Abstract
The microRNA(miRNA)-34a is a key regulator of tumor suppression. It controls the expression of a plethora of target proteins involved in cell cycle, differentiation and apoptosis, and antagonizes processes that are necessary for basic cancer cell viability as well as cancer stemness, metastasis, and chemoresistance. In this review, we focus on the molecular mechanisms of miR-34a-mediated tumor suppression, giving emphasis on the main miR-34a targets, as well as on the principal regulators involved in the modulation of this miRNA. Moreover, we shed light on the miR-34a role in modulating responsiveness to chemotherapy and on the phytonutrients-mediated regulation of miR-34a expression and activity in cancer cells. Given the broad anti-oncogenic activity of miR-34a, we also discuss the substantial benefits of a new therapeutic concept based on nanotechnology delivery of miRNA mimics. In fact, the replacement of oncosuppressor miRNAs provides an effective strategy against tumor heterogeneity and the selective RNA-based delivery systems seems to be an excellent platform for a safe and effective targeting of the tumor.
Keywords: apoptosis, cancer, cell cycle, delivery, DNA damage, microRNA, miR-34a, multiple myeloma, nanotechnology, p53
Introduction
MicroRNAs (miRNAs) are a class of small non-coding RNAs, which regulate gene expression at the post-transcriptional level and may conceivably play a key role in tumorigenesis. From the first demonstration in 2002.1 to the present, the involvement of miRNAs in human cancer has led to extensive research.2,3,4 MiRNAs are dysregulated in almost all solid and hematological malignancies, and specific signatures allow the characterization of poorly differentiated tumors5 as well as harbor relevant clinical implications.6,7,8,9 MiRNAs, which are upregulated in cancer cells and contribute to carcinogenesis by inhibiting tumor suppressor genes, are considered oncogenic miRNAs (OncomiRs),10 while downregulated miRNAs, that normally prevent cancer development by inhibiting the expression of proto-oncogenes, are known as tumor suppressor miRNAs.11 Silencing Oncomirs with miRNA inhibitors or replacing tumor suppressor miRNAs with synthetic miRNA mimics is demonstrating a valuable experimental strategy for the treatment of solid and hematological malignancies.12,13,14,15,16,17,18,19,20 MiRNAs with epigenetic or antiangiogenetic or modulatory activity within the microenvironment as well as with synergistic activity with chemotherapeutic agents are particularly promising in translational therapeutic approaches.21,22,23,24,25,26,27,28,29
The miR-34 family consists of three members: miR-34a, miR-34b, and miR-34c; miR-34a is encoded by its own transcript, whereas miR-34b and miR-34c share a common primary transcript. In mice, miR-34a is ubiquitously expressed with the highest expression in brain, whereas miR-34b/c is mainly expressed in lung tissues. Moreover, miR-34a is expressed at higher levels than miR-34b/c, with the exception of the lung.30 Therefore, the two miR-34 genes have presumably tissue-specific functions, even though they are anyway direct transcriptional targets of the onco-suppressor p53, whose expression is greatly affected by DNA damage and oncogenic stress. miR-34 family, thus, contributes to p53 downstream effects on proliferation arrest and induction of apoptosis, by targeting c-MYC, CDK6, and c-MET.30
MiRNAs are originally transcribed as long hairpin molecules (pri-miRNAs) that are subsequently processed over several steps until DICER cuts them intoduplexes of their final 22–23 nt length. RNase III, human DROSHA, is the core nuclease that executes the initiation step of pri-miRNAs processing into stem-loop precursors of approximately 70 nucleotides (pre-miRNAs). DICER, a member of the RNase III superfamily of bidentate nucleases, mediates the cytoplasmic processing of pre-miRNAs into mature miRNAs. Thus, the two RNase III proteins, DROSHA and DICER, have key roles in miRNA-mediated gene regulation in processes such as development and differentiation. As a last step, one strand of the miRNA duplex (“mature strand”) is incorporated into the RNA-induced silencing complex (RISC) while the other is supposedly degraded. Once integrated into the RISC, miRNAs guide this protein complex to partially complementary binding sites located in the 3′ untranslated region (UTR) of target mRNAs and inhibit their expression either by interfering with translation or by destabilizing the target mRNA.31 BRCA1 triggers miRNA biogenesis in collaboration with DROSHA microprocessor and SMADs/p53. In details, BRCA1 has been shown to trigger expression of let-7a-1, miR-16-1, miR-145, and miR-34a.32 It has lately been shown that DICER1 depleted colorectal cancer cells revealed a notably higher expression of cancer stem cell markers such as Sox9, Sox2, Lgr5, CD44, and Nanog. Moreover, ectopic re-expression of miR-34a in both primary and tumor-derived cell lines was put in correlation with cycle arrest, apoptosis and cell growth inhibition.11 Di Martino et al. have recently explored the molecular effects induced by enforced expression of miR-34a on multiple myeloma (MM) cells, showing a time-dependent modulation of several signaling pathways involved in the control of cell proliferation and apoptosis. The last group was the first one to have demonstrated a role of miR-34a in the pathogenesis of MM finding it downregulated in a wide series of MM samples.31,32 The involvement of miR-34a in this haematological neoplasia can open a new scenario in understanding the molecular pathways involved in the generation of this disease from preneoplastic lesions such as monoclonal gammopathy. One of the most affected was the Erk/Akt-dependent pathway.31 The authors demonstrated that miR-34a induces sequential down modulation of both Erk and Akt activity, which is followed by pro-caspase-6 and -3 cleavage and apoptosis induction in MM cells. Previously, the same authors have also showed the potential of miR-34a treatment in downregulating both Bcl-2 and NOTCH1 and in the induction of apoptosis both in vitro and in a xenograft mouse model.32 In addition, downregulation of DICER in cancer cells was found to correlate with the promotion of metastasis. Intriguingly, DICER1 deficient colon cancer cells showed lower expression of EpCAM, indicating invasive potential, and significant over-expression of CD44 and other cancer stem cell markers. Increased metastatic potential in DICER1-impaired cells associated with the defective production of the miRNAs that regulate the pathways involved in this process, such as miR-34a, miR-126, and the miR-200 family.33 MiR-34a has been also associated with regulation of cancer stem cells function in various cancer types such as prostate cancer,34 pancreatic cancer,35 medulloblastoma,36 glioblastoma.37 Furthermore, miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44, indicating the direct role of CD44 and miR-34a in cancer development and progression.34 Consistently, Shi et al. described that overexpression of miR-34a inhibited CD44hi NSCLC cell growth in vitro and in xenograft tumors, demonstrating that miR-34a impairs tumors regeneration by negatively regulation of stem-like NSCLC.38 In lung cancers, miR-34a has been evaluated as a replacement therapy candidate; in fact, exogenous miR-34a-mimics delivery was found to substantially reduce the tumor growth.39 In addition, a relative loss of miR-34a expression was considered a key etiologic factor in contributing to the aggressive behavior of lung cancer stem cells (CSC), and thus those features were mitigated by exogenous delivery and restoration of miR-34a activity.40
DNA Damage Regulation
MiRNAs are actively involved in the regulation of genes that are related to DNA damage and repair; therefore, changes in miRNA biogenesis and maturation processes are often associated with the response to these mechanisms. Recent studies show that transcription of miRNA can be directly affected by DNA damage. We have already underlined the critical role played by the p53 gene in this regulation and the p53-dependent modulation of miR-34a in response to DNA injury.30 Several studies have found that DNA damage-induced miR-34a expression was dependent on p53, and that this was followed by induction of cell cycle arrest, promotion of apoptosis, and DNA repair.36 Wild-type p53 expressing glioblastoma cell lines have been shown to respond to radiation and there was significantly higher DNA damage in post-irradiated cancer cells. Mechanistically, it has been shown that miR-34a-mediated negative regulation of p53-binding protein 1(53BP1) resulted in suppression of genomic instability in tumor cells.41 p53 can induce expression of miR-34a also in irradiated mice. Moreover, upregulation of miR-34a in response to genotoxic agent exposure is observed in different biological systems.38 When DNA damage activates the p53 gene, p53 protein binds to the promoter of miR-34a and upregulates miRNA expression (Figure 1). MiR-34a is, in fact, a direct transcriptional target of p53 because the promoter region of miR-34a contains a canonical p53 binding site.39 The p53 network suppresses tumor formation through the coordinated activation of multiple transcriptional targets, and miR-34 may act in concert with other effectors to inhibit inappropriate cell proliferation. DNA damage signaling also affects the miRNA maturation biogenesis processes through the activation of the p53 gene. In fact, p53 binding to DROSHA facilitates the processing of pri-miRNAs in pre-miRNAs and mutation in the DNA-binding domain of p53 negatively affects this processing, thus reducing the expression of the related miRNAs. Moreover, in silico analyses, reveal that all three components of the p53 tumor suppressor, p53, p63, and p73, can regulate the major components of miRNA processing, such as DROSHA-DGCR8, DICER-TRBP2, and Argonaute proteins. In addition to being a direct transcriptional target of p53, miR-34a indirectly upregulates p53-dependent apoptosis through another intermediate protein, SIRT1,39 which has a well-conserved NAD-dependent sirtuin core domain, and catalyzes NAD-dependent deacetylation of several targets. Once activated, SIRT1 deacetylates also a variety of non-histone target proteins, such as p53, the retinoblastoma protein (Rb), FoxO transcription factors, Ku70, NFκB and PGC-1alpha. SIRT1-mediated deacetylation of p53 decreases the transcriptional activation and consequent protein expression of p53 downstream targets, such as p21 and PUMA.39 Therefore, SIRT1 mediates the survival of cells during periods of severe stress through the inhibition of apoptosis. MiR-34 inhibition of SIRT1 leads to an increase in p53 acetylation and p53 activity. As a result, miR-34 suppression of SIRT1 strengthens p53-mediated apoptosis. Simultaneously, increased p53, heightens miR-34a production, thus enhancing p53 stabilization and completing a positive feedback loop that protects cells from cellular oxidative stress and DNA damage.
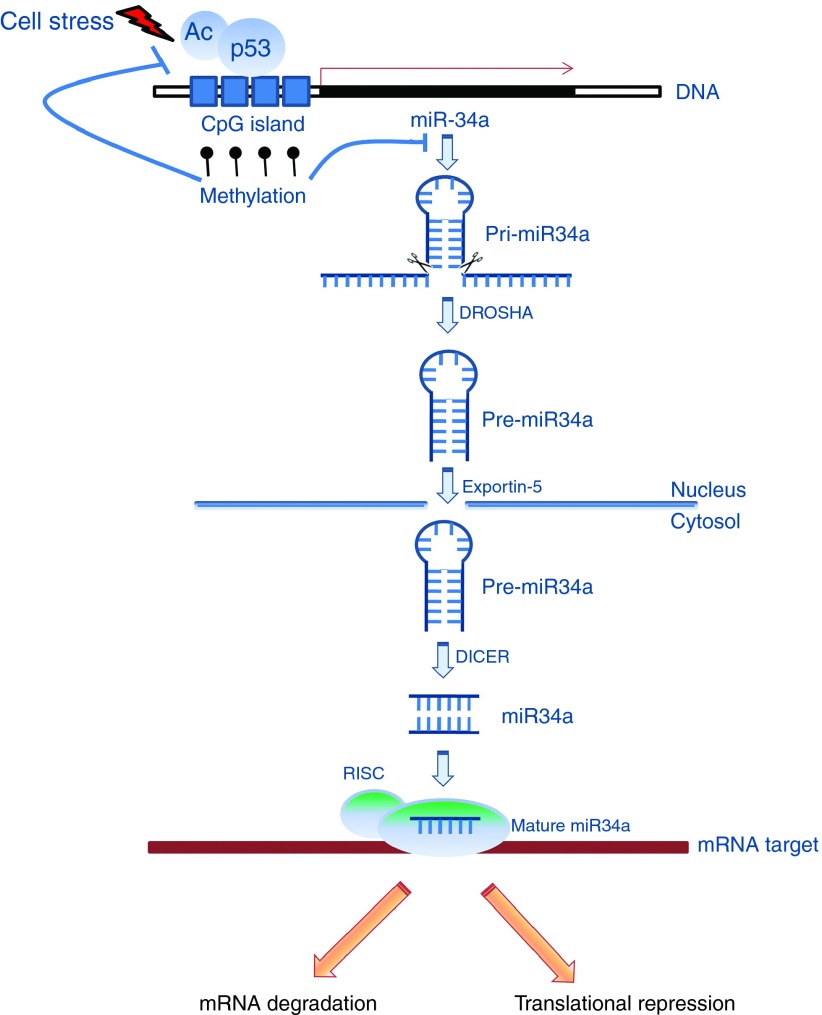
Figure 1.
MiR-34a biogenesis. DNA damage activates the p53 gene, that binds to the miR-34a promoter and up-regulates miRNA expression. Mutation in the DNA-binding domain of p53 negatively affects this processing, thus reducing the expression of the related miRNAs. CpG island miR-34a promoter hypermethylation induces a miR-34a silencing that is dominant over its transactivation by p53 after DNA damage. MiR-34a is originally transcribed as long hairpin molecule (pri-miRNA) that is subsequently processed by the human RNase III DROSHA, into a stem-loop precursor of approximately 70 nucleotides (pre-miRNAs). Exportin-5 mediates the miR-34a traslocation from the nucleus to the cytoplasm. Another human RNase III, DICER, through several steps, cuts miR-34a into duplexes with final 22–23nt length. As a last step, one strand of the miRNA duplex (‘‘mature strand'') is incorporated into the RNA-induced silencing complex (RISC) while the other is supposedly degraded. Once integrated into the RISC, miR34a guide this protein complex to partially or totally complementary binding sites located in the 3' untranslated region (UTR) of target mRNAs and inhibit their expression. In detail, the perfect alignment causes mRNA degradation, while the partial alignment interfers with mRNA translation.
