Abstract
Mutations in BRCA2 increase susceptibility to breast, ovarian and prostate cancers. The product of human BRCA2, BRCA2 protein, plays a key role in the repair of DNA double strand breaks and interstrand crosslinks by RAD51-mediated homologous recombination. Here, we present a biochemical and structural characterization of full length (3,418 amino acid) BRCA2, alone and in complex with RAD51. We show that BRCA2 facilitates nucleation of RAD51 filaments at multiple sites on single-stranded DNA. Three-dimensional electron microscopy reconstructions revealed that BRCA2 exists as a dimer and that two oppositely-oriented sets of RAD51 molecules bind the dimer. Single stranded DNA binds along the long axis of BRCA2, such that only one set of RAD51 monomers can form a productive complex with DNA and establish filament formation. Our data define the molecular mechanism by which this tumor suppressor facilitates RAD51-mediated homologous recombinational repair.
Introduction
Many individuals carry genetic mutations that confer a predisposition to early onset breast or ovarian cancers1. Of these familial cases, 30-50% are associated with mutations in the breast cancer susceptibility genes BRCA1 or BRCA22,3. The cumulative incidence of breast cancer in germ-line BRCA2 mutant carriers approaches 80%, leading many women with familial history to make difficult choices relating to radical surgical approaches that can reduce the risk.
Genomic DNA suffers tens of thousands of endogenous DNA damage events each day, due to the presence of metabolic by-products, toxic chemicals and UV-irradiation4. Unrepaired lesions caused by these agents are a contributing factor to genome instability and cancer development. One of the most serious forms of damage is a DNA double-strand break (DSB), as unrepaired or improperly repaired breaks can lead to mutation, chromosome translocation and deletion – all of which can contribute to cancer development - or to cell death5.
BRCA2 encodes BRCA2 protein (384 kDa), which is involved in the repair of DSBs by homologous recombination (HR)6,7. A key step in HR is the recruitment of RAD51 recombinase to exposed single-stranded DNA (ssDNA) overhangs generated by the resection of DSB termini. RAD51 assembles on the ssDNA to form highly ordered helical nucleoprotein filaments that subsequently mediate the search for homologous sequences on the undamaged sister chromatid. The homology search is then followed by DNA strand exchange leading to the formation of recombination intermediates in which interacting DNAs become covalently linked.
Cells defective for BRCA2 are sensitive to DNA inter-strand crosslinking agents, exhibit a decreased efficiency of DSB repair, and an inability to promote the efficient restart of stalled replication forks8-10. They also fail to promote the nuclear relocalization of RAD51 following exogenous DNA damage11,12. The inability to target RAD51 to sites of repair indicates that BRCA2 acts as a mediator for the formation of RAD51 filaments, a proposal supported by several other observations: (i) BRCA2 interacts with RAD51 through a cluster of eight conserved motifs, the BRC repeats, and an additional unrelated C-terminal RAD51 interaction domain13-18, (ii) mutations within the BRC repeats confer DNA damage sensitivity19, (iii) purified BRCA2 stimulates the formation of RAD51 nucleoprotein filaments in vitro20-22, and (iv) over-expression of RAD51 suppresses the HR defects observed in BRCA2-depleted mouse cells23.
Apart from the BRC repeats that interact with RAD51, we have limited knowledge relating to the functions of other regions of this large protein: The N-terminal region interacts with PALB2 (partner and localizer of BRCA2)24-27, a central domain interacts with the meiotic recombinase DMC128, and the C-terminal region contains an alpha helical domain followed by three OB (oligonucleotide-oligosaccharide binding) folds (residues 2480-3190) that bind ssDNA29.
Due to the large size of BRCA2, which hinders structural studies, its precise functions in promoting RAD51-mediated HR remain poorly defined. Here, we set out to use biochemical and structural approaches to provide mechanistic insights into how full-length BRCA2 interacts with RAD51 and ssDNA to promote RAD51 nucleoprotein filament formation. Our results show that BRCA2 stimulates the nucleation of RAD51 filaments and facilitates unidirectional 3′-5′ filament growth, two processes essential for successful DSB repair and/or replication fork restart.
