Abstract
• Premise of the study: Variation in the distribution of methylated CpG (methyl-CpG) in genomic DNA (gDNA) across the tree of life is biologically interesting and useful in genomic studies. We illustrate the use of human methyl-CpG-binding domain (MBD2) to fractionate angiosperm DNA into eukaryotic nuclear (methyl-CpG-rich) vs. organellar and prokaryotic (methyl-CpG-poor) elements for genomic and metagenomic sequencing projects.
• Methods: MBD2 has been used to enrich prokaryotic DNA in animal systems. Using gDNA from five model angiosperm species, we apply a similar approach to identify whether MBD2 can fractionate plant gDNA into methyl-CpG-depleted vs. enriched methyl-CpG elements. For each sample, three gDNA libraries were sequenced: (1) untreated gDNA, (2) a methyl-CpG-depleted fraction, and (3) a methyl-CpG-enriched fraction.
• Results: Relative to untreated gDNA, the methyl-depleted libraries showed a 3.2–11.2-fold and 3.4–11.3-fold increase in chloroplast DNA (cpDNA) and mitochondrial DNA (mtDNA), respectively. Methyl-enriched fractions showed a 1.8–31.3-fold and 1.3–29.0-fold decrease in cpDNA and mtDNA, respectively.
• Discussion: The application of MBD2 enabled fractionation of plant gDNA. The effectiveness was particularly striking for monocot gDNA (Poaceae). When sufficiently effective on a sample, this approach can increase the cost efficiency of sequencing plant genomes as well as prokaryotes living in or on plant tissues.
Keywords: endophyte, enrichment, genome sequencing, metagenome, microbiome, methyl-binding domain
Studies focused on genomic and metagenomic questions often struggle to obtain the nucleotide sample(s) ideally suited for successful investigations. For example, organellar genome sequencing projects have employed a variety of techniques to obtain sequence data enriched with the genome of interest. These include laborious centrifugation prior to DNA extraction (e.g., Atherton et al., 2010), genome skimming approaches (e.g., Straub et al., 2012) that rely on the sheer scale of sequencing, long-range PCR (e.g., Uribe-Convers et al., 2014), and hybrid sequence capture techniques (reviewed by Cronn et al., 2012). Nuclear genome sequencing projects face the opposite problem, often wasting valuable sequencing efforts by redundantly sequencing small multicopy organellar genomes or prokaryotic contaminants. Studies focused on metagenomic communities (microbiomes) in eukaryotes deal with an even greater problem, plant nuclear DNA potentially eclipsing small prokaryotic genomes that may occur in low titer. As a result of these issues, the development of approaches that allow one to extract total genomic DNA (gDNA) and subsequently partition DNA from different genomic compartments into enriched fractions has considerable potential for numerous lines of research.
One means by which such fractionation has been done in animal systems involves the use of a DNA methyl-binding domain (Gebhard et al., 2006; Feehery et al., 2013). Various classes of DNA methylation are well known from all forms of life. In eukaryotic nuclear genomes, methylated CpG (methyl-CpG) sites have been particularly well studied, representing a eukaryote-specific form of methylation that is associated with critical epigenetic signaling and gene expression (e.g., Deaton and Bird, 2011). Whereas vertebrate nuclear genomes appear to largely use methyl-CpG (up to 80%; Feng et al., 2010), angiosperm genomes reveal more complex patterns of methylation. The latter also possess extensive cytosine methylation (Feng et al., 2010), which includes methyl-CHG and methyl-CHHG in addition to methyl-CpG (e.g., Feng et al., 2010; Zemach et al., 2010). Furthermore, plant nuclear genomes encode a more complex array of methyl-binding domains (e.g., Springer and Kaeppler, 2005; Feng et al., 2010). In contrast to methylation of nuclear genomes, the prokaryote-derived organellar genomes of eukaryotes have received less attention. In fact, a study has only recently overturned the commonly held view that human mitochondrial DNA (mtDNA) contains methyl-CpG (Hong et al., 2013). Similar work on Arabidopsis thaliana (L.) Heynh., Populus trichocarpa Torr. & A. Gray, and Oryza sativa L. suggests that angiosperm plastomes have ca. 1–1.5% total methylation and that these include less than 0.75% methyl-CpG compared to 22–59% in the same nuclear genomes (Feng et al., 2010). If similar patterns characterize angiosperm mtDNA, then the nuclear and organellar genomes of these organisms appear to be well differentiated in their methyl-CpG composition.