MiR-34a Targets
Bioinformatic approaches and cellular experiments have allowed identifying several mRNAs as direct miR-34a targets (Table 1). Most of them encode factors required for G1/S transition (c-MYC, E2F, CDK4, CDK6), anti-apoptotic proteins (Bcl2, SIRT1), and proteins involved in invasion (c-MET).30 MiRNAs regulate their targets via association of a 7 nucleotide stretch, the so-called seed-sequence, located in their 5′-portion with a complementary sequence in the 3′-UTR of the target mRNA.31 Additional base pairing may occur via nucleotides in the middle and 3′-portion of the miRNA. Since the relatively short seed region is the primary determinant of target recognition, a single miRNA presumably regulates dozens or even hundreds of target mRNAs. Bioinformatic predictions suggest that several hundred mRNAs contain matches to the miR-34a seed sequence. Recently, proteomics analysis revealed early targets of miR-34a involved in neuroblastoma tumorigenesis.42,43 However, the main miR-34a targets also confirmed by cellular experiments are summarized below and illustrated in the Figure 2.
Table 1. Main miR-34a targets. Sequence alignment based on bioinformatic predictions.
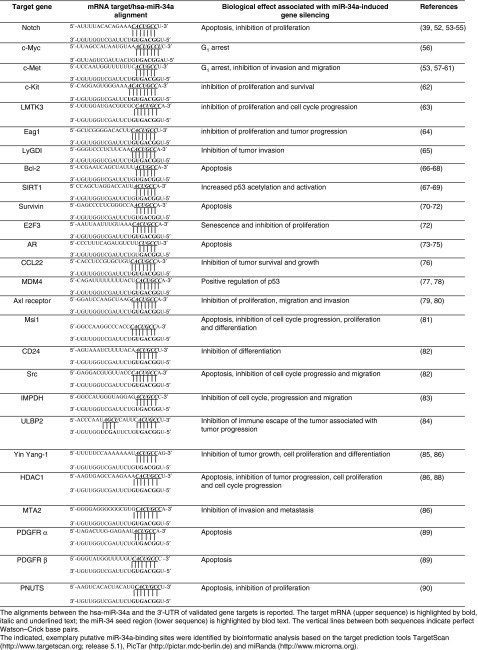
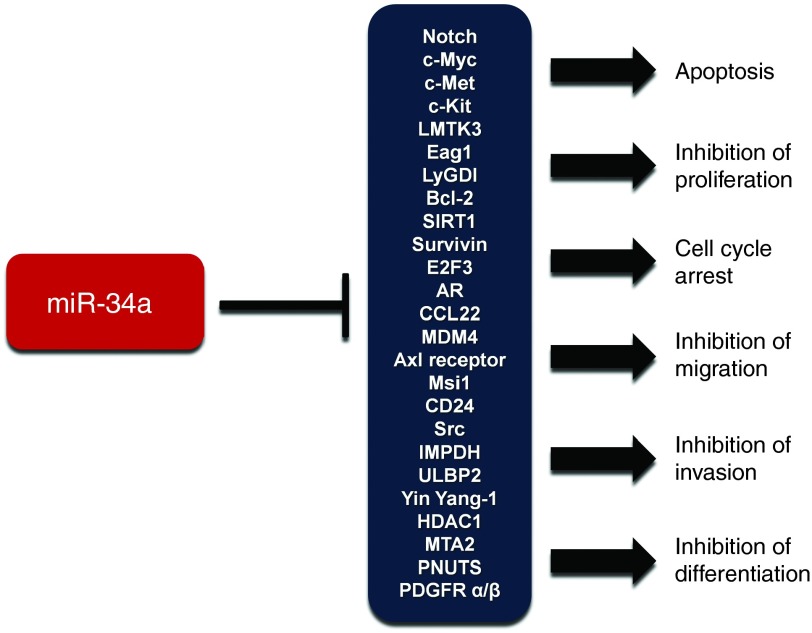
Figure 2.
Cellular outcomes associated with miR-34a-induced gene silencing. Representation of the main miR-34a target mRNAs, and biological effects associated with their repression. MiR-34a can antagonize many different oncogenic processes.
Notch
Notch signaling pathway participates in a variety of cellular processes, including cell fate specification, differentiation, proliferation, apoptosis, adhesion, epithelial-mesenchymal transition, migration, and angiogenesis. Activation of Notch signaling contributes to the pathogenesis of several human neoplasms and its inhibition may be an effective therapeutic approach. Li et al., put for the first time in relation the augmented levels of Notch with the miR-34a downregulation in glioblastoma and medulloblastoma, showing that miR-34a targeted both Notch-1 and Notch-2.44 Furthermore, in cervical cancer and trophoblast cell lines, miR-34a was found to regulate the expression of Jagged-1 and its receptor Notch-1; enforced expression of miR-34a was related with the reduction of the invasion capacity Jagged-1 and Notch-1–mediated.45 It is well acclaimed that Delta-like ligand-1 (Dll1) is a known ligand of the Notch-1 and Notch-2 receptors. It has been shown that Dll1 is negatively regulated by miR-34a. Medulloblastoma cells reconstituted with miR-34a displayed remarkably reduced expression of Dll1 that resulted in activation of Notch-1 mediated signaling, as evidenced by Notch-1 intracellular domain (NICD1) protein after 12 hours. Also, one of the target genes of Notch-1 signaling, HEY1 was noted to be increased. Interestingly, Notch-2 mediated signaling was inhibited in miR-34a reconstructed cancer cells, as evidenced by downregulation of NICD2 and of its known target: the Hairy and enhancer of split 1 (Hes1).46 Notably, since the Notch pathway is a critical regulator of asymmetric division of stem cells, Bu et al. described that well-differentiated colorectal cancer may perform either self-renewal or differentiated division depending on miR-34a levels. Specifically, low mir-34a levels upregulate notch signaling and promote symmetric division to produce colon cancer stem cells (CCSC), while asymmetric division and production of differentiated non-CCSC is controlled by high miR-34a levels that depress Notch signaling.47,48
c-Myc
c-Myc is one of the major factors involved in tumor development. Thus, its inhibition by miR-34a binding in 3′ UTR region is thought to be a significant function of miR-34a. In primary renal cell carcinoma, miR-34a reduces the c-Myc–Miz–Skp2 transcriptional complex, which induces RhoA transcription and inhibits cell invasion. MiR-34a also directly suppresses formation of c-Myc–P-TEFb complex involved in the control of the elongation phase of transcription by Polymerase II. MiR-34a was also shown to strongly inhibit cell proliferation, in vivo xenograft tumor growth and cell invasion. Moreover, C-Myc resulted negatively regulated by miR-34a in kidney cancer cells and ChIP assay revealed that miR-34a suppressed the recruitment of c-Myc to the RhoA promoter.49
c-Met
A series of studies carried out in parallel with those about Notch, revealed that also c-Met is a known target of miR-34a and correlates to the metastasis potential of tumors. Enforced miR-34a expression downregulated c-Met oncogene and inhibited proliferation and metastasis of osteosarcoma cells,50 of cervical carcinoma and choriocarcinoma cells,45 and of human hepatocellular carcinoma cells.51 In HepG2 cells, ectopic expression of miR-34a potently reduced both mRNA and protein levels of c-Met, inhibited tumor cell migration and invasion in a c-Met-dependent manner and decreased c-Met-induced phosphorylation of extracellular signal-regulated kinases 1 and 2 (ERK1/2).51 Interestingly, transfection of mesothelial cells with miR-34a inhibitors considerably enhanced cell proliferation and invasive potential via upregulation of c-Met, phosphorylated c-MET, and Bcl-2 proteins.52 Moreover, proliferation of UCH1 chordoma cells was effectively inhibited after miR-34a transfection and apoptosis was inducted by c-Met downregulation following a direct targeting by miR-34a.53 Finally, Menges et al. demonstrated that malignant mesotheliomas are characterized by enhanced presence of CSC population associated with p53/miR-34a-dependent activation of c-Met.54
c-Kit
c-Kit, which is also known as CD117 or stem cell factor receptor, is a type III receptor tyrosine kinase (RTK) and an important mediator/initiator of several signaling cascades. Deregulated expression and activation of c-Kit contributes to several types of cancers, such as leukemia, glioblastoma, melanoma, lung and breast cancer. c-Kit, as well as various stemness markers (CD44, Lgr5, and BMI-1), is over-expressed in colorectal cancer cells. A recent report by Siemens et al., shows that miR-34 directly targets the c-Kit mRNA and thereby mediates repression of c-Kit expression. Accordingly, miR-34 activation negatively regulates c-Kit mediated signaling events and cell transformation, and stimulates the chemosensitization.55
LMTK3
Another recent issue identifies in lemur tyrosine kinase 3 (LMTK3), an important regulator of estrogen receptor alpha (ERα), a novel functional target of miR-34a involved in cell proliferation and cell cycle progression.56 Treating ERα-positive breast cancer cell line MCF-7 with estrogen, results in downregulation of miR-34a. However, enforced expression of miR-34a, using lentiviral vector, causes downregulation of LMTK3 and reduces Akt phosphorylation. This study reveals that, according to the mRNA sequence, LMTK3 is a predicted target of miR-34a and this is confirmed by dual luciferase reporter assay and by the decrease of LMTK3 mRNA and protein levels after over-expression of miR-34a.56
Ether à go-go 1 channel
Ether à go-go 1 (Eag1) channel is constitutively overexpressed in osteosarcoma. Enforced expression of miR-34a in MG-63 and Saos-2 cells mediated downregulation of Eag1, via a negative feed-forward mechanism. Moreover, cancer cells reconstructed with miR-34a remarkably induced regression of growth of osteosarcoma cells in nude mice.57
LyGDI
There are multiple lines of evidence showing that LyGDI expression is related to tumor invasion and altered radio-sensitivity. LyGDI is one of the key regulators of Rho GTPases based on its ability to bound Rac1, Rac3, and CDC42 in their inactive state. Duan et al. found that ectopic expression of miR-34a downregulated LyGDI expression, and thus promoted constitutive activation and membrane translocation of Rac1, dramatically reducing expression of cyclooxygenase-2 and thus restoring radiation induced apoptosis in non-small cell lung cancer cells. Furthermore, these investigators predicated by TargetScan and further confirmed by both real-time RT-PCR and protein expression studies, that LyGDI is a new miR-34a target gene.58
Bcl-2 and SIRT1
Several investigational studies have detected Bcl-2 and SIRT1 as direct targets of miR-34a, identifying also an inverse correlation between the miR-34a level and both Bcl-2 and SIRT1 expression. Recently, hepatocellular carcinoma (HCC) tissues with lower miR-34a expression were found to express higher levels of Bcl-2 protein than those with elevated expression of miR-34a. Bioinformatics and luciferase reporter assays revealed that miR-34a binds the 3′-UTR of the Bcl-2 mRNA and represses its translation. Western blotting analysis and qRT-PCR confirmed that Bcl-2 is inhibited by miR-34a overexpression.59 Remarkably, it has been demonstrated that transfection of HCC cells with oncolytic adenovirus co-expressing IL-24 and miR-34a was an effective antitumor strategy and Bcl-2 and SIRT1 were noted be direct miR-34a targets.60 In breast cancer cells, restoration of miR-34a induced apoptosis and suppressed both proliferation and invasion through the simultaneous inhibition of Bcl-2 and SIRT1, and this effect was significantly amplified by the combination with 5-Fluorouracil (5-FU) treatment.61 Moreover, it has been shown that the treatment with the DNA damaging agent adriamycin induced expression of miR-34a in p53 wild-type A549 human lung cancer cell line, and this effect was not observed in mutant p53 SBC-5 cells. Sirtuins have emerged as multifunctional regulators reported to deacetylate histones and several transcriptional regulators in the nucleus and SIRT1 have been shown to be quantitatively controlled by miR-34a and miR-199a. It has also been shown that gene silencing of SIRT1 in lung cancer cells considerably improved sensitivity to cisplatin.62
Survivin and E2F3
Survivin has been shown to be an ideal target for cancer gene therapy based on its strong antiapoptotic effect. Shen et al. explored the relationship between miR-34a and survivin in larynx squamous cell carcinoma (LSCC). They found a direct negative regulation of gene expression, explored by using quantitative real-time PCR, Western blot and cell cycle analyses after transfection of miR-34a mimic in LSCC. MiR-34a expression levels were correlated with tumor differentiation, lymphatic metastasis, clinical stages, and survival rates.63 Transfection of gastric cancer cells with miR-34a mimic resulted in inhibition of proliferation and invasion and miR-34a inhibitory effect was regulated by survivin targeting.64 More recently, the same authors have related miR-34a re-expression with improved sensitivity of gastric cancer cells against cisplatin-based chemotherapies, with PI3K/AKT/survivin signaling pathway possibly involved in this mechanism.64 Furthermore, the enforced expression of miR-34a in head and neck squamous cell carcinoma cells extensively reduced cell proliferation, colony formation and migration via targeting of E2F3 and survivin. Cancer cells reconstituted with E2F3 restored expression of survivin in miR-34a expressing cells.65
Androgen receptor
Androgen Receptor (AR) is a key determinant for the molecular changes required for apoptosis resistance and for driving prostate cancer cells from an androgen-dependent to an androgen-independent or androgen depletion-independent (ADI-castration resistant) state. Therefore, downregulation of AR expression should be considered the main strategy for the treatment of ADI prostate cancer. Enforced expression of miR-34a in prostate cancer cells considerably inhibits self-renewal capacity and exerts a direct negative control on AR, PSA and Notch-1.66 It has been recently shown that the cooperation between miR-34a and 34c plays an important role in AR-dependent p53-mediated apoptosis in prostate cancer. Active AR contributes to the activation of some kinases that after treatment with DNA damaging agents enhance p53 activity and its downstream targets. Accordingly, knocking down of AR prevents activation of kinases that upregulate p53. Concurrently, active AR inhibits calcium/calmodulin-dependent kinase II (CaMKII), while AR inhibition activates CaMKII that partially restores p53 activation.