Results
Structural organization of the BRCA2 dimer
To provide mechanistic insights into the role of BRCA2 in recombinational repair, we set out to provide a structural characterization of full-length BRCA2 and the BRCA2-RAD51 complex. To do this, purified BRCA220 (Supplemental Fig. 1a), was visualized by electron microscopy (EM). Negatively stained EM images of BRCA2 showed a range of particle sizes with relatively low abundance (Supplemental Fig. 2), precluding them from high-resolution cryo-EM 3D reconstructions. However, a total of 8873 negatively stained particles were selected and subjected to multivariate statistical analysis and classification. Selected class averages were used to generate a 3D reconstruction using angular reconstitution methodologies30, and the 3D structure was refined iteratively in IMAGIC31. The initial eigen-images and 3D reconstructions showed a two-fold symmetry suggestive of a dimer (Supplemental Fig. 3). After several cycles of refinement without imposed symmetry, subsequent refinement was carried out with two-fold symmetry imposed. The reprojections from the 3D model were in excellent agreement with the corresponding class averages (Supplemental Fig. 3c) and the final 3D reconstruction incorporated the vast majority of the dataset at a resolution of 18.5 Å using 0.5 Fourier shell correlation (FSC) criterion (Supplemental Fig. 4). The 3D reconstruction was validated using tilt-pair and tilt-series analyses32,33 (Supplemental Fig. 4).
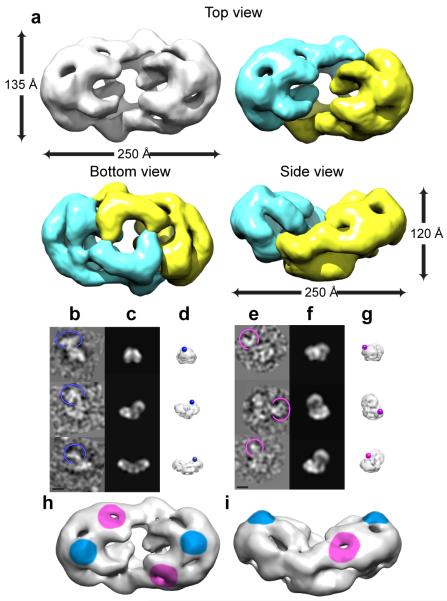
The 3D reconstruction of BRCA2 revealed a slightly curved elongated molecule of ~ 250 Å × 135 Å × 120 Å (Fig. 1a). Looking from the concave face (top view), the elliptic density contains two halves that enclose a central channel, with the main density forming a smooth arc at the far end and with the two halves being connected by elongated density at the outer rim of the channel. From the side view, the molecule displayed a kidney bean shape with a groove in the middle and a relatively flat base at the bottom. When viewed from the bottom, the central channel was enclosed by distinct regions of density arranged in a rectangular shape.
Figure 1. 3D reconstruction of BRCA2 and identification of domains.
(a) Surface view of the 3D reconstruction. Two halves were colored yellow and cyan, representing two potential monomers although the exact boundary is unknown. (b-g) Antibody labeling against C-terminal Flag tag (b-d) and BRC repeats (e-g). In (b) and (e), raw particles with antibody are circled. (c) and (f) Reprojections along the same orientations of (b) and (e). (d) and (g) 3D reconstructions viewed along the same directions as (b) and (e), with antibody locations represented by spheres. (h) and (i) Top and side views of BRCA2 with antibody locations colored (Flag tag- blue, BRC – magenta). Magnification bars in all single particle images represent 100 Å.
