Abstract
CCR5 disruption by zinc finger nucleases (ZFNs) is a promising method for HIV-1 gene therapy. However, successful clinical translation of this strategy necessitates the development of a safe and effective method for delivery into relevant cells. We used non-integrating lentivirus (NILV) for transient expression of ZFNs and pseudotyped the virus with HIV-envelope for targeted delivery to CD4+ T cells. Both activated and resting primary CD4+ T cells transduced with CCR5-ZFNs NILV showed resistance to HIV-1 infection in vitro. Furthermore, NILV transduced resting CD4+ T cells from HIV-1 seronegative individuals were resistant to HIV-1 challenge when reconstituted into NOD-scid IL2rγc null (NSG) mice. Likewise, endogenous virus replication was suppressed in NSG mice reconstituted with CCR5-ZFN–transduced resting CD4+ T cells from treatment naïve as well as ART-treated HIV-1 seropositive patients. Taken together, NILV pseudotyped with HIV envelope provides a simple and clinically viable strategy for HIV-1 gene therapy.
Keywords: CCR5 gene editing, HIV-1 therapy, humanized mice, non-integrating lentivirus, resting CD4+ T cells, zinc finger nucleases
Introduction
HIV-1 infection still remains a global epidemic with significant morbidity and mortality. Although anti-retroviral therapy (ART) has extended the lives of HIV-1-infected individuals, it has several limitations including the need for life long therapy, high cost and side effects associated with the drugs.1 Moreover, the virus is not completely eradicated with this approach and rebounds rapidly upon interruption of ART.2,3 Hence, there has been interest in pursuing alternative strategies that are aimed at rendering cells intrinsically resistant to HIV-1.
Zinc finger nucleases (ZFNs) have been widely used for genome editing in various types of cells and living organisms4,5,6,7,8,9,10 and is also a highly promising strategy for HIV-1 therapy aimed at disrupting specific host genes important for virus replication,11,12,13,14,15,16,17,18 or to eliminate provirus in latently infected cells.19 There has been a renewed interest in CCR5 disruption as a strategy for HIV-1 gene therapy because of the so-called “Berlin Patient”, who was completely cured of HIV-1 infection after receiving bone marrow stem cells transplanted from a donor homozygous for the Δ32 mutation (a 32-bp deletion in the single coding exon of the gene) in the CCR5 gene which leads to blockade of HIV-1 entry via this co-receptor.20,21 Unlike conventional gene knockdown by RNAi, which requires the continued presence of the effector siRNA moiety for silencing to be maintained, transient ZFN expression results in permanent and heritable gene editing so that the CCR5-modified cells can confer long-term HIV-1 resistance in patients. CCR5 disruption mediated by ZFNs has been tested in primary CD4+ T cells as well as in CD34+ stem cells.11,12,13,14,15,16,17,18 Reconstitution of NSG mice with activated CD4+ T cells transduced with adenovirus encoding CCR5-ZFNs resulted in resistance to HIV-1 challenge.22 Promising new data have been reported from one of the ongoing clinical trials in which CCR5 gene modified autologous CD4+ T cells were used for adoptive therapy in HIV infected subjects.23 The therapy was found to be safe and well tolerated. Moreover, increased levels of CD4+ T cells were seen following treatment interruption, which attest to the feasibility of the approach. However, large-scale expansion of gene-edited cells for personal therapy is labor intensive and requires a specialized laboratory set up. Alternative simplified regimens may facilitate the dissemination of the therapeutic approach for large-scale clinical application.
Lentiviral vectors are among the most efficient gene delivery vehicles for primary cells, including T cells, making them attractive vectors for ZFN delivery for HIV-1 therapy. The third generation vectors currently used in gene therapy trials have undergone extensive modifications to ensure safety.24,25,26 Nevertheless, insertional mutagenesis could pose a potential risk for gene therapy with integrating lentiviral vectors.27,28 On the other hand, non-integrating lentivirus (NILV) generated by packaging vectors with a mutated integrase gene minimizes vector integration into the host genome, which makes them ideal for gene editing applications, where only transient expression of the transgene is needed.29 Indeed, CCR5 disruption using a non-integrating lentiviral vector in human stem cells has been attempted in vitro,11 but the applicability of the approach for selective delivery to primary CD4+ T cells for inducing HIV-1 resistance remains untested.
In this work, we tested transduction efficiency and CCR5 disruption frequency after ZFN delivery to cell lines and primary CD4+ T cells via a non-integrating lentiviral vector. In previous studies using adenoviral vector-based transduction, activated CD4+ T cells were gene-modified and expanded in vitro before transfusion.12,23 However, in addition to being time-consuming and expensive, extended culture after T-cell activation may result in an altered cell surface phenotype and exhaustion of the cells, which may have an impact on the efficacy of gene therapy in the long-term.30,31,32 Although conventional adenoviral and lentiviral vectors efficiently transduce a multitude of cell types, including primary cells, they are inefficient in transducing resting T cells.32,33 Here, we used NILV pseudotyped with HIV-1 envelope to enable transduction of unstimulated CD4+ T cells from healthy or HIV-infected donors. This allowed selective transduction of the cells within unfractionated PBMCs, which could then be infused into NSG mice (Hu/PBL model) without further in vitro manipulation and then evaluated for reconstitution and resistance to HIV-1 infection. These efforts are anticipated to lead to the development of a simple approach for generating HIV-resistant cells for cellular therapy in HIV-1 infected patients.
Results
Construction and characterization of non-integrating CCR5-ZFN encoding lentivirus
Non-integrating lentivirus has been demonstrated to successfully infect various types of cell lines, as well as stem cells without integration of viral DNA into the cellular genome.29,34 For nucleolytic activity, two arms of Zinc finger nucleases are designed to bind to 9–18 nucleotides on the left and right hand side of the targeted gene on the sense and antisense strands respectively. To improve the disruption efficiency, we expressed the ZFN pair targeting a specific site within the CCR5 gene in a single construct by connecting them via a FMDV 2A sequence (Supplementary Figure S1a, for the ZFN protein sequence see Supplementary Figure S1c). The sequence was cloned into the lentiviral vector, pLVX-ZsGreen (a bicistronic IRES vector to allow simultaneous expression of ZFNs and ZsGreen from the same RNA transcript, enabling easy confirmation of ZFNs expression by monitoring ZsGreen expression) under the CMV promoter to produce a fusion protein containing both arms (Supplementary Figure S1b). However, this fusion protein is expected to be cleaved soon after translation, because of self-cleavage activity of the 2A sequence,35 yielding two functional units that simultaneously target their respective DNA binding sites. To derive the NILV, the ZFN vector was co-transfected with a packaging plasmid and an envelope (either VSV-G env or HIV-1 env) encoding plasmid into 293 T cells and the supernatants collected after 48 hours of culture. In order to achieve transient ZFN expression, we used an intergrase-defective packaging plasmid, vTK939, to package the lentivirus. In vitro and in vivo studies have demonstrated that the resulting lentivirus shows much lower levels of illegitimate integration.36 We first tested the transduction efficiency of the VSV-G pseudotyped non-integrating vector compared with the integrating lentiviral vector in HEK 293 T cells. The results show that 48 hours after lentiviral infection, ZsGreen expression was comparable between the two vectors (Supplementary Figure S1d). To evaluate whether NILV expression was more transient compared to conventional integrating LV, we transduced 293 T cells with both vectors at a multiplicity of infection (MOI) of 10 (20 ng of p24 for 1 × 105 cells) and then monitored the GFP expression over time. In the 10 day period of observation, the GFP fluorescence was progressively lost in NILV transduced cells, declining to baseline level by day 8 whereas the percentage of GFP-positive cells remained stable after transduction with the WT integrating vector (Supplementary Figure S2).