CCL22
A wealth of information suggests that, in tumor microenvironment, FoxP3 expressing regulatory T cells (Treg) are co-opted by tumor cells to tactfully escape from immune surveillance. Hepatitis B virus has been shown to potentiate TGF signaling in liver microenvironment, thus suppressing miR-34a and restoring the expression of chemokine CCL22. To better understand the role of miR-34a and CCL22 in regulation of tumor growth, Hepa cells were inoculated subcutaneously into C57BL/6J mice and tumor growth was noted. Enforced expression of miR-34a significantly reduced tumor growth in immunocompetent mice, while the removal of miR-34a target sequence from CCL22 mRNA, inhibited miR-34a mediated negative regulation of CCL2 expression.67
MDM4
Among many reported putative miR-34 targets, the MDM4 gene (also referred to as HDM4 in humans) has emerged as a prominent candidate due to its important functions as one major negative regulator of p53. It is a well-established that MDM4 binding to p53 unable the triggering of its target genes. MDM4 is negatively regulated by MDM2 via MDM2 mediated degradation. Recent emerging evidence has started to shed light on the MDM4 negative regulation by miR-34a, showing that it is likely not mediated by the 3′ untranslated region (UTR), but rather by a miR-34a site in the coding region of the last exon of MDM4 (exon 11).68 Okada et al., demonstrated a positive feedback loop between p53 and miR-34a that is at least in part due to the miR-34a-mediated repression of the negative p53 regulator, MDM4. While p53 induces the transcription of miR-34a, this miRNA in turn represses post-transcriptionally MDM4, in parallel with the repression of SIRT1, to enhance p53 transcription activity and decrease p53 protein turnover. This positive feedback loop between p53 and miR-34 further strengthens the p53 activity upon p53 activation, conferring a robust tumor suppression response.69
Axl receptor tyrosine kinase
Axl is a tyrosine kinase receptor involved in the induction of proliferation, migration and invasion. In triple negative breast cancer cells, MDA-MB-231, transfection of miR-34a resulted in suppression of Axl receptor, that has been first identified as a putative miR-34a target using multiple miRNA/target prediction algorithms and further confirmed by reporter assays. Moreover, it was observed a notable decline in Akt phosphorylation and phenotypic effects on cell migration in miR-34a transfected cancer cells.70 Luciferase-reporter assays confirmed the specificity of miR-34a targeting on the 3′-UTR of Axl. An inverse correlation between Axl protein and miR-34a expression in a panel of non-small cell lung cancer (NSCLC), colorectal cancer (CRC), and breast cancer (BRC) cell lines was also observed. Moreover, pre-miR34a transfection inhibited in vitro migration and invasion and, in vivo, reduced the number of distant lung- or liver-metastases.71
Msi1
Musashi1 (Msi1) is an evolutionarily conserved RNA-binding protein that has been connected to the development of multiple tumor types such as glioblastoma multiforme, medulloblastoma, cervical carcinoma, lung and colon cancers. It can interfere with the expression of specific gene subsets involved in cell cycle control, proliferation, apoptosis and differentiation, and results to be negatively regulated by miR-34a. Msi1 long 3′-UTR region was proven to be targeted by several tumor suppressor miRNAs including miR-34a. It has been reported that cells reconstituted with miR-34a displayed a decrease in Msi1 expression profile and a reduced cell proliferation. Furthermore, cell proliferation inhibition induced by the tumor suppressor miRNAs was partially rescued by Msi1 transgenic expression.72
CD24 and Src
CD24 is a glycosylphosphatidylinositol (GPI)-anchored membrane protein and has been shown to activate Src. Src is well known to induce the activation of AP-1 transcription factors via MAPK mediated phosphorylation of c-Jun. Therefore, CD24 mediate expression of miR-21 via AP-1 transcription factors. Transient transfection of CD24 in low CD24-expressing Rko and HCT-116 cell lines triggers upregulation of miR-21. 3′-UTR of CD24 or Src were individually co-transfected together with pre-miR-34a into the HT-29 and Geo cell lines and the resulting data revealed that cancer cells displayed notably reduced expression of CD24 and/or Src. These findings suggest that miR-34a inhibited expression of miR-21 via negative regulation of CD24 and Src. Moreover, reversion of the epigenetic silencing of miR-34a was assumed to be therapeutically beneficial for colorectal cancer patients.73
Inosine 5′-monophosphate dehydrogenase
MiR-34a has also been shown to quantitatively control the expression of inosine 5′-monophosphate dehydrogenase (IMPDH), a rate-limiting enzyme of de novo GTP biosynthesis. MiR-34a mediates negative control of IMPDH, resulting in dramatic suppression of GTP-dependent Ras signaling pathway.74
ULBP2
Natural killer cell immunoreceptor ligands NKG2DL are present on surface of tumor cells and sensitize them to killing by NK cells and cytotoxic T lymphocytes. ULBP2 is a NKG2DL and strong prognostic marker in human malignant melanoma regulated by miR-34a/c and miR-449a/miR-449c. Bioinformatic analyses revealed that both miRNAs directly target the 3′-UTR region of ULBP2 mRNA. Accordingly, transfection of miR-34a in melanoma cells notably reduced ULBP2 expression and NK cells-mediated recognition of tumor cells.75
Yin Yang-1, HDAC1, and MTA2
Overexpression of miR-34a in GBM cells (U251) resulted in suppression of EGFR protein. It was mechanistically shown that miR-34a mediates targeting of Yin Yang-1 (YY1), a transcription factor that can stimulate the expression of EGFR, thus repressing the expression of cell-cycle proteins and EGFR. Moreover, both deletion of miR-34a and amplification of EGFR were associated with a drastic reduction of overall survival in glioblastoma (GBM) patients.76 Kaller et al., confirmed miR-34a targeting of YY1 mRNA and identified additional miR-34a targets known to be negative regulators of p53 such as MTA2 and HDAC1.77 MTA2 and HDAC1 are components of the NURD complex, which mediates deacetylation and destabilization of the p53 protein. YY1, in addition to stimulating the expression of EGFR, is also able to modulate MDM2-mediated ubiquitination of p53. The repressing activity of YY1, MTA2 and HDAC1 is therefore one of the different mechanisms by which miR-34a exerts its growth inhibitory effects.77 TP53-positive and TP53-negative cancer cells displayed notable miR-34-mediated inhibitory effect on cancer cell growth. It has been shown that HDAC1 represses expression of p21 in cancer cells. However, cancer cells reconstituted with miR-34 displayed an increase in p21 expression via HDAC1 mediated negative regulation.78
PDGFR α/β
Data obtained from five different NSCLC cell lines with p53 WT, mutant or null indicated that miR-34a, -34b, and -34c expression was downregulated with respect to normal cells. Furthermore, expression of PDGFRα and PDGFRβ was found to be notably upregulated. The most intriguing feature was an inverse correlation between PDGFRα/β and miR-34a/c. Overexpression of miR-34a/c resulted in the downregulation of PDGFRα/β and in the consequent restoration of TRAIL-induced apoptosis in NSCLC cell lines.79
PNUTS
Xenografting nude mice with CD44+CD133+ gallbladder cancer tumor stem-like cells resulted in establishment of tumor. It was astonishing to note that lower miR-34a expression and longer telomere length were observed in gallbladder adenocarcinoma tissues. Enforced expression of miR-34a dramatically reduced tumor-forming capacity and increased sensitivity to radiation via negative regulation of PNUTS, thus providing an effective antitumor strategy. PNUTS has been shown to be constitutively overexpressed in gallbladder cancer cells, where it is able to promote telomere lengthening. Therefore miR-34a inhibitory effect was also mediated by negative regulation of telomere lengthening.80
Regulators Involved in The Modulation of MiR-34a
MiRNA maturation is controlled by the DROSHA microprocessor complex. Recently, additional molecules have been shown to be involved in miRNA maturation; e.g., the TGF-β signal transducer SMADS promotes miR-21 maturation, and the tumor suppressor p53 enhances the processing of miR-16, miR-143, miR-145, and miR-206,81 whereas estrogen receptor α attenuates maturation of pri-miRNAs into pre-miRNAs.82 However, only a limited number of miR-34a modulators are currently known and these are listed and discussed below.
CD95
It is interesting to note that CD95 is a target gene of p53 and p53 knockout in HCT116 cells induced significant reduction of CD95 levels. Surprisingly, it has been shown that overexpressing CD95 in CD95 low expressing MCF7 cells caused dramatic upregulation of miR-34a in chemotherapeutic drug treated cancer cells. Moreover, miR-34a expression was correlated with the sensitivity of cells to CD95-mediated apoptosis. MiR-34a was also identified as a marker of cancer cells that are more sensitive to CD95-mediated apoptosis, and as a sensitizer to apoptosis mediated by CD95. Another exciting piece of information related to CD95-mediated signaling is the negative regulation of let-7 expression. In details, it was discovered a p53-regulated network in which CD95 maintains p53 expression and, indirectly, also the expression of miR-34a, providing a substantial protective axis against the loss of let-7.83
PPAR-γ
Recent evidence has demonstrated the underlying mechanisms of miR-34a induction in TP53 (biallelically altered) cell lines treated with PPAR-γ ligand.84
PAR2
Proteinase activated receptor 2 (PAR2) was found to be broadly expressed in cancer and was positively correlated with tumor progression in colorectal cancer. It has recently been shown that PAR2 triggers the expression of Cyclin D1 via suppressing miR-34a in colon cancer cells. In addition, it was shown that PAR2 modulates upregulation of TGF-β that also suppresses miR-34a expression.85
Myc
Myc mediates downregulation of miR-34a in diffuse large B-cell lymphoma (DLBCL) cells and enhances FoxP1-regulated cell proliferation. Cells transfected with miR-34a considerably reduced proliferation via the targeting of FoxP1.86
SNAIL and ZEB1
MiR-34a downregulation is now well studied during EMT in TGF treated cancer cells. SNAIL and ZEB1 trigger expression of miR-34a/c via binding to E-boxes in the miR-34a/b/c promoters. MiR-34a has also been shown to negatively regulate SNAIL and ZEB1. Therefore, enforced expression of miR-34a resulted in reversal of mesenchymal to epithelial transition.87 Consistently, EMT-mediated invasion and metastasis was described after IL-6R/STAT3/miR-34a feedback loop activation in CRC primary tumors.88 The authors demonstrated that the exposure of CRC cells to IL-6 activates the transcription factor STAT3 that directly repress miR-34a gene and that IL-6R is a miR-34a target.