Due to BRCA2’s large size, and difficulty in obtaining sufficient amounts of protein, we were unable to use internal labeling and nanogold particles to identify functional domains within the reconstruction. We therefore used antibodies (which have a distinct Y-shape in negative stain EM) against BRCA2’s C-terminal Flag tag to visualize this region of the protein. BRCA2 particles, with antibodies attached to them (Fig. 1b, circle indicates the antibody), were then assigned Euler angles using the BRCA2 3D reconstruction. Based on Euler angle assignments of individual particles and the locations of the antibody, the 3D position of the antibody was obtained through triangulation (Supplemental Fig. 3d), indicating that the C-terminus was located at the top and near the far end of the BRCA2 dimer (Fig. 1b-d and h-i, blue surface). Similarly, the location of the BRC repeats was identified using an antibody against a region between BRC5 and BRC6 (Fig. 1e-g), and was located at the outer rim and towards the top of the 3D structure (Fig. 1h and i, purple surface).
Structure of the BRCA2-RAD51 complex
Previously, it was shown that BRC repeats 1-4 exhibit a higher affinity for RAD51 than BRC repeats 6-816,17,34. Accordingly, four to six RAD51 molecules are bound by the BRCA2 monomer21. To investigate the spatial arrangement of RAD51 within the context of BRCA2, an excess of purified RAD51 was mixed with BRCA2 and the BRCA2-RAD51 complex was affinity purified (Supplemental Fig. 1a). The complex was then visualized by EM and the 3D structure determined. We obtained images of 6877 particles and performed independent image processing, without using the BRCA2 3D reconstruction as references for alignment or angular assignment, in order to avoid any potential model bias. Interestingly, the particle sizes were again consistent with BRCA2 existing as dimers, indicating that RAD51 binding does not dissociate the dimer. Initial eigen-images and class averages revealed two-fold symmetry and elongated particles, except that the particles appeared wider than the BRCA2 particles (Supplemental Fig. 3 and 5).
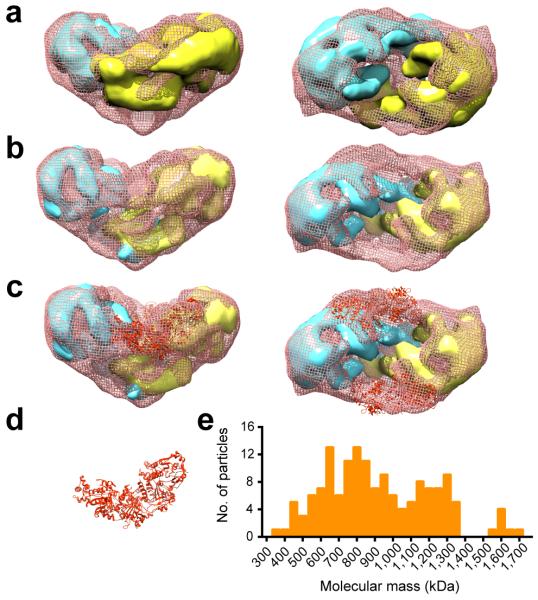
The final 3D reconstruction using angular reconstitution followed by iterative cycles of refinement was at 19.5 Å resolution and incorporated the vast majority of the dataset (Fig. 2). The structure was validated using similar approaches to those used for the BRCA2 reconstruction, and again showed two halves connected by an elongated density along the outer rim of a central cage (Fig. 2a and b). The structure is longer (265 Å vs. 250 Å), wider (165 Å vs. 135 Å) and taller (165 Å vs. 120Å) than BRCA2 alone. In addition, in the BRCA2-RAD51 complex, we did not find a cavity at the bottom of the BRCA2 dimer, and when viewed from the side the reconstruction displayed a more triangular shape (Fig. 2b) that contrasted with the BRCA2 reconstruction, which had a clear groove at the top and a relatively flat base at the bottom (Fig. 1a). Other notable features were the extra density along the sides of the central cavity region towards the top of the BRCA2-RAD51 complex, contributing to the increased width and the loss of the clear groove (compare Figs. 1 and 2).
Figure 2. 3D reconstruction of BRCA2-RAD51 complex.