The MTS cell viability assay did not reveal any overt cytotoxic effects of NILV-mediated ZFN expression in 293T cells (Supplementary Figure S3). In silico genome-wide analysis with PROGNOS software for potential off target gene modification by the ZFN sequences used in this study (obtained from Sigma-Aldrich, St Louis, MO) did not reveal any off-target sites when no mismatch was allowed but showed 32 potential off-target sites when two mismatches per ZFN half-site were allowed.37 However, deep sequencing data with K562 cells treated with the same ZFN pair by another group revealed no detectable off-target activity.38
We next evaluated the efficiency of CCR5 disruption after NILV-mediated ZFN delivery in 293T cells. CCR5 disruption was quantified by PCR amplification across the CCR5 locus, heating and re-annealing of PCR products, and digestion with T7 endonuclease, which is able to cleave mismatched dsDNA caused by non-homologous end joining (NHEJ).39 At an MOI of 30 (60 ng of p24 for 1 × 105 cells), the transduction efficiency was 63.8%, for HEK293T cells, with a corresponding CCR5 disruption frequency of 60% (Figure 1a).
Figure 1.
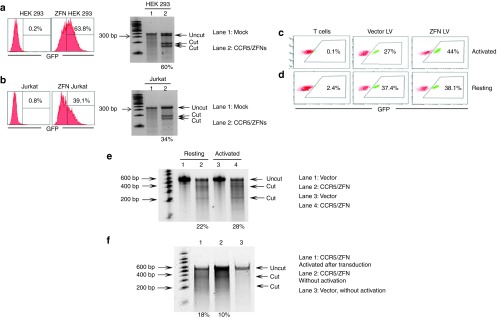
ZFN-expressing NILV-mediated disruption of CCR5 gene in cell lines and primary human CD4+ T cells. (a) Transduction efficiency and CCR5 disruption efficiency of HEK 293 cells. HEK 293 cells were transduced with CCR/ZFNs NILV pseudotyped with VSV-G at an MOI of 30 and examined for ZsGreen expression by flow cytometry after 48 hours. The genomic DNA extracted from the transduced cells was tested for CCR5 disruption by T7 endonuclease assay using a Tris-Acetate PAGE gel. Empty vector (GFP only) was used as control. (b) Transduction efficiency and CCR5 disruption efficiency in Jurkat cells after ZFN delivery with HIV envelope pseudotyped NILV. (c) PHA-activated CD4+ T cells were transduced with CCR5/ZFN NILV pseudotyped with HIV envelope at an MOI of 30 and ZsGreen-positive cells were enumerated by flow cytometry. (d) Resting CD4+ T cells were transduced as in c, and then activated with PHA 4 hours after transduction. For both activated and resting CD4+ T cells transduction assays, the empty NILV (without CCR5/ZFN) was used as control. (e) The frequency of CCR5 disruption in activated and resting CD4+ T cells (activated 4 hours after transduction) was assessed by T7 endonuclease assay. (f) The frequency of ZFN-mediated CCR5 disruption in resting CD4+ T cells with or without activation after transduction. Purified resting CD4+ T cells were transduced with NILV at an MOI of 15. The frequency of CCR5 disruption assessed by T7 nuclease assay is indicated as percentage of total population, calculated by the formula: 100 × [1 − (1 − fraction of cleavage)1/2]. Representative data from experiments performed at least three times are shown.
HIV envelope pseudotyped NILV can deliver ZFNs to resting and activated CD4+ T cells to efficiently disrupt the CCR5 gene
Findings from multiple studies indicate poor transduction of unstimulated but not stimulated CD4+ T cells with VSV-G pseudotyped lentiviral particles.32,40 As CXCR-4 tropic HIV envelope pseudotyping has been shown to allow efficient lentiviral transduction of quiescent CD4+ T cells,32 we used this approach for NILV-mediated delivery of ZFNs to HIV-1-susceptible resting and activated CD4+ T cells. We first tested whether the HIV envelope pseudotyped NILV could deliver ZFNs to CD4 expressing Jurkat cells. As shown in Figure 1b, at an MOI of 30 (60 ng of p24 for 1 × 105 cells), the transduction efficiency was 39.1% with a corresponding CCR5 disruption frequency of 34%.
We next compared NILV transduction in resting and activated primary T cells with conventional VSV-G or HIV env pseudotyped NILV, using ZsGreen to track expression. In agreement with previous data,32 we found equivalent ZsGreen expression with both HIV env and VSV-G in CD4+ T cells activated with phytohemagglutinin (PHA) prior to transduction. On the other hand, in unstimulated CD4+ T cells, there was negligible ZsGreen expression with VSV-G, whereas with HIV-env pseudotyped NILV the expression level was comparable to that in activated T cells (Supplementary Figure S4).
We also evaluated whether HIV env pseudotyping could be used for selective ZFN delivery to CD4+ T cells within a mixed PBMC population. PBMCs isolated from a normal donor were transduced with NILV at an MOI of 10 (20 ng of p24 for 1 × 105 cells) and after 48 hours of culture, ZsGreen expression on CD8, CD4, and CD19 gated cells was evaluated by flow cytometry. As shown in Supplementary Figure S5, after transduction, ZsGreen was expressed only in CD4+ T cells and not in CD8+ T cells or in B cells (CD19+).
CCR5 editing efficiency was also compared between PHA activated and resting primary CD4+ T cells transduced with ZFN-expressing HIV-env pseudotyped NILV at an MOI of 30. Resting cells were stimulated with PHA 4 hours after transduction. After 2 days of culture, the CCR5 disruption frequency was found to be 28% for prior activated and 22% for resting CD4+ T cells with a corresponding transduction efficiency of 44% and 38.1% respectively (Figure 1c–e).
In the previous experiment, the resting cells were activated following NILV transduction, so it was important to directly test whether ZFN-cleavage was activation dependent or could occur even in the absence of any stimulation. To test this, resting CD4+ T cells transduced with NILV at an MOI of 15 (30 ng p24 for 1 × 105 cells) were cultured for 2 days with or without PHA activation and T7 endonuclease assay was performed with genomic DNA extracted from the cells. The results show that the CCR5 locus was successfully disrupted with an efficiency of 10% in unstimulated, compared with 18% in CD4+ T cells that were activated 4 hours after transduction (Figure 1f). The transduction efficiency assessed by ZsGreen expression was 13% and 24% with or without activation respectively (data not shown), so the proportion of ZFN cleavage in the transduced cells was similar in both cases (about 75% of the transduced population).