PDGF
MiR-34a expression was downregulated in response to PDGF treatment in NIH-3T3 cell line harboring a unique fusion protein (KP) composed of the extracellular domain of KDR (VEGF receptor II) and the intracellular domain of PDGFRA. It was further shown that PI3K/Akt/MDM2 signaling axis did not reduce the expression of p53 in KP-expressing NIH-3T3 cells; therefore, the regulation of miR-34a expression by PDGF signaling likely operates through a p53-independent mechanism. These findings thus identify a reciprocal negative feedback loop influencing oncogenic receptor tyrosine kinase (RTK) signaling.89
Modulation of Drug Response in Cancer Cells
Several works have established a role for miR-34a in modulating responsiveness to chemotherapy. This effect is at least in part mediated by the direct post-transcriptional repression of the oncogenic substrates of miR-34a. The altered modulation of drug response mediated by miR-34a has been reported in a number of cellular models including NSCLC, HCC, breast cancer, bladder cancer, colorectal cancer, prostate cancer, and head and neck squamous cell carcinoma cell lines. In NSCLC cells with primary and acquired erlotinib resistance, therapeutic miR-34a treatment induced tumor growth inhibition by the replacement of the sensitization to the drug. Moreover, a strong synergistic interaction between erlotinib and miR-34a mimics was also observed in a panel of HCC known to be refractory to erlotinib.90 Li et al. showed that enforced expression of miR-34a in MCF-7 breast cancer cells was able to restore sensitivity to adriamycin and this effect was partially reversed by the use of miR-34a inhibitor. It was also found an inverse association between Notch1 and miR-34a expression; thus the acquired adriamycin resistance of breast cancer was at least in part attributed to the targeting of Notch1.91 Similarly, miR-34a transfection increased the sensitivity to cisplatin for lung cancer cells, and this effect could be reverted by the miR-34a inhibition.62 Several studies have demonstrated that perturbation of the p53-Rb signaling axis is linked to muscle-invasive transitional cell carcinoma of the bladder (MI-TCC) progression following cystectomy. Based on this premise, and considering the ability of miR-34a to target multiple points of the p53-Rb pathway, Vinall et al. investigated whether increased miR-34a expression could sensitize bladder cancer cell lines to treatment with cisplatin. They demonstrated that transfection with pre-miR-34a increased chemosensitivity to cisplatin through inhibition of Cdk6 and SIRT1. Analysis of miR-34a expression in TCC patient samples taken before chemotherapy revealed that many patients express low levels of miR-34a and that lower miR-34a expression may be a useful predictor of response to chemotherapy.92 Another study compared miR-34a the levels of human colorectal cancer DLD-1 cells 5-fluorouracil (5-FU)-resistant with the parental counterpart of DLD-1 cells. It was not only found that 5-FU-resistant cells expressed significant higher levels of miR-34a, but also that the introduction of miR-34a into resistant cells significantly inhibited cell growth and induced a considerable attenuation of 5-FU-resistance, which was accompanied by reduced expression of SIRT1 and E2F family proteins. These findings suggest that the resistance to 5-FU in DLD-1 cells was at least in part mediated by miR-34a/SIRT1/E2F3 cascade.93 Furthermore, also paclitaxel resistance resulted overcome by miR-34a over-expression and SIRT1 knockdown in hormone-refractory PC3, as demonstrated by Kojima et al.94 Moreover, cis-diamminedichloroplatinum resistant head and neck squamous cell carcinoma cell line revealed downregulated miR-34a. In addition, it is relevant to mention that a significant association was found between decreased expression of miR-34a and poor disease specific survival (P = 0.0015), poor disease free survival (P = 0.0019), and poor local control rates (P = 0.017) in patients with nasal squamous cell carcinomas treated with intra-arterial infusion of cis-diamminedichloroplatinum. Furthermore, multivariate analyses demonstrated significant associations between miR-34a expression and the hazard ratios of disease free survival.95 Therefore, there are excellent premises for the use of miR-34a as prognostic biomarker in patients with nasal squamous cell carcinoma. Another study underlined the important advance in diagnostic and prognostic settings represented by the use of miRNA signatures. In details, Nakatani et al.96 investigated miRNAome expression in 34 Ewing's sarcoma (EWS) patients, validating the expression of the most significant miRNAs by qRT-PCR. The resulting data, with the support of in vitro studies attesting the increased chemo-sensitivity and the decreased aggressiveness of cancer cells after enforced expression of miR-34a, lead to consider miR-34a as the most significant predictor of outcome.
Nutrigenomics
There are several evidences regarding the influence of phytochemicals and nutrients such as amino acids, carbohydrates, fatty acids, and vitamins on miRNA gene expression in animals and on the consequent physiological consequences. For this reason, it was also explored the potential use of miRNAs as new therapeutic agents for the dietary-derived disorders and the putative effect of food-derived miRNAs to influence host gene expression. More in details, as regards the effects on miR-34a expression, the lack of folic acid (vitamin B 9) in the nutrient medium of cell cultures caused changes in its expression levels, which returned to the basal extent after the re-addition of folate to the growth medium. It was demonstrated that rats fed with a diet deficient in methyl donors (folate, methionine, and choline) developed hepatocellular carcinoma97 and that miR-16, -17–92, -34a, and -127 presented a decreased expression in hepatocellular carcinoma induced by methyl deficiency.98 Interestingly, changes in the expression of these miRNAs and protein levels of their targets, resulted a prominent event during development of nonalcoholic steatohepatitis (NASH) induced by methyl deficiency, and this strongly suggests that severity of NASH and susceptibility to NASH may be determined by variations in miRNA expression response. Recently, it was found a synergism between curcumin (a biphenyl compound derived from rhizome) and emodin (an active component isolated from the root and rhizome of Rheum palmatum) in breast cancer cells growth inhibition, suggesting a new and potentially useful approach for breast cancer therapy. Interestingly, breast cancer cell lines, including MDA-MB-231 and MDA-MB-435, show low expression of miR-34a; however, emodin and curcumin treatment revealed an increase in miR-34a levels and a concomitant negative regulation of miR-34a-dependent anti-apoptotic genes, including Bcl-2 and Bmi-1.99 CDF is a curcumin analog with an improved bioavailability. It is now well known that instead of HCT116 colon cancer cell line, SW620 colon cancer cell line is positive for CpG methylation of miR-34a promoter. 5-aza-2′-deoxycytidine, a methyltransferase inhibitor, effectively induced expression of miR-34a in SW620 cells. Likewise, CDF also triggered upregulation of miR-34a in SW620 cells.100 The action of other natural compounds such as Rhamnetin, a polyphenol, and Cirsiliol, a 5, 3′, 4′-trihydroxy 6,7-dimethoxyflavone, was studied for the treatment of non-small cell lung cancer cells (NSCLC) cells. Upon the drug combination, cells displayed an increase in miR-34a expression and a Notch-1 suppression, with consequent notable downregulation of the nuclear factor-κB pathway.101 In terms of cancer prevention and treatment, the tocotrienols belonging to the Vitamin E family, seem to be able to both stop angiogenesis and promote apoptosis. Delta-tocotrienols appear to be the most effective in inducing apoptosis in cancer cells by inhibiting multiple signaling pathways such as the EGFR, NF-κB, MAPK, and PI3K/AKT pathways. Experimental evidence showed that delta-tocotrienol inhibited Notch-1 signaling, cell proliferation, invasion and induced apoptosis in NSCLC cells via alteration of miR-34a expression.102 The induction of apoptosis via the indirect targeting of Notch-1, through the induction of miR-34a upregulation, was also found in pancreatic cancer cells treated with the soy isoflavone Genistein,103 that was reported to be involved also in the regulation of prostate cancer.104 The treatment of DU145 prostate cancer cells (Mutant RB) with flavone substantially reduced cdk4 and cdk6 and concomitantly enhanced Cdk inhibitors p21/p27. Moreover, flavones-mediated increase in miR-34a expression, caused the negative regulation of E2F1 and E2F3.104 It has subsequently been shown that Genistein was able to regulate HOTAIR. HOX transcript antisense RNA (HOTAIR) gene is located within the Homeobox C (HOXC) gene cluster on chromosome 12 and encodes lncRNA molecule. Emerging evidence has started to shed light on HOTAIR interaction with both PRC2 and lysine specific demethylase 1 (LSD1) complexes. HOTAIR interaction with PRC2 and LSD1 mediated histone H3 lysine 27 methylation and lysine 4 demethylation for epigenetic silencing of different genes. Astonishingly, Genistein induced upregulation of miR-34a and mediated negative regulation of HOTAIR in PC3 and DU145 prostate cancer cells inhibiting their cell growth.105 Oral administration of 3,6-dihydroxyflavone (3,6-DHF) to BALB/c nude mice inoculated with breast cancer cells induced in vivo regression of tumor growth as well as a reversion of the global upregulation of miR-21 and the downregulation of miR-34a, markedly promoting pro-apoptotic effects.106 It has recently been shown that treatment of malignant neuroblastoma SH-SY5Y and SK-N-DZ cells with epigallocatechin-3-gallate (EGCG) notably reduced expression of miR-92, miR-93, and miR-106. However, miR-7-1, miR-34a, and miR-99a were found to be upregulated.107 Moreover, also the combined treatment of malignant neuroblastoma SK-N-BE2 and IMR-32 cells with N-(4-hydroxyphenyl) retinamide (4-HPR) and EGCG, induced an increase in the expression of miR-34a.108
Animal Model Studies
The biological activity of miR-34a studied in cell models suggests the potential ability to also suppress tumor growth in vivo. A common approach to achieve miRNA expression in vivo is based on vectoral systems that function similarly to those used for traditional gene therapy.109,110 These can be used either on conventional xenografts, before transplantation, or systemically as nanoparticles, by parenteral administration. One example of systemic delivery is provided by Di Martino et al., who explored the clinical translatability of their experimental findings examining the antitumor activity of miR-34a in murine xenograft models of human MM. In details, the authors demonstrated the successful delivery of miR-34a mimics in MM xenografts in SCID mice and in a recently established novel SCID-synth-hu model111,112 via NLE, a novel lipid-based delivery vehicle, which overcomes many of the most important limitations of other vehicles. Importantly, these findings supported the development of formulated miR-34a as new experimental agent for the treatment of MM.32 In order to explore the potential clinical applications of miR-34a, Li et al. incorporated it into a targeting expression plasmid based on an engineered expression vector, the T-VISA-miR-34a system, which can enhance cancer-specific promoter activity by 100-fold and prolong the duration of gene expression. T-VISA-miR-34a was found to induce both robust and persistent expression of miR-34a, dramatically inhibiting tumor growth and prolonging survival, did not inducing systemic toxicity, in an orthotopic mouse model of breast cancer.113 It is noteworthy that the therapeutically resistant KrasLSL-G12D/+; Trp53LSL-R172H/+ mouse lung cancer model demonstrated downregulation of miR-34a and heightened expression of miR-34 target genes, such as Met and Bcl-2,as evidenced by qRT-PCR. Moreover, animals bearing lung-specific transgene activation infected with mir-34a-expressing lentivirus, showed little or no evidence of tumorigenesis and reduced progression of pre-formed tumors, thus supporting the use of miR-34 as a lung tumor-preventative and tumor-static agent.114 Based on the use of cell culture-based and mouse models, Iqbal et al. have recently investigated on miR-34a involvement in p53-independent blockade of cell cycle. They defined a new role for miR-34a, providing an essential link between p19Arf and its capacity to block cell proliferation driven by PDGFRβ.115 Substantial fraction of information has been added into the biology of EMT through the use of an experimental mouse model of human lung adenocarcinoma metastasis driven by ZEB1. This model has hallowed to establish a strong link between high levels of EMT activators and loss of cell polarity, reduced expression of basement membrane components, and increased propensity for metastasis. In details, ZEB1 was found to modulate the expression of numerous oncogenic and tumor-suppressive miRNAs, including miR-34a, whose ectopic re-expression decreased tumor cell invasion and metastasis, by the inhibition of pro-migratory cytoskeletal structures, and suppressed activation of the RHO GTPase family; moreover, miR-34a replacement regulated gene expression signature enriched in cytoskeletal functions and predictive of outcome in human lung adenocarcinomas. These findings demonstrated that ZEB1 drives pro-metastatic actin cytoskeletal remodeling by downregulating miR-34a expression and provided the rationale to develop miR-34a as a therapeutic agent in lung cancer patients.116 The in vivo tumor suppressor function of miR-34a as well as its therapeutic activity was also demonstrated by Wiggins et al. that used a synthetic miR-34a mimic in an effort to restore a loss of function in cancer. They developed a therapeutic formulation based on chemically synthesized miR-34a and a lipid-based delivery vehicle able to block tumor growth in mouse models of non-small cell lung cancer. This formulation was effective and well tolerated when administered either locally or systemically and the anti-oncogenic effects were accompanied by an accumulation of miR-34a in the tumor tissue and by a downregulation of the direct miR-34a targets.117 Similar results were obtained in a genetically engineered mouse model of lung cancer based on the constitutively active KRAS mutant G12D.118 Ten weeks after the activation of the KRAS-G12D mutant and the initiation of orthotopic lung tumors, miR-34a mimics conjugated with NLE were systemically introduced every other day for a total of eight injections at a concentration of 1 mg/kg each time. Systemic treatment led to a significant decrease in tumor burden. Specifically, mice treated with miR-34a displayed a 60% reduction in tumor area compared to mice treated with a miRNA control. This result correlated with a significant elevation of miR-34a levels in lung tissue, reduced expression of Ki67 and an increase in TUNEL-positive cells. Another example of systemic delivery is provided by the intravenous delivery of miR-34a in polycationic liposome-hyaluronic acid nanoparticles modified with a GC4 single-chain antibody fragment (scFv).119 The authors hypothesized that the antibody moiety GC4 scFv could facilitate the targeting of the nanoparticles to tumor metastasis. To test the therapeutic utility, the formulation was administered on two consecutive days (0.3 mg/kg) to mice that had lung metastases induced by the B16F10 melanoma xenograft. Eleven days after the first treatment, the tumor burden was reduced by approximately 50% as assessed by luminescence imaging of metastatic lesions. In a separate experiment, miR-34a activity was confirmed by an increase of miR-34a-induced apoptosis and the downregulation of survivin protein levels in lung metastases. The therapeutic potential of miR-34a was also investigated in subcutaneous and orthotopic pancreas MiaPaca-2 xenografts, were miR-34a was delivered systemically using a 100 nm-lipid-based nanoparticle that contains a miR-34a-encoding vector. The tumors from mice treated with miR-34a were significantly smaller and showed signs of necrosis and apoptosis. Moreover, orthotopic and subcutaneous tumors displayed reduced levels of Ki67 and SIRT1 proteins, respectively. Also CD44 and ALDH mRNA levels were reduced in tumor tissues of miR-34a-treated animals.120
In summary, the preclinical data from animal tumor models demonstrate the therapeutic usefulness of developing novel miR-34a formulations designed to achieve the clinical trial.
