Abstract
Aptamer ligands for myelin basic protein (MBP) were obtained using the systematic evolution of ligand by exponential enrichment (SELEX) method. Two clones were isolated from a pool of oligonucleotides and tested for MBP targeting. Using purified MBP, we demonstrated the binding activity of the aptamers and we also showed the affinity of MBP for oligonucleotides of specific length. Moreover, one selected aptamer competitively inhibited the binding of an MBP-specific antibody to MBP and the aptamer was found more sensitive than a commercial antibody. In addition, we showed the ability of the aptamer to detect myelin-rich regions in paraffin-embedded mouse brain tissue. Therefore, the MBP-binding activity of the selected oligonucleotide may prove useful as a tool for life science and medical research for myelin detection and might be a good lead for testing it in autoimmune diseases such as multiple sclerosis.
Keywords: aptamers, histochemistry, multiple sclerosis, myelin basic protein
Introduction
The discovery of new chemical entities for medical research, diagnostics or basic research is being obtained by the screening of large chemical libraries. These libraries are composed of related/unrelated molecules and constitute a potential source of new leads for the medical field. A relatively new type of chemical libraries includes the aptamers. Aptamers are short single-stranded nucletodic molecules (ssDNA or ssARN) with the ability to fold in a stable three-dimensional structure that allows them to interact sterically and through electrostatic interactions with a target molecule. In general, they are 15–80 nucleotides long. Aptamers are identified from synthetic ssDNA or ssRNA libraries through a process termed SELEX (Systematic Evolution of Ligands by EXponential enrichment).1 They have been selected to bind a plethora of targets including small molecules, peptides, proteins, and cells.2 So far, the accumulated data on aptamer functionality have demonstrated that antibodies are no longer the only entities that may be developed to bind to targets with high affinity and specificity.
Although aptamer technology is not robust yet, continued efforts are being made toward its consolidation given its numerous advantages over current technologies.3 The application of aptamer technology for the detection of myelin basic protein (MBP) has not fully been exploited and a MBP-specific aptamer could lead to a potential therapeutic drug for multiple sclerosis (MS) or a novel reagent for myelin detection.
MBP is the most widely studied myelin protein in MS and the second most abundant myelin protein after the proteolipid protein4 that constitutes the myelin sheath, which is necessary for the normal activity of the vertebrate nervous system. MBP has various isoforms generated by alternative splicing, which include the classical 18.5 and 21.5 kDa isoforms. These proteins are highly basic; however, various post-translational modifications can generate different charge isomers.5 MBP is considered to be an intrinsically unstructured protein and its conformation is defined by its environment or upon association with a ligand.6 While the presence of disordered sequences makes it difficult to solve the protein crystal structure, this characteristic and the basicity of the protein render it very attractive for aptamer-targeting.
Much evidence suggests that MBP may be an autoantigen candidate in MS.7 In particular, MBP-specific T cells have been isolated from MS patients and healthy individuals.8,9,10,11,12,13,14 Hence, MBP seems to play an important role in MS progression and could therefore be an interesting drug target. Published data suggest that in MS patients, autoantibodies recognize MBP and recruit inflammatory cells to focal areas, thereby targeting the central nervous system myelin components and affecting its stability. Moreover, T cells in MS patients are able to recognize MBP peptides exposed at the surface of antigen-presenting cells, which triggers an immune response against MBP.
We hypothesized that a molecule able to protect MBP from autoantibodies may be a good neuroprotective agent. Additionally, life science researchers and clinicians may benefit from the identification of an aptamer able to detect MBP in tissue sections. This would indeed provide the possibility of a one-step staining of myelin under physiological conditions or permit the detection of myelin-rich regions using different types of dyes.
The present study reports the selection process of an aptamer specific for MBP. We demonstrate its ability to detect MBP in different in vitro assays and mouse brain tissue sections as well as its capacity to compete with a specific anti-MBP antibody. We also discuss the potential use of this molecule as a therapeutic agent, diagnostic agent and research tool.
Results
Aptamer selection
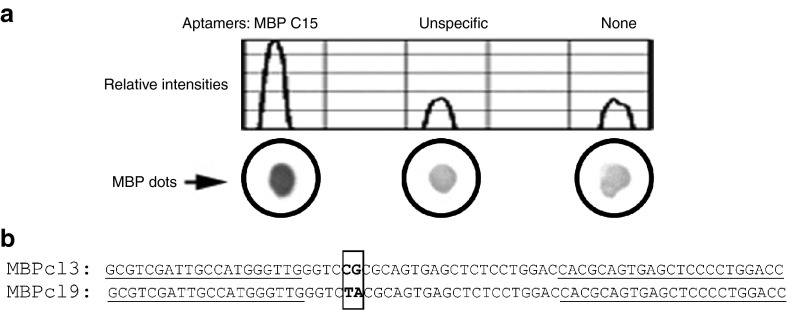
Single-stranded DNA molecules exhibiting high affinity for mouse MBP were obtained by an in vitro aptamer selection procedure, named SELEX. Our initial ssDNA library was designed to contain a randomized sequence region of 35 nucleotides, and be flanked by fixed primer sites. High biases in composition of the nucleotide library may lead to an unsuccessful SELEX, such that equal representation of four nucleotides is preferred. To test for the nucleotide composition, our initial pool of aptamers was cloned out and 20 individual clones were sequenced, resulting in a composition of 27% A, 29.4% C, 19.7% G, and 23.4% T. Several variables were modified during the successive SELEX rounds in order to increase the stringency of the selection (Table 1). The SELEX was started with a library of ~3 × 1014 distinct ssDNA molecules. Aptamers obtained at the 15th cycle (MBP C15) were analyzed using a radio isotope-free dot-blot assay (Figure 1a), which has become a common technique for protein detection, and may be performed by most research laboratories due to its simplicity and speed. Figure 1a shows the MBP-binding activity of the selected pool of aptamers compared to the activity of another pool of aptamers that had been selected to an unrelated protein. Whereas the binding activity of the unspecific pool of aptamers is comparable to a control without aptamers, MBP C15 showed a higher binding activity suggesting that the pool of aptamers was enriched for molecules that have affinity for MBP. This aptamer pool was therefore cloned out and 15 individual clones were sequenced. Sequences were clustered into groups based on their nucleotide content, which revealed two prominent families with highly conserved sequences, represented by the clones MBPcl3 and MBPcl9 (Figure 1b). Interestingly, the length of the randomized region was shortened in both sequences. Indeed, while the original ssDNA library was designed to contain ssDNA molecules with a 35-nucleotide long randomized region, the aptamer obtained after 15 rounds of SELEX displayed a 26-nucleotide long randomized region. 5′ biotinylated versions of MBPcl9, MBPcl3, and of the unselected random library were generated by solid-phase synthesis for further studies in enzyme-linked immunosorbent assay (ELISA)-like assays. These assays followed the basic principles of an ELISA with the difference that the first antibody is replaced by an aptamer conjugated to a reporter for detection with a secondary horseradish peroxidase (HRP)-conjugated antibody. Our ELISA-like assay also differs from the enzyme-linked oligonucleotide assay,15 which only makes use of oligonucleotides for detection.
Table 1. Protein and aptamer quantity, washing stringency and incubation time for each SELEX.