CCR5 disruption protects primary CD4+ T cells from HIV-1 infection in vitro
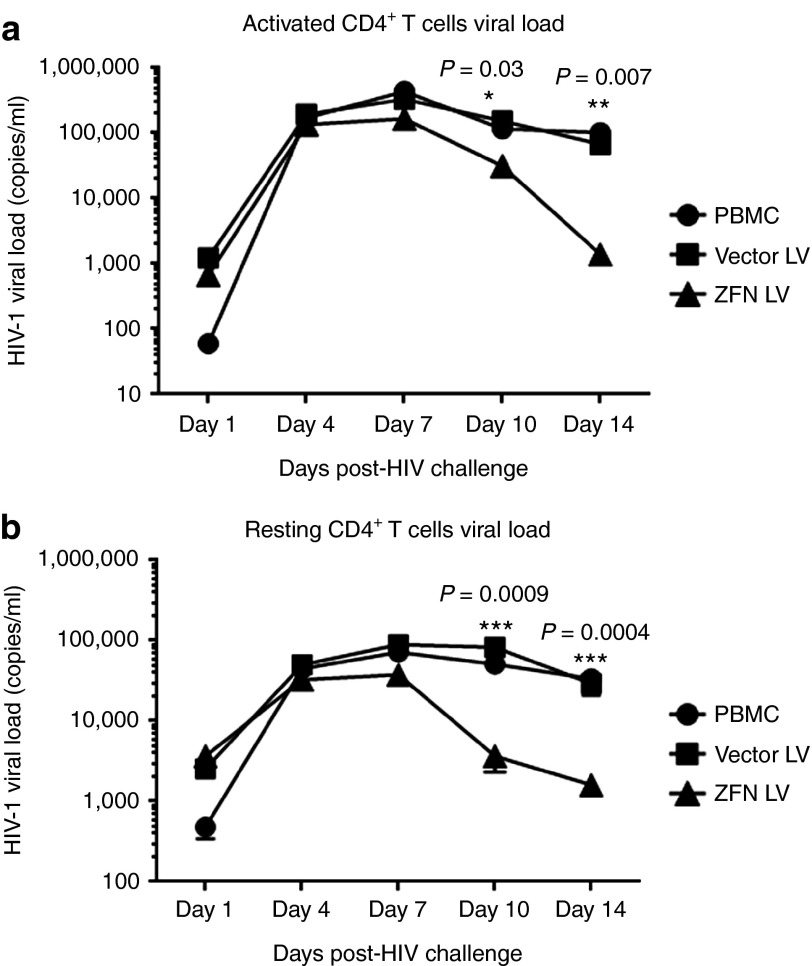
To investigate whether CCR5 disruption by CCR5-ZFNs NILV can suppress HIV-1 infection, primary CD4+ T cells were transduced with HIV-pseudotyped NILV at an MOI of 30 (60 ng of p24 for 1 × 105 cells) and then challenged with CCR5 tropic HIV-1 isolate HIVBaL at an MOI of 0.1 (200 pg p24 for 1 × 105 cells). Activated cells were transduced with NILV and then challenged with HIV-1 48 hours after transduction, whereas resting CD4+ T cells were first transduced with NILV, activated with PHA for 2 days and then subjected to viral challenge. Supernatants were collected from the cultures on days 1, 4, 7, 10, and 14 for viral load determination by qRT-PCR. With mock and control vector transduction, the response pattern to HIV-1 challenge was very similar in both activated and resting CD4+ T cells in that the viral load peaked at day 7, and then dropped two- to fourfolds after day 14. We reason that this decrease in viral load is caused by the decreasing availability of CD4+ T cells for further de novo infection. By contrast, in both resting and activated CD4+ T cells transduced with CCR5-ZFNs, the viral load was substantially lower than either non-transduced or vector-transduced CD4+ T cells on day 7, and became nearly undetectable by 14 days after infection. These results suggest that CCR5 disruption protects against HIV-1 infection and thus provides survival advantage to the cells (Figure 2a,b).
Figure 2.
CCR5-ZFN-expressing NILV transduced activated and resting CD4+ T cells resist HIV infection in vitro. (a) activated or (b) resting CD4+ T cells were challenged with HIVBAL 48 hours after transduction and the supernatants collected at different time points were tested by LRT qRT-PCR to determine the viral titers. PBMC and empty NILV were used as controls. All the samples for qRT-PCR were in triplicates and plotted as average copy numbers. One way analysis of variance was used to compare the differences between groups at different time points. Significance levels are shown in the figures as *P < 0.05; **P < 0.01; and ***P < 0.001.
CCR5 disruption in resting CD4+ T cells from a seronegative donor confers HIV-1 resistance in Hu/PBL mice
We used the Hu/PBL model to assess whether CCR5-ZFNs-transduced resting CD4+ T cells can be reconstituted in humanized mice, and whether the gene modified cells are able to resist HIV-1 infection in vivo. CD4+ T cells isolated from a HIV-1 seronegative donor, were transduced with HIV-pseudotyped NILV for 2 hours at an MOI of 10 (200 ng of p24 per 2 × 106 cells) and injected into the animals (5 × 105 CD4+ cells/mouse) along with 0.5 million unmanipulated PBMCs to facilitate engraftment. Non-transduced and empty NILV vector-transduced cells were also similarly injected in separate groups of mice as negative controls. All mice were challenged with 100 ng p24 of R5-tropic virus, HIVBaL 5 days after cell infusion. The CD4+ T cell profile (% CD3+CD4+) was monitored on days 7, 21, 34, and 42 (Figure 3a). The transduction efficiencies for vector and CCR5-ZFNs NILV were 10.1% and 16.7% respectively (Figure 3b). On day 7 after infusion, similar levels of CD4 reconstitution (around 50–70%) were observed in all groups. However, by day 21 after challenge, the control LV-transduced and non-transduced groups exhibited a dramatic decrease in CD3+CD4+ cells, compared to the CCR5-ZFN group, where five out of eight animals showed a significantly higher percentages of CD3+CD4+ cells. Furthermore, the CD4+ T cell levels were maintained over time in CCR5-ZFN group as assessed on days 21, 34, and 42, whereas the levels consistently remained low in both non-transduced group and vector-transduced groups (Figure 3c). We also investigated whether transfer of ZFN modified CD4+ T cells could control plasma viremia by determining the p24 levels by ELISA in sera collected after sacrificing the animals 7 weeks after challenge. The results revealed that HIV-1 was at a very low level in most of the animals in the ZFN group (around 10 pg/ml in four mice and 50 pg/ml in one mouse), whereas the control groups showed a high viral load, ranging from 100–900 pg/ml (Figure 3d). The three animals in the ZFN group that failed to maintain their CD4 counts showed viral loads similar to the control mice. The result of p24 ELISA was also corroborated by analysis of HIV-1 cDNA late reverse transcripts (LRTs) by qRT-PCR.41 Even here, in the CCR5-ZFN transduced group, the viral loads were very low in the same five protected animals (1,000–2,500 copies/ml), whereas both non-transduced and vector transduced groups presented a significantly higher viral load (average around 10,000 copies/ml) (Figure 3e). Overall, these results suggest that transient ZFN expression in resting CD4+ T cells using HIV envelope pseudotyped NILV can generate HIV-1-resistant cells that efficiently engraft and expand in humanized mice.
Figure 3.