Variation in patterns of DNA methylation between major phyla and genomic compartments therefore offer unique opportunities to partition total gDNA into different source elements. Prior research on animal tissues has shown that the human methyl-CpG-binding domain (MBD2) can be used to partition fragments of DNA into those with methyl-CpG present vs. those lacking appreciable CpG methylation (Gebhard et al., 2006; Feehery et al., 2013). Through this technique, Feehery et al. (2013) demonstrated that gDNA extracted from human and fish tissues can be readily fractionated into methyl-CpG-rich and methyl-CpG-poor elements representing nuclear host and prokaryotic parasite or endosymbiont components, respectively.
We reasoned that an available MBD2 construct might be applicable to fractionate gDNA extracted from plant tissues into high-methyl-CpG nuclear chromosomal fragments vs. low-methyl-CpG elements. The latter classes that we considered include plastid DNA and mtDNA in addition to DNA from prokaryotic parasites or endophytes. If successful, such an approach could reduce the relative cost and time invested in a variety of studies on plant genomes as well as those focused on plant metagenomic communities.
METHODS
To test the potential utility of methyl-CpG capture on plant systems, we sequenced gDNA from A. thaliana, Glycine max (L.) Merr., Leucaena leucocephala (Lam.) de Wit, O. sativa, and Zea mays L. For each sample, three gDNA libraries were sequenced using a MiSeq instrument (Illumina, San Diego, California, USA): (1) untreated gDNA, (2) a fraction depleted of methyl-CpG, and (3) a fraction enriched for methyl-CpG. The input DNA sources for the latter two libraries represent the supernatant and bead fractions derived from the application of the NEBNext Microbiome DNA Enrichment Kit (#E2612S; New England Biolabs, Ipswich, Massachusetts, USA). This approach uses the IgG1 fused to the human methyl-CpG-binding domain (together “MBD2-Fc”) to pull down a methyl-CpG-enriched fraction in the bead-associated element, leaving a methyl-depleted fraction in the supernatant.
Seeds for G. max (Burpee Be Sweet 292), L. leucocephala, and Z. mays (Plantation Products, Norton, Massachusetts, USA) were grown under standard greenhouse or growth chamber conditions in Las Cruces, New Mexico. Voucher information is provided in Appendix 1. Total gDNA was obtained from young leaf material through standard cetyltrimethylammonium bromide (CTAB) extraction (Doyle and Doyle, 1987). Using these standard approaches, DNA from L. leucocephala was highly viscous and difficult to work with, as were standard column-based plasmid-type DNA extractions (e.g., QIAGEN). Therefore, we extracted DNA using Norgen Biotek’s new Plant/Fungal DNA Isolation Kit (#26200; Norgen Biotek, Thorold, Ontario, Canada), which yielded a clean sample of L. leucocephala. Arabidopsis thaliana and O. sativa DNA was purchased directly from BioChain Institute (Newark, California, USA).
Due to the critical nature of gDNA quality as well as the quantity of gDNA to MBD2-Fc ratio for effective methyl-CpG capture, DNA quality and quantity were checked using two different methods. NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, Delaware, USA) was used to calculate purity of DNA by A260/A280 as well as concentration. In addition, a double-stranded DNA (dsDNA) assay was also used to calculate concentration using the Qubit dsDNA BR Assay Kit (#Q32850; Life Technologies, Grand Island, New York, USA). Differences in concentration between NanoDrop and Qubit are likely due to single-stranded RNA contamination in genomic preps or other contaminants absorbing at A260. As a result, we used Qubit-derived estimates whenever there was a discrepancy. Enrichments followed manufacturer’s recommendations (#E2612S; New England Biolabs). In short, we used a ratio of 1 μg of gDNA for 160-μL protein A magnetic beads and 16-μL MBD2-Fc protein. First, 160-μL protein A magnetic beads were prebound to 16-μL MBD2-Fc protein by incubating with rotation for 10 min at room temperature. Protein A/MBD2-Fc complex was pulled down by magnetic field and washed twice with 1 mL of ice-cold 1× wash/bind buffer to remove unbound MBD2-Fc protein. The beads were resuspended in 160 μL of ice-cold 1× wash/bind buffer. The gDNA was added to the protein A/MBD2-Fc beads and incubated with rotation for 15 min at room temperature. The beads were pulled down by magnetic field and the methyl-CpG-depleted supernatant carefully removed without disturbing the beads. The bead pellets were washed once with ice-cold 1× wash/bind buffer while sitting on a magnetic stand, resuspended in 150 μL 1× TE buffer (pH 8.0), and then treated with Proteinase K to release methyl-CpG-enriched DNA. After proteinase K treatment at 65°C for 20 min, the beads were pulled down, and the supernatant containing the methyl-CpG-enriched fraction was recovered. Both the methyl-CpG-enriched and methyl-CpG-depleted fractions were purified using 1.8× volume of AMPure beads (#A63880; Beckman Coulter, Brea, California, USA) and eluted in 60 μL of 1× TE buffer (pH 8.0).
