Abstract

We describe a microfluidic Western blot assay (μWestern) using a Tris tricine discontinuous buffer system suitable for analyses of a wide molecular mass range (6.5–116 kDa). The Tris tricine μWestern is completed in an enclosed, straight glass microfluidic channel housing a photopatterned polyacrylamide gel that incorporates a photoactive benzophenone methacrylamide monomer. Upon brief ultraviolet (UV) light exposure, the hydrogel toggles from molecular sieving for size-based separation to a covalent immobilization scaffold for in situ antibody probing. Electrophoresis controls all assay stages, affording purely electronic operation with no pumps or valves needed for fluid control. Electrophoretic introduction of antibody into and along the molecular sieving gel requires that the probe must traverse through (i) a discontinuous gel interface central to the transient isotachophoresis used to achieve high-performance separations and (ii) the full axial length of the separation gel. In-channel antibody probing of small molecular mass species is especially challenging, since the gel must effectively sieve small proteins while permitting effective probing with large-molecular-mass antibodies. To create a well-controlled gel interface, we introduce a fabrication method that relies on a hydrostatic pressure mismatch between the buffer and polymer precursor solution to eliminate the interfacial pore-size control issues that arise when a polymerizing polymer abuts a nonpolymerizing polymer solution. Combined with a new swept antibody probe plug delivery scheme, the Tris tricine μWestern blot enables 40% higher separation resolution as compared to a Tris glycine system, destacking of proteins down to 6.5 kDa, and a 100-fold better signal-to-noise ratio (SNR) for small pore gels, expanding the range of applicable biological targets.
Western blotting comprises an indispensable analytical tool for both research and clinical laboratories.1−6 In conventional Western blots, slab-gel electrophoresis forms the basis for protein sizing. Antibody probing is conducted after transfer of protein bands from the small-pore-size polyacrylamide gel (e.g., 19 nm to 140 nm pores for 3.5–10.5% total monomer (%T) and 0.5–10% cross-linker (%C) gels)7 to a larger-pore-size polymer membrane (e.g., PVDF or nitrocellulose with 200–450 nm size pores).8 Proteins are immobilized on the membrane via hydrophobic interactions.9 Immobilization of protein bands on a large-pore-size membrane facilitates antibody-based probing of the immobilized species with large-molecular-mass antibodies.1 In effect, the design of conventional Western blotting decouples pore-size demands required for effective molecular sieving during polyacrylamide gel electrophoresis (PAGE) from specifications for effective probing.
Microfluidic design affords faster assay times, smaller sample volumes, and easier integration with automation systems than conventional slab-gel systems. Building on these advantages, protein separations from capillaries,10 microfluidic chips,11 and a microarrayer12 have been interfaced to blotting membranes for probing. In another approach to full integration, a capillary system supports the protein separation (mass or pI) and subsequent photocapture and immunoprobing of antigens on the capillary wall.13,14 The commercial capillary Western blot completes in 3 h. While an advance in integration, the assay sees low protein immobilization efficiencies (∼0.01%) and substantial hardware (fluid pumping, high voltage, and robotic control for multiplexing).13 In an alternate approach using a planar glass microdevice, protein separation and probing steps are integrated on chip yielding assay times as fast as 3 min.15−18 A microchamber patterned with functionalized polymers forms the basis for assay integration. Nevertheless, the approach requires complex fabrication protocols that limit throughput and multiplexing capabilities.
Using a planar microfluidic device, our group reported on a single microchannel Western technique that unifies protein separation and probing in a single microfluidic channel. The simple single channel design provides a basis for scaleup and multiplexing. In order to integrate the assay stages and obtain high immobilization efficiencies, the μWestern makes dual use of the separation axis: the axis forms the molecular sieving dimension during PAGE and also forms the path for antibody probe introduction during probing (i.e., antibody is introduced through the protein separation gel).
While the in-channel probing strategy underpins completion of Western blotting in a single microchannel, the approach poses a particular challenge for small-molecular-mass species. The gel must both effectively sieve small proteins during PAGE and also allow large antibodies to electromigrate through the gel pores during probing. As such, the standard Tris glycine microchannel Western blot is limited to analyses of proteins larger than ∼21 kDa,19 as smaller species remain stacked between the leading and terminating electrolyte in the 7.5%T gel. While the use of a higher %T gel allows destacking of smaller-molecular-mass species, the small-pore-size gel traps antibodies at the separation gel interface, thus substantially reducing immunoprobing signals. To expand the applicability of microfluidic Western blotting to smaller molecular mass proteins, we describe a new fabrication technique that reduces confounding interactions of the gel with large antibody probes, as well as transition to assay conditions relevant to smaller species.
Materials and Methods
In-Channel Gel Fabrication
Microchannel designs are completed in-house, then fabricated using standard wet glass etching at a glass foundry (Perkin Elmer).20 Separation channels are 1 cm long, 10 μm deep, and 70 μm wide. Each well pair is connected by three parallel separation channels for technical triplicates. Prior to gel fabrication, channel walls are functionalized with acrylate monomers, as previously described,21 to enable gel cross-linking to the channel walls.