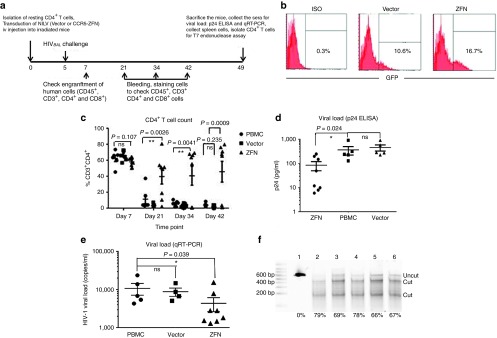
Reconstitution with ZFN-treated CD4+ T cells from a normal donor protects Hu-PBL mice from HIV challenge. (a) Schematic of study design. (b) NILV transduction efficiency in resting CD4 T cells used for infusion into Hu-PBL mice. (c) CD4 T-cell counts at different time points in HIV infected Hu-PBL mice infused with 0.5 × 106 CD4 cells treated with CCR5-ZFN- (8 mice), empty vector- (6 mice, 1 mouse died on day 21), or left untreated (6 mice, 1 mouse died on day 28) along with 0.5 × 106 unfractionated PBMCs. One-way ANOVA was used to compare the difference between these three groups. ns = P > 0.05; *P < 0.05; **P < 0.01; and ***P < 0.001. (d,e) Viral load measured (d) by p24 ELISA or (e) by qRT-PCR using sera collected on Day 49. Multiple T test was used to test the significances between groups. ns = P > 0.05; *P < 0.05. (f) T7 endonuclease assay with human CD4+ T cells isolated from spleen on day 49 from 5 ZFN-treated and 1 vector control-treated mice. Lane 1: Vector control; Lanes 2–6: ZFN group. The frequency of CCR5 gene disruption is indicated as percentage, calculated by the formula: 100 × [1 − (1 − fraction of cleavage)1/2].
The increase in the proportion of CD4+ T cells over time in the CCR5-ZFN group suggests that CCR5 disruption provides survival advantage to the gene modified relative to the non-transduced CD4+ T cells, which became infected and died. To confirm CCR5 disruption in this population, genomic DNA extracted from purified CD4+ T cells was subjected to T7 endonuclease assay. For many of the unprotected mice, the T7 endonuclease assay could not be carried out because of a paucity of CD4+ T cells after HIV-1 challenge. By contrast, the CCR5 ZFN-protected showed an average disruption frequency of 71.8%, confirming the enrichment of protected cells (Figure 3f).
Suppression of endogenous virus in Hu/PBL mice engrafted with CCR5 modified resting CD4+ T cells from HIV-1-seropositive subjects
Infusion of autologous CCR5 gene modified CD4+ T cells is being tried as a potential therapy for HIV-1 infection. Towards this end, we tested the protective efficacy of ZFN-mediated CCR5 disruption in Hu/PBL mice reconstituted with PBMCs from HIV-1+ donors. As HIV pseudotyped NILV is expected to selectively target CD4+ T cells, we directly transduced 3 × 106 PBMCs from the patients in the absence of CD4+ T-cell purification. PBMCs transduced at an MOI of 10 (300 ng of p24 for 3 × 106 PBMCs) without prior activation were infused into NSG mice. The CD4+ T-cell levels were monitored over time and sera collected at the time of sacrifice were used for viral load analysis.
We first tested the strategy with PBMCs from two treatment-naïve HIV-1+ subjects with high viral loads (for patient information see Supplementary Table S1). We used only four mice for each group (mock and ZFN transduced) because of limitation of the number of cells available. PBMCs from only one of the two subjects showed successful engraftment in NSG mice, with three out of four animals in both control and test groups showing adequate human cell reconstitution. The results showed that the CD4 count was maintained in the CCR5 ZFN-transduced group (CD3+CD4+ levels between 49–57%), whereas in the mock-transduced group, the CD4+ T cells dropped from a mean of 59% on day 7 to a mean of 20% on day 42 (Figure 4a). Protection was also assessed by measuring the viral load by p24 ELISA in sera collected on day 49 after transplantation. The viral load was very much reduced in the ZFN group (average 25 pg/ml blood, with undetectable virus in one animal) compared with the mock treated group (averaging 360 pg/ml) (Figure 4b). Consistent with the p24 ELISA results, a qRT-PCR assay also revealed that the average viral load was around 1,500 copies/ml blood in CCR5 ZFN group, whereas in the mock treated group it was about fivefold higher (Figure 4c).
Figure 4.
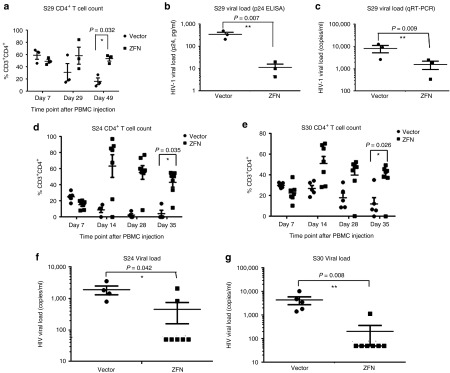
Direct ZFN transduction of whole PBMCs from treated or untreated seropositive individuals confers protection from endogenous virus in Hu-PBL mice. Whole PBMCs (3 x 106) from indicated subjects were mock-transduced or transduced with HIV env pseudotyped NILV and injected iv into NSG mice. CD4 T-cell counts over time (a) viral loads measured by (b) p24 ELISA, and (c) qRT-PCR in sera collected on day 49 in Hu-PBL mice infused with PBMCs from an ART negative HIV-1+ individual. Multiple T test was used to compare the differences between Vector or ZFN treated groups (three mice each group), significance values are shown in the figures. *P < 0.05, **P < 0.01. (d–g) CD4 T-cell counts over time (d,e) and viral load measured by qPCR in sera collected on day 35 (f,g) in Hu-PBL mice infused with CD4 T cells from two ART treated HIV positive individuals. Multiple t-test was used to compare the differences between Vector (n = 4 for S24 and n = 5 for S30) and ZFN (N = 7 for both S24 and S30) treated groups. *P < 0.05, **P < 0.01.
Next, we used the same Hu/PBL model to test HIV-1 resistance mediated by CCR5 disruption in resting PBMCs from 2 HIV-1+ subjects on ART, whose viral loads were undetectable (Subject 24, S24) or at very low level (Subject 30, S30). In both cases, good human cell engraftment was observed in all mice (Figure 4d,e). The CD4 levels on day 7 were slightly higher for subject S30 (30% for the vector-treated and 23% for ZFN-treated group) than for subject S24 (24% for the vector-treated and 16% for ZFN-treated group), which is consistent with the CD4+ T-cell counts in the input PBMCs (40% in S30 and 27% in S24). Although the CD4+ T-cell counts were slightly higher in the control group compared with the CCR5/ZFNs group on day 7, CD4+ T cells in this group gradually declined for both subjects (dropping from an average of 24% on day 7 to 12.7% on day 35 for S30 and from 23% to 3% for S24) (Figure 4,e). By contrast, in the CCR5/ZFN group, the CD4+ T-cell levels increased with time (from ~23% to ~45% for S30, and from 16% to 50% for S24).