DNA was sheared to 150–200-bp fragments using a Covaris S2 ultrasonicator (Covaris, Woburn, Massachusetts, USA) and directly used to prepare libraries with the NEBNext Ultra DNA Library Prep Kit (#E7370S; New England Biolabs) and multiplex barcodes (#E7335S and #E7500S; New England Biolabs) with nine rounds of PCR. To keep the amplification of libraries constant, 3–5% of adapter-ligated DNA was used as a template in a 50-μL PCR reaction for untreated (350 ng) and methyl-CpG-enriched fractions, whereas a third of adapter-ligated DNA was used as a template for the methyl-CpG-depleted fraction. The indexed libraries were multiplexed and sequenced (paired-end) on various runs of an Illumina MiSeq instrument using the 300-cycle reagent kit (version 2). Raw FASTQ files were generated using MiSeq Control Software version 2.2.0 (Illumina).
Paired-end sequencing reads were mapped to available relevant organellar genomes (Table 1) for each species using Bowtie 2 version 2.1.0 (Langmead and Salzberg, 2012). A conservative mapping approach was applied, requiring each read to: (1) stringently map (–score-min L,0,−0.6), (2) have its pair occur within a reasonable expected distance (-X 700), and (3) have each pair be properly oriented relative to one another (BamTools version 2.3.0, option “–isProperPair true”; Barnett et al., 2011). The percentage of reads mapping to a genome was calculated as the fraction of read-pairs mapped to the total read-pairs in the library. In addition, for the modest-sized high-quality nuclear genome for A. thaliana (TAIR 9; Arabidopsis Genome Initiative, 2000) the data were mapped to the nuclear, chloroplast, and mitochondrial genomes simultaneously using the same approach. Geneious (version 6.8 created by Biomatters [http://www.geneious.com]) was used for graphical representation of the evenness of organellar genome coverage for Arabidopsis (TAIR 9) and Oryza (build 4; Zhao et al., 2004).
Table 1.
Plant species, genome accessions, and genome sizes used in this study.
| Species | Plastome / Mitochondrial DNA genome accession | Plastome / Mitochondrial genome size (bp) |
| Arabidopsis thaliana | NC_0009321.1 / NC_001284.2 | 154,478 / 366,924 |
| Glycine max | DQ317523.1 / NC_020455.1 | 152,218 / 402,558 |
| Leucaena leucocephalaa | Hernandez et al., personal communication | 164,692a / 1,013,450a |
| Oryza sativa | NC_008155.1 / NC_007886.1 | 134,496 / 491,515 |
| Zea mays | NC_001666.2 / AY506529.1 | 140,384 / 569,630 |
Draft genomes made available by Hernandez et al. for read mapping Leucaena libraries.
RESULTS
The DNA extracts used here included high-molecular-weight samples that comigrated with lambda DNA marker (#N3019L; New England Biolabs) on an agarose gel and that had a 260/280 ratio of at least 1.80 (Table 2). Hereafter each library sequenced will be referred to as “UT” (untreated DNA), “E” (methyl-enriched), or “D” (methyl-depleted). The resulting Illumina library characteristics and percentage of read-pairs appropriately mapping to the plastome and mitochondrial genome for each library are presented in Table 3 and Fig. 1, and the primary data are available through the National Center for Biotechnology Information Sequence Read Archive (SUB581050).
Table 2.
Starting genomic DNA characteristics for species in the study.
| Species | 260/280 ratio | NanoDrop (ng/μL) | Qubita (ng/μL) |
| Arabidopsis thaliana | 1.92 | 160 | 85 |
| Glycine max | 1.85 | 232 | 68 |
| Leucaena leucocephala | 1.99 | 168 | 52 |
| Oryza sativa | 1.85 | 454 | 116 |
| Zea mays | 1.80 | 66 | 70 |
Qubit values are based on the dsDNA BR Assay Kit (Life Technologies).
Table 3.