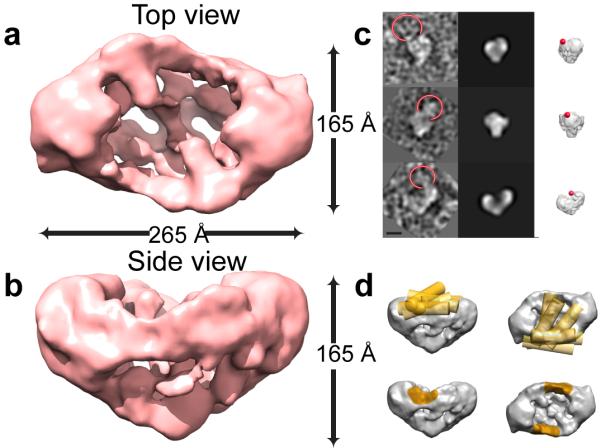
(a) and (b) Side and top surface views of the 3D reconstruction. (c) Antibody labeling against RAD51. Left: individual particles with antibody circled. Middle: corresponding reprojections from the BRCA2-RAD51 reconstruction. Right: surface view along the same direction with antibody locations indicated with spheres. (d) Cylinders representing antibody locations defined from individual particles (upper). These intersect at the density regions on the outer rim connecting the two halves (lower, orange surface).
Due to the observed changes in the overall shape between the BRCA2 and BRCA2-RAD51 reconstructions, it was not immediately obvious which density region contained RAD51. However, the location of the BRC repeats (Fig. 1) suggested that RAD51 binds to the sides and between the two halves of the arc-like globular density. To determine if this was the case, we probed the complex with an antibody against RAD51, and found that it bound to the region consistent with the location of the BRC repeats (Fig. 2c and d, orange surface). These results support the view that RAD51 is located at the outer rim of the central cage region between the two arc-shaped halves of the reconstruction towards the top of the molecule (viewed from the side).
Conformational changes of BRCA2 upon RAD51 binding
Since RAD51 binds towards the top of the BRCA2 dimer, the altered shape observed at the bottom of the BRCA2-RAD51 reconstruction, compared with that of BRCA2 alone, indicated that RAD51 binding induced structural rearrangements (Fig. 3a). Clear differences were the steep slope of the side view, resulting in a pointed tip at the base of the reconstruction and the disappearance of the channel at the bottom (Figs. 2a and 3a). The slope indicated a tilt of each monomer towards the vertical axis relative to the dimer plane. In BRCA2, the channel at the bottom is formed by two L-shaped densities, and we propose that each L-shape is contributed by a BRCA2 monomer (Fig. 1a, bottom view). The closure of the channel in the BRCA2-RAD51 complex indicated that the L-shaped density was reorganized and became more closely associated in an intercalated arrangement. Using the segmentation command implemented in Chimera35, we separated the BRCA2 density into two halves (Fig. 1a and Fig. 3, yellow and cyan density regions), and fitted the two halves independently into the BRCA2-RAD51 density (Fig. 3b). This resulted in an increase in the correlation coefficient between BRCA2 and BRCA2-RAD51 reconstructions from 0.64 to 0.78. Although there are some uncertainties in the exact boundary of the BRCA2 monomers, the rearrangement mainly involved a rotation and tilt of the two monomers (Fig. 3b) to accommodate the steep slope on the side of the molecule. This rearrangement resulted in extra density at the top that connects the two halves of the dimer (Fig. 3b), consistent with the RAD51 antibody labeling data (Fig. 2d). This extra density region could accommodate approximately four RAD51 monomers on each side, arranged in a filament like fashion (Fig. 3c and d).
Figure 3. RAD51 binding to BRCA2.
(a) Overlay of BRCA2 dimer (yellow and cyan) and BRCA2-RAD51 (pink mesh) highlighting the differences in their shape. (b) Rearranged BRCA2 dimer fitted into the BRCA2-RAD51 complex. (c) as in (b). Four RAD51 monomers (orange ribbon) were fitted into the additional density in BRCA2-RAD51 not accounted for by BRCA2 density. (d) Four RAD51 monomers, arranged as in filaments. (e) Histogram of mass measurement of BRCA2-RAD51 complex using STEM, showing peaks at 800 kD and 1200 kD, corresponding to BRCA2 dimer and BRA2 dimer binding to 8-10 RAD51.