The separation gel precursor solution is composed of acrylamide/bis(acrylamide) at a ratio of 37.5:1 (Sigma–Aldrich, No. A3699) diluted to a final %T between 8 and 12, as indicated in the text. To enable protein photocapture, 1.5 mM N-[3-[(4-benzoylphenyl)formamido]propyl]methacrylamide (BPMAC, C21H22N2O3, 350.2 g/mol) is added from a stock solution of 100 mM in DMSO. BPMAC is synthesized in-house.22 The gel precursor buffer is 500 mM Tris HCl titrated to pH 8.45 for the Tris tricine discontinuous buffer system and 375 mM Tris HCl titrated to pH 8.8 for the Tris glycine discontinuous buffer system, consistent with the Tris HCl pH typically used in slab gels for each respective buffer system.23 These components are degassed in a sonicator under vacuum for 3 min. After degassing, sodium dodecyl sulfate (SDS) (0.1% final concentration) and Triton X-100 (0.1%) are added along with the initiators riboflavin 5′-monophosphate (0.0006%) (No. F1392, Sigma–Aldrich), TEMED (0.05% vol/vol) (No. T9281, Sigma–Aldrich), and ammonium persulfate (0.015%) (No. A3678, Sigma–Aldrich). Gel precursor is applied to one well, with capillary action wicking the solution into the dry microchannels. In the original protocol,19 glass chips are submerged in a Petri dish with gel buffer precursor solution with SDS (0.1%) and Triton X-100 (0.1%) after gel precursor loading to prevent flow in the channels and remove gel precursor from the wells. In the alternate protocol utilized here (unless otherwise noted), the gel precursor in the well is replaced with buffer solution after gel loading but before submerging the chip, so as to generate a buffer/gel precursor interface partway through the channel (detailed in the Results and Discussion section). For the comparison of antibody plug to antibody front probe approaches, additional buffer is added to the opposite well to subsequently migrate the gel precursor back toward the center of the channel and establish a buffer/gel precursor interface on both ends of the gel in the channel. While submerged, chips are photopolymerized using a collimated blue LED source (470 nM, No. M470L2, Thorlabs) at 300 lm (Sper Scientific 840022 Advanced Light Meter) for 6 min. After fabrication, the chip is stored in gel buffer solution with 0.1% SDS and 0.1% Triton X-100 until use. In the case of the DHEBA gel, the acrylamide/bis(acrylamide) solution is replaced with acrylamide monomer and an N,N′-(1,2-dihydroxyethylene)bis(acrylamide) (DHEBA) cross-linker (No. 294381, Sigma–Aldrich) at a molar ratio equivalent to the 37.5:1 bis/acrylamide for a final concentration of 12%T, 3.5%C acrylamide/DHEBA.
Sample and Antibody Preparation
A protein ladder is used to optimize the assay, with the ladder consisting of Alexa Fluor 488 conjugated proteins: β-galactosidase (116 kDa) (No. G8511, Sigma–Aldrich) (labeled in-house), bovine serum albumin (BSA) (66 kDa) (No. A13100, Life Technologies, prelabeled), ovalbumin (OVA) (45 kDa) (No. 34781, Life Technologies, prelabeled), c-reactive protein (CRP) (25 kDa) (labeled in-house), and aprotinin (AP) (6.5 kDa) (No. sc-3595, Santa Cruz Biotechnology) (labeled in-house). Proteins are labeled using an Alexa Fluor 488 Protein Labeling Kit (No. A10235, Life Technologies), following the package protocol.
Prior to Western blotting, the sample is prepared in a buffer consisting of 2% SDS and 100 mM of the reducing agent dithiothreitol (DTT) then heated for 3 min at 90 °C. Finally, 500 mM Tris HCl pH 6.8 is added to bring the sample to 50 mM Tris HCl pH 6.8 prior to analysis. Target sample consists of purified prostate specific antigen (PSA) (No. 539834, Calbiochem) at either 300 nM or 600 nM concentration, as indicated in the text, and probed using a polyclonal PSA antibody (No. AF1344, Fisher Scientific). Antibodies are labeled using Alexa Fluor 568 (No. A10238, Life Technologies), following the package protocol.
Sample Separation
Voltage is applied to the chip using platinum electrodes attached to a custom-built, eight-channel high-voltage power supply with current/voltage feedback control. To load sample onto the chip, 2.3 μL of sample is pipetted into a well and electrophoresed into the channel at 1.5 μA (∼11 V) (or 1.0 μA, ∼11 V for Tris glycine) for 80 s. The well is then washed out with the terminating electrolyte run buffer consisting of 0.1% Triton X-100, 0.1% SDS, 3% DMSO with either 1X Tris glycine (25 mM Tris, 192 mM glycine Bio-Rad 161-0734) or 1X Tris tricine (100 mM Tris, 100 mM tricine, No. T1165, Sigma–Aldrich) and a fixed current applied across the channel to stack the injected plug via transient isotachophoresis (ITP) and then size the sample species. Sizing uses a fixed current of 1.5 μA for Tris tricine and 1.0 μA for Tris glycine systems, both of which result in a voltage ramp of ∼25–55 V/cm during separation. For Tris glycine separations, 0.3 μA is applied during stacking (∼4–8 V/cm ramp), followed by 1 μA for separation once the proteins enter the gel, as this yields slightly improved stacking and separation performance compared to applying a continuous current of 1 μA. For the Tris tricine system, a 1.5 μA current is applied during both the stacking (∼12–25 V/cm ramp) and sizing phases.
Protein Blotting
Protein capture on the photoactive gel is performed using a Hamamatsu Lightening Cure LC5 UV source through a light guide, with the gel exposed for 30–45 s at 100% intensity. After photocapture, unconjugated proteins are electrophoresed out of the channels by applying a reverse voltage for 10 min at 100 V/cm. During this step, both wells are filled with Tris tricine SDS buffer. The Tris tricine SDS buffer is then replaced with Tris glycine buffer (no SDS) in both wells for an additional 10 min washout at 100 V/cm in the same direction.