DNA sequence and read mapping results. For each species, three libraries were sequenced: untreated DNA (UT), methyl-enriched DNA (E), and methyl-depleted DNA (D). The total reads per library and the number of reads mapping (2× the read-pairs), and percentage of read-pairs mapping for each are listed.
| Chloroplast | Mitochondrial | ||||
| Library | Reads per library (million reads) | cpDNA read matches (thousand reads) | Percent cpDNA read-pairs | Mitochondrial read matches (thousand reads) | Percent mitochondrial read-pairs |
| UT-Arabidopsis | 3.47 | 602 | 17.36 | 56 | 1.62 |
| E-Arabidopsis | 4.49 | 33 | 0.74 | 24 | 0.55 |
| D-Arabidopsis | 3.72 | 2070 | 55.79 | 287 | 7.72 |
| UT-Oryza | 3.21 | 207 | 6.46 | 77 | 2.41 |
| E-Oryza | 4.37 | 41 | 0.94 | 18 | 0.43 |
| D-Oryza | 3.86 | 2410 | 62.52 | 877 | 22.69 |
| UT-Glycine | 3.12 | 95 | 3.04 | 19 | 0.63 |
| E-Glycine | 4.38 | 73 | 1.68 | 20 | 0.47 |
| D-Glycine | 4.40 | 856 | 19.46 | 159 | 3.61 |
| UT-Zea | 2.69 | 166 | 6.20 | 63 | 2.36 |
| E-Zea | 2.91 | 8 | 0.30 | 5 | 0.18 |
| D-Zea | 3.38 | 2350 | 69.55 | 908 | 26.82 |
| UT-Leucaena | 5.47 | 496 | 9.07 | 251 | 4.60 |
| E-Leucaena | 4.85 | 14 | 0.29 | 7 | 0.16 |
| D-Leucaena | 4.13 | 1260 | 30.56 | 648 | 15.69 |
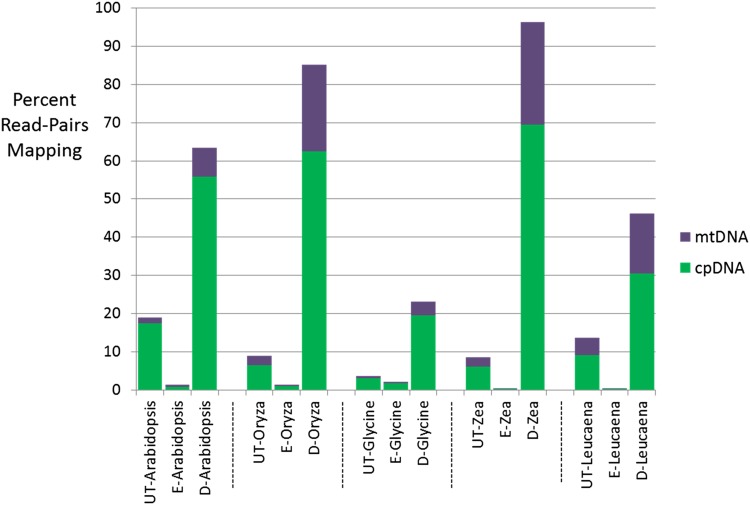
Fig. 1.
Percentage of read-pairs mapping to plastome and mitochondrial genomes using Bowtie 2. UT = untreated genomic DNA (gDNA) library; E = methyl-enriched gDNA library; D = methyl-depleted gDNA library.
Relative to untreated gDNA, the methyl-depleted libraries resulted in a 3.2–11.2-fold and 3.4–11.3-fold increase in chloroplast DNA (cpDNA) and mtDNA, respectively. Conversely, the methyl-enriched fraction resulted in a 1.8–31.3-fold and 1.3–29.0-fold decrease in cpDNA and mtDNA reads, respectively.