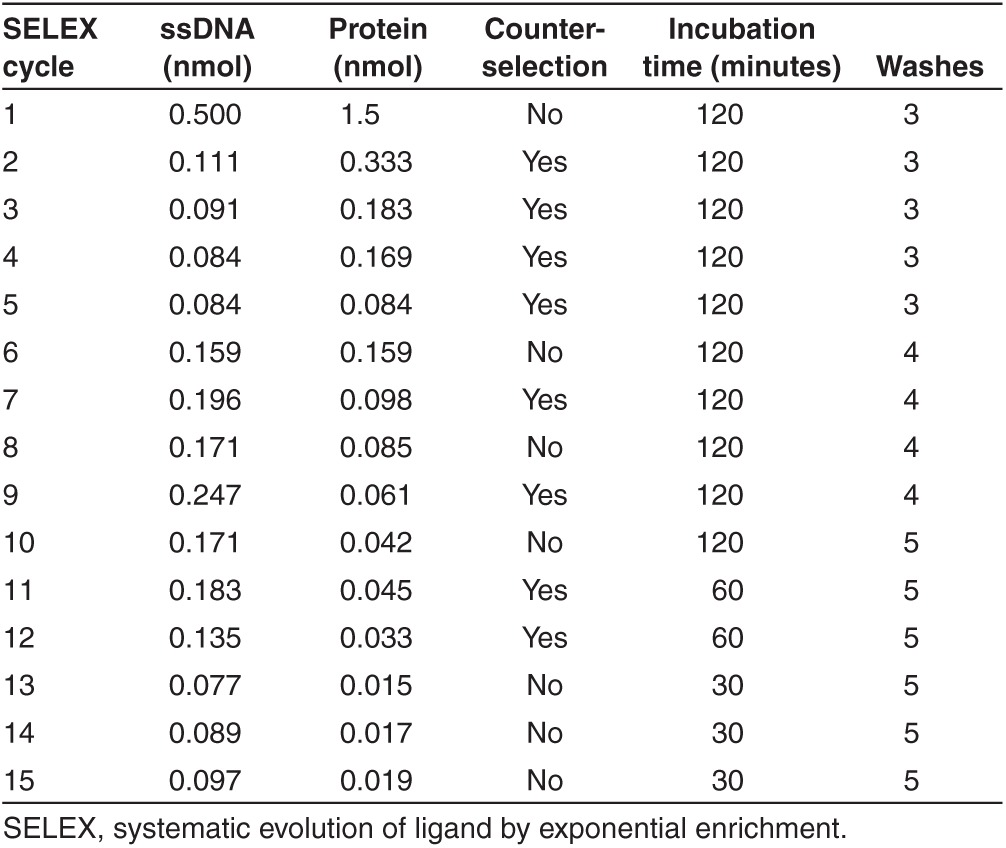
Figure 1.
Dot-blot analysis of the selected library after 15 rounds of systematic evolution of ligand by exponential enrichment (SELEX). (a) 1 µg of pure myelin basic protein (MBP) was spotted onto nitrocellulose membranes and incubated with equal nmol of different biotinylated aptamers. MBP C15 is the selected library against MBP; “unspecific” is another pool of aptamers selected against an unrelated target; “none” is without aptamers. Bound aptamers were detected with an alkaline phosphatase-conjugated anti-biotin antibody and developed with NBT/BCIP substrates. Relative intensities were measured with the ImageJ software. (b) Sequence alignment of the two most representative clones found in the enriched library selected for its affinity for MBP. After the 15th round of selection, the aptamer pool was cloned out and sequenced. MBPcl3 had the highest frequency among all nucleotide sequences followed by MBPcl9. Sequences corresponding to the constant primer regions are underlined. Letters in bold show differences between the two clones.
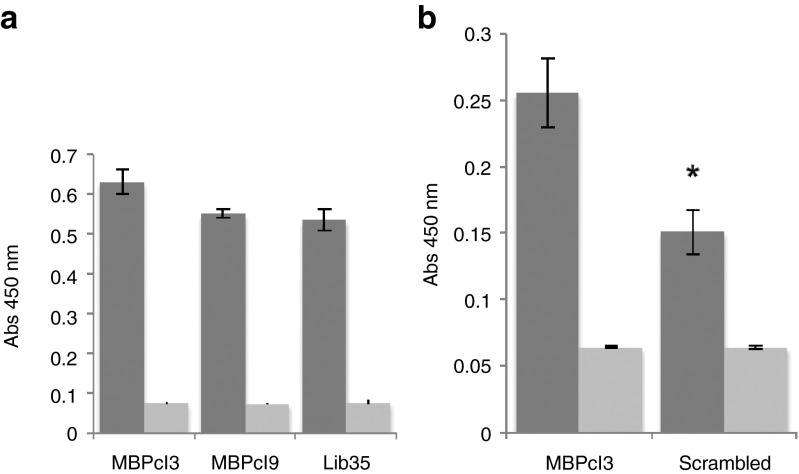
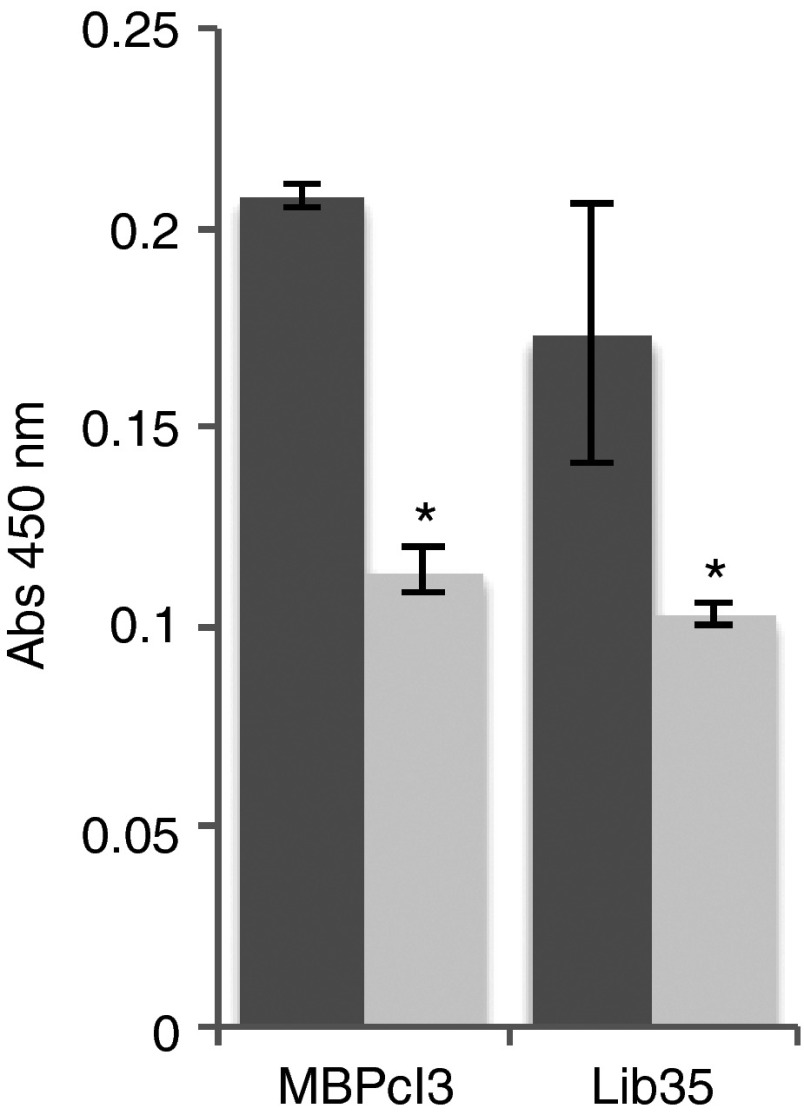
The MBP-binding activity of MBPcl3 and MBPcl9 was compared and no statistical difference was found (Figure 2a). Interestingly, the unselected aptamer pool also showed a strong interaction with immobilized MBP suggesting that the protein binds to nucleic acids irrespective of their sequence due to its highly positive net charge at physiological pH. However, it is also possible that the binding activity resides in part within the fixed reverse primer-annealing region of the aptamer. Sequence analysis of both aptamer clones revealed a highly conserved nucleotide sequences between the randomized region and the constant reverse primer-annealing region of the aptamers. To determine if the binding activity of MBPcl3 was sequence specific, a scrambled version of MBPcl3 was generated by randomizing its nucleotide sequence and its MBP binding activity was assessed. Figure 2b clearly shows that the integrity of MBPcl3 sequence is necessary for its binding to the target molecule.