Antibody Probing
Following the second wash step, electrophoresis is used to drive probe antibodies (500 nM in Tris glycine (no SDS)) through the protein decorated gel. Two antibody probing schemes are utilized: a plug of antibody (a “top hat” concentration distribution) and a front of antibody (a “step function” concentration distribution). In the antibody plug scheme, the concentration distribution is defined by first electrophoretically loading an antibody concentration front into the channel (7 min at 200 V/cm) from one well. To create the plug, the electric field is set to zero and the loading well is thoroughly washed with Tris glycine buffer via gentle aspiration. After the well is devoid of antibodies, the electric field is reapplied (200 V/cm), defining the back of the antibody plug, which is then allowed to migrate along the separation axis. In the continual antibody loading scheme, antibodies are electrophoretically loaded (200 V/cm) along the separation axis until the axial signal is uniform (∼54 min). Both wells are then thoroughly washed with Tris glycine buffer via gentle aspiration and a reverse polarity voltage is applied to electromigrate unbound antibodies out of the channel.
For both probing schemes, fluorescence images are collected every 2 min using automated time lapse imaging controlled via Metamorph. Monitoring allows determination of the wash time that yields a maximum signal-to-noise ratio (SNR).
Imaging and Image Analysis
Chips are imaged on an inverted epi-fluorescence microscope (Olympus IX-50) using a 10× objective (Olympus UPlanFLN, NA 0.3) with CCD camera (CoolSNAP HQ2, Photometrics), filter cubes (XF102-2 and XF100-3 (Omega Optical, Brattleboro, VT), and automated x–y stage.
Background values are calculated by taking the raw image values and subtracting the autofluorescence signal from regions adjacent to the channel of interest. Prior to antibody loading, the fluorescence intensity in the channel is approximately equal to the surrounding glass, so channel intensity higher than the surrounding glass is likely due to nonspecific antibody adsorption in the channel. To estimate the standard deviation of the background signal in the channel, we measure the signal standard deviation in two 50-pixel (192 μm) regions. The location of the two regions is more than one peak width away from the probe signal in both channel directions.
The separation resolution (SR) is defined as the distance between peak maxima divided by 4 times the average standard deviation of two neighboring ladder protein concentration distributions, as per convention. The concentration distribution metrics are calculated via least-squares fitting to assumed Gaussian distributions (MATLAB).
Results and Discussion
Conducted in a single enclosed microchannel, the μWestern (Figure 1) is a multistage assay comprised of polyacrylamide gel electrophoresis (PAGE), protein blotting via photocapture, and probing with antibodies. The present study addresses three central performance considerations necessary to optimize a single-channel microfluidic Western blotting assay for low-molecular-mass proteins.
Figure 1.

Low-molecular-mass μWestern. Conducted in an enclosed microchannel filled with photoactive PA gel, the assay is comprised of three steps: (1) protein sizing after transient isotachophoresis, (2) immobilization of proteins on gel via UV photocapture (blotting), and (3) in situ antibody probing via electrophoresis. Optimization for low-molecular-mass species focuses on the separation and probing stages (bold labels). The applied electric potential is indicated by plus (+) and minus (−) symbols. Arrows indicate the direction of species electromigration under the conditions used.
First, to expand the molecular mass applicability of in-channel microfluidic Western blotting to include low-molecular-mass species, we implement a discontinuous Tris tricine buffer system. Originally developed by Schägger and Von Jagow,23 the Tris tricine system offers improved destacking of small-molecular-weight species in a given pore-size PAGE gel, as compared to the Laemmli Tris glycine system.24 The basis for the extended molecular mass range of the Tris tricine system stems from the higher electrophoretic mobility of the tricine terminating electrolyte, as compared to the lower mobility of the commonly used glycine terminating electrolyte.
Second, to mitigate the reduced effective PA gel pore size often observed at a gel/buffer or gel/gel interface, we introduce a new gel fabrication method. Importantly, careful control of the interfacial gel pore size is essential to avoid the accumulation of material (i.e., antibody probe or sample) at the interface. We also consider an alternative hydrophilic cross-linker (DHEBA) that offers flexibility in forming and cleaving cross-links. To assess the performance across these systems, we utilize immunoprobing of PSA, a 28 kDa protein,25 which is important to prostate cancer screening diagnostics.
Third, to reduce background signal on the gel, unwanted accumulation of antibody material at the gel interface, and total assay duration in the antibody probing step, we implement antibody probing, using a swept antibody plug (along the separation axis), compared to earlier approaches that relied on continuous introduction of a concentration front.
Separation Step: ITP for Low-Molecular-Mass Proteins
Prior to PAGE, transient isotachophoresis affords both sample enrichment and low injection dispersion.26 A discontinuous buffer system establishes the ITP protein stack. A discontinuous pore-size gel (step change from large to small pore-size) transitions the assay from ITP to PAGE. As the ITP sample stack enters the separation gel, proteins slow, relative to the terminating electrolyte; thus, proteins destack and separate in the molecular sieving gel.26 In the separation gel, any proteins small enough to migrate faster than the terminating electrolyte will remain stacked, which can prevent effective separation of small-molecular-weight species, particularly in large-pore-size (low %T) gels.