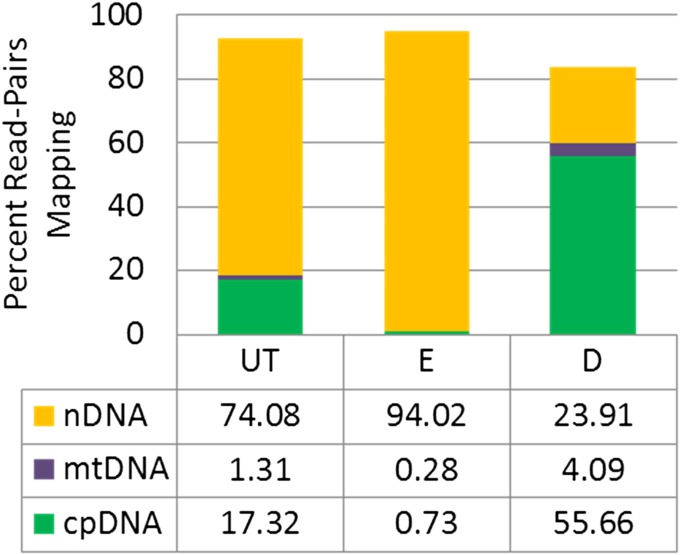
The mapping of reads to a reference genome that contained both nuclear and organellar DNA sequences for A. thaliana provided additional insight into the characteristics of the methyl-CpG-depleted and methyl-CpG-enriched fractions (Fig. 2). The percentage of reads mapped to the plastome and mitochondrial genomes using this approach (Fig. 2), and the aforementioned mapping only to the organellar genomes (Fig. 1), was essentially identical. Considering the nuclear genome, 74% of the untreated DNA read-pairs mapped to the genome while 94% and 23% mapped for the methyl-enriched and methyl-depleted fractions, respectively. The combined percentage of reads mapping to the three genomes was similar for the starting DNA and methyl-enriched library; however, the methyl-depleted library had ca. 10% fewer read-pairs mapping to any one of the three Arabidopsis genomes (see Discussion).
Fig. 2.
Arabidopsis thaliana read-pairs mapping to any of the three genomes (mitochondrial DNA [mtDNA], chloroplast DNA [cpDNA], and nuclear DNA [nDNA]). UT = untreated genomic DNA (gDNA) library; E = methyl-enriched gDNA library; D = methyl-depleted gDNA library.
To investigate whether one or more of these classes of libraries (UT, E, or D) was prone to bias in sequencing coverage across genomes, we mapped the A. thaliana and O. sativa libraries to their respective organellar genomes (cpDNA presented in Appendix S1 (1.6MB, pdf) ) and chromosome 1 (selected arbitrarily) using Geneious for easy visualization. We were particularly interested in identifying whether multiple-kilobase segments had been excluded, likely a result of the method failing to pull down the large complete segments through the enrichment process. The resulting percentages of read-pairs mapping were consistent between the Geneious and aforementioned Bowtie 2 approach, and overall read coverage appears to be uniform for all three classes of libraries. For simplicity, the range of findings is presented in the following examples, focusing on the easily visualized plastome (Appendix S1 (1.6MB, pdf) ). The O. sativa cpDNA genome showed uniform coverage across the plastome for all libraries. For the methyl-CpG-depleted fraction, the lower coverage (mean 31×) likely explains slightly lower uniformity (Appendix S1A (1.6MB, pdf) ). With A. thaliana, there is nearly uniform read coverage for the UT library and D library except in three AT-rich zones showing lower coverage (Appendix S1B (1.6MB, pdf) ). This lower coverage is unlikely to be due to the enrichment process, as problems with Illumina coverage in low-complexity AT- and GC-rich regions are a known issue (e.g., Oyola et al., 2012). As with O. sativa, the A. thaliana E library showed the expected decrease in plastome coverage (mean 22×) and expected increase in variation in overall coverage as a result. The methyl-enriched libraries mapped to chromosome 1 revealed only small gaps (typically <500 bp) in coverage across the length (data not shown), which suggests that this approach has not excluded large regions due to failed capture.
DISCUSSION
The objective of this study was to investigate whether the MBD2-Fc construct can be used on plant samples to effectively partition total gDNA into methyl-rich and methyl-poor elements that better reflect their genomic origin than untreated DNA extractions. Using the relative representation of organellar (likely methyl-CpG-poor) and nuclear DNA (known methyl-CpG-rich) from DNA extracts of high purity (Table 2) and high molecular weight, the approach showed the predicted enrichment and depletion of methyl-CpG fractions in all cases. However, the changes relative to untreated DNA varied by species, library type, and major lineage. Results from our two monocot samples, both representatives of the economically important Poaceae, showed the greatest fold increase (9.3–11.3-fold) in the representation of organellar DNA in methyl-poor libraries (Table 3). These samples also showed considerable depletion of organellar DNA in the methyl-rich fraction (5.6–20.3-fold). Among the eudicot samples, the results were more variable by library and/or organelle. The Arabidopsis and Leucaena samples had the greatest starting percentage of cpDNA, and the methyl-rich fraction revealed a 23–31-fold decrease in cpDNA contamination. Although the starting concentration of mtDNA reads in any of the samples was low (all <8%), depletion of mtDNA reads in the methyl-rich fraction ranged from just 1.34-fold in Glycine to 29-fold in Leucaena.