Figure 2.
Myelin basic protein (MBP)-binding activity of MBPcl3 aptamer using an ELISA-like assay. 0.5 µg of pure MBP was incubated in a microtiter plate with equal nmol of an unselected aptamer library (Lib35), MBPcl3 or MBPcl9 (a) or with a scrambled MBPcl3 aptamer whose entire nucleotide sequence was randomly rearranged (b). Bound aptamers were detected with horseradish peroxidase (HRP)-conjugated avidin and developed with 3,3',5,5'-tetramethylbenzidine (TMB) substrate (dark gray). Wells without MBP were used as controls (light gray). Experiments were performed in triplicate. The asterisk indicates a significant difference in comparison with MBPcl3 (P < 0.05, Student's t-test). Data are shown as mean ± SD.
Structural analysis
To further investigate whether aptamer sequence can be minimized, structural analysis of MBPcl9 and MBPcl3 was performed with Mfold16 to predict the most likely single-stranded DNA secondary structures (Figure 3a–c). MBPcl9 structure displayed an inverted L-shape (Figure 3a) with two long stems, each followed by a pair of loops, and contained several unpaired 3′ terminal nucleotides. On the other hand, two optimal structures were shown for MBPcl3 with comparable free energies. One of these structures showed an inverted L-shaped figure, similar to MBPcl9 (Figure 3b). The second structure (Figure 3c) contained a higher number of unpaired nucleotides. The difference between MBPcl3 and MBPcl9 resided in the nucleotides at position 25 and 26 corresponding to CG and TA, respectively. While the TA nucleotides are part of the long stem 5 in MBPcl9, the CG pair in MBPcl3 is either included in the loop E or among the unpaired nucleotides, suggesting that these two nucleotides allow MBPcl3 to adopt different conformational structures. This result allowed to define distinct domains that contain stems and loops such that shorter versions of the MBP-specific aptamers were generated by solid-phase synthesis and tested for their affinity for MBP (Figure 3d). Aptamer domain AF comprised stem 5 with loops A and F, domain BC included stem 8 and loops B and C, while domain AE contained stem 7 with loops A and E (Figure 3a–c). Interestingly, all three shorter versions of aptamer MBPcl3 and MBPcl9 were found to lose the MBP-binding activity, suggesting that a full length version of MBPcl3 and MBPcl9 is necessary for their MBP-binding activity.
Figure 3.
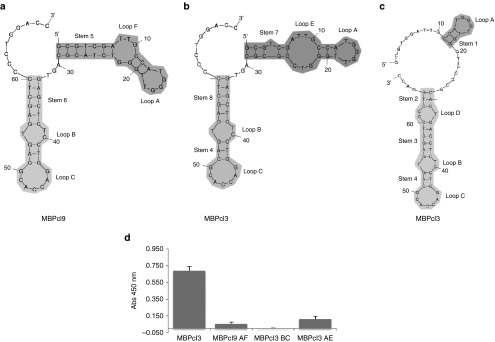
Aptamer conformations and their binding activities. Upper panel shows schematic representations of secondary structures of MBPcl3 and MBPcl9 as predicted by the mfold program. (a) is the predicted most favorable structure for MBPcl9; (b) and (c) are two different possible conformations for MBPcl3 with similar free energies. The myelin basic protein (MBP)-binding activity of three different MBPcl3 domains was determined by an enzyme-linked immunosorbent assay (ELISA)-like assay (d). 0.5 µg of pure MBP was immobilized onto wells of a microtiter plate and incubated with MBPcl3 or equal nmol of MBPcl3 or MBPcl9 truncated products. Truncation was performed to retain a stem-loop structure. MBPcl9 AF includes Stem 5, Loop A and F; MBPcl3 BC includes stem 4 and 8 and loops B and C; MBPcl3 AE includes stem 7 with loops A and E. Experiments were performed in duplicate. The asterisks indicate a significant difference in comparison with MBPcl3 (P < 0.05, Student's t-test). Data are shown as mean ± SD.
MBP aptamer specificity
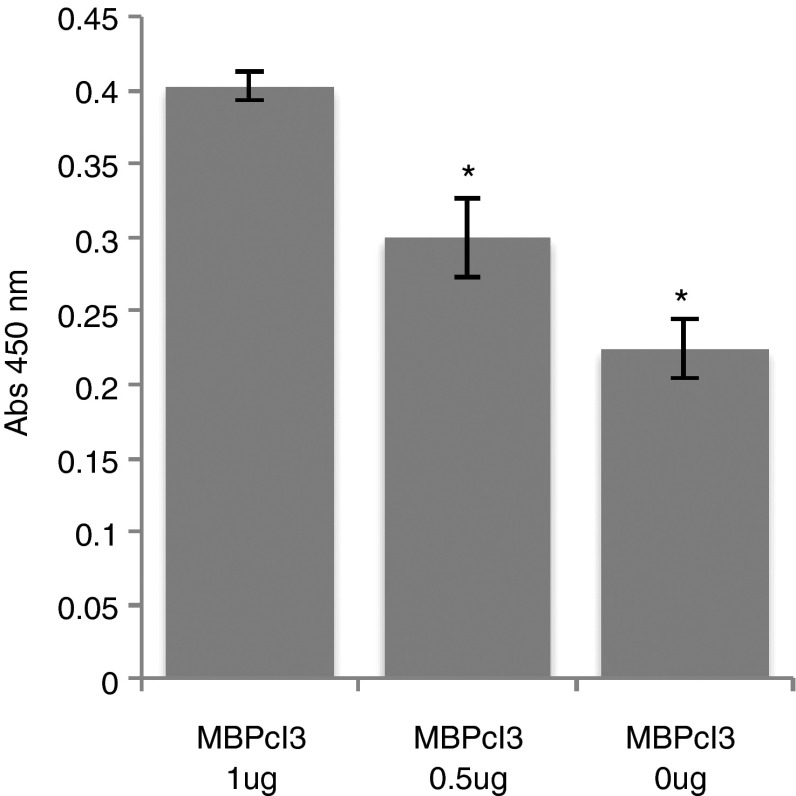
In order to analyze whether the MBPcl3 aptamer is able to detect MBP in a complex mixture of proteins, 1 or 0.5 µg of MBP was immobilized onto wells of a microtiter plate and a cleared Hela cells lysate was added to the wells. Wells were then coated with bovine serum albumin (BSA) to block the remaining free surface. In consequence, if there was a high unspecific binding activity of MBPcl3, a higher absorbance should be expected in wells containing 0.5 µg of MBP due to a higher surface availability for the binding of soluble Hela cell proteins. The absorbance measured in wells containing 0.5 µg of MBP was much lower than in wells originally coated with 1 µg of MBP, reflecting a reduced protein–aptamer interaction in the former, thereby indicating that MBPcl3 specifically detected MBP in a mixture of proteins (Figure 4).