To confirm that BRCA2 remained as a dimer upon binding to RAD51, we carried out mass measurements of the BRCA2-RAD51 complex using Scanning Transmission Electron Microscopy (STEM). The histogram showed two major populations at ~800 kD and ~1200 kD respectively, in agreement with the presence of BRCA2 dimers and their complex with 8-10 RAD51 molecules (Fig. 3e). Our data therefore indicate that the BRCA2 dimer binds to two sets of RAD51 molecules, and that each set can accommodate approximately four or five RAD51 monomers that bind in between the two BRCA2 monomers towards the top of the molecule (Fig. 3c).
DNA binding of BRCA2 and the BRCA2-RAD51 complex
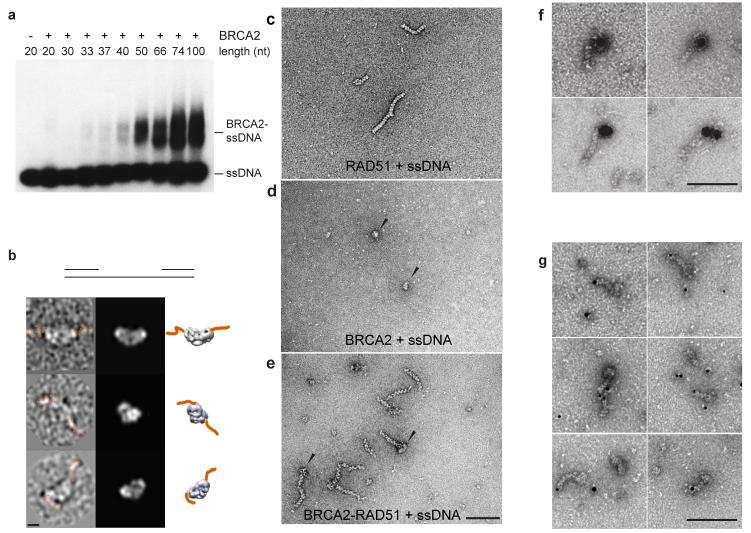
An important property of BRCA2 is its preference for binding to single rather than double stranded DNA (dsDNA)20,21. This DNA-binding specificity of BRCA2 is critical for the association of RAD51 with ssDNA, as RAD51 alone binds to both ssDNA and dsDNA with similar affinities20,36-38. To understand how BRCA2 binds ssDNA and recruits RAD51, we determined the minimum length of ssDNA required for BRCA2 binding. Using 32P-labeled oligos of defined length (20-100 nucleotides), electromobility shift assays (EMSA) revealed that the minimum binding size for the formation of a stable BRCA2-ssDNA complex was approximately 50 nucleotides, although longer oligos (>66 nucleotides) were required for maximal binding (Fig. 4a). Little or no binding was observed with ssDNAs shorter than 40 nucleotides. In contrast, previous studies using only the DNA binding domain of BRCA2 showed that it exhibits weak binding to 12-20 nucleotides whereas stronger binding was observed with oligos that were 36 nucleotides in length39. Our results are therefore consistent with the notion that the stable association of BRCA2 with ssDNA may require simultaneous binding to both subunits of the dimer.
Figure 4. ssDNA binding of BRCA2 and BRCA2-RAD51 complex.
(a) Gel-shift assay showing the binding of BRCA2 to 5′-32P-labeled ssDNA substrates ranging from 20 to 100 nt. DNA was detected by autoradiography. (b) Images of individual particles of BRCA2 bound to gapped DNA (duplex arms are indicated in orange). Magnification bars represent 100 Å. (c-e) Electron microscopic visualization of RAD51-ssDNA filaments, BRCA2-ssDNA complexes, and BRCA2-RAD51-ssDNA complexes, as indicated. (f) Localization of BRCA2 in BRCA2-RAD51-ssDNA complexes by immunogold labeling. (g) Visualization of BRCA2-RAD51 filaments formed with 5′-gold particle labeled ssDNA. Magnification bars represent 100 nm.