Human Studies
The involvement of miR-34a in cancer, as well as its precise role during carcinogenesis is still being dissected. MiR-34a has been found to be deregulated in cancer by DNA copy number changes and epigenetic alterations, as well as by altered processing by the miRNA biogenesis machinery and through altered transcriptional activation. Although it is clear that miR-34a can acts as tumor suppressor by regulating processes such as proliferation, cell cycle, apoptosis, invasion and metastasis, it is however still under investigation the precise correlation between miR-34a expression levels and the metastatic cancer stage. Overall, the expression of miR-34a was reduced in many types of cancers. To evaluate the potential role of miR-34 family in human epithelial ovarian cancer (EOC), it has determined its expression level in a panel of 83 cancer tissues.121 MiR-34a expression was reduced in 100% of EOC with p53mutation, and miR-34b/c in 72%; at the same time, miR-34a was downregulated in 93% of tumors with wild-type p53. In all cases, decreased miR-34a levels were associated with metastatic clinical stage. Furthermore, miR-34 re-expression in p53 mutant EOC cancer cells correlated with reduced proliferation, motility and invasion, and decreased expression of receptor protein tyrosine kinase c-MET.121 Downregulation of miR-34a was reported in neuroblastoma tumors harboring 1p36.3 loss122 and in chronic lymphocytic leukemia patients carrying 17p13/TP53 deletions,123 compared with patients without such deletions. Loss of 1p36, the genomic interval harboring miR-34a, is common in diverse human cancers, but one of the other mechanisms responsible for decrease of miR-34 family expression levels seems to be CpG island hypermethylation. miR-34a promoter methylation was reported in 3 of 23 cases of colon cancer, 19 out of 24 primary prostate carcinomas, 6 out 24 breast carcinomas, 7 out 24 lung carcinomas, 3 out 14 kidney carcinomas, 1 out 5 bladder carcinomas, 3 out 19 pancreatic carcinomas, as well as in 20 out 32 primary melanomas.124 As a result of this methylation, it was observed a concomitant loss of miR-34a expression and cancer development. Interestingly, another contemporary study revealed miR-34a promoter methylation in 45.1% of the samples and a strong association with colon cancer metastasis.125 Accordingly, miR-34 family was found to be epigenetically silenced in 9 of 9 cell lines examined and in 101 of 111 primary colorectal carcinomas (CRCs), but they were not in normal colonic epithelium. After treatment with demethylating agents, their expression was restored. As a result, it was reported a severe inhibition of both tumor motility and metastasis formation.126 Moreover, miR-34a expression was found to be downregulated in the 36% of human CRCs, compared with the counterpart of normal tissues, and this was put in relation with the subsequent upregulation of the E2F pathway, and the downregulation of the p53/p21 pathway, thus leading to aberrant cell proliferation and colon cancer development.127 In human HCCs, there were discrepant reports on the expression of miR-34a. Pineau et al.128 observed by microarray that miR-34a increased in HCC and this was linked to disease progression from normal liver through cirrhosis to full-blown HCC. On the other hand, Li et al.51 reported that downregulation of miR-34a expression was highly significant in 19 of 25 (76%) human HCC tissues compared with adjacent normal tissues, using real time RT-qPCR. The different source of the samples and the various assays might partly explain the discordance of the different results. A recent report showed a significantly decreased miR-34a expression in eighty-three cases of HCC formalin-fixed paraffin-embedded (FFPE) tissues, compared to that in the adjacent liver tissues. Moreover, functional experiments showed that HCC cells transfected with miR-34a mimic displayed remarkable decrease in pERK1/2 and pSTAT-5, as evidenced by western blot assay, as well as suppression of cell growth, migration and invasion and a simultaneous increase of cellular apoptosis and caspase activity.129 In non-small-cell lung cancer (NSCLC) patients treated only with curative surgery, low levels of miR-34a in tumor samples correlated with a high rate of relapse. Moreover, miR-34a levels were modulated by methylation of the promoter region of the miR-34a gene. The frequency of p53 mutations was significantly higher in patients with low miR-34a expression, and the group of patients with both p53 mutations and low miR-34a expression had a very poor prognosis, indicating a potential synergism for these two factors.130 However, a recent report suggested upregulated miR-34a (P < 0.05) in advanced NSCLC patients treated with pemetrexed, compared with healthy controls. No significant association with clinical outcome was recorded for miR-34a.131 Peurala et al., focused on the expression of miR-34a in an extensive series of human breast carcinomas and on its relationship with tumor phenotype and prognosis. In details, low expression of miR-34a was found in about 32% and high expression in about 25% of the tumors, with the remaining tumors showing intermediate expression levels. Unexpectedly, high miR-34a expression was correlated with an aggressive phenotype of hormone receptor negative tumors, p53-immunopositive, high tumor grade and high proliferation rate of the tumors, indicating miR-34a expression activation per se as a marker for an aggressive breast tumor. However, it was found to exert an independent effect for a lower risk of recurrence or death from breast cancer.132 Noteworthy, there is another conflicting report that indicated increased amounts of miR-34a in ER− and ER−/PR− breast carcinoma patients.133 Surprisingly, a recent report indicated, for breast tumors, a downregulation of miR-34a independent of the two main regulating mechanisms, such as mutant p53 or epigenetic silencing. Furthermore, downregulation of miR-34a was significantly associated with metastasis, while there was a significant correlation between upregulation of miR-34a and non-metastatic condition, indicating a protective role for miR-34a against more invasive disease.134 Recent blood-based miRNA reports, clearly show that malignancy alters miRNA levels in the circulation, although it is still unclear how tumor associated miRNAs make their way into the bloodstream. Significantly reduced circulating miR-34a levels in both colorectal and breast cancer patients, compared to healthy individuals, highlights the potential of these molecules as novel non-invasive biomarkers for cancer.135 Overall, miR-34a seems to represent an attractive candidate for gene therapy approaches, rather than siRNA approaches, because of its capacity of regulating cell proliferation and cell death, as well as cell migration and invasion, through the targeting of several target genes. For this reason, there are several ongoing studies aimed at better understanding the usefulness of the miR-34a replacement, in view to improve the clinical management of cancer patients.
Nanotechnology to Deliver MiR-34A In Cancer
Despite the interest on the use of miR-34a as new anticancer agent, biopharmaceutical issues, i.e., rapid degradation in biological fluids, poor uptake into cells and not specific distribution into the body, hamper the use of wild-type non-coding RNA by systemic administration.136 Chemical modifications of the RNA backbone can increase the in vivo half-life,6 although the interaction with the intracellular target could be affected. In the light of these considerations, the use of delivery systems represents a valid tool to facilitate miRNA passage from the bench to the bed-side. Ideally, a delivery system should be safe, stable, effective and tumor-specific. Nanotechnology have been largely investigated for drug delivery and different nanocarriers have been proposed in oligonucleotide-based therapies. Nanotechnology-based formulations are in advanced phase of clinical study to deliver non-coding RNA.137 The use of nanocarriers is especially advantageous in cancer, due to the possibility to passively accumulate drug in tissues (i.e., tumors) characterized by highly permeable endothelium of the capillaries and by a reduced lymphatic function (the so called enhanced and permeability and retention (EPR) effect).138 The possibility to modify the nanocarrier surface with different molecules (i.e., ligands binding specific receptor or monoclonal antibody) have been explored to target cells overexpressing receptors or antigens characteristic of tumor phenotype (active targeting).139
Different biomaterials have been investigated to deliver nucleic acid, and more specifically non-coding RNA, in cancer. Lipid-based nanocarriers are certainly the most investigated systems for nucleic acids delivery.140 In details, cationic liposomes are commonly used as transfection reagents in vitro. They are able to interact with negatively charged nucleic acid forming colloidal nanoparticles, also named lipoplexes that are efficiently up taken into the cells by endocytosis. However, lipoplexes aggregate in serum and their use in vitro requires serum-free cell culture media; moreover, safe and efficacious delivery in vivo is rarely achieved due to toxicity, non-specific uptake, and unwanted immune response.141 To overcome these concerns, formulations based on cationic lipids have been modified with different approaches. For instance, cationic liposomes were associated to hyaluronic acid to deliver miR-34a in an animal model of lung metastasis.119 This delivery system consisted on hyaluronic acid and miR-34a firstly complexed with protamine; this complex was then encapsulated in cationic liposomes by charge interaction. The PEGylation of the formulation increased its stability in serum, while the surface modification with tumor-targeting single-chain antibody fragment (scFv) allowed to target lung metastasis cells. The efficient inhibition of the tumor growth was associated with a very limited systemic toxicity.119 Cationic liposomes have also been used to deliver plasmid DNA, namely pMSCV-puro vectors expressing corresponding miR-34a.120 In this study, the growth of orthotopic pancreatic cancer xenografts was significantly inhibited by replacement of miR-34a through systemic delivery.
The development of the stable nucleic acid lipid particles (SNALPs) represents a successful strategy to reduce both physical instability in serum and toxicity of liposomes, due to the cationic charge of the lipid, maintaining the high encapsulation efficiency of nucleic acids. These nanoparticles are characterized by ionizable lipids that assure high miR-34a entrapment in the protonated form and higher stability in serum upon charge neutralization on the particle surface.31 These systems were rapidly assessed on mammalian primates for the delivery of small interfering RNA and different clinical trials are ongoing. SNALPs were successfully used to deliver miR-34a in vitro in medulloblastoma cells, showing equal effects to those achieved with adenovirus miR-34a cell infection.36 SNALPs were also used to deliver a miR-34a mimic in MM cells with a significant change of gene expression with relevant effects on multiple signal transduction pathways. The same system was then tested in vivo in a MM xenograft. In these experiments, a significant tumor growth inhibition and a prolonged mice survival were observed. These findings were associated to an efficient miR-34a delivery into the tumor cells and to reduced NOTCH1 expression.33 In order to additionally improve the delivery of miR-34a in MM cells overexpressing transferrin (Tf) receptor, SNALPs were conjugated with Tf.142 In this study, a miR-34a with two different chemical backbones was used. The use of a completely 2′-O-methylated miR-34a resulted in higher oligonucleotide encapsulation into the vesicles. In an experimental model of MM, all the animals treated with SNALPs encapsulating miR-34a showed a significant inhibition of the tumor growth. However, SNALPs conjugated with Tf and encapsulating 2′-O-methylated miR-34a caused the highest increase of mice survival. A different lipid-based approach consists on the use of neutral phospholipids formulated in nanoemulsion prepared by extrusion immediately before use. This formulation has been successfully used to deliver miR-34a in different experimental tumor models. MiR-34a was formulated with this neutral emulsion and locally or systemically administered in a murine model of NSCLC.39 In both cases, miR-34a accumulation into the tumor was found, although higher levels were found in the case of the locally injected formulation. In both cases, a therapeutic effect was found, suggesting that only minimal amounts of miR-34a are needed to elicit the pharmacological activity. In the case of systemic delivery, miR-34a displayed a 60% reduction in tumor area compared to mice treated with a naked miRNA.120 Finally, the administration of formulated miR-34a was well tolerated, confirming that pathways regulated by the miRNA mimic are already activated by the endogenous miRNA in healthy cells. The same formulation was investigated in an experimental model of MM in which a comparison with the stable transfection with miR-34a lentivirus was carried out. In this study, similar inhibition of tumor formation was found by comparing the effect of miR-34a lentivirus and nanocarriers encapsulating miR-34a.39 The observed downregulation of miR-34a validated targets at both mRNA and protein levels additionally supported the therapeutic potential of miR-34a mimics formulated with a neutral lipid emulsion.34
Active targeting of CD44 molecule, an integral membrane glycoprotein over-expressed on the surface of various tumor cells, has been used to specifically deliver miR-34a to tumor cells. To do this, nanoparticles “decorated” with hyaluronic acid have been developed. In particular, chitosan/hyaluronic acid based nanoparticles encapsulating miR-34a, alone or associated with doxorubicin, have been prepared. These nanoparticles were stable in different media and protected miRNA against enzymatic degradation. Doxorubicin and miR-34a were released from the nanoparticle at a different release rate that was accelerated in acidic pH (characteristic of such solid tumors). These nanoparticles demonstrated higher antitumor activity in vitro and in vivo likely mediated by a CD44 receptor endocytosis pathway.143 The same authors proposed a easier CD44-targeting system consisting on hyaluronic acid/protamine sulfate interpolyelectrolyte complexes encapsulating miR-34a.144 Antibody specifically binding GD2, a glycolipid highly expressed on the cell surface of neuroblastoma and several other cancers, was conjugated on silica nanoparticles to restore miR-34a in tumor cell. In this study, none of the organs exhibited any increase in mature miR-34a transcript levels subsequent to treatment; adverse effects on kidney and/or liver function were also not observed. The use of these nanoparticles resulted in a significant inhibition of neuroblastoma tumor growth in a murine orthotopic disease model.145 In a different approach, a tumor-homing and -penetrating bifunctional peptide was physically combined with the cationic β-cyclodextrin-polyethylenimine conjugates for effective miR-34a delivery in tumor cells.146 These nanoparticles induced migration suppression and displayed antiproliferative activity in vitro and in vivo. In a recent study, nanoparticles based on inorganic materials, namely calcium/phosphate, were also proposed for miR34a delivery. The nucleic acid was chemically modified with a spacer to improve entrapment efficiency into calcium phosphate nanoparticles. The nanoparticles have also been modified with linear polyethylenimine, a cationic polymer used as powerful transfection agent. These nanosized particles showed great homogeneity and were successfully delivered into cancer cell in vitro. Starting from these evidences, nanotechnology-based approach should be taken into account when designing a new therapy involving miR-34a restoration. In this light, miRNA therapeutics screened multiple existing delivery systems in pre-clinical development or already in clinical trials. This evaluation encompassed an assessment of efficacy in appropriate mouse models of cancer, miRNA biodistribution, and preliminary safety. From these tests, liposomes based on ionizable lipids (NOV340 technology—SMARTICLES) were selected for the following phase of the study. In May 2013, miRNA Therapeutics announced the beginning of the phase 1 clinical study MRX34, bringing the first miRNA (miR-34a) in humans. The study is, at the moment, ongoing.