Figure 4.
Myelin basic protein (MBP)-binding activity of the MBPcl3 aptamer in a complex mixture of proteins. A cleared lysate of Hela cells was immobilized onto a microtiter plate containing 1 µg, 0.5 µg, or nothing of pure MBP. Wells were then coated with 3% BSA such that wells with lesser amount of MBP contained a higher amount of either the Hela cell lysate or the BSA protein. Therefore, the observed difference between the two treatments may only be explained by the difference in the amount of MBP. Experiments were performed in triplicate. The asterisk indicates a significant difference when compared to MBPcl3 1 µg (P < 0.05, Student's t-test). Data are shown as mean ± SD.
MBP-binding activity of an anti-MBP antibody compared to MBPcl3
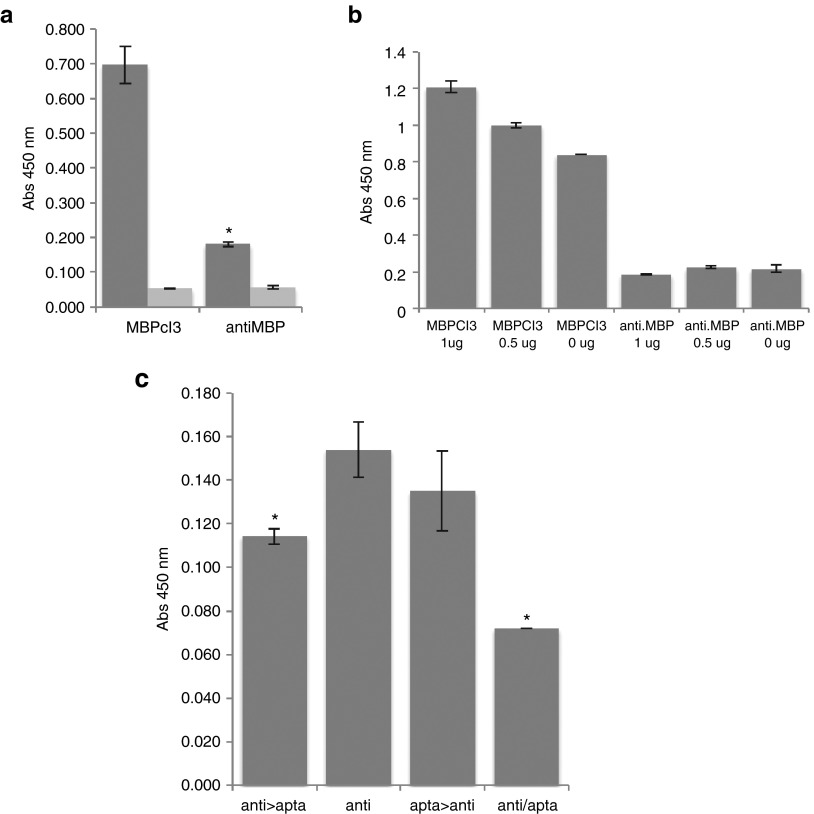
MBPcl3-binding activity was also compared with a commercial polyclonal anti-MBP antibody. For this purpose, equal molar concentrations of MBPcl3 and anti-MBP antibody were incubated with 0.5 µg of MBP (Figure 5a). Interestingly, MBPcl3 was found to bind MBP almost fivefolds more effectively than the antibody. In addition, MBPcl3 was also compared to the anti-MBP antibody in a complex mixture of proteins as described before (Figure 5b). For MBP detection, 7 pmol of either MBPcl3 or anti-MBP antibody was used. Figure 5b shows that MBPcl3 was able to detect MBP, while the anti-MBP antibody did not. MS is a disease where autoimmune antibodies play a key role. The recognition of MBP by autoantibodies induces the recruitment of lymphocytes to MBP sites and could participate in the degradation of the myelin protein. In order to test whether MBPcl3 could inhibit the activity of an MBP-specific antibody, a competition assay was performed (Figure 5c). MBPcl3 and the anti-MBP antibody were incubated at different times or together in Microlon ELISA microplates that contained previously immobilized MBP. Figure 5c shows that the presence of MBPcl3 blocked the binding of the antibody to MBP and that this effect was greater when MBPcl3 and the anti-MBP antibody were incubated together.
Figure 5.
The myelin basic protein (MBP)-binding activity of an antibody compared to MBPcl3. (a) Equal nmol of MBPcl3 and of the antibody were incubated with the MBP that was first immobilized on a microtiter plate. Wells without MBP were used as controls (light gray). (b) A cleared lysate of Hela cells was immobilized on a microtiter plate containing 1 or 0.5 µg of pure MBP. Wells were then blocked with 3% BSA and after washing 7 pmol of aptamer or antibody was added. (c) 0.5 µg of pure MBP was immobilized onto a microtiter plate and incubated with 0.028 nmol of MBPcl3, 0.010 nmol of anti-MBP antibody or with both at different times. Columns from left to right: (1) MBP incubated with anti-MBP antibody, washed and then with the MBPcl3 aptamer. (2) MBP incubated with the antibody only. (3) MBP incubated with MBPcl3, washed and then with anti-MBP antibody. (4) anti-MBP and MBPcl3 aptamer incubated together. For (a), (b), and (c), fraction of bound antibody was detected with an horseradish peroxidase (HRP)-conjugated anti-rabbit antibody and developed with 3,3',5,5'-tetramethylbenzidine (TMB) substrate, while for (a) and (b), fraction of bound aptamer was detected with an HRP-conjugated avidin. Experiments were performed in triplicate. The asterisks indicate a significant difference in comparison with MBPcl3 (a) or with “anti” (b) (P < 0.05, Student's t-test). Data are shown as mean ± SD.
MBP peptide binding of MBPcl3 aptamer
The recognition by T-cell receptors of antigenic peptides exposed on the surface of antigen presenting cells is considered an important step in MS progression. Thus, an MBP peptide comprising residues 90–106 (MBP90106) was synthesized and the MBP90106-binding activity of MBPcl3 was tested in an ELISA-like assay. Interestingly, MBPcl3 was able to bind MBP90106 as well as the unselected aptamer pool (Figure 6).
Figure 6.
Detection of a myelin basic protein (MBP) peptide with MBPcl3 aptamer. Two micrograms of pure MBP peptide were incubated in a microtiter plate with equal nmol of an unselected aptamer library (Lib35) or MBPcl3. Bound aptamers were detected with HRP-conjugated avidin and developed with 3,3',5,5'-tetramethylbenzidine (TMB) substrate (dark gray). Wells without MBP peptide were used as controls (light gray). Experiments were performed in triplicate. One asterisk indicates a significant difference in comparison with MBPcl3 and two asterisks indicates a significant difference in comparison with Lib35 (P < 0.05, Student's t-test). Data are shown as mean ± SD.