Although generally effective, there are several potential causes for the apparent variation in the overall efficacy of MBD2-Fc to partition methyl-rich and methyl-poor genomic regions in our genomic extracts. First, divergence in MBD proteins has been documented between animal and plant systems as well as between monocots and eudicots (e.g., Feng et al., 2010). As a result, MBD2, for some unknown reason, may not bind plant methyl-CpG as effectively as with vertebrate methyl-CpG sites. This issue could be confounded if one or more of the samples was “contaminated” by native plant methyl-binding domains, not removed by DNA purification, that impeded access and binding of MBD2-Fc. Furthermore, limited existing data suggest that there may be variation in the frequency of CpG methylation between monocots and eudicots. Feng et al. (2010) found that chromosomal DNA of the eudicots A. thaliana and P. trichocarpa contained 22–42% methyl-CpG, but their single monocot sample, O. sativa, displayed 59%. Further research on patterns of plant DNA methylation will identify whether there are lineage-specific differences between monocots and eudicots and indicate whether these may be influencing the results presented here. In addition, Gebhard et al. (2006) discussed variation in binding sensitivity associated with salt concentration, which is worth future consideration in the application of this and similar approaches.
Based on our preliminary results (not shown), it is clear that clean high-molecular-weight DNA is important for the application of MBD2-Fc. With high-molecular-weight DNA, a large fragment with even one CpG island along its length may be bound and pulled down and available for library prep even though other regions may lack methyl-CpG. Without high-molecular-weight DNA, variation in methyl-CpG could easily lead to unintentional partitioning into different fractions from a collinear segment of DNA (e.g., a chromosome). Although such findings could be helpful for studies on methylated vs. unmethylated collinear regions, they were not the objective of the current study.
The visualization of reads mapping to A. thaliana and O. sativa (Appendix S1 (1.6MB, pdf) ) illustrated that sequencing of either the methyl-enriched or methyl-depleted fractions provided relatively even coverage of sequence, without signs of large regions lacking coverage in any of the three genomes. There appears to be a slight decrease in coverage across low-complexity AT-rich regions in the A. thaliana methyl-depleted library (Appendix S1B (1.6MB, pdf) ). This result is most likely a slight exacerbation of a known Illumina AT sequencing issue (e.g., Oyola et al., 2012) rather than a representation issue in the actual libraries being sequenced.
Read mapping to the nuclear and organellar genomes of A. thaliana revealed an intriguing result. The total number of read-pairs mapping to any one of these genomes was ca. 10% less in the methyl-depleted library than the untreated or methyl-enriched libraries (Fig. 2). While this may represent stochastic variation among libraries, our preliminary studies of various libraries showed little variation in relative genomic representation between independently generated and sequenced libraries (data not shown). We suspected that the decrease in read mapping with methyl-depleted libraries is, at least in part, due to enrichment of “contaminating” prokaryotic sequences, which would be expected for this approach. This would be consistent with the findings of Feehery et al. (2013), who used this approach to better characterize prokaryotic species inhabiting animal tissues. To roughly investigate this idea, we ran each of the three A. thaliana libraries through MetaPhlAn (Segata et al., 2012), which focuses primarily on human-associated prokaryotic genomes. Running reads that were unmapped to the A. thaliana genomes, we applied the “Sensitive-Local” setting in MetaPhlAn and found nine, two, and 23 prokaryotic genome hits in the UT, E, and D libraries, respectively. The decreased representation in the E library and increase in the D library, from the same starting DNA sample, is consistent with the method having partitioned the prokaryotic contaminants into the D library and suggests that the protocol may be particularly important in plant metagenomic studies (e.g., Zheng et al., 2014).