As demonstrated by Schägger and Von Jagow,23 a Tris tricine terminating electrolyte ITP system allows effective destacking of smaller-molecular-weight proteins in a given pore-size gel. The isoelectric point of tricine is lower than that of glycine, resulting in tricine having a higher mobility for a given gel pH. Proteins that electromigrate faster than the terminating electrolyte will remain stacked. Consequently, a higher-mobility terminating electrolyte enables destacking of smaller-molecular-weight species in a gel with a given pore size. In this system, we seek to optimize PAGE for a protein ladder spanning a mass range of 6.5–116 kDa. Previous work with our in-channel μWestern performed well across a 21–116 kDa mass range in a 7.5%T, 2.7%C acrylamide/bis(acrylamide) gel and utilized the conventional Tris glycine trailing electrolyte.19
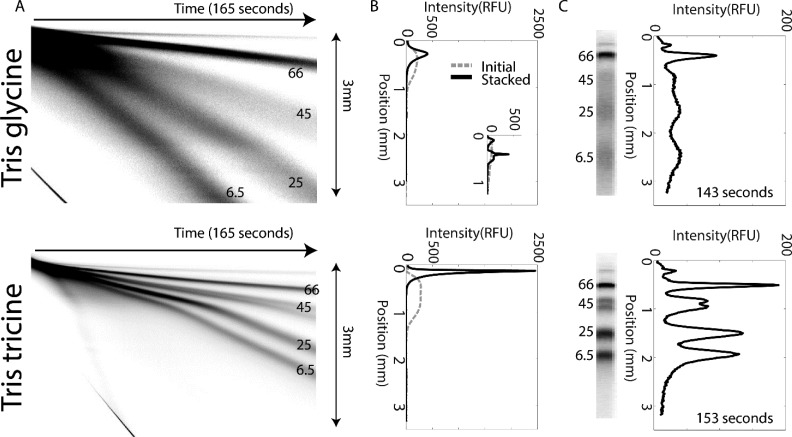
We first compare the stacking and separation performance of the Tris glycine and Tris tricine discontinuous buffer systems, both with Tris HCl as leading electrolyte (Figure 2A). ITP stacking is conducted in an open channel (free solution) region abutting a 12%T discontinuous gel for PAGE. For the conventional Tris glycine system, we observe a 440 μm (CV = 1.6%, n = 3) ITP stack (Figure 2B) in free solution and a 200 μm (CV = 3.4%, n = 3) stack at the gel interface. The Tris tricine system presents an ITP stack of 70 μm (CV = 9.1%, n = 3) in free solution and a 50 μm stack at the gel interface (CV = 7.8%, n = 3). For Tris glycine, the injected plug is stacked at a ratio of 3.6, based on comparison of full width at half-maximum (fwhm) for the loaded sample plug width to the minimum stacked fwhm (CV = 2.6%, n = 3). In contrast, the Tris tricine system yields a stacking ratio of 17.8 (CV = 5.6%, n = 3). In the case of the Tris glycine system, a minimum free solution stack width is achieved within 35 s in the first 1.1 mm of the channel. With the Tris tricine system, the free solution stack width continues to decrease as the sample migrates toward the gel interface. With a gel interface located 4.4 mm from the well inlet, a minimum free solution stack width is achieved in 100 s and at a location just before entering the gel.
Figure 2.
Optimization of discontinuous buffer system for low-molecular-mass PAGE. (A) PAGE kymograph of Tris glycine (top) and Tris tricine (bottom) discontinuous buffer systems in a 12%T discontinuous gel; PAGE is operated under a fixed current of 1.5 μA for Tris tricine and 1 μA for Tris glycine, yielding a voltage ramp of ∼25–55 V/cm during each separation. (B) ITP sample stacking intensity profiles for protein ladder stack in open-channel regions for both the Tris glycine (upper) and Tris tricine (lower) systems at an initial sample loading and minimum sample width. During stacking, a 1.5 μA fixed current is applied for Tris tricine (∼12–25 V ramp) and a 0.3 μA fixed current (∼4–8 V ramp) for Tris glycine (as lower current yielded better stacking). Inset shows ITP stacking in a 4%T stacking gel for the Tris glycine system, added to reduce putative EOF-induced dispersion. (C) Inverted fluorescence micrographs and corresponding intensity profiles of sizing in the Tris glycine (top, open-channel loading, no 4%T gel) and Tris tricine (bottom) systems. In both cases, the 25 kDa ladder protein is observed at the 1.5 mm separation distance position.
We hypothesize that the larger stack width observed in the Tris glycine system may be due to the greater difference in conductivity between Tris glycine and the leading electrolyte, Tris HCl, as compared to the Tris tricine system. The buffer conductivities are measured as 1.3 mS/cm for the Tris tricine run buffer, 0.47 mS/cm for the Tris glycine run buffer, 5.1 mS/cm for the 500 mM Tris HCl pH 8.45 gel buffer in the Tris tricine system, and 3.1 mS/cm for the 375 mM Tris HCl pH 8.8 gel buffer in the Tris glycine system. We hypothesize that the enhanced conductivity difference measured for the Tris glycine system, as compared to the Tris tricine system, may result in a substantial mismatch in electro-osmotic flow (EOF) between the leading and terminating electrolyte. Consequently, EOF generated in the open-channel region during ITP may be contributing dispersion to the Tris glycine system.27
To test the EOF dispersion hypothesis, we replace the open channel region with a 4%T stacking gel. The presence of even a large pore-size gel should reduce EOF and any associated dispersion. In the modified Tris glycine system (Figure 2B), we observe an ITP stack width of 31 μm (CV = 13.1%, n = 3). The ratio of the change in fwhm (from injected sample plug to the stacked plug) for OVA and CRP (BSA destacks in stacking gel) is 23.2 (CV = 9.7%, n = 3) with Tris glycine and 4%T loading gel, notably higher than the factor of 5.2 stacking previously observed with the Tris glycine and no loading gel (i.e., free solution) system for the same proteins. The Tris glycine system with stacking gel offers stacking similar to that observed in the Tris tricine system (factor of 27.9 stacking for BSA, OVA, CRP) in free solution. These observations suggest that EOF-induced dispersion in the open channel/PA gel Tris glycine system may reduce the ITP stacking capability.