Histochemistry
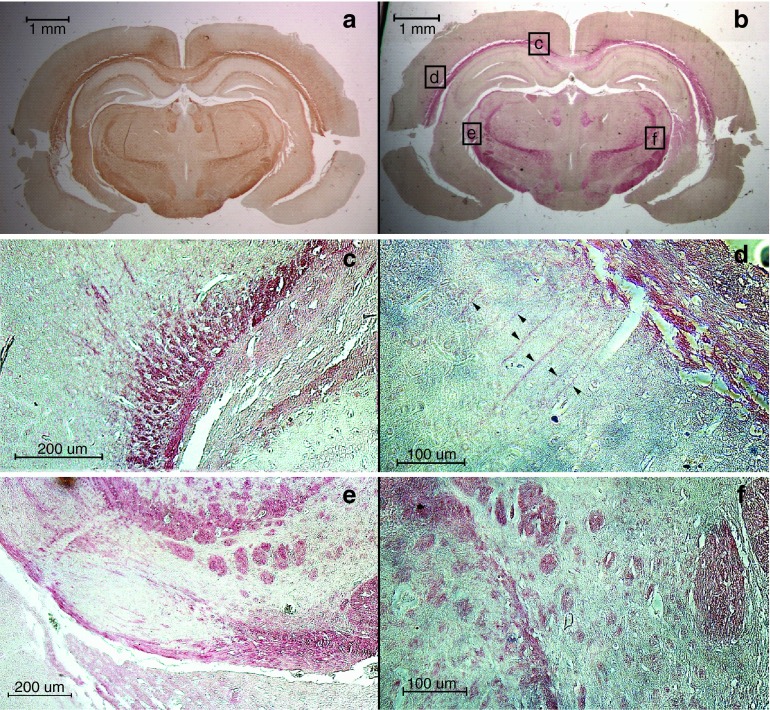
In order to test the ability of MBPcl3 to detect MBP in tissues, we used mouse brain sections for histochemical reaction. MBPcl3 was able to recognize myelin-rich regions in the mouse brain (Figure 7). Brain areas known to contain neural fibers including the corpus callosum, cerebral penduncle, thalamic radiation, fasciculus retroflexus, and premammilary nucleus (Figure 7b) were reactive to MBPcl3. The pattern of MBP detection through MBPcl3 was comparable to that observed with anti-MBP antibody (Figure 7a).
Figure 7.
Detection of myelin-rich regions in mouse brain sections through immunostaining with anti-myelin basic protein (MBP) polyclonal antibody (a) and oligo-stained with the MBPcl3 aptamer (b–f). (c–f) are higher magnifications of squared areas in (b). (d) Arrowheads indicate axons near the corpus callosum.
Discussion
SELEX is a proven methodology to obtain active nucleic acids with the ability to bind specific targets (for reviews, see refs. 2,17). Essentially SELEX consists in repeated binding tests, selections and amplifications of aptamers from an initial library until enrichment of specific molecules is obtained. Aptamers are being tested for different purposes such as protein purification18 or for protein-function regulation in an antagonistic or agonistic manner.19,20,21
In this study, we obtained specific aptamers against pure MBP from mice containing mainly the 18.5 kDa isoform. Whereas the aptamer library was designed to contain a randomized region of 35 nucleotides, two selected clones, namely MBPcl3 and MBPcl9, showed a shorter randomized region of 26 nucleotides that mediated the MBP-binding activity in nitrocellulose membranes and in ELISA microtiter plates. Interestingly, the unselected library of aptamers was found to also bind MBP suggesting a natural affinity of MBP for nucleic acids. Nevertheless, the MBP-binding activity of a scrambled version of MBPcl3 aptamer was not as good as the unmodified MBPcl3, showing that binding was sequence-specific and not simply due to physicochemical properties of the protein. MBP binding of the unselected aptamer pool might, therefore, arise from the primer-annealing region present at both the aptamer and the library. The analysis of MBPcl3 and cl9 randomized region showed (sequence) similarity to the reverse primer-annealing region that might drive MBP binding. On the other hand, MBP contains a strong positive net charge at physiological pH, and its nucleic acid binding activity could be driven mainly by electrostatic interaction. Indeed, it is well known that MBP binds negatively charged molecules such as the polar head of acidic lipids22,23 and fluorescently labeled triphosphate nucleotides.24 Even more, Smith et al.25 have proposed the presence of a nontraditional nuclear localization signal on MBP that allows to translocate the protein to the nucleus. Hence, our results add new insight on the ability of MBP to bind nucleic acids in vitro and it is likely that binding activity might occur in vivo.
The secondary structure of MBPcl3 and MBPcl9 aptamers was predicted with mFold16 to rationally detect possible active domains. Shorter versions of both aptamers were synthesized. MBPcl9 AF and MBPcl3 BE domains contained the forward primer-priming region, while MBPcl3 BC contained part of the reverse primer-priming region. However, none of them was able to equal the activity of the full-length MBPcl3 suggesting that the whole nucleotide sequence is required for the binding activity of the aptamer. Similarly, chemokine platelet factor 4 (PF4) is another highly positive charged-protein with the ability to bind many negatively charged polyanions and recently nucleic acids were added to the list.26 Interestingly, PF4 was able to bind RNA and DNA of different length, sequence and structural configurations that regulated its activity. Jaax et al. showed that longer constructs (45mer) are more potent regulators than shorter ones (10-21mer). However, many of the conclusions observed by Jaax et al. were based on an experimental assay that was not indicative of the presence of nucleic acids bound to PF4 but rather to the ability of PF4 to bind platelets in the presence of nucleic acids. In addition, an aptamer that binds to myelin, named 3064, has been recently raised using a suspension of crude murine myelin.27 It was shown that not only aptamer 3064 had the ability to bind to myelin but also the aptamer control. In agreement with this, our results also suggest an intrinsic ability of MBP to bind nucleic acids, although it is not fully understood whether MBP is able to bind any kind of oligonucleotide or its binding is length-, sequence-, and/or structure-related. Nastasijevic et al.27 shed a little more light in this issue showing the inability of a 40mer oligodeoxythymidylate molecule to bind myelin. Thus, further effort should be directed toward identifying the affinity constant of MBPcl3 aptamer and understanding the effect nucleic acids length, sequence and structural configuration on the binding to myelin.
MBPcl3 was also able to detect MBP in a complex mixture of proteins in a concentration-dependent manner. In addition, MBPcl3 binding activity was compared to that of an anti-MBP polyclonal antibody. Using equivalent amounts of both the aptamer and the antibody, we observed a higher binding activity with MBPcl3. Moreover, while 7 pmol of aptamer was sufficient to detect MBP in a complex mixture of proteins, the same amount of anti-MBP antibody was not, suggesting that MBPcl3 has a higher affinity for MBP than the antibody.