When comparing the application of MBD2-Fc to other options for genome and microbiome sequencing, there are several issues to consider. These include project objectives, ease of use, cost, time, reproducibility, required starting material, and results. Careful consideration of all these elements should help narrow in on the most logical method for a given study (e.g., Jansen et al., 2005). Whole-genome sequencing of any genomic compartment using unperturbed total gDNA has been the standard for nuclear genome sequencing and even for organellar genomes. This rapid approach is appropriate for many applications and starting materials (e.g., Straub et al., 2012), but a considerable percentage of reads can be wasted on genomic compartments of lesser or no interest to the parent study. The isolation of entire genomic compartments (e.g., nuclei, mitochondria, and/or chloroplasts) prior to DNA extraction from one or more fractions is also a widely applied approach. However, the process is laborious, requires copious fresh starting material (e.g., 20 g in Shi et al., 2012), and the results with plant material have varied, occasionally resulting in little effective change in the total concentration of different fractions (noted by Atherton et al., 2010). In addition, the coverage presented by Shi et al. (2012) is far less uniform (see their Fig. 4) than what is presented for the MBD2-Fc (Appendix S1 (1.6MB, pdf) ). PCR-based approaches offer a powerful option that can be based on low-quantity and/or low-quality starting material. Nonetheless, coverage tends to be highly variable (e.g., Uribe-Convers et al., 2014), and numerous problems can presumably result from the coamplification of nuclear-encoded organellar genomic elements (e.g., Vieira et al., 2014). Hybridization approaches continue to rise in popularity and can be exceptionally cost-effective, particularly when sequencing a portion of the nuclear genome or most or all of organellar genomes for large numbers of individuals, populations, or species (e.g., Cronn et al., 2012; Stull et al., 2013; Mandel et al., 2014; Mariac et al., 2014; Weitemier et al., 2014). However, when the hybridization resources have not already been developed, the initial setup cost and time may be prohibitive, and variability in probe specificity can lead to limited uniformity of coverage. In addition, hybridization approaches do not differentiate between nuclear-encoded organellar elements vs. their organellar sequence (as is an issue with the PCR approach), but they should not create chimeric sequences that are more commonly associated with long-range PCR.
The methyl-CpG capture approach presented here requires intact high-molecular-weight starting DNA for optimal results, but it does not require large quantities of DNA (we have used as little as 500 ng of starting material for the enrichment and generation of libraries). Our results indicate a high degree of fractionation in many samples and uniform sequencing coverage across genomes. If one is concerned about the relative success of enrichments, real-time quantitative PCR can be used to test for shifts in well-developed cpDNA loci in the different fractions (untreated, methyl-rich, and methyl-poor) prior to costly library prep and sequencing. At ca. US$30 per enrichment, we see the use of MBD2-Fc as a fast and useful approach for modest numbers of samples in sequencing projects of organellar genomes (i.e., when the initial setup of hybridization approaches is not justified) and nuclear genome sequencing projects on plant groups with moderate to high organellar contamination that can lead to substantial losses of genomic sequence data on nontarget genomes. Furthermore, the deep sequencing of methyl-depleted and methyl-enriched gDNA libraries derived from plant tissues may give a more complete picture of the unobserved prokaryotic and eukaryotic coinhabitants. Libraries selectively depleted of plant nuclear DNA should be rich in plant organellar DNA as well as prokaryotic coinhabitants. These libraries may also be enriched for organellar DNA from microscopic eukaryotic coinhabitants (e.g., fungal or algal). In contrast, the methyl-enriched libraries not only should include a greater representation of plant nuclear DNA, but also may contain more nuclear DNA derived from microscopic eukaryotic coinhabitants (e.g., fungal or algal). Thus the use of MBD2-Fc shows considerable potential in plant biology and suggests that future research on the use of plant-specific MBDs is warranted for even greater efficiency of plant DNA partitioning in genomic and metagenomic studies.
Supplementary Material
Appendix 1.
Voucher information for seed-grown samples. The Arabidopsis thaliana and Oryza sativa data were derived from commercially available DNA and lack physical vouchers.
| Species | Seed origin | Herbarium | Herbarium acquisition no. |
| Glycine max | “Burpee Be Sweet” seed packet | NMC | 84564 |
| Leucaena leucocephala | Honolulu, HI | NMC | 84563 |
| Zea mays | “Hybrid Sweet” seed packet | NMC | 84565 |
Note: NMC = New Mexico State University herbarium.