Separation Step: PAGE for Low-Molecular-Mass Proteins
We next compare the PAGE performance of the Tris tricine to the conventional Tris glycine system (Figure 2C). We first observe a total separation time of 153 s for Tris tricine and 143 s for Tris glycine. PAGE assay completion is defined as the time from when the sample first enters the separation gel to the arrival of the 25 kDa ladder protein at the 1.5 mm position on the separation axis. Second, the SNR for the Tris glycine system is notably lower than that of the Tris tricine system, as expected given the transient ITP behaviors described in the previous section. We attribute the slightly lower protein peak area under the curves (AUCs) for the Tris glycine system (as compared to the Tris tricine system) to ∼25% less sample material loaded in the Tris glycine system.
Lastly, the Tris tricine system is observed to provide ∼40% more SR than the Tris glycine system, under otherwise similar conditions. The SR between the two lowest-molecular-mass species (CRP and AP) is SRCRP–AP = 0.66 (CV = 2.3%, n = 3) for Tris glycine and SRCRP–AP = 0.91 (CV = 3.0%, n = 3) for Tris tricine. Considering the larger proteins, SRBSA-OVA = 0.78 for Tris glycine (CV = 2.4%, n = 3) and SRBSA-OVA = 1.04 for Tris tricine (CV = 3.3%, n = 3). Whereas SROVA-CRP = 0.63 (CV = 0.9%, n = 3) for Tris glycine and SROVA-CRP = 1.2 (CV = 3.0%, n = 3) for Tris tricine. In summary, our observations suggest that both the greater sample preconcentration and lower injection dispersion attained with the Tris tricine system affords higher PAGE separation performance for the molecular mass range considered here.
Blotting: Protein Immobilization Efficiency
In lieu of physical sample transfer from a gel to a hydrophobic blotting membrane, the μWestern uses UV photocapture of proteins in the channel via a benzophenone-functionalized polyacrylamide gel.19 While protein is not physically transferred per se, mass is indeed immobilized: this is the critical aspect of the blotting step. For a UV exposure of 45 s in the Tris tricine system, we observe a BSA capture efficiency (ηBSA) of 65.7% (CV = 8.1%, n = 3). For the Tris glycine system, we observe ηBSA = 51.6% (CV = 6.8%, n = 3). Capture efficiencies for photoimmobilization in the Tris tricine system are observed to be species-dependent, as reported previously:22 ηOVA = 48.9% (CV = 4.8%, n = 3), ηCRP = 35.3% (CV = 11.5%, n = 3), and ηAP = 63.0% (CV = 14.1%, n = 3). Raw data and intensity profiles used to calculate these values are shown in Figure S-1 in the Supporting Information.
Antibody Probing: Discontinuous Gel Interface
As the μWestern is completed in a single enclosed microchannel, introduction of probing antibodies makes use of directed electromigration of antibody down the separation axis. An implication of this scheme is that the large probe antibody electromigrates through the molecular sieving gel (which, here, has been optimized for a wide molecular mass range separation that, importantly, includes low-molecular-mass species). To evaluate the impact of probe introduction into the 12%T sieving gel after sizing, we utilize a fluorescently labeled polyclonal antibody against prostate specific antigen (PSA) (see Figure 3A). A plug of antibody is loaded into the channel for 7 min and then electromigrated along the separation axis (both at 200 V/cm).
Figure 3.

A larger pore-size gradient at the open-channel/gel interface reduces unwanted size-exclusion effects during probing. (A) Inverted fluorescence micrographs show antibody probing across a gel with smaller pore sizes at the interface19 (left) and for a gel with a gradient to larger pore sizes at the interface (right), both with 12%T gels utilizing DHEBA cross-linker and 600 nM purified PSA sample. Gel interface is marked with black arrow; expected location of the PSA major isoform is indicated with an asterisk (*). (B) Inverted fluorescence kymographs of a 116–6.5 kDa ladder separation in an 8%T (top) and 12%T gel (bottom) with a Tris tricine discontinuous buffer. Right panel shows the ladder when the 25 kDa marker is 1.5 mm into the gel. In the 8%T gel, the small 6.5 kDa marker migrates faster than the stack and so rejoins the stack a short distance into the gel. 12%T enables destacking and separation of full 116–6.5 kDa ladder. (C) Schematic depicting fabrication protocol yielding a short larger-than-bulk to bulk pore-size gradient at the separation gel interface.
Using this directed electromigration of probe to immobilized antigen, we observe interfering antibody accumulation near the open-channel/12%T gel interface, when using the original fabrication protocol.19 The background signal near the gel/free solution interface is higher than the probe signal at the immobilized PSA, resulting in an SNR of <3, even with a sample containing 600 nM of PSA. High antibody background at the interface occurs regardless of directionality of antibody loading relative to the interface. In contrast, a fabrication method enabling the generation of larger pore sizes at the interface relative to the bulk of the gel generates a clear antiPSA probe signal for the 28 kDa primary isoform. We also observe additional minor peaks at a lower and higher MW. We attribute the smaller MW peak to known biological cleavage PSA isoforms28−31 and/or sample degradation and the larger MW peak to proPSA28−30 and/or PSA aggregates (Figure 3A). Validation of the μWestern was completed via conventional Western blot. We observed signal at both larger and smaller MW positions, relative to the 28 kDa PSA peak position (see Figure S-2 in the Supporting Information), consistent with the μWestern blot results for the same sample. The lower-molecular-mass PSA cleavage isoforms present as a single peak in the μWestern, because of the short separation distance used compared to conventional slab gel (2 mm vs 8 cm).