Conclusion
MiRNA-based therapy has a great potential to be a more powerful tool in tumor treatment by the simultaneous modulation of multiple genes involved in distinct tumor-related signaling networks. MiR-34a is, to date, one of the most characterized tumor suppressor miRNAs in a variety of tumors. As evidenced by the current literature, it is lost or expressed at reduced levels in numerous tumor tissues, and the re-introduction of miR-34 mimics was found to inhibit cancer cell growth both in vitro and in vivo. Therefore, miR-34a has proved to be a tumor suppressor in cancer cells and an ideal therapeutic tool to combat metastasis, chemoresistance and tumor recurrence. However, several obstacles, including degradation and inactivation by nucleases, poor intracellular delivery, rapid plasma elimination, and both renal and dose-limiting hemodynamic toxicities, hamper the development of therapies based on miRNAs. Hence, there is a growing interest for the development of ad hoc delivery systems aimed to lead miR-34a toward the transition from bench to bedside. In the last decade, more than one technology has been developed to establish a promising clinically relevant candidate as antitumor therapeutic agent. Recently, it has been developed a SNALP-based delivery system of miR-34a that has been also modified through the use of two strategies to enhance miR-34a delivery in cancer.142 In another study,143 it was reported a successful application of hyaluronic acid-chitosan (HA-CS) as a dual nanocarrier system to simultaneous delivery of the chemotherapeutic drug Doxorubicin (DOX) and the tumor suppressive miR-34a, exploiting their synergistic effect and the enhanced anti-cancer chemotherapeutic efficacy after intracellular internalization of co-loaded nanoparticles. Another chance associated to the development of new strategies for the use of miR-34a as a therapeutic weapon in cancer is the combination of miR-34a replacement in combination with other anticancer agents. In this light, it was recently reported that Mir-34a regulates interferon (IFN)-β protein expression in human cells147 and we have also reported the regulation of interferon genes and pathways in MM cells.31 Moreover, IFN-β seems to have a role in the pathogenesis and therapy of pancreatic cancer.148,149,150 Based upon these results, it should be suggested the combined use of IFN-β and miR-4a replacement as an interesting strategy in order to treat pancreatic cancer, a neoplasm almost orphan of available medical treatments. Other important considerations have to be done regarding he general strategy of treat human cancers with miRNAs. First, miRNAs have multiple intracellular targets and thus they can induce pleiotropic intracellular effects on multiple signal transduction pathways involved in the maintenance of the neoplastic phenotype. In this light, anticancer treatment based upon miRNAs resembles that one based on multi kinase inhibitors. The latter, differently from kinase inhibitors raised against a specific molecular target, have contributed to give important advancements in the therapy of several neoplasms such as renal cell cancer and gastro-intestinal stromal tumors that were before almost without any active medical treatment. Based upon these considerations, miRNAs have a great therapeutic potential in human cancers. Second, it is widely demonstrated that different neoplasms derived from different histotypes or also from the same type of tumor can have a different miRNA signature that is important on the choice of the “best miRNA target” to be used for therapeutic purposes. This consideration discloses the interest on the molecular characterization of the tumors also for the specific miRNAs found to be altered if compared with the normal counterparts. Moreover, miRNAs themselves can be mutated in tumor tissues and this mutation can have a role in cell transformation. Therefore, as already occurred for monoclonal antibodies or kinase inhibitors also in the case of miRNAs it can be predicted the importance of the molecular diagnostic study of the tumor in order to drive the clinician in the choice of the most promising strategy. Finally, miRNAs are differently involved in the maintenance of the homeostasis of cancer and normal tissues. Therefore, it cannot be excluded that the same miRNA that has advantageous effects on cancer cells can, on the other hand, have negative side effects on normal tissues. This phenomenon is known with the name of “off target” effects. Based on this last consideration, it becomes more evident the need to deliver the miRNAs specifically in cancer tissues sparing the normal ones. Based on these considerations, chemical modifications, aimed to improve the biological stability of miR-34a, could be successfully combined with targeting approaches based upon the active delivery of the nanotechnological devices, in order to finally optimize these models for the subsequent translation in the clinical setting.
Acknowledgments
This work has been supported by funds of Italian Association for Cancer Research (AIRC), PI: PT. “Special Program Molecular Clinical Oncology–5 per mille” n. 9980, 2010/15. The authors declare no conflict of interest.
References
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti M, Musto P, Di Martino MT, Fabris S, Agnelli L, Todoerti K, et al. Biological and clinical relevance of miRNA expression signatures in primary plasma cell leukemia. Clin Cancer Res. 2013;19:3130–3142. doi: 10.1158/1078-0432.CCR-12-2043. [DOI] [PubMed] [Google Scholar]
- Negrini M, Cutrona G, Bassi C, Fabris S, Zagatti B, Colombo M, et al. microRNAome Expression in Chronic Lymphocytic Leukemia: Comparison with Normal B-cell Subsets and Correlations with Prognostic and Clinical Parameters. Clin Cancer Res. 2014;20:4141–4153. doi: 10.1158/1078-0432.CCR-13-2497. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Dimopoulos K, Gimsing P, Grønbæk K. Aberrant microRNA expression in multiple myeloma. Eur J Haematol. 2013;91:95–105. doi: 10.1111/ejh.12124. [DOI] [PubMed] [Google Scholar]
- Tagliaferri P, Rossi M, Di Martino MT, Amodio N, Leone E, Gulla A, et al. Promises and challenges of MicroRNA-based treatment of multiple myeloma. Curr Cancer Drug Targets. 2012;12:838–846. doi: 10.2174/156800912802429355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio N, Di Martino MT, Neri A, Tagliaferri P, Tassone P. Non-coding RNA: a novel opportunity for the personalized treatment of multiple myeloma. Expert Opin Biol Ther. 2013;13 (suppl. 1):S125–S137. doi: 10.1517/14712598.2013.796356. [DOI] [PubMed] [Google Scholar]
- Rossi M, Amodio N, Di Martino MT, Caracciolo D, Tagliaferri P, Tassone P. From target therapy to miRNA therapeutics of human multiple myeloma: theoretical and technological issues in the evolving scenario. Curr Drug Targets. 2013;14:1144–1149. doi: 10.2174/13894501113149990186. [DOI] [PubMed] [Google Scholar]
- Rossi M, Amodio N, Di Martino MT, Tagliaferri P, Tassone P, Cho WC. MicroRNA and Multiple Myeloma: from Laboratory Findings to Translational Therapeutic Approaches. Curr Pharm Biotechnol. 2014;15:459–467. doi: 10.2174/1389201015666140519104743. [DOI] [PubMed] [Google Scholar]
- Tutar L, Tutar E, Tutar Y. MicroRNAs and Cancer; an Overview. Curr Pharm Biotechnol. 2014;15:430–437. doi: 10.2174/1389201015666140519095304. [DOI] [PubMed] [Google Scholar]
- Rolfo C, Fanale D, Hong DS, Tsimberidou AM, Piha-Paul SA, Pauwels P, et al. Impact of MicroRNAs in Resistance to Chemotherapy and Novel Targeted Agents in Non-Small Cell Lung Cancer. Curr Pharm Biotechnol. 2014;15:475–485. doi: 10.2174/1389201015666140519123219. [DOI] [PubMed] [Google Scholar]
- Thorsen SB, Obad S, Jensen NF, Stenvang J, Kauppinen S. The therapeutic potential of microRNAs in cancer. Cancer J. 2012;18:275–284. doi: 10.1097/PPO.0b013e318258b5d6. [DOI] [PubMed] [Google Scholar]
- Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovell LC, Putcha BD, Samuel T, Manne U. Clinical implications of microRNAs in cancer. Biotech Histochem. 2013;88:388–396. doi: 10.3109/10520295.2013.788735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio N, Leotta M, Bellizzi D, Di Martino MT, D'Aquila P, Lionetti M, et al. DNA-demethylating and anti-tumor activity of synthetic miR-29b mimics in multiple myeloma. Oncotarget. 2012;3:1246–1258. doi: 10.18632/oncotarget.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio N, Di Martino MT, Foresta U, Leone E, Lionetti M, Leotta M, et al. miR-29b sensitizes multiple myeloma cells to bortezomib-induced apoptosis through the activation of a feedback loop with the transcription factor Sp1. Cell Death Dis. 2012;3:e436. doi: 10.1038/cddis.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi M, Pitari MR, Amodio N, Di Martino MT, Conforti F, Leone E, et al. miR-29b negatively regulates human osteoclastic cell differentiation and function: implications for the treatment of multiple myeloma-related bone disease. J Cell Physiol. 2013;228:1506–1515. doi: 10.1002/jcp.24306. [DOI] [PubMed] [Google Scholar]
- Leone E, Morelli E, Di Martino MT, Amodio N, Foresta U, Gullà A, et al. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin Cancer Res. 2013;19:2096–2106. doi: 10.1158/1078-0432.CCR-12-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino MT, Gullà A, Cantafio ME, Lionetti M, Leone E, Amodio N, et al. In vitro and in vivo anti-tumor activity of miR-221/222 inhibitors in multiple myeloma. Oncotarget. 2013;4:242–255. doi: 10.18632/oncotarget.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio N, Bellizzi D, Leotta M, Raimondi L, Biamonte L, D'Aquila P, et al. miR-29b induces SOCS-1 expression by promoter demethylation and negatively regulates migration of multiple myeloma and endothelial cells. Cell Cycle. 2013;12:3650–3662. doi: 10.4161/cc.26585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leotta M, Biamonte L, Raimondi L, Ronchetti D, Martino MT, Botta C, et al. A p53-dependent tumor suppressor network is induced by selective miR-125a-5p inhibition in multiple myeloma cells in vitro. J Cell Physiol. 2014;229:2106–2116. doi: 10.1002/jcp.24669. [DOI] [PubMed] [Google Scholar]
- Raimondi L, Amodio N, Di Martino MT, Altomare E, Leotta M, Caracciolo D, et al. Targeting of multiple myeloma-related angiogenesis by miR-199a-5p mimics: in vitro and in vivo anti-tumor activity. Oncotarget. 2014;5:3039–3054. doi: 10.18632/oncotarget.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Maruyama R, Yamamoto E, Kai M. DNA methylation and microRNA dysregulation in cancer. Mol Oncol. 2012;6:567–578. doi: 10.1016/j.molonc.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- Di Martino MT, Campani V, Misso G, Gallo Cantafio ME, Gullà A, Foresta U, et al. In vivo activity of miR-34a mimics delivered by stable nucleic acid lipid particles (SNALPs) against multiple myeloma. PLoS ONE. 2014;9:e90005. doi: 10.1371/journal.pone.0090005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin Cancer Res. 2012;18:6260–6270. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliou MS, da Silva-Diz V, Carmona FJ, Ramalho-Carvalho J, Heyn H, Villanueva A, et al. Impaired DICER1 function promotes stemness and metastasis in colon cancer. Oncogene. 2014;33:4003–4015. doi: 10.1038/onc.2013.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Kelnar K, Liu B, Chen X, Calhoun-Davis T, Li H, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nat Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls D, Tang SN, Rodova M, Srivastava RK, Shankar S. Targeting epigenetic regulation of miR-34a for treatment of pancreatic cancer by inhibition of pancreatic cancer stem cells. PLoS ONE. 2011;6:e24099. doi: 10.1371/journal.pone.0024099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankevicins L, Almeida da Silva AP, Ventura Dos Passos F, Dos Santos Ferreira E, Menks Ribeiro MC, G David M, et al. MiR-34a is up-regulated in response to low dose, low energy X-ray induced DNA damage in breast cells. Radiat Oncol. 2013;8:231. doi: 10.1186/1748-717X-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guessous F, Zhang Y, Kofman A, Catania A, Li Y, Schiff D, et al. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle. 2010;9:1031–1036. doi: 10.4161/cc.9.6.10987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Branham WS, Dial SL, Wang Y, Guo L, Shi L, et al. Genomic analysis of microRNA time-course expression in liver of mice treated with genotoxic carcinogen N-ethyl-N-nitrosourea. BMC Genomics. 2010;11:609. doi: 10.1186/1471-2164-11-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakuchi M, Lowenstein CJ. MiR-34, SIRT1 and p53: the feedback loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- Basak SK, Veena MS, Oh S, Lai C, Vangala S, Elashoff D, et al. The CD44(high) tumorigenic subsets in lung cancer biospecimens are enriched for low miR-34a expression. PLoS ONE. 2013;8:e73195. doi: 10.1371/journal.pone.0073195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofman AV, Kim J, Park SY, Dupart E, Letson C, Bao Y, et al. microRNA-34a promotes DNA damage and mitotic catastrophe. Cell Cycle. 2013;12:3500–3511. doi: 10.4161/cc.26459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Antonellis P, Carotenuto M, Vandenbussche J, De Vita G, Ferrucci V, Medaglia C, et al. Early Targets of miR-34a in Neuroblastoma. Mol Cell Proteomics. 2014;13:2114–2131. doi: 10.1074/mcp.M113.035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothe K, Lin H, Lin KB, Leung A, Wang HM, Malekesmaeili M, et al. The core autophagy protein ATG4B is a potential biomarker and therapeutic target in CML stem/progenitor cells. Blood. 2014;123:3622–3634. doi: 10.1182/blood-2013-07-516807. [DOI] [PubMed] [Google Scholar]
- Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009;69:7569–7576. doi: 10.1158/0008-5472.CAN-09-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang RT, Leung CO, Ye TM, Liu W, Chiu PC, Lam KK, et al. MicroRNA-34a suppresses invasion through downregulation of Notch1 and Jagged1 in cervical carcinoma and choriocarcinoma cells. Carcinogenesis. 2010;31:1037–1044. doi: 10.1093/carcin/bgq066. [DOI] [PubMed] [Google Scholar]
- de Antonellis P, Medaglia C, Cusanelli E, Andolfo I, Liguori L, De Vita G, et al. MiR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS ONE. 2011;6:e24584. doi: 10.1371/journal.pone.0024584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu P, Chen KY, Chen JH, Wang L, Walters J, Shin YJ, et al. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell. 2013;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winton DJ. miR-34a sets the “sweet spot” for notch in colorectal cancer stem cells. Cell Stem Cell. 2013;12:499–501. doi: 10.1016/j.stem.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Yamamura S, Saini S, Majid S, Hirata H, Ueno K, Deng G, et al. MicroRNA-34a modulates c-Myc transcriptional complexes to suppress malignancy in human prostate cancer cells. PLoS ONE. 2012;7:e29722. doi: 10.1371/journal.pone.0029722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Gao J, Yang T, Ma Q, Qiu X, Fan Q, et al. MicroRNA-34a inhibits the proliferation and metastasis of osteosarcoma cells both in vitro and in vivo. PLoS ONE. 2012;7:e33778. doi: 10.1371/journal.pone.0033778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Fu H, Tie Y, Hu Z, Kong W, Wu Y, et al. miR-34a inhibits migration and invasion by down-regulation of c-Met expression in human hepatocellular carcinoma cells. Cancer Lett. 2009;275:44–53. doi: 10.1016/j.canlet.2008.09.035. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Toyooka S, Soh J, Tsukuda K, Shien K, Furukawa M, et al. Downregulation of microRNA-34 induces cell proliferation and invasion of human mesothelial cells. Oncol Rep. 2013;29:2169–2174. doi: 10.3892/or.2013.2351. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Schiff D, Park D, Abounader R. MicroRNA-608 and microRNA-34a regulate chordoma malignancy by targeting EGFR, Bcl-xL and MET. PLoS ONE. 2014;9:e91546. doi: 10.1371/journal.pone.0091546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges CW, Kadariya Y, Altomare D, Talarchek J, Neumann-Domer E, Wu Y, et al. Tumor suppressor alterations cooperate to drive aggressive mesotheliomas with enriched cancer stem cells via a p53-miR-34a-c-Met axis. Cancer Res. 2014;74:1261–1271. doi: 10.1158/0008-5472.CAN-13-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens H, Jackstadt R, Kaller M, Hermeking H. Repression of c-Kit by p53 is mediated by miR-34 and is associated with reduced chemoresistance, migration and stemness. Oncotarget. 2013;4:1399–1415. doi: 10.18632/oncotarget.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Guo J, Li D, Jia C, Yin W, Sun R, et al. MicroRNA-34a suppresses cell proliferation by targeting LMTK3 in human breast cancer mcf-7 cell line. DNA Cell Biol. 2013;32:699–707. doi: 10.1089/dna.2013.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhong D, Gao Q, Zhai W, Ding Z, Wu J. MicroRNA-34a inhibits human osteosarcoma proliferation by downregulating ether à go-go 1 expression. Int J Med Sci. 2013;10:676–682. doi: 10.7150/ijms.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan W, Xu Y, Dong Y, Cao L, Tong J, Zhou X. Ectopic expression of miR-34a enhances radiosensitivity of non-small cell lung cancer cells, partly by suppressing the LyGDI signaling pathway. J Radiat Res. 2013;54:611–619. doi: 10.1093/jrr/rrs136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Li QJ, Gong ZB, Zhou L, You N, Wang S, et al. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol Cancer Res Treat. 2014;13:77–86. doi: 10.7785/tcrt.2012.500364. [DOI] [PubMed] [Google Scholar]
- Lou W, Chen Q, Ma L, Liu J, Yang Z, Shen J, et al. Oncolytic adenovirus co-expressing miRNA-34a and IL-24 induces superior antitumor activity in experimental tumor model. J Mol Med. 2013;91:715–725. doi: 10.1007/s00109-012-0985-x. [DOI] [PubMed] [Google Scholar]
- Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13:109–117. doi: 10.1007/s10238-012-0186-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Dong K, Gao P, Long M, Lin F, Weng Y, et al. microRNA-34a sensitizes lung cancer cell lines to DDP treatment independent of p53 status. Cancer Biother Radiopharm. 2013;28:45–50. doi: 10.1089/cbr.2012.1218. [DOI] [PubMed] [Google Scholar]
- Shen Z, Zhan G, Ye D, Ren Y, Cheng L, Wu Z, et al. MicroRNA-34a affects the occurrence of laryngeal squamous cell carcinoma by targeting the antiapoptotic gene survivin. Med Oncol. 2012;29:2473–2480. doi: 10.1007/s12032-011-0156-x. [DOI] [PubMed] [Google Scholar]
- Cao W, Yang W, Fan R, Li H, Jiang J, Geng M, et al. miR-34a regulates cisplatin-induce gastric cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour Biol. 2014;35:1287–1295. doi: 10.1007/s13277-013-1171-7. [DOI] [PubMed] [Google Scholar]
- Kumar B, Yadav A, Lang J, Teknos TN, Kumar P. Dysregulation of microRNA-34a expression in head and neck squamous cell carcinoma promotes tumor growth and tumor angiogenesis. PLoS ONE. 2012;7:e37601. doi: 10.1371/journal.pone.0037601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashat M, Azzouz L, Sarkar SH, Kong D, Li Y, Sarkar FH. Inactivation of AR and Notch-1 signaling by miR-34a attenuates prostate cancer aggressiveness. Am J Transl Res. 2012;4:432–442. [PMC free article] [PubMed] [Google Scholar]
- Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-ß-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandke P, Wyatt N, Fraser J, Bates B, Berberich SJ, Markey MP. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS ONE. 2012;7:e42034. doi: 10.1371/journal.pone.0042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X, et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 2014;28:438–450. doi: 10.1101/gad.233585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackiewicz M, Huppi K, Pitt JJ, Dorsey TH, Ambs S, Caplen NJ. Identification of the receptor tyrosine kinase AXL in breast cancer as a target for the human miR-34a microRNA. Breast Cancer Res Treat. 2011;130:663–679. doi: 10.1007/s10549-011-1690-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudduluru G, Ceppi P, Kumarswamy R, Scagliotti GV, Papotti M, Allgayer H. Regulation of Axl receptor tyrosine kinase expression by miR-34a and miR-199a/b in solid cancer. Oncogene. 2011;30:2888–2899. doi: 10.1038/onc.2011.13. [DOI] [PubMed] [Google Scholar]
- Vo DT, Qiao M, Smith AD, Burns SC, Brenner AJ, Penalva LO. The oncogenic RNA-binding protein Musashi1 is regulated by tumor suppressor miRNAs. RNA Biol. 2011;8:817–828. doi: 10.4161/rna.8.5.16041. [DOI] [PubMed] [Google Scholar]
- Muppala S, Mudduluru G, Leupold JH, Buergy D, Sleeman JP, Allgayer H. CD24 induces expression of the oncomir miR-21 via Src, and CD24 and Src are both post-transcriptionally downregulated by the tumor suppressor miR-34a. PLoS ONE. 2013;8:e59563. doi: 10.1371/journal.pone.0059563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HR, Roe JS, Lee JE, Hwang IY, Cho EJ, Youn HD. A p53-inducible microRNA-34a downregulates Ras signaling by targeting IMPDH. Biochem Biophys Res Commun. 2012;418:682–688. doi: 10.1016/j.bbrc.2012.01.077. [DOI] [PubMed] [Google Scholar]
- Heinemann A, Zhao F, Pechlivanis S, Eberle J, Steinle A, Diederichs S, et al. Tumor suppressive microRNAs miR-34a/c control cancer cell expression of ULBP2, a stress-induced ligand of the natural killer cell receptor NKG2D. Cancer Res. 2012;72:460–471. doi: 10.1158/0008-5472.CAN-11-1977. [DOI] [PubMed] [Google Scholar]
- Yin D, Ogawa S, Kawamata N, Leiter A, Ham M, Li D, et al. miR-34a functions as a tumor suppressor modulating EGFR in glioblastoma multiforme. Oncogene. 2013;32:1155–1163. doi: 10.1038/onc.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaller M, Liffers ST, Oeljeklaus S, Kuhlmann K, Röh S, Hoffmann R, et al. Genome-wide characterization of miR-34a induced changes in protein and mRNA expression by a combined pulsed SILAC and microarray analysis. Mol Cell Proteomics. 2011;10:M111.010462. doi: 10.1074/mcp.M111.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Lammers P, Torrance CJ, Bader AG. TP53-independent function of miR-34a via HDAC1 and p21(CIP1/WAF1.) Mol Ther. 2013;21:1678–1686. doi: 10.1038/mt.2013.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Jeon YJ, Nuovo GJ, Middleton J, Secchiero P, Joshi P, et al. MiR-34a/c-Dependent PDGFR-a/ß Downregulation Inhibits Tumorigenesis and Enhances TRAIL-Induced Apoptosis in Lung Cancer. PLoS ONE. 2013;8:e67581. doi: 10.1371/journal.pone.0067581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Xiang Y, Tang J, Wu G, Li J, Xiao H, et al. miR-34 is associated with poor prognosis of patients with gallbladder cancer through regulating telomere length in tumor stem cells. Tumour Biol. 2014;35:1503–1510. doi: 10.1007/s13277-013-1207-z. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata K, Fujiyama S, Ito S, Ueda T, Murata T, Naitou M, et al. Maturation of microRNA is hormonally regulated by a nuclear receptor. Mol Cell. 2009;36:340–347. doi: 10.1016/j.molcel.2009.08.017. [DOI] [PubMed] [Google Scholar]
- Hau A, Ceppi P, Peter ME. CD95 is part of a let-7/p53/miR-34 regulatory network. PLoS ONE. 2012;7:e49636. doi: 10.1371/journal.pone.0049636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rücker FG, Russ AC, Cocciardi S, Kett H, Schlenk RF, Botzenhardt U, et al. Altered miRNA and gene expression in acute myeloid leukemia with complex karyotype identify networks of prognostic relevance. Leukemia. 2013;27:353–361. doi: 10.1038/leu.2012.208. [DOI] [PubMed] [Google Scholar]
- Ma Y, Bao-Han W, Lv X, Su Y, Zhao X, Yin Y, et al. MicroRNA-34a mediates the autocrine signaling of PAR2-activating proteinase and its role in colonic cancer cell proliferation. PLoS ONE. 2013;8:e72383. doi: 10.1371/journal.pone.0072383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig VJ, Cogliatti SB, Imig J, Renner C, Neuenschwander S, Rehrauer H, et al. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117:6227–6236. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens H, Jackstadt R, Hünten S, Kaller M, Menssen A, Götz U, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- Rokavec M, Öner MG, Li H, Jackstadt R, Jiang L, Lodygin D, et al. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J Clin Invest. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Jacobsen A, Ozawa T, Harinath G, Pedraza A, Sander C, et al. miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS ONE. 2012;7:e33844. doi: 10.1371/journal.pone.0033844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kelnar K, Bader AG. In-depth analysis shows synergy between erlotinib and miR-34a. PLoS ONE. 2014;9:e89105. doi: 10.1371/journal.pone.0089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Ji MH, Zhong SL, Zha QB, Xu JJ, Zhao JH, et al. MicroRNA-34a modulates chemosensitivity of breast cancer cells to adriamycin by targeting Notch1. Arch Med Res. 2012;43:514–521. doi: 10.1016/j.arcmed.2012.09.007. [DOI] [PubMed] [Google Scholar]
- Vinall RL, Ripoll AZ, Wang S, Pan CX, deVere White RW. MiR-34a chemosensitizes bladder cancer cells to cisplatin treatment regardless of p53-Rb pathway status. Int J Cancer. 2012;130:2526–2538. doi: 10.1002/ijc.26256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akao Y, Noguchi S, Iio A, Kojima K, Takagi T, Naoe T. Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 2011;300:197–204. doi: 10.1016/j.canlet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- Kojima K, Fujita Y, Nozawa Y, Deguchi T, Ito M. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate. 2010;70:1501–1512. doi: 10.1002/pros.21185. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Saiki Y, Shiga K, Chen N, Fukushige S, Sunamura M, et al. miR-34a is downregulated in cis-diamminedichloroplatinum treated sinonasal squamous cell carcinoma patients with poor prognosis. Cancer Sci. 2012;103:1737–1743. doi: 10.1111/j.1349-7006.2012.02338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani F, Ferracin M, Manara MC, Ventura S, Del Monaco V, Ferrari S, et al. miR-34a predicts survival of Ewing's sarcoma patients and directly influences cell chemo-sensitivity and malignancy. J Pathol. 2012;226:796–805. doi: 10.1002/path.3007. [DOI] [PubMed] [Google Scholar]