MBPcl3 was also used to detect myelin-rich areas in mouse brain tissue sections. Currently, there are only a few methods for myelin staining in tissue sections, which include Luxor Fast Blue described by Kluver and Barrera in 1953,28 Black Gold II (BKII) commercialized by Merck and the classical antibody-mediated immune reaction. Luxor Fast Blue and BKII are the cheapest approaches but they are not only time-consuming methodologies but also involve processing the tissue in nonphysiological conditions. Antibodies constitute the most expensive alternative although they allow researchers to work under physiological conditions, with the possibility of selecting a variety of dye-conjugated antibodies and stain tissues in a fewer steps. On the contrary, MBPcl3 results from a relatively cheap and easy approach to synthesize molecules with multiple advantages over antibodies, Luxor Fast Blue and BKII methods. MBPcl3 may be synthesized by solid-phase and is more amenable to modifications during its manufacture such that virtually any type of dye, enzyme, or marker may be added during this process. Furthermore, staining can be performed under physiological conditions and the number of staining steps may be reduced to only one (e.g., MBPcl3 conjugated to an alkaline phosphatase or horseradish peroxidase). The finding that MBPcl3 is able to identify MBP in tissue sections suggests that it may become a great research tool for histochemical staining of the central nervous system and opens up a promising technical output of aptamers for molecule detection and cell localization.
Finally, MBPcl3 could be also a great therapeutic lead and it would be very interesting to try in the near future its activity in different animal models of MS. MS is an inflammatory disease that affects the central nervous system and leads to substantial disabilities through deficits of sensation and impaired motor, autonomic, and neurocognitive functions. At present, no cure exists for MS and current treatments aim to arrest or slow the progression of the disease and alleviate the symptoms. MS is currently considered to be a CD4+ Th1-mediated autoimmune disease.7,29 This assumption is based on the cellular composition of brain and cerebrospinal fluid-infiltrating cells and data obtained from the experimental autoimmune encephalomyelitis model.30 Experimental autoimmune encephalomyelitis is induced in susceptible rats and mice by the injection of myelin components, promoting a CD4+-mediated autoimmune disease that shares similarities with MS.7,30 Despite clear indications of CD4+ Th1 cells key role in the disease, its etiology remains unknown. Slowing down, halting the disease progression or reversing its effects remains an unmet objective for patients with MS. MBP constitutes one of the major components of the myelin sheath31 and is found throughout the whole myelin sheath.32 Previous works confirmed that many MS patients carry MBP autoantibodies and suggest that these antibodies may be responsible for the loss of myelin in neuronal axons.7,33,34,35 Moreover, it has been proposed that the antigenic epitope of MBP was located between residues 61 and 110.36 Hence, targeting the MBP protein may result in an immune-protective therapy by blocking either the interaction between autoantibodies and the endogenous MBP or by blocking the activation immunoreactive T-cells that recognize MBP peptides displayed on the surface of antigen-presenting cells. Indeed, this study showed that the MBPcl3 aptamer inhibited the MBP-binding activity of an MBP-specific antibody and that MBPcl3 was also able to interact with an MBP peptide comprising residues 90–106. Thus, it would be very interesting to analyze in the future whether MBPcl3 aptamer is able to block MBP-mediated T-cell activation.
In summary, this is the first report to show an aptamer capable to detect MBP in tissue sections and demonstrate the potential of MBPcl3 as a medical research tool. This work also opens the possibility for the development of novel aptamer-based therapeutic approaches to pursue better drugs in diseases such as MS. However, more efforts should be directed towards understanding the nature of the MBP-oligonucleotides interaction.
Materials and methods
Materials. Mouse myelin basic protein, Bovine Serum Albumin fraction V, alkaline phosphatase-conjugated monoclonal anti-biotin, and anti-MBP polyclonal antibodies were purchased from Sigma Aldrich (St Luis, MO). Yeast tRNA was purchased from Biodynamics (BA, Argentina). Taq Polymerase and dideoxynucleotides were obtained from Fermentas (Vilnius, Lithuania). Alkaline phosphatase substrate NBT/BCIP and HRP substrate 3,3',5,5'-tetramethylbenzidine were purchased from Promega (Madison, WI). HRP-conjugated avidin was purchased from Pierce (Rockford, IL). High binding Microlon ELISA plates were obtained from Greiner Bio-One (Monroe, NC). All oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). Nitrocellulose Hybond ECL membranes were obtained from GE Healthcare (Piscataway, NJ). Salmon sperm DNA and Sybr Gold were purchased from Life Technologies (Gran Island, NY). All other reagents and chemicals were of the highest purity available and were obtained from commercial sources.
SELEX. The ssDNA library was designed to contain a randomized sequence region of 35 nucleotides, and to be flanked by constant primer annealing regions (5′-GCGTCGATTGCCATGGGTTG(N)35 cacgcagtgagctcccctggacc-3′). A biased molar mixture (30% A, 30% C, 20% G, and 20% T) of the four phosphoramidites was used to compensate for their incorporation efficiencies and to generate a truly randomized region. The first round of selection was initiated with 0.5 nmol (~3 × 1014 molecules) of ssDNA. Prior to selections, ssDNAs were diluted in selection buffer (SB; 20 mmol/l Tris-HCl pH 7.6, 150 mmol/l NaCl, 5 mmol/l MgCl2), denatured at 95 °C for 5 minutes and cooled on ice for another 5 minutes. Affinity selections were done as follows: MBP was immobilized onto a nitrocellulose membrane disc and blocked in blocking buffer (SB containing 2% BSA) for 60 minutes. After several washes with SB, MBP was incubated with the ssDNA library at 37 °C for a fixed period of time (Table 1). Counter-selections of aptamers were performed by incubating the ssDNA library with nitrocellulose membrane discs that had been blocked with blocking buffer for 60 minutes at SELEX cycle 2, 3, 4, 5, 7, 9, 11, and 12. Molar ratio of MBP to aptamers, washing stringency, and incubation time were modified over the SELEX cycles (Table 1). After washing with SB, the aptamers bound to MBP were eluted in distilled water by heating the complex at 95 °C for 10 minutes. Asymmetric polymerase chain reaction (PCR) was performed with eluted aptamers in 100 µl reaction mix containing 2 µmol/l forward primer 5′-GCGTCGATTGCCATGGGTTG-3′; 0.02 µmol/l reverse primer 5′- GGTCCAGGGGAGCTCACTGCGTG-3′; 200 µmol/l of each dNTP; 10 mmol/l Tris-HCl (pH 8.8 at 25 °C), 50 mmol/l KCl, 0.08% (v/v) Nonidet P40 and 0.8 mmol/l MgCl2. Thermal cycling consisted of one cycle at 95 °C for 10 minutes and multiple cycles at 95 °C for 30 seconds, 58 °C for 40 seconds, and 72 °C for 40 seconds. The number of PCR cycles was estimated empirically at each round of SELEX to avoid the amplification of PCR products of incorrect size by analyzing an aliquot of the PCR product on a 3% agarose gel electrophoresis every 10 PCR cycles containing sybr gold for visualization. Following PCR amplification, the ssDNA library was ethanol-precipitated and pelleted by centrifugation. The pellet was dried under vacuum and resuspended in distilled water for the next cycle of selection.
Cloning and sequencing. Aptamers from the 15th round of selection were asymmetrically PCR-amplified, purified, and an aliquot was used as a template for a standard PCR. dsDNA PCR products were cloned using the pGEM-T Easy kit (Promega) and individual clones of transformed bacteria were screened for the presence of aptamers with the correct size. Plasmid DNA extracted from single colonies was sequenced at the Instituto Nacional de Tecnología Agropecuaria (Castelar, Argentina). Sequence alignments were performed with CLUSTAL W.