The preferential accumulation of antibody probe material at the open-channel/gel interface is attributed primarily to size exclusion at the interface. The size exclusion is exacerbated by the difficulty in controlling the pore size of the gel at an open channel interface.32,33 Briefly, monomer and cross-linker from unpolymerized regions of the microfluidic channel diffuse into the polymerizing region during fabrication, thus establishing smaller pore sizes at the gel/free solution interface, relative to the pore size in the bulk of the gel.32,33 The small pore size occurs when either photomasking or oxygen inhibition from the wells is used to create the gel interface. Effective preconcentration and separation requires a region of free solution (or much larger pore sizes) adjacent to a separation gel to enable good sample stacking and destacking. The smaller effective pore size at the open-channel/gel interface presents an issue for probing, particularly for higher %T gels, because it is this accumulation of high antibody background at the interface that obscures the probe signal from the antigen, greatly reducing assay sensitivity. Simply utilizing a lower %T gel does not allow for destacking of smaller-molecular-weight species, thus limiting the assay to larger proteins. As shown in Figure 3B, an 8%T gel results in ineffective destacking of the small molecular mass protein aprotinin (7 kDa with label), with aprotinin migrating faster than the stack, thus rejoining the stack a short distance into the gel. The lower %T gel also results in lower SR between the other destacked ladder proteins at a given distance into the gel.
To overcome antibody probe size exclusion at the open-channel/gel interface and enable immunoprobing even in high %T gels, we seek to eliminate the interfacial small pore-size artifact by establishing a short gradient of larger-than-bulk to bulk pore-sizes at the interface. We hypothesized that eliminating the unpolymerized gel precursor abutting the polymerizing region would inherently eliminate the source of additional monomer and cross-linker that yield smaller pore sizes at the interface than the bulk of the gel. To achieve the configuration prior to blue light photopolymerization, the channel is first filled with gel precursor solution from a well (Figure 3C). Next, gel precursor solution is removed from the well, thus leaving two empty wells and a channel filled with gel precursor solution. One well is then filled with buffer solution (3 μL). Because of the hydrostatic pressure mismatch generated between the wells, the buffer then flows into the channel. After a fixed duration (ca. 15 s), the entire chip is submerged in buffer, thus eliminating the hydrostatic pressure head and the resultant flow in the channel. The entire chip is illuminated with a blue LED for 6 min to photopolymerize the gel. The duration between buffer loading into the well and subsequent submersion of the chip in buffer establishes the location of the interface gradient in the channel. The time interval between establishing multifluid configuration in the channel and photopolymerization establishes the pore-size gradient characteristics (i.e., pore-size gradient length and steepness), because of diffusion of the gel precursor into the free solution region. Longer durations between chip submersion and photopolymerization would lead to a longer region of the larger pore size gradient at the interface. For the examples here, the chip is illuminated with blue light immediately (<1 min) after submerging to minimize diffusion and generate a relatively sharp interface. As shown in Figure 2, this fabrication method enables excellent stacking and separation performance when using the Tris tricine discontinuous buffer system.
Using the fabrication technique, we observe an interface located ∼4.4 mm from the edge of the input well for an equilibration time of 15 s (i.e., duration between buffer loading into the well and subsequent submersion of the chip in buffer). Variation in the position of the gel interface between triplicate channels sharing a well is 1.1% (n = 4 triplicates) and the CV of the gel interface position between lanes not sharing a well on a chip is 4.8% (n = 4 triplicates). By observing the location of the buffer with a fluorescent dye tracer included, the position of the fluid/fluid interface within the channel is observed to be linearly dependent on time (R2 = 0.999). The fluid interface migrates at the same velocity (<1% difference), regardless of whether the well is filled with buffer or gel precursor, indicating that the position is dictated by hydraulic pressure alone and differences in viscosity and surface tension are negligible between the buffer and the gel precursor. Furthermore, we note that fabrication of a stacking gel by one-step UV exposure is also possible using this approach, simply by loading a 4%T, 2.7%C gel precursor solution into the well instead of buffer solution.
This fabrication method yields the antibody probe performance shown on the right-hand side of Figure 3A, which provides a clear antiPSA probe signal for the PSA isoforms. For both Figure 3A results, the use of a DHEBA cross-linker, in place of bis(acrylamide), is utilized as a proof of concept. DHEBA is a more hydrophilic cross-linker that is cleavable in highly acidic or basic conditions.34,35 Further development with DHEBA may enable greater design flexibility in the μWestern blot system by allowing for an increase in pore size between the separation and probe stages of the assay.
Antibody Probing: Swept Plug Scheme
Our broad molecular mass range μWestern19 utilizes electromigration to drive a front of primary antibodies into and then along the entire separation axis to achieve probing. In practice, this front introduction method is applied for 20 min to fill the channel with antibodies, followed by a 20 min antibody washout in the reverse direction in a 7.5%T, 2.7%C gel. We seek to establish and validate an alternative transport approach for probing to (i) reduce assay time and complexity, (ii) provide uniform antibody incubation times across a gel, (iii) enable a consistent protocol regardless of gel length or pore size, and (iv) reduce nonspecific antibody background in the gel, which increases substantially with increasing %T gels, when using an antibody front probe approach.19 To achieve these goals, we investigate electrophoretic introduction of a well-defined plug of 500 nM antibody into the gel, then along the entire separation axis (7 min, E = 200 V/cm) (see Figure 4A).