- Kutay H, Bai S, Datta J, Motiwala T, Pogribny I, Frankel W, et al. Downregulation of miR-122 in the rodent and human hepatocellular carcinomas. J Cell Biochem. 2006;99:671–678. doi: 10.1002/jcb.20982. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pogribny IP, Tryndyak VP, Ross SA, Beland FA. Differential expression of microRNAs during hepatocarcinogenesis induced by methyl deficiency in rats. Nutr Rev. 2008;66 (suppl. 1):S33–S35. doi: 10.1111/j.1753-4887.2008.00064.x. [DOI] [PubMed] [Google Scholar]
- Guo J, Li W, Shi H, Xie X, Li L, Tang H, et al. Synergistic effects of curcumin with emodin against the proliferation and invasion of breast cancer cells through upregulation of miR-34a. Mol Cell Biochem. 2013;382:103–111. doi: 10.1007/s11010-013-1723-6. [DOI] [PubMed] [Google Scholar]
- Roy S, Levi E, Majumdar AP, Sarkar FH. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J Hematol Oncol. 2012;5:58. doi: 10.1186/1756-8722-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Kim E, Kim W, Seong KM, Youn H, Kim JW, et al. Rhamnetin and cirsiliol induce radiosensitization and inhibition of epithelial-mesenchymal transition (EMT) by miR-34a-mediated suppression of Notch-1 expression in non-small cell lung cancer cell lines. J Biol Chem. 2013;288:27343–27357. doi: 10.1074/jbc.M113.490482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Wang Z, Geamanu A, Goja A, Sarkar FH, Gupta SV. Delta-tocotrienol suppresses Notch-1 pathway by upregulating miR-34a in nonsmall cell lung cancer cells. Int J Cancer. 2012;131:2668–2677. doi: 10.1002/ijc.27549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Duan Q, Ahmad A, Bao B, Banerjee S, Shi Y, et al. Genistein inhibits cell growth and induces apoptosis through up-regulation of miR-34a in pancreatic cancer cells. Curr Drug Targets. 2012;13:1750–1756. doi: 10.2174/138945012804545597. [DOI] [PubMed] [Google Scholar]
- Tomosugi M, Sowa Y, Yasuda S, Tanaka R, te Riele H, Ikawa H, et al. Retinoblastoma gene-independent G1 phase arrest by flavone, phosphatidylinositol 3-kinase inhibitor, and histone deacetylase inhibitor. Cancer Sci. 2012;103:2139–2143. doi: 10.1111/cas.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, Majid S, et al. Genistein inhibits prostate cancer cell growth by targeting miR-34a and oncogenic HOTAIR. PLoS ONE. 2013;8:e70372. doi: 10.1371/journal.pone.0070372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui C, Yujie F, Lijia Y, Long Y, Hongxia X, Yong Z, et al. MicroRNA-34a and microRNA-21 play roles in the chemopreventive effects of 3,6-dihydroxyflavone on 1-methyl-1-nitrosourea-induced breast carcinogenesis. Breast Cancer Res. 2012;14:R80. doi: 10.1186/bcr3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M, Ai W, Banik NL, Ray SK. Overexpression of miR-7-1 increases efficacy of green tea polyphenols for induction of apoptosis in human malignant neuroblastoma SH-SY5Y and SK-N-DZ cells. Neurochem Res. 2013;38:420–432. doi: 10.1007/s11064-012-0936-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M, Khandkar M, Banik NL, Ray SK. Alterations in expression of specific microRNAs by combination of 4-HPR and EGCG inhibited growth of human malignant neuroblastoma cells. Brain Res. 2012;1454:1–13. doi: 10.1016/j.brainres.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–1017. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Friedman JM, Liang G. Creating a flexible multiple microRNA expression vector by linking precursor microRNAs. Biochem Biophys Res Commun. 2011;411:276–280. doi: 10.1016/j.bbrc.2011.06.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calimeri T, Battista E, Conforti F, Neri P, Di Martino MT, Rossi M, et al. A unique three-dimensional SCID-polymeric scaffold (SCID-synth-hu) model for in vivo expansion of human primary multiple myeloma cells. Leukemia. 2011;25:707–711. doi: 10.1038/leu.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone P, Neri P, Burger R, Di Martino MT, Leone E, Amodio N, et al. Mouse models as a translational platform for the development of new therapeutic agents in multiple myeloma. Curr Cancer Drug Targets. 2012;12:814–822. doi: 10.2174/156800912802429292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Xie X, Luo J, Liu M, Xi S, Guo J, et al. Targeted expression of miR-34a using the T-VISA system suppresses breast cancer cell growth and invasion. Mol Ther. 2012;20:2326–2334. doi: 10.1038/mt.2012.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinski AL, Slack FJ. miRNA-34 prevents cancer initiation and progression in a therapeutically resistant K-ras and p53-induced mouse model of lung adenocarcinoma. Cancer Res. 2012;72:5576–5587. doi: 10.1158/0008-5472.CAN-12-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal N, Mei J, Liu J, Skapek SX. miR-34a is essential for p19(Arf)-driven cell cycle arrest. Cell Cycle. 2014;13:792–800. doi: 10.4161/cc.27725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, et al. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest. 2012;122:3170–3183. doi: 10.1172/JCI63608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D, et al. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010;70:5923–5930. doi: 10.1158/0008-5472.CAN-10-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trang P, Wiggins JF, Daige CL, Cho C, Omotola M, Brown D, et al. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhu X, Zhang X, Liu B, Huang L. Nanoparticles modified with tumor-targeting scFv deliver siRNA and miRNA for cancer therapy. Mol Ther. 2010;18:1650–1656. doi: 10.1038/mt.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik D, Campbell NR, Karikari C, Chivukula R, Kent OA, Mendell JT, et al. Restitution of tumor suppressor microRNAs using a systemic nanovector inhibits pancreatic cancer growth in mice. Mol Cancer Ther. 2011;10:1470–1480. doi: 10.1158/1535-7163.MCT-11-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin Cancer Res. 2010;16:1119–1128. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg-Gorenshtein G, Avigad S, Jeison M, Halevy-Berco G, Mardoukh J, Luria D, et al. Reduced levels of miR-34a in neuroblastoma are not caused by mutations in the TP53 binding site. Genes Chromosomes Cancer. 2009;48:539–543. doi: 10.1002/gcc.20662. [DOI] [PubMed] [Google Scholar]
- Dijkstra MK, van Lom K, Tielemans D, Elstrodt F, Langerak AW, van ‘t Veer MB, et al. 17p13/TP53 deletion in B-CLL patients is associated with microRNA-34a downregulation. Leukemia. 2009;23:625–627. doi: 10.1038/leu.2008.264. [DOI] [PubMed] [Google Scholar]
- Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, et al. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7:2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- Siemens H, Neumann J, Jackstadt R, Mansmann U, Horst D, Kirchner T, et al. Detection of miR-34a promoter methylation in combination with elevated expression of c-Met and ß-catenin predicts distant metastasis of colon cancer. Clin Cancer Res. 2013;19:710–720. doi: 10.1158/1078-0432.CCR-12-1703. [DOI] [PubMed] [Google Scholar]
- Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68:4123–4132. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:264–269. doi: 10.1073/pnas.0907904107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Luo D, Rong M, Chen G. Underexpression of miR-34a in hepatocellular carcinoma and its contribution towards enhancement of proliferating inhibitory effects of agents targeting c-MET. PLoS ONE. 2013;8:e61054. doi: 10.1371/journal.pone.0061054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo E, Navarro A, Viñolas N, Marrades RM, Diaz T, Gel B, et al. miR-34a as a prognostic marker of relapse in surgically resected non-small-cell lung cancer. Carcinogenesis. 2009;30:1903–1909. doi: 10.1093/carcin/bgp219. [DOI] [PubMed] [Google Scholar]
- Franchina T, Amodeo V, Bronte G, Savio G, Ricciardi GR, Picciotto M, et al. Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non-small cell lung cancer. J Cell Physiol. 2014;229:97–99. doi: 10.1002/jcp.24422. [DOI] [PubMed] [Google Scholar]
- Peurala H, Greco D, Heikkinen T, Kaur S, Bartkova J, Jamshidi M, et al. MiR-34a expression has an effect for lower risk of metastasis and associates with expression patterns predicting clinical outcome in breast cancer. PLoS ONE. 2011;6:e26122. doi: 10.1371/journal.pone.0026122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelser C, Flesch-Janys D, Chang-Claude J, Pantel K, Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin Chem. 2013;59:1489–1496. doi: 10.1373/clinchem.2013.205161. [DOI] [PubMed] [Google Scholar]
- Javeri A, Ghaffarpour M, Taha MF, Houshmand M. Downregulation of miR-34a in breast tumors is not associated with either p53 mutations or promoter hypermethylation while it correlates with metastasis. Med Oncol. 2013;30:413. doi: 10.1007/s12032-012-0413-7. [DOI] [PubMed] [Google Scholar]
- Nugent M, Miller N, Kerin MJ. Circulating miR-34a levels are reduced in colorectal cancer. J Surg Oncol. 2012;106:947–952. doi: 10.1002/jso.23174. [DOI] [PubMed] [Google Scholar]
- Tabernero J, Shapiro GI, LoRusso PM, Cervantes A, Schwartz GK, Weiss GJ, et al. First-in-humans trial of an RNA interference therapeutic targeting VEGF and KSP in cancer patients with liver involvement. Cancer Discov. 2013;3:406–417. doi: 10.1158/2159-8290.CD-12-0429. [DOI] [PubMed] [Google Scholar]
- Lee JM, Yoon TJ, Cho YS. Recent developments in nanoparticle-based siRNA delivery for cancer therapy. Biomed Res Int. 2013;2013:782041. doi: 10.1155/2013/782041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H, Wu J, Sawa T, Matsumura Y, Hori K. Tumor vascular permeability and the EPR effect in macromolecular therapeutics: a review. J Control Release. 2000;65:271–284. doi: 10.1016/s0168-3659(99)00248-5. [DOI] [PubMed] [Google Scholar]
- Basile L, Pignatello R, Passirani C. Active targeting strategies for anticancer drug nanocarriers. Curr Drug Deliv. 2012;9:255–268. doi: 10.2174/156720112800389089. [DOI] [PubMed] [Google Scholar]
- De Rosa G, De Stefano D, Galeone A. Oligonucleotide delivery in cancer therapy. Expert Opin Drug Deliv. 2010;7:1263–1278. doi: 10.1517/17425247.2010.527942. [DOI] [PubMed] [Google Scholar]
- Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Scognamiglio I, Di Martino MT, Campani V, Virgilio A, Galeone A, Gullà A, et al. Transferrin-conjugated SNALPs encapsulating 2'-O-methylated miR-34a for the treatment of multiple myeloma. Biomed Res Int. 2014;2014:217365. doi: 10.1155/2014/217365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Cao M, Zhang J, Hu K, Yin Z, Zhou Z, et al. Hyaluronic acid-chitosan nanoparticles for co-delivery of MiR-34a and doxorubicin in therapy against triple negative breast cancer. Biomaterials. 2014;35:4333–4344. doi: 10.1016/j.biomaterials.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Wang S, Cao M, Deng X, Xiao X, Yin Z, Hu Q, Zhou Z, Zhang F, Zhang R, Wu Y, Sheng W, Zeng Y . (2014) Degradable Hyaluronic Acid/Protamine Sulfate Interpolyelectrolyte Complexes as miRNA-Delivery Nanocapsules for Triple-Negative Breast Cancer Therapy. Adv Healthc Mater. 9. [DOI] [PubMed]
- Tivnan A, Orr WS, Gubala V, Nooney R, Williams DE, McDonagh C, Prenter S M, Harvey H, Domingo-Fernández R, Bray IM, Piskareva O, Ng CY, Lode HN, Davidoff AM, Stallings RL. Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PLoS One. 2012;7:e38129. doi: 10.1371/journal.pone.0038129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu QL, Jiang QY, Jin X, Shen J, Wang K, Li YB, Xu FJ, Tang GP, Li ZH. Cationic microRNA-delivering nanovectors with bifunctional peptides for efficient treatment of PANC-1 xenograft model. Biomaterials. 2013;34:2265–76. doi: 10.1016/j.biomaterials.2012.12.016. [DOI] [PubMed] [Google Scholar]
- Witwer KW, Sisk JM, Gama L, Clements JE. MicroRNA regulation of IFN-beta protein expression: rapid and sensitive modulation of the innate immune response. J Immunol. 2010;184:2369–2376. doi: 10.4049/jimmunol.0902712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicitore A, Caraglia M, Gaudenzi G, Manfredi G, Amato B, Mari D, et al. Type I interferon-mediated pathway interacts with peroxisome proliferator activated receptor-? (PPAR-?): at the cross-road of pancreatic cancer cell proliferation. Biochim Biophys Acta. 2014;1845:42–52. doi: 10.1016/j.bbcan.2013.11.003. [DOI] [PubMed] [Google Scholar]
- Vitale G, Zappavigna S, Marra M, Dicitore A, Meschini S, Condello M, et al. The PPAR-? agonist troglitazone antagonizes survival pathways induced by STAT-3 in recombinant interferon-ß treated pancreatic cancer cells. Biotechnol Adv. 2012;30:169–184. doi: 10.1016/j.biotechadv.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Vitale G, van Eijck CH, van Koetsveld Ing PM, Erdmann JI, Speel EJ, van der Wansem Ing K, et al. Type I interferons in the treatment of pancreatic cancer: mechanisms of action and role of related receptors. Ann Surg. 2007;246:259–268. doi: 10.1097/01.sla.0000261460.07110.f2. [DOI] [PMC free article] [PubMed] [Google Scholar]