We next visualized the BRCA2-ssDNA complex in order to determine the path of the DNA across BRCA2. Because ssDNA is difficult to see by negative staining, whereas dsDNA can be observed, we used gapped duplex DNA in which ssDNA regions (70 or 100 nucleotides in length) were flanked by duplex arms of 65 or 50 base pairs. The gaps were large enough to accommodate a BRCA2 dimer for maximal DNA binding, while the duplex arms allowed us to define the path of the DNA (Fig. 4b). Binding to the gapped duplex DNA was confirmed by EMSA (Supplemental Fig.1c). We analyzed particles in which we could see DNA bound to BRCA2 by projection-matching using the BRCA2 3D reconstruction. The observed relative positions of the dsDNA arms were consistent with a model in which ssDNA binds along the long axis of the BRCA2 dimer (Fig. 4b). These results are in agreement with the location of the C-terminus of BRCA2, as determined by antibody labeling, as the C-terminus is close to the DNA binding domain (Fig. 1h and i).
To address how BRCA2 recruits RAD51 to ssDNA, we visualized the assembly of BRCA2-RAD51-ssDNA on linear φX174 ssDNA. RAD51-ssDNA, BRCA2-ssDNA and BRCA2-RAD51-ssDNA complexes, formed at short incubation times, were adsorbed onto carbon grids, negatively stained with uranyl acetate, and images acquired for analysis (Fig. 4c-e). As expected, BRCA2 formed discrete particles that could be visualized quite easily due to their large size (Fig. 4d, arrowed). Striking results, however, were obtained when we viewed the BRCA2-RAD51-ssDNA samples, as we observed many examples in which BRCA2 particles were positioned at only one end of the RAD51-ssDNA filaments (Fig. 4e, arrowed). Because BRCA2 carries a C-terminal Flag-tag, we confirmed that these terminal particles were indeed BRCA2 using a monoclonal anti-Flag antibody coupled to 20 nm gold particles (Fig. 4f). At longer reaction time points, BRCA2 was observed only infrequently indicating that it may dissociate once nucleoprotein filament formation has been initiated.
The visualization of BRCA2-RAD51-ssDNA filaments indicated unidirectional growth of the RAD51 filament from BRCA2. To determine the polarity of filament growth, we generated linear ssDNA molecules that were gold-labeled either at their 5′- or 3′-termini. EM analysis revealed that RAD51 filaments extended from BRCA2 to the 5′-gold label (Fig. 4g). In contrast, we failed to detect similar complexes oriented in the opposite direction (5′-3′) using 3′-gold labeled ssDNAs (data not shown). Our interpretation of these results is that RAD51 filaments grow from BRCA2 in the 3′-5′ direction, consistent with the 3′-5′ polarity of strand transfer relative to bound ssDNA40,41.
BRCA2 promotes increased nucleation of RAD51-ssDNA filaments
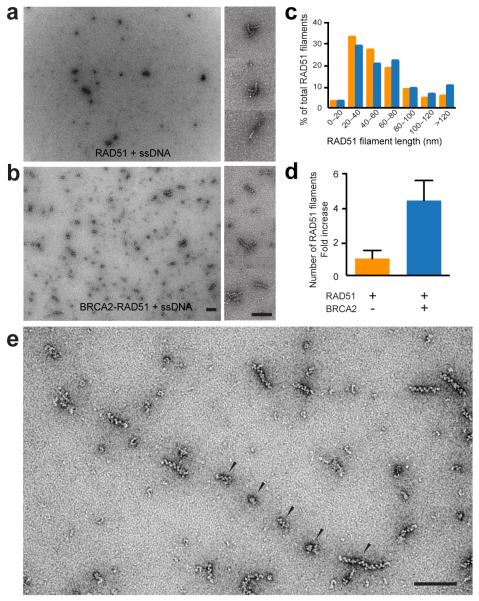
We next determined whether BRCA2 influences the nature of the filaments formed. We found that only short filaments (40 - 100 nm) were observed in the presence or absence of BRCA2 (Fig. 5a and b). Quantification of the average filament length revealed little or no change in length as a consequence of BRCA2 inclusion (Fig. 5c). The presence of BRCA2, however, led to a significant increase in the number of RAD51 filaments. To confirm this, we investigated the initial nucleation events and found that approximately 4x more RAD51-ssDNA nucleation events occurred in the presence of BRCA2 than in its absence (Fig. 5a and b, quantified in Fig. 5d, and Supplemental Fig. 1b). Interestingly, we also observed multiple BRCA2 particles and short RAD51 filaments aligned in a linear fashion (Fig. 5e), presumably along a single ssDNA molecule, suggesting that BRCA2 (and therefore BRCA2-RAD51) can bind to multiple sites along the ssDNA and initiate multiple nucleation events.