Dot blotting. Biotinylated aptamers were generated by asymmetric PCR with a 5′ biotinylated forward primer and purified. Aptamers were heated at 95 °C for 5 minutes and then cooled on ice for another 5 minutes. Pure MBP protein was spotted onto 0.45 µm pore-size nitrocellulose membranes and air-dried in a BioRad biodot system. Membranes were blocked with blocking buffer or Tris buffer saline (TBS; 20 mmol/l Tris-HCl pH 7.5, 500 mmol/l NaCl) with 2% BSA for 1 hour at room temperature. Membranes were then washed three times with SB or TBS and incubated with 0.014 nmol of aptamers in SB or TBS containing 0.1 mg/ml of yeast tRNA for 2 hours at 37 °C. Then, membranes were washed three times with SB or TBS and incubated for 1 hour at room temperature with alkaline phosphatase -conjugated anti-biotin monoclonal antibody in SB or TBS containing 2% BSA. Membranes were washed three times and biotinylated aptamers were detected with NBT/BCIP alkaline phosphatase substrate.
ELISA-like binding assays. A high binding Microlon microtiter plate was coated overnight with MBP or a MBP peptide comprising residues 90–106 (Anaspec, cat. 60977. CA) in 100 µl of 0.05 mol/l sodium carbonate pH 9.6. Wells were washed three times with phosphate buffer saline containing Tween 20 (PBS-T; 1 mmol/l KH2PO4, 10 mmol/l Na2HPO4, 137 mmol/l NaCl, 2.7 mmol/l KCl, 0.05% Tween 20) and blocked with PBS-T containing 3% BSA for 1 hour. Then, wells were washed three times with SB and incubated with aptamers in SB supplemented with 0.1 mg/ml of yeast tRNA or salmon sperm DNA. Before addition to the wells, aptamers were heated at 95 °C for 5 minutes and cooled on ice for another 5 minutes. Wells containing MBP and incubated without aptamers and wells without MBP and incubated with aptamers were used as internal controls. Wells were washed three times with SB and incubated with HRP-avidin in SB containing 2% BSA for 1 hour. Then, wells were washed three times with SB and 3,3',5,5'-tetramethybenzidine was used as HRP substrate. Absorbance was measured at 450 nm in a Biotek µQuant microplate spectrophotometer.
Activity of MBPcl3 in cell lysates. Three 10 cm Ø cell culture plates of Hela cells (ATCC CCL-2.1) cultured to confluence were washed twice with PBS and lysed in 400 µl of radio immunoprecipitation assay buffer (50 mmol/l Tris-HCl pH 7.4, 150 mmol/l NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 0.2 mmol/l phenylmethylsulfonyl fluoride, 10 nmol/l leupeptin, and 10 pg/ml aprotinin). Lysates were centrifuged at 15,000× g in a table centrifuge at 4 °C for 15 minutes. High binding Microlon microtiter plates were prepared with MBP as described above and 75 µl of cell lysate was added to each well containing different amounts of MBP for 1 hour. Wells without MBP were used as internal controls. After three washes with PBS-T, wells containing MBP protein and cell lysates were blocked with PBS-T containing 3% BSA for 1 hour. Addition of aptamers and detection followed as described earlier (see ELISA-like binding assays) with the difference that aptamers were synthesized with nucleotide analogues containing phosphorothioate interlinkages.
Histochemistry. Brains from adult BALB/c mice were fixed in cold 4% paraformaldehyde (PFA) in 0.1 mol/l phosphate buffer (pH 7.4) for 72 hours, dehydrated through a graded series of ethanol, and embedded in paraffin. Paraffin-embedded tissues were sectioned at 5 µm thickness. Histochemical identification of myelin was performed as follows: tissue sections were deparaffinized in xylene, rehydrated through a decreasing series (100, 95, 70, and 50%) of ethanol, washed in distilled water for 5 minutes, and stored in SB 1X until further use. For myelin detection with aptamer MBPcl3, tissue sections were incubated in PBS containing 4% PFA and 0.3% Triton for 1 minute, washed in SB 1X, and blocked in SB 1X containing 2% BSA for 30 minutes. Tissue sections were then washed and incubated with 5 mmol/l of 5′ biotinylated MBPcl3 aptamer for 2 hours at 37 °C in SB 1X containing 4 µg of salmon sperm gDNA. Sections were then washed, incubated with an anti-biotin alkaline phosphatase-conjugated antibody for 1 hour in SB 1X containing 2% BSA and alkaline phosphatase activity was revealed using SIGMA's Magenta kit. Examination was performed with a Nikon Eclipse E200 upright microscope. For myelin detection with a commercial anti-MBP antibody, tissue sections were incubated in PBS containing 4% PFA and 0.3% Triton for 1 minute, washed in PBS, and treated with 0.3% H2O2 in water to quench endogenous peroxidase activity. Sections were then washed in PBS, blocked in PBS containing 2% BSA for 30 minutes, washed again in PBS and incubated with a rabbit anti-MBP antibody (1 × 102 dilution) in PBS containing 1% BSA for 1 hour at room temperature. Sections were then washed and incubated with an anti-rabbit IgG HRP-conjugated antibody for 1 hour in PBS containing 1% BSA and staining was performed with the Vectastatin Elite ABC kit rabbit (Rabbit IgG) from Vector Laboratories (CA). Examination was performed with a Nikon Eclipse E200 upright microscope or a Nikon T1 confocal microscope equipped with transmission light and a Nikon Digital Sight DS-Fi1 camera.
Statistical Analysis. Results from at least two independent experiments in high binding Microlon microtiter plates were analyzed and the P values were calculated with the Student's t-test. Differences were considered statistically significant when P < 0.05.
Acknowledgments
This work was supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET, Argentina); the Agencia Nacional de Promoción Científica y Tecnológica through PICT-Start Up 2011-1840 granted to ADV; and Universidad Maimónides. The authors thank Sophie Adjalley for critical reading of the manuscript, and Veronica Dorfman, Martín Radrizzani, Ana Adamo and Germán Roth for their advice. Rozenblum G. and Vitullo A.D. have a patent pending on “Aptámeros contra la proteína básica de mielina como agentes neuroprotectores”, Instituto Nacional de la Propiedad Industrial, Argentina. Application number 20130104294. The other author declares no conflict of interest.