Figure 4.

Antibody probing scheme impacts background signal. (A) Inverted fluorescence micrographs for electrophoresis of antibody probe via swept plug introduction (top) and continuous front loading (bottom). Loading images use an exposure time of 50 ms; washout images use an exposure time of 300 ms. The antibody loading concentration is 500 nM; E = 200 V/cm. (B) Inverted fluorescence micrograph showing the protein ladder and final probe results for PSA for each method. The PSA primary isoform is indicated with an asterisk (*).
Selection of the 7 min antibody loading time for the plug scheme is informed by results of a kinetic model, indicating that >94% of immobilized antigen would be bound assuming kon = 2 × 104 M–1 s–1 and koff = 5 × 10–4 s–1 for an antiPSA mAb.36 For the configuration under study, the antibody plug requires ∼22 min to traverse a 4.5 mm long 12%T, 2.7%C gel with (E = 200 V/cm). We observe maximum SNR after an additional 50 min of antibody washout via electrophoresis of clear buffer into the channel (E =200 V/cm).
During antibody probing, we observed both the major antibody probe peak and two low-concentration, large-molecular-mass peaks electromigrating along the microchannel axis. Since the low-concentration peaks continually migrate during the time lapse imaging, we hypothesize that the peaks are contamination in the stock antibody probe solution (e.g., aggregation37). As the spurious peaks are continuously mobile, whereas the probe antibody is immobile at the target peak location, mobility is used to differentiate the signal sources. We suggest a filtration step to remove aggregates from the stock antibody solution, which would reduce assay time by minimizing the probe washout duration.
With a sweep of antibody plug along the separation axis, we observe an SNR = 263 (CV = 11.4%, n = 3) and average background intensity in the gel of 24 RFU (CV = 3.2%, n = 3). In comparison, continual antibody loading yields an SNR of 131 (CV = 18.8%, n = 3) and a more than 2-fold higher average antibody background intensity in the gel of 54 RFU (CV = 16.3%, n = 3) for the same sample, which contains 300 nM of PSA (Figure 4B). Thus, the swept antibody plug technique results in higher SNR due to a lower background and lower standard deviation of the background, compared to continually loading antibodies into the gel.
We further note that the swept antibody plug scheme offers uniformity in antibody incubation time with target for all targets, regardless of molecular mass. In other words and in contrast to a continuous antibody front introduction approach, high-molecular-mass proteins immobilized near the head of the separation channel will be incubated with the antibody probe for the same duration as small proteins immobilized near the end of the separation axis. Varying the loading time of the swept plug would allow optimization for different antibody–antigen binding kinetics by increasing or decreasing the incubation time.
The μWestern channel design also supports optimization to meet a desired separation resolution and/or a desired assay time. For example, using a longer separation length would enable improved separation resolution, with the tradeoff of a longer assay time, since the antibodies must traverse the full separation length for probing. Conversely, using a gel length even shorter than the 5-mm gel presented here would reduce the total assay time.
Conclusions
To advance μWestern blotting to detection of low molecular mass species, we report on optimization of the assay by: (i) Tris tricine discontinuous buffer, (ii) fabrication to generate larger interfacial pores, and (iii) antibody plug probe. The Tris tricine discontinuous buffer system enables improved separation resolution and the destacking of smaller-molecular-weight species for a given pore size gel. Control over gel interfacial pore size through fabrication optimization is critical for enabling both good stacking for high separation resolution as well as good antibody transport for high SNR probing. Furthermore, utilization of an antibody plug sweep probe approach reduces antibody consumption while improving assay time and SNR. Through continued future development with more hydrophilic and cleavable cross-linkers such as DHEBA, with the effective separation and probing initially demonstrated here, we plan to further push the bounds of sample molecular weight, sensitivity, and assay speed. The approaches presented here expand the applicability of the μWestern to a wider variety of diagnostic and basic biological research applications.
Acknowledgments
This work was supported by an NIH New Innovator Award (No. DP2OD007294, to A.E.H.). A.E.H. is an Alfred P. Sloan Foundation Research Fellow in Chemistry. R.E.G. was supported with a National Science Foundation Graduate Research Fellowship. Elisabet Rosas assisted with running the traditional Western blot slab gel shown in the Supporting Information.