Figure 5. Nucleation of RAD51 filaments by BRCA2.
(a) and (b) Effect of BRCA2 on the number of RAD51-ssDNA nucleation events, as determined by electron microscopy. Inserts show enlargements of RAD51 filaments. (c-d) Quantification of RAD51-ssDNA filament length (c) and nucleation events (d) in the presence (blue) or absence (orange) of BRCA2, as determined by measurement of images shown in Fig. 4c (n = 314) and 4e (n = 332), n = number of RAD51 filaments. In total, 204 (RAD51-ssDNA) and 149 (BRCA2-RAD51-ssDNA) randomly collected grid areas were quantified. P-values (P < 0.0001) were determined using a two-tailed t test, error bars represent SD. (e) BRCA2-RAD51-ssDNA complexes visualized as multiple distinct filament nucleation sites on the same ssDNA molecule (arrowed). Magnification bars represent 100 nm.
Taken together, our data show that BRCA2 facilitates the interaction of RAD51 with ssDNA by creating nucleation sites from which RAD51 self polymerizes with a 3′-5′ directionality. Consistent with previous data42, filament assembly involves the formation of multiple nucleation clusters and subsequent gap filling, rather than the progressive elongation of RAD51 filaments from just a small number of nucleation sites. The extension of these short nascent filaments into a single extended filament capable of efficient homologous pairing and strand exchange may require the actions of other RAD51 mediator proteins including PALB2, RAD52, RAD54 and/or the RAD51 paralogs.
Discussion
Our results reveal new insights into the role that BRCA2 plays in driving RAD51 filament formation. Specifically, we report the first detailed view of the structure of full-length BRCA2, and show that (i) a BRCA2 dimer recruits two sets of RAD51 molecules arranged in opposing orientations, (ii) ssDNA binds across one surface of the BRCA2 dimer, (iii) BRCA2 increases the frequency, but not the length, of nascent RAD51 filament nucleation events, and (iv) that it facilitates unidirectional 3′-5′ filament growth from each nucleation site.
Our data show that purified BRCA2 predominately exists in dimeric form and that dimers bind RAD51 and ssDNA without dissociation. Approximately four or five RAD51 monomers bind into each BRCA2 subunit. Consistent with this, four or five RAD51 monomers represent the minimal nucleation length that enables filament growth49. It is known that each BRC repeat can mimic the RAD51 inter-subunit elements required for filament/ring contacts, supporting the concept that the RAD51 monomers are aligned by BRC binding in preparation for nucleation on ssDNA. Due to the intrinsic symmetry imposed by the BRCA2 dimer, the two sets of RAD51 molecules bound to the dimer are oriented in opposing directions (Fig. 6). Upon ssDNA binding, which imposes polarity, only one set of RAD51 molecules will be in the correct polarity for interaction with ssDNA (Fig. 6). Dimers therefore offer certain mechanistic advantages as they allow BRCA2 to bind ssDNA in either orientation and still present one set of RAD51 molecules in the correct orientation for productive filament formation. However, we cannot exclude the possibility that upon interaction with other partners, such as PALB2 or BRCA1, or the introduction of DNA damage or cell cycle mediated post-translational modifications, a dimer to monomer transition occurs. Such a mechanism has been proposed for Ustilago maydis Brh2