As the viral load was below detection by p24 ELISA in all groups (data not shown), we used the more sensitive qRT-PCR assay to evaluate plasma viremia. The results showed that the virus remained below the limit of detection (<60 copies/ml) in the sera of 6/7 mice (for S30) and 5/7 mice (for S24) in the CCR5-ZFN group, whereas for both subjects, a significantly higher viral load was detected in all the control mice (Figure 4f,g).
Taken together, these results demonstrate that direct transduction of PBMCs from HIV-1 seropositive subjects with HIV-1-pseudotyped NILV is a feasible approach for CCR5 gene editing; and that after infusion into Hu/PBL mice, the gene modified cells are protected from HIV-1 infection.
Discussion
Here, we used a non-integrating lentiviral system for transient expression of ZFNs to edit the CCR5 gene in primary CD4+ T cells. More importantly, pseudotyping NILV with HIV envelope allowed targeting of resting CD4 T cells, obviating the need for CD4+ T-cell isolation from PBMCs or prior activation. Using the Hu/PBL mouse model, we also showed that the gene modified resting CD4+ T cells from normal, as well as HIV-1+ donors, could engraft and provide protection from HIV-1 infection in vivo.
Recently, multiple studies have demonstrated the efficacy of ZFN-mediated CCR5 disruption as a therapeutic strategy to block HIV-1 infection.11,12,13,17,22 Adenoviral vector has been the preferred vehicle for ZFN delivery because of its ability to transduce hard to transfect primary cells.42 Despite a similar advantage, conventional lentiviral vectors have not been utilized because of their propensity to integrate into the host genome, which poses the obvious risk of insertional mutagenesis. In addition, long-term expression of ZFN from an integrated transgene can also increase the risks of off target effects.43,44,45 Tracking of ZsGreen expression revealed that NILV expression in cultured cells decreased to a nearly undetectable level by day 8, which is in agreement with published data showing that NILV expression through cell division reaches baseline levels by day 10.29 These properties of NILV make it well suited for transient ZFN expression to confer stable CCR5 gene modification and thereby induce HIV-1 resistance in susceptible cells.
Integrase defective vectors are known to induce only transient accumulation of episomal DNA in the nucleus as they retain a very weak integration capacity compared with WT vectors.46,47 NILVs lacks the integrase protein that facilitates integration, but background integrations due to illegitimate IN-independent insertions still occurs in transduced cells, the reported rate of which has varied considerably.48 With the D64E mutant used in this study, the residual integration was demonstrated to be ~0.2% in NILV-transduced 293T cells.36 Suwanmanee et al. have reported a 35-fold reduction in integration frequencies in liver tissue when the same D64E vector used in this study was injected in vivo to correct factor IX deficiency in a mouse model of hemophilia.49 Thus, although very low, some residual illegitimate integrations do occur with NILV, which poses a definite safety concern for clinical use of NILV. Recent studies have shown that the frequency of such residual integration can be reduced to minimal levels by using a PPT deleted vector.36 Another potential risk for use of NILV for HIV-1 therapy is the possibility that in HIV-1 infected cells NILVs could be complemented in trans by an active integrase of similar specificity. As suggested by Lombardo et al. incorporating cis-acting mutations in the integrase attachment sites of the vector may prevent such a rescue.11
The ZFNs employed for CCR5 gene targeting in this study were obtained from a commercial source (Sigma-Aldrich) and differ in sequence from the one from Sangamo Biosciences that has been used in multiple studies for CCR5 gene editing in hematopoietic stem cells and primary T cells.12,13,18 In silico genome-wide analysis of the sequences targeted by the ZFN pair used in this study did not reveal any off-target sites. Although allowing two mismatches per ZFN half-site for in silico analysis identifies 32 potential off target sites, deep sequencing data published by Badia et al. found no off target effects in human K562 cells transfected with the same ZFN pair.38 However, the recent demonstration that NILV can be captured at sites of double stranded breaks induced by ZFNs by nonhomologous end joining suggests that such illegitimate integrations could also contribute to off target toxicity.50,51 Thus, for actual clinical use, the genome wide impact of NILV-mediated ZFN delivery in primary T cells has to be understood. The potential for insertional mutagenesis is somewhat mitigated by studies showing that mature CD4+ T cells are relatively resistant to oncogenic transformation in vivo even after transduction with integrating retroviral vector expressing potent T-cell oncogenes.52
Conventionally, VSV-G is used for pseudotyping lentiviruses because of its broad tropism for many different cell types. However, cells in the G0 stage of the cell cycle, such as resting CD4+ T cells are highly recalcitrant to transduction with VSV-G pseudotyped lentivirus. On the other hand, unlike VSV-G, CXCR4-tropic HIV envelope has been shown to mediate fusion of lentiviral particles in both unstimulated and stimulated CD4+ T cells.32 Our study also shows that lentivirus packaged with X4-tropic envelope from LAI can efficiently deliver ZFNs for CCR5 disruption into resting CD4+ T cells. This makes it possible to selectively target HIV-1 susceptible CD4+ T cells within unfractionated PBMC population. Furthermore, as ex vivo transduction is the only external manipulation required, after ZFN modification, the PBMCs can be immediately infused back into the patient, which would simplify ZFN-based therapy for wider clinical application.
CCR5 disruption has been the goal in most studies evaluating ZFNs for HIV-1 therapy. This is because the vast majority of newly transmitted HIV-1 strains use the co-receptor and CCR5 tropic viruses predominate during the early stages of infection.53,54,55 Nevertheless, over time, there is a significant increase in quasispecies that utilize CXCR4 as an alternative co-receptor for infection.55,56,57,58,59 Thus, in addition to CCR5, CXCR4 disruption may also be needed, particularly for the treatment of chronically infected patients.
Although a majority of animals transplanted with CCR5 ZFNs modified CD4 T cells were protected from HIV-1 challenge, we also observed that some animals (3 out of 8 animals receiving PBMCs from a normal donor and 2 out of 14 animals receiving patient PBMCs) failed to show protection. Although the reason for this is not clear, another study also found that a fraction of animals treated with CCR5 ZFN-modified T cells remained unprotected.12 As only a small number of xenoreactive cells are expected to expand after infusion of resting CD4+ T cells, it is possible that paucity of gene modified cells within this fraction in some of the animals could explain the lack of protection. Thus, increasing the infusion size may be necessary to show complete antiviral efficacy in humanized mice.
Taken together, our study demonstrates that NILV can serve as a potentially useful alternative vector for ZFN-mediated CCR5 alteration. HIV envelope pseudotyping of NILV allows unfractionated PBMCs from HIV-1 patients to be directly transduced ex vivo and reinfused back into the patients without extensive ex vivo manipulation. As no T-cell activation or large-scale expansion of gene modified cells are involved, the impact on immune homeostasis is likely to be minimal. The simplicity of the delivery approach used here is likely to broaden the scope of ZFNs-mediated CCR5 disruption for HIV-1 gene therapy.
Materials and Methods
Cells and cell culture. HEK 293T cells and TZM-bl cells were cultured in complete DMEM medium (Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS), and penicillin-streptomycin-glutamine (PSG, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mmol/l L-glutamine). Jurkat cells were cultured in complete RPMI medium (RPMI 1640 medium supplemented with 10% FBS and PSG). Human Peripheral blood mononuclear cells (PBMCs) were purified from normal human donor blood by Ficoll density gradient centrifugation. Primary CD4+ T cells were purified from either healthy donors or patients' PBMCs by two rounds of negative selection using the EasySep Human CD4+ T-cell enrichment kit (Stemcell Technologies, Vancouver, BC, Canada) according to the manufacturer's instructions.