References
- Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Nimjee SM, Rusconi CP, Sullenger BA. Aptamers: an emerging class of therapeutics. Annu Rev Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- Benjamins JA, Morell P. Proteins of myelin and their metabolism. Neurochem Res. 1978;3:137–174. doi: 10.1007/BF00964057. [DOI] [PubMed] [Google Scholar]
- Ridsdale RA, Beniac DR, Tompkins TA, Moscarello MA, Harauz G. Three-dimensional structure of myelin basic protein. II. Molecular modeling and considerations of predicted structures in multiple sclerosis. J Biol Chem. 1997;272:4269–4275. doi: 10.1074/jbc.272.7.4269. [DOI] [PubMed] [Google Scholar]
- Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Farès C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron. 2004;35:503–542. doi: 10.1016/j.micron.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Martin R, McFarland HF, McFarlin DE. Immunological aspects of demyelinating diseases. Annu Rev Immunol. 1992;10:153–187. doi: 10.1146/annurev.iy.10.040192.001101. [DOI] [PubMed] [Google Scholar]
- Burns J, Rosenzweig A, Zweiman B, Lisak RP. Isolation of myelin basic protein-reactive T-cell lines from normal human blood. Cell Immunol. 1983;81:435–440. doi: 10.1016/0008-8749(83)90250-2. [DOI] [PubMed] [Google Scholar]
- Richert JR, McFarlin DE, Rose JW, McFarland HF, Greenstein JI. Expansion of antigen-specific T cells from cerebrospinal fluid of patients with multiple sclerosis. J Neuroimmunol. 1983;5:317–324. doi: 10.1016/0165-5728(83)90052-8. [DOI] [PubMed] [Google Scholar]
- Chou YK, Vainiene M, Whitham R, Bourdette D, Chou CH, Hashim G, et al. Response of human T lymphocyte lines to myelin basic protein: association of dominant epitopes with HLA class II restriction molecules. J Neurosci Res. 1989;23:207–216. doi: 10.1002/jnr.490230211. [DOI] [PubMed] [Google Scholar]
- Martin R, Jaraquemada D, Flerlage M, Richert J, Whitaker J, Long EO, et al. Fine specificity and HLA restriction of myelin basic protein-specific cytotoxic T cell lines from multiple sclerosis patients and healthy individuals. J Immunol. 1990;145:540–548. [PubMed] [Google Scholar]
- Pette M, Fujita K, Kitze B, Whitaker JN, Albert E, Kappos L, et al. Myelin basic protein-specific T lymphocyte lines from MS patients and healthy individuals. Neurology. 1990;40:1770–1776. doi: 10.1212/wnl.40.11.1770. [DOI] [PubMed] [Google Scholar]
- Ota K, Matsui M, Milford EL, Mackin GA, Weiner HL, Hafler DA. T-cell recognition of an immunodominant myelin basic protein epitope in multiple sclerosis. Nature. 1990;346:183–187. doi: 10.1038/346183a0. [DOI] [PubMed] [Google Scholar]
- Olsson T, Zhi WW, Höjeberg B, Kostulas V, Jiang YP, Anderson G, et al. Autoreactive T lymphocytes in multiple sclerosis determined by antigen-induced secretion of interferon-gamma. J Clin Invest. 1990;86:981–985. doi: 10.1172/JCI114800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drolet DW, Moon-McDermott L, Romig TS. An enzyme-linked oligonucleotide assay. Nat Biotechnol. 1996;14:1021–1025. doi: 10.1038/nbt0896-1021. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keefe AD, Pai S, Ellington A. Aptamers as therapeutics. Nat Rev Drug Discov. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romig TS, Bell C, Drolet DW. Aptamer affinity chromatography: combinatorial chemistry applied to protein purification. J Chromatogr B Biomed Sci Appl. 1999;731:275–284. [PubMed] [Google Scholar]
- Jellinek D, Green LS, Bell C, Janjic N. Inhibition of receptor binding by high-affinity RNA ligands to vascular endothelial growth factor. Biochemistry. 1994;33:10450–10456. doi: 10.1021/bi00200a028. [DOI] [PubMed] [Google Scholar]
- Adachi H, Ishiguro A, Hamada M, Sakota E, Asai K, Nakamura Y. Antagonistic RNA aptamer specific to a heterodimeric form of human interleukin-17A/F. Biochimie. 2011;93:1081–1088. doi: 10.1016/j.biochi.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Pestourie C, Tavitian B, Duconge F. Aptamers against extracellular targets for in vivo applications. Biochimie. 2005;87:921–930. doi: 10.1016/j.biochi.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Boggs JM, Moscarello MA, Papahadjopoulos D. Phase separation of acidic and neutral phospholipids induced by human myelin basic protein. Biochemistry. 1977;16:5420–5426. doi: 10.1021/bi00644a003. [DOI] [PubMed] [Google Scholar]
- Sankaram MB, Brophy PJ, Marsh D. Selectivity of interaction of phospholipids with bovine spinal cord myelin basic protein studied by spin-label electron spin resonance. Biochemistry. 1989;28:9699–9707. doi: 10.1021/bi00451a024. [DOI] [PubMed] [Google Scholar]
- Wang C, Neugebauer U, Bürck J, Myllykoski M, Baumgärtel P, Popp J, et al. Charge isomers of myelin basic protein: structure and interactions with membranes, nucleotide analogues, and calmodulin. PLoS ONE. 2011;6:e19915. doi: 10.1371/journal.pone.0019915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GS, Seymour LV, Boggs JM, Harauz G. The 21.5-kDa isoform of myelin basic protein has a non-traditional PY-nuclear-localization signal. Biochem Biophys Res Commun. 2012;422:670–675. doi: 10.1016/j.bbrc.2012.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaax ME, Krauel K, Marschall T, Brandt S, Gansler J, Fürll B, et al. Complex formation with nucleic acids and aptamers alters the antigenic properties of platelet factor 4. Blood. 2013;122:272–281. doi: 10.1182/blood-2013-01-478966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nastasijevic B, Wright BR, Smestad J, Warrington AE, Rodriguez M, Maher LJ., 3rd Remyelination induced by a DNA aptamer in a mouse model of multiple sclerosis. PLoS ONE. 2012;7:e39595. doi: 10.1371/journal.pone.0039595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KLUVER H, BARRERA E. A method for the combined staining of cells and fibers in the nervous system. J Neuropathol Exp Neurol. 1953;12:400–403. doi: 10.1097/00005072-195312040-00008. [DOI] [PubMed] [Google Scholar]
- Hafler DA. Multiple sclerosis. J Clin Invest. 2004;113:788–794. doi: 10.1172/JCI21357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamvil SS, Steinman L. The T lymphocyte in experimental allergic encephalomyelitis. Annu Rev Immunol. 1990;8:579–621. doi: 10.1146/annurev.iy.08.040190.003051. [DOI] [PubMed] [Google Scholar]
- Deber CM, Reynolds SJ. Central nervous system myelin: structure, function, and pathology. Clin Biochem. 1991;24:113–134. doi: 10.1016/0009-9120(91)90421-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner C, Lassmann H, Waehneldt TV, Matthieu JM, Linington C. Differential ultrastructural localization of myelin basic protein, myelin/oligodendroglial glycoprotein, and 2',3'-cyclic nucleotide 3'-phosphodiesterase in the CNS of adult rats. J Neurochem. 1989;52:296–304. doi: 10.1111/j.1471-4159.1989.tb10930.x. [DOI] [PubMed] [Google Scholar]
- Allegretta M, Nicklas JA, Sriram S, Albertini RJ. T cells responsive to myelin basic protein in patients with multiple sclerosis. Science. 1990;247:718–721. doi: 10.1126/science.1689076. [DOI] [PubMed] [Google Scholar]
- Uccelli A, Giunti D, Salvetti M, Ristori G, Fenoglio D, Abbruzzeseet al. (1998A restricted T cell response to myelin basic protein (MBP) is stable in multiple sclerosis (MS) patients Clin Exp Immunol 111186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielekova B, Sung MH, Kadom N, Simon R, McFarland H, Martin R. Expansion and functional relevance of high-avidity myelin-specific CD4+ T cells in multiple sclerosis. J Immunol. 2004;172:3893–3904. doi: 10.4049/jimmunol.172.6.3893. [DOI] [PubMed] [Google Scholar]
- Warren KG, Catz I, Steinman L. Fine specificity of the antibody response to myelin basic protein in the central nervous system in multiple sclerosis: the minimal B-cell epitope and a model of its features. Proc Natl Acad Sci USA. 1995;92:11061–11065. doi: 10.1073/pnas.92.24.11061. [DOI] [PMC free article] [PubMed] [Google Scholar]