Supporting Information Available
Additional information is available as noted in the text. This material is available free of charge via the Internet at http://pubs.acs.org/.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Soundy P.; Harvey B. In Medical Biomethods Handbook; Walker J. M., Rapley R., Eds.; Humana Press: Totowa, NJ, 2005; pp 43–62. [Google Scholar]
- Allain J.-P.; Paul D.; Laurian Y.; Senn D. Members of the AIDS–Haemophilia French Study Group. Lancet 1986, 328, 1233–1236.2878129 [Google Scholar]
- Shayesteh L.; Lu Y.; Kuo W.-L.; Baldocchi R.; Godfrey T.; Collins C.; Pinkel D.; Powell B.; Mills G. B.; Gray J. W. Nat. Genet. 1999, 21, 99–102. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S.; Huh W.-K.; Bower K.; Howson R. W.; Belle A.; Dephoure N.; O’Shea E. K.; Weissman J. S. Nature 2003, 425, 737–741. [DOI] [PubMed] [Google Scholar]
- Dalmau J.; Furneaux H. M.; Gralla R. J.; Kris M. G.; Posner J. B. Ann. Neurol. 1990, 27, 544–552. [DOI] [PubMed] [Google Scholar]
- Ida N.; Hartmann T.; Pantel J.; Schröder J.; Zerfass R.; Förstl H.; Sandbrink R.; Masters C. L.; Beyreuther K. J. Biol. Chem. 1996, 271, 22908–22914. [DOI] [PubMed] [Google Scholar]
- Holmes D. L.; Stellwagen N. C. Electrophoresis 1991, 12, 612–619. [DOI] [PubMed] [Google Scholar]
- MacPhee D. J. J. Pharmacol. Toxicol. Methods 2010, 61, 171–177. [DOI] [PubMed] [Google Scholar]
- Lin W.; Kasamatsu H. Anal. Biochem. 1983, 128, 302–311. [DOI] [PubMed] [Google Scholar]
- Anderson G. J.; M. Cipolla C.; Kennedy R. T. Anal. Chem. 2011, 83, 1350–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S.; Anderson G. J.; Kennedy R. T. Anal. Chem. 2013, 85, 6073–6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaccio M. F.; Wagner J. P.; Chuu C.-P.; Lauffenburger D. A.; Jones R. B. Nat. Methods 2010, 7, 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill R. A.; Bhamidipati A.; Bi X.; Deb-Basu D.; Cahill L.; Ferrante J.; Gentalen E.; Glazer M.; Gossett J.; Hacker K.; Kirby C.; Knittle J.; Loder R.; Mastroieni C.; MacLaren M.; Mills T.; Nguyen U.; Parker N.; Rice A.; Roach D.; Suich D.; Voehringer D.; Voss K.; Yang J.; Yang T.; Horn P. B. V. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 16153–16158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan A. C.; Deb-Basu D.; Orban M. W.; Gotlib J. R.; Natkunam Y.; O’Neill R.; Padua R.-A.; Xu L.; Taketa D.; Shirer A. E.; Beer S.; Yee A. X.; Voehringer D. W.; Felsher D. W. Nat. Med. 2009, 15, 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.; Karns K.; Tia S. Q.; He M.; Herr A. E. Anal. Chem. 2012, 84, 2533–2540. [DOI] [PubMed] [Google Scholar]
- Hou C.; Herr A. E. Analyst 2013, 138, 158. [DOI] [PubMed] [Google Scholar]
- He M.; Herr A. E. Anal. Chem. 2009, 81, 8177–8184. [DOI] [PubMed] [Google Scholar]
- Tia S. Q.; He M.; Kim D.; Herr A. E. Anal. Chem. 2011, 83, 3581–3588. [DOI] [PubMed] [Google Scholar]
- Hughes A. J.; Herr A. E. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 21450–21455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousse L.; Mouradian S.; Minalla A.; Yee H.; Williams K.; Dubrow R. Anal. Chem. 2001, 73, 1207–1212. [DOI] [PubMed] [Google Scholar]
- Apori A. A.; Herr A. E. Anal. Chem. 2011, 83, 2691–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A. J.; Lin R. K. C.; Peehl D. M.; Herr A. E. Proc. Natl. Acad. Sci. U.S.A. 2012, 109, 5972–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H.; von Jagow G. Anal. Biochem. 1987, 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Nature 1970, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Bélanger A.; van Halbeek H.; Graves H. C.; Grandbois K.; Stamey T. A.; Huang L.; Poppe I.; Labrie F. Prostate 1995, 27, 187–197. [DOI] [PubMed] [Google Scholar]
- Ornstein L. Ann. N.Y. Acad. Sci. 1964, 121, 321–349. [DOI] [PubMed] [Google Scholar]
- Burgi D. S.; Chien R. L. Anal. Chem. 1991, 63, 2042–2047. [Google Scholar]
- Jung K.; Reiche J.; Boehme A.; Stephan C.; Loening S. A.; Schnorr D.; Hoesel W.; Sinha P. Clin. Chem. 2004, 50, 2292–2301. [DOI] [PubMed] [Google Scholar]
- Balk S. P.; Ko Y.-J.; Bubley G. J. J. Clin. Oncol. 2003, 21, 383–391. [DOI] [PubMed] [Google Scholar]
- Tabarés G.; Jung K.; Reiche J.; Stephan C.; Lein M.; Peracaula R.; de Llorens R.; Hoesel W. Clin. Biochem. 2007, 40, 343–350. [DOI] [PubMed] [Google Scholar]
- Wang T. J.; Linton H. J.; Sokoloff R. L.; Grauer L. S.; Rittenhouse H. G.; Wolfert R. L. Tumor Biol. 1999, 20, 79–85. [DOI] [PubMed] [Google Scholar]
- Herr A. E.; Throckmorton D. J.; Davenport A. A.; Singh A. K. Anal. Chem. 2005, 77, 585–590. [DOI] [PubMed] [Google Scholar]
- Hou C.; Herr A. E. Anal. Chem. 2010, 82, 3343–3351. [DOI] [PubMed] [Google Scholar]
- O’Connell P. B. H.; Brady C. J. Anal. Biochem. 1976, 76, 63–73. [DOI] [PubMed] [Google Scholar]
- Gelfi C.; Alloni A.; de Besi P.; Righetti P. G. J. Chromatogr. A 1992, 608, 343–348. [Google Scholar]
- Kapil M. A.; Herr A. E. Anal. Chem. 2014, 86, 2601–2609. [DOI] [PubMed] [Google Scholar]
- Mahler H.-C.; Friess W.; Grauschopf U.; Kiese S. J. Pharm. Sci. 2009, 98, 2909–2934. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.