Lentiviral vector. A CCR5-ZFN plasmid pair encoding the left and right ZFN arms was purchased from Sigma-Aldrich (St Louis, MO). Lentiviral vector pLVX-ZsGreen was purchased from Clontech Laboratory (Mountain View, CA). The following primer pairs were used to amplify ZFN1 and ZFN2 fragments from CCR5-ZFNs plasmids: zfn1 forward: cgctctgaattcgccaccatggactacaaagaccatgac (which includes a kozak sequence and the start codon) and zfn1 reverse: gcccctcgagagatctgaagttgatctcgc; zfn2 forward: gcccctcgagggatccggagccacgaacttctctctgttaaagcaagcaggagacgtggaag aaaaccccggtcctatgagatctgactacaaagac (which contains a 2A sequence from foot-and-mouth disease virus) and zfn2 reverse: ccgacatctagattatcagaagttgatctcgccg. After PCR amplification of zfn fragments, the zfn1 fragment was digested with EcoR I/Xho I and zfn2 with Xho I/Xba I. pLVX-ZsGreen vector was digested with EcoR I/Xba I and the three digested fragments were ligated together at a molar ratio of 1:1:1 and transformed into E. coli DH5α. The resulting recombinant plasmid (pLVX-ZFN1-2A-ZFN2) which co-expresses both ZFN arms was used to transfect 293T cells to generate lentivirus.
Generation and titration of lentivirus. 1 × 107 293 T cells were co-transfected with 20 μg of vector plasmid (pLVX-ZFN1-2A-ZFN2, or pLVX as control), 16 μg of packaging plasmid and 10 μg of envelope plasmid using Polyethylenimine (PEI, Polysciences, Warrington, PA).60 For integrating lentivirus, pHR'8.9ΔVPR was used as packaging plasmid (which expresses functional integrase)61; while vTK939 (which expresses D64E-mutant integrase) was used in a similar manner as when making non-integrating lentivirus.36 The plasmids pCMV-VSV-G or pHIV LAI (generously provided by Dr Una O'Doherty at the University of Pennsylvania) were used for pseudotyping the lentiviruse with VSV-G and HIV-1 envelopes respectively. After 30 minutes of transfection, DMEM supplemented with 10% FBS and PSG was added and the cells were incubated at 37 °C under 5% CO2. Two days later, the supernatants were collected, filtered (low binding filter, PVDF membrane, Millipore) and concentrated by ultracentrifugation at 25,000g at 4 °C using an SW 40 Ti rotor for 2.5 hours and the lentivirus pellets were dissolved in PBS. To determine the viral titers, 293 T or TZM-bl cells were transduced with VSV-G or HIV-1 pseudotyped lentivirus respectively. Briefly, 1 × 105 cells were spin-transduced with serially diluted lentivirus preparations in serum free DMEM in the presence of 7 μg/ml polybrene (Sigma-Aldrich) at 1,200 g for 2 hours, and then incubated at 37 °C for another 2 hours. After changing medium to complete DMEM, the transduced cells were incubated at 37 °C for 48 hours. The cells were then fixed with 1% formaldehyde. The transduction efficiency was quantified by evaluating the infected cells for GFP expression by flow cytometry.
T7 endonuclease assay. Genomic DNA was extracted using the GenElute Mammalian Genomic DNA miniprep kit (Sigma-Aldrich) according to the manufacturer's instructions. The frequency of gene modification by NHEJ was evaluated as described previously.16 Briefly, the purified genomic DNA was used as a template to amplify a fragment of the CCR5 gene using the specific primers (CCR5 forward: 5′-TTTTGTGGGCAACATGCTGGTCATCCT-3′ and CCR5 reverse: 5′-TGTAGGGAGCCCAGAAGAGA-3′), to yield a 346 bp amplicon. To amplify the CCR5 gene fragment in CD4+ T cells from humanized mice the primer pairs were: CCR5 forward: 5′-GGTCATCCTCATCCTGAT-3′ and CCR5 reverse: 5′-TGTAGGGAGCCCAGAAGAGA-3′) which results in a 605 bp amplicon. The PCR products were gel purified, denatured and re-annealed to form heteroduplex DNA, followed by treatment with T7 endonuclease. Since T7 endonuclease is mismatch-sensitive, the insertions and deletions caused by NHEJ could be detected by running a 7% Tris-Acetate PAGE gel and quantitated by densitometry using Quantity One 4.6.6 software. The disruption frequency was defined as 100 × [1-(1-fraction cleaved)1/2], wherein the fraction cleaved is the density sum of the cleaved bands divided by the density sum of the cleaved bands and un-cleaved band.
Viral load determination. HIV-1 viral loads in cell culture or in the sera were determined by HIV-1 cDNA Late Reverse Transcripts (LRTs) qRT-PCR and/or p24 ELISA. LRTs PCR was performed as described elsewhere26 but with a small modification. Briefly, for cell culture supernatants, the supernatants were treated with 10% Triton-100, then directly used for qRT-PCR. For mice sera, RNAs were isolated according to the manufacturer's instructions (QIAamp viral RNA mini kit, Qiagen, Germantown, MD), and used as PCR templates. The PCR mixtures were formulated according to the manufacturer's instructions (Taqman Fast Virus 1-Step Master Mix kit, Life Technologies, Grand Island, NY). The primers (Sigma-Aldrich) were as follows: MH531 (5′ TGTGTGCCCGTCTGTTGTGT 3′), MH532 (5′ GAGTCCTGCGTCGAGAGAGC 3′), and fluorescent probe LRT-P (5′ FAM-CAGTGGCGCCCGAACAGGGA-TAMRA 3′). The PCR mixtures were incubated at 95 °C for 5 minutes, followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. A sample quantitated by the 4th generation Roche Cobas Ampliprep/TaqMan method at the Laboratory of Clinical and Biological studies, Miller School of Medicine, University of Miami was used as standard. A standard curve generated using dilutions of this standard to calculate the absolute copy numbers of copies. The lower limit of detection of the assay was determined to be 50 copies/ml. The Alliance HIV-1 ELISA kit from PerkinElmer (Waltham, MA) was used to perform p24 ELISA according to the manufacturer's instructions.
Lentivirus transduction and HIV-1 challenge in vitro. 293 T cells, Jurkat cells and primary CD4+ T cells were used for lentiviral transduction. CD4+ T cells were purified from PBMCs of HIV-1-seronegative donors by two rounds of negative selection. CD4+ T cells were transduced either directly (resting) or after activation with PHA (1 µg/ml) in the presence of 50 U/ml rIL-2 for 2 days. In brief, 1 × 105 cells were spin-transduced with VSV-G or HIV-pseudotyped lentivirus (ZFN or control virus) at indicated multiplicity of infection (MOI). Resting CD4 T cells were activated with PHA following transduction for 2 days before HIV-1 challenge. Cells were then infected with the CCR5 tropic strain HIVBAL (stocked in our laboratory) at an MOI of 0.1 (200 pg of p24 for 1 × 105 cells). Supernatants collected at various time points were assayed for viral titers.