LITERATURE CITED
- Arabidopsis Genome Initiative. 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Atherton R., McComish B., Shepherd L., Berry L., Albert N., Lockhart P. 2010. Whole genome sequencing of enriched chloroplast DNA using the Illumina GAII platform. Plant Methods 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett D. W., Garrison E. K., Quinlan A. R., Strömberg M. P., Marth G. T. 2011. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics (Oxford, England) 27: 1691–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronn R., Knaus B. J., Liston A., Maughan P. J., Parks M., Syring J. V., Udall J. 2012. Targeted enrichment strategies for next-generation plant biology. American Journal of Botany 99: 291–311. [DOI] [PubMed] [Google Scholar]
- Deaton A. M., Bird A. 2011. CpG islands and the regulation of transcription. Genes & Development 25: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. J., Doyle J. L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemistry Bulletin 19: 11–15. [Google Scholar]
- Feehery G. R., Yigit E., Oyola S. O., Langhorst B. W., Schmidt V. T., Stewart F. J., Dimalanta E. T., et al. 2013. A method for selectively enriching microbial DNA from contaminating vertebrate host DNA. PLoS ONE 8: e76096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Cokus S. J., Zhang X., Chen P.-Y., Bostick M., Goll M. G., Hetzel J., et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proceedings of the National Academy of Sciences 107: 8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhard C., Schwarzfischer L., Pham T. H., Andreesen R., Mackensen A., Rehli M. 2006. Rapid and sensitive detection of CpG-methylation using methyl-binding (MB)-PCR. Nucleic Acids Research 34: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E. E., Okitsu C. Y., Smith A. D., Hsieh C.-L. 2013. Regionally-specific and genome-wide analyses conclusively demonstrate the absence of CpG methylation in human mitochondrial DNA. Molecular and Cellular Biology 33: 2683–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R. K., Raubeson L. A., Boore J. L., dePamphilis C. W., Chumley T. W., Haberle R. C., Wyman S. K., et al. 2005. Methods for obtaining and analyzing whole chloroplast genome sequences. In E. A. Zimmer and E. Roalson [eds.], Molecular evolution: Producing the biochemical data, 348–384. Methods in enzymology, vol. 395. Academic Press, Waltham, Massachusetts, USA. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel J. R., Dikow R. B., Funk V. A., Masalia R. R., Staton S. E., Kozik A., Michelmore R. W., Rieseberg L. H., Burke J. M. 2014. A target enrichment method for gathering phylogenetic information from hundreds of loci: An example from the Compositae. Applications in Plant Sciences 2(2): 1300085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariac C., Scarcelli N., Pouzadou J., Barnaud A., Billot C., Faye A., Kougbeadjo A., et al. 2014. Cost-effective enrichment hybridization capture of chloroplast genomes at deep multiplexing levels for population genetics and phylogeography studies. Molecular Ecology Resources 14: 1103–1113. [DOI] [PubMed] [Google Scholar]
- Oyola S., Otto T., Gu Y., Maslen G., Manske M., Campino S., Turner D., et al. 2012. Optimizing Illumina next-generation sequencing library preparation for extremely AT-biased genomes. BMC Genomics 13: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Waldron L., Ballarini A., Narasimhan V., Jousson O., Huttenhower C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature Methods 9: 811–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C., Hu N., Huang H., Gao J., Zhao Y.-J., Gao L.-Z. 2012. An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS ONE 7: e31468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer N. M., Kaeppler S. M. 2005. Evolutionary divergence of monocot and dicot methyl-CpG-binding domain proteins. Plant Physiology 138: 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub S., Parks M., Weitemeyer K., Fishbein M., Cronn R. C., Liston A. 2012. Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. American Journal of Botany 99: 349–364. [DOI] [PubMed] [Google Scholar]
- Stull G. W., Moore M. J., Mandala V. S., Douglas N. A., Kates H.-R., Qi X., Brockington S. F., et al. 2013. A targeted enrichment strategy for massively parallel sequencing of angiosperm plastid genomes. Applications in Plant Sciences 1(2): 1200497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uribe-Convers S., Duke J. R., Moore M. J., Tank D. C. 2014. A long PCR–based approach for DNA enrichment prior to next-generation sequencing for systematic studies. Applications in Plant Sciences 2(1): 1300063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira L. N., Faoro H., Fraga H. P. F., Rogalski M., de Souza E. M., de Oliveira Pedrosa F., Nodari R. O., Guerra M. P. 2014. An improved protocol for intact chloroplasts and cpDNA isolation in conifers. PLoS ONE 9: e84792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitemier K., Straub S., Cronn R. C., Fishbein M., Schmickl R., McDonnell A., Liston A. 2014. Hyb-Seq: Combining target enrichment and genome skimming for plant phylogenomics. Applications in Plant Sciences 2(9): 1400042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A., McDaniel I. E., Silva P., Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328: 916–919. [DOI] [PubMed] [Google Scholar]
- Zhao W., Wang J., He X., Huang X., Jiao Y., Dai M., Wei S., et al. 2004. BGI-RIS: An integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Research 32: D377–D382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z., Deng X., Chen J. 2014. Whole-genome sequence of “Candidatus Liberibacter asiaticus” from Guangdong, China. Genome Announcements 2: e00273-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.