Testing HIV-1 resistance in Hu-PBL mice. All work with animals was approved by the Institutional Review Board of the Texas Tech University Health Sciences Center.
NOD.cg PrkdcscidIL2rgtm/Wjl/Sz (NSG) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Hu-PBL mice were essentially generated as described.62 PBMCs from HIV-1 seronegative or seropositive individuals on or off ART were used. Freshly prepared PBMCs or negatively isolated CD4+ T cells (2 × 105) were transduced with HIV-pseudotyped lentivirus as described earlier. The cells were injected intravenously (iv) in a volume of 0.2 ml PBS into irradiated (180 rads) 4–6 week-old mice. Transduced CD4+ T cells were infused after mixing with 5 × 105 untreated PBMCs (as a source of antigen presenting cells to promote engraftment). Cell engraftment was tested 5 days after transplantation by flow cytometric analysis of PBMCs stained with human CD45, CD3, CD4, and CD8 antibodies. Animals receiving PBMCs from HIV-1-seronegative donors were infected with 100 ng p24 of HIVBAL by intraperitoneal (ip) route 5–7 days after human cell transfer. Blood samples from the animals were periodically assayed for viral load and CD4+ T cell changes. After 7 weeks blood and spleen from euthanized animals were harvested for final analysis.
Statistical analysis. Statistical analyses were performed with GraphPad Prism version 6. Data between groups were compared using one-way analysis of variance with Bonferroni correction, or multiple t-tests. P < 0.05 was considered significant and P < 0.01 was considered extremely significant.
SUPPLEMENTARY MATERIAL Figure S1. Generation of non-integrating lentivirus encoding ZFNs for CCR5 gene editing. Figure S2. ZsGreen expression over time after transduction of 293 T cells with non-integrating (red) or wild type integrating (blue) lentiviruses. Figure S3. MTS assay to test the toxicity of CCR5-ZFNs non-integrating lentivirus. Figure S4. Transduction efficiencies of CCR5-ZFN-expressing NILV pseudotyped with HIV envelope or VSV-G envelope in both activated and resting CD4+ T cells. Figure S5. Selective transduction CD4+ T cells with HIV envelope pseudotyped NILV. Table S1. Clinical information on HIV seropositive subjects.
Acknowledgments
We thank the PBMC donors. We also thank Una O'Doherty at the University of Pennsylvania for providing us the HIVLAI envelope plasmid for lentiviral pseudotyping. This work was supported by the following grants: R21HL116268 (P.S.,) from NIH/NHLBI and RO1 AI071882 (P.S.,) and R01-DK058702-10 (T.K.) from NIH/NIAID.
Supplementary Material
References
- Carr A. Toxicity of antiretroviral therapy and implications for drug development. Nat Rev Drug Discov. 2003;2:624–634. doi: 10.1038/nrd1151. [DOI] [PubMed] [Google Scholar]
- Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature. 2009;459:437–441. doi: 10.1038/nature07992. [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, et al. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob HJ, Lazar J, Dwinell MR, Moreno C, Geurts AM. Gene targeting in the rat: advances and opportunities. Trends Genet. 2010;26:510–518. doi: 10.1016/j.tig.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt N, Wang J, Kim K, Friedman G, Wang X, Taupin V, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28:839–847. doi: 10.1038/nbt.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilen CB, Wang J, Tilton JC, Miller JC, Kim KA, Rebar EJ, et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog. 2011;7:e1002020. doi: 10.1371/journal.ppat.1002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Wang J, Crain K, Fearns C, Kim KA, Hua KL, et al. Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4(+) T cell resistance and enrichment. Mol Ther. 2012;20:849–859. doi: 10.1038/mt.2011.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Ousterout DG, Gersbach CA. Advances in targeted genome editing. Curr Opin Chem Biol. 2012;16:268–277. doi: 10.1016/j.cbpa.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone D, Kiem HP, Jerome KR. Targeted gene disruption to cure HIV. Curr Opin HIV AIDS. 2013;8:217–223. doi: 10.1097/COH.0b013e32835f736c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Krymskaya L, Wang J, Henley J, Rao A, Cao LF, et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21:1259–1269. doi: 10.1038/mt.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu X, Wang P, Ding D, Li L, Wang H, Ma L, et al. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 2013;41:7771–7782. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allers K, Hütter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, et al. Evidence for the cure of HIV infection by CCR5?32/?32 stem cell transplantation. Blood. 2011;117:2791–2799. doi: 10.1182/blood-2010-09-309591. [DOI] [PubMed] [Google Scholar]
- Hütter G, Zaia JA. Allogeneic haematopoietic stem cell transplantation in patients with human immunodeficiency virus: the experiences of more than 25 years. Clin Exp Immunol. 2011;163:284–295. doi: 10.1111/j.1365-2249.2010.04312.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier DA, Brennan AL, Jiang S, Binder-Scholl GK, Lee G, Plesa G, et al. Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum Gene Ther. 2013;24:245–258. doi: 10.1089/hum.2012.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flight MH. Trial watch: Clinical trial boost for lentiviral gene therapy. Nat Rev Drug Discov. 2013;12:654. doi: 10.1038/nrd4111. [DOI] [PubMed] [Google Scholar]
- Biffi A, Montini E, Lorioli L, Cesani M, Fumagalli F, Plati T, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- Aiuti A, Biasco L, Scaramuzza S, Ferrua F, Cicalese MP, Baricordi C, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagen FH, Rademaker HJ, Fallaux FJ, Hoeben RC. Insertion vectors for gene therapy. Gene Ther. 2000;7:271–272. doi: 10.1038/sj.gt.3301121. [DOI] [PubMed] [Google Scholar]
- Cesana D, Ranzani M, Volpin M, Bartholomae C, Duros C, Artus A, et al. Uncovering and dissecting the genotoxicity of self-inactivating lentiviral vectors in vivo. Mol Ther. 2014;22:774–785. doi: 10.1038/mt.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkis C, Philippe S, Mallet J, Serguera C. Non-integrating lentiviral vectors. Curr Gene Ther. 2008;8:430–437. doi: 10.2174/156652308786848012. [DOI] [PubMed] [Google Scholar]
- Marktel S, Magnani Z, Ciceri F, Cazzaniga S, Riddell SR, Traversari C, et al. Immunologic potential of donor lymphocytes expressing a suicide gene for early immune reconstitution after hematopoietic T-cell-depleted stem cell transplantation. Blood. 2003;101:1290–1298. doi: 10.1182/blood-2002-08-2351. [DOI] [PubMed] [Google Scholar]
- Qasim W, King D, Buddle J, Verfuerth S, Kinnon C, Thrasher AJ, et al. The impact of retroviral suicide gene transduction procedures on T cells. Br J Haematol. 2003;123:712–719. doi: 10.1046/j.1365-2141.2003.04672.x. [DOI] [PubMed] [Google Scholar]
- Agosto LM, Yu JJ, Liszewski MK, Baytop C, Korokhov N, Humeau LM, et al. The CXCR4-tropic human immunodeficiency virus envelope promotes more-efficient gene delivery to resting CD4+ T cells than the vesicular stomatitis virus glycoprotein G envelope. J Virol. 2009;83:8153–8162. doi: 10.1128/JVI.00220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham TJ, Lee GM, Titus JA, Sconocchia G, Bakács T, Kovesdi I, et al. Targeted adenovirus-mediated gene delivery to T cells via CD3. J Virol. 1997;71:7663–7669. doi: 10.1128/jvi.71.10.7663-7669.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe S, Sarkis C, Barkats M, Mammeri H, Ladroue C, Petit C, et al. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc Natl Acad Sci USA. 2006;103:17684–17689. doi: 10.1073/pnas.0606197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felipe P, Martín V, Cortés ML, Ryan M, Izquierdo M. Use of the 2A sequence from foot-and-mouth disease virus in the generation of retroviral vectors for gene therapy. Gene Ther. 1999;6:198–208. doi: 10.1038/sj.gt.3300811. [DOI] [PubMed] [Google Scholar]
- Kantor B, Bayer M, Ma H, Samulski J, Li C, McCown T, et al. Notable reduction in illegitimate integration mediated by a PPT-deleted, nonintegrating lentiviral vector. Mol Ther. 2011;19:547–556. doi: 10.1038/mt.2010.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine EJ, Cradick TJ, Zhao CL, Lin Y, Bao G. An online bioinformatics tool predicts zinc finger and TALE nuclease off-target cleavage. Nucleic Acids Res. 2014;42:e42. doi: 10.1093/nar/gkt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badia R, Riveira-Muñoz E, Clotet B, Esté JA, Ballana E. Gene editing using a zinc-finger nuclease mimicking the CCR5?32 mutation induces resistance to CCR5-using HIV-1. J Antimicrob Chemother. 2014;69:1755–1759. doi: 10.1093/jac/dku072. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frecha C, Costa C, Nègre D, Gauthier E, Russell SJ, Cosset FL, et al. Stable transduction of quiescent T cells without induction of cycle progression by a novel lentiviral vector pseudotyped with measles virus glycoproteins. Blood. 2008;112:4843–4852. doi: 10.1182/blood-2008-05-155945. [DOI] [PubMed] [Google Scholar]
- Yoder KE, Fishel R. Real-time quantitative PCR and fast QPCR have similar sensitivity and accuracy with HIV cDNA late reverse transcripts and 2-LTR circles. J Virol Methods. 2008;153:253–256. doi: 10.1016/j.jviromet.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manjunath N, Yi G, Dang Y, Shankar P. Newer gene editing technologies toward HIV gene therapy. Viruses. 2013;5:2748–2766. doi: 10.3390/v5112748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Händel EM, Cathomen T. Zinc-finger nuclease based genome surgery: it's all about specificity. Curr Gene Ther. 2011;11:28–37. doi: 10.2174/156652311794520120. [DOI] [PubMed] [Google Scholar]
- Ramalingam S, Kandavelou K, Rajenderan R, Chandrasegaran S. Creating designed zinc-finger nucleases with minimal cytotoxicity. J Mol Biol. 2011;405:630–641. doi: 10.1016/j.jmb.2010.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Ramirez CL, Linder SJ, Pattanayak V, Shoresh N, Ku M, et al. In silico abstraction of zinc finger nuclease cleavage profiles reveals an expanded landscape of off-target sites. Nucleic Acids Res. 2013;41:e181. doi: 10.1093/nar/gkt716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yáñez-Muñoz RJ, Balaggan KS, MacNeil A, Howe SJ, Schmidt M, Smith AJ, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- Rahim AA, Wong AM, Howe SJ, Buckley SM, Acosta-Saltos AD, Elston KE, et al. Efficient gene delivery to the adult and fetal CNS using pseudotyped non-integrating lentiviral vectors. Gene Ther. 2009;16:509–520. doi: 10.1038/gt.2008.186. [DOI] [PubMed] [Google Scholar]
- Banasik MB, McCray PB. Integrase-defective lentiviral vectors: progress and applications. Gene Ther. 2010;17:150–157. doi: 10.1038/gt.2009.135. [DOI] [PubMed] [Google Scholar]
- Suwanmanee T, Hu G, Gui T, Bartholomae CC, Kutschera I, von Kalle C, et al. Integration-deficient lentiviral vectors expressing codon-optimized R338L human FIX restore normal hemostasis in Hemophilia B mice. Mol Ther. 2014;22:567–574. doi: 10.1038/mt.2013.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Olsen PA, Gelazauskaite M, Randøl M, Krauss S. Analysis of illegitimate genomic integration mediated by zinc-finger nucleases: implications for specificity of targeted gene correction. BMC Mol Biol. 2010;11:35. doi: 10.1186/1471-2199-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newrzela S, Cornils K, Li Z, Baum C, Brugman MH, Hartmann M, et al. Resistance of mature T cells to oncogene transformation. Blood. 2008;112:2278–2286. doi: 10.1182/blood-2007-12-128751. [DOI] [PubMed] [Google Scholar]
- Margolis L, Shattock R. Selective transmission of CCR5-utilizing HIV-1: the ‘gatekeeper' problem resolved. Nat Rev Microbiol. 2006;4:312–317. doi: 10.1038/nrmicro1387. [DOI] [PubMed] [Google Scholar]
- Grivel JC, Shattock RJ, Margolis LB. Selective transmission of R5 HIV-1 variants: where is the gatekeeper. J Transl Med. 2011;9 Suppl 1:S6. doi: 10.1186/1479-5876-9-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicenzi E, Liò P, Poli G. The puzzling role of CXCR4 in human immunodeficiency virus infection. Theranostics. 2013;3:18–25. doi: 10.7150/thno.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naif HM. Pathogenesis of HIV Infection. Infect Dis Rep. 2013;5 Suppl 1:e6. doi: 10.4081/idr.2013.s1.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorry PR, Ancuta P. Coreceptors and HIV-1 pathogenesis. Curr HIV/AIDS Rep. 2011;8:45–53. doi: 10.1007/s11904-010-0069-x. [DOI] [PubMed] [Google Scholar]
- Scarlatti G, Tresoldi E, Björndal A, Fredriksson R, Colognesi C, Deng HK, et al. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- Connor RI, Sheridan KE, Ceradini D, Choe S, Landau NR. Change in coreceptor use correlates with disease progression in HIV-1–infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Kutner RH, Bazan NG, Reiser J. Simplified lentivirus vector production in protein-free media using polyethylenimine-mediated transfection. J Virol Methods. 2009;157:113–121. doi: 10.1016/j.jviromet.2008.11.021. [DOI] [PubMed] [Google Scholar]
- Lee SK, Dykxhoorn DM, Kumar P, Ranjbar S, Song E, Maliszewski LE, et al. Lentiviral delivery of short hairpin RNAs protects CD4 T cells from multiple clades and primary isolates of HIV. Blood. 2005;106:818–826. doi: 10.1182/blood-2004-10-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata H, Maeda K, Miyakawa T, Shibayama S, Matsuo M, Takaoka Y, et al. Potent anti-R5 human immunodeficiency virus type 1 effects of a CCR5 antagonist, AK602/ONO4128/GW873140, in a novel human peripheral blood mononuclear cell nonobese diabetic-SCID, interleukin-2 receptor gamma-chain-knocked-out AIDS mouse model. J Virol. 2005;79:2087–2096. doi: 10.1128/JVI.79.4.2087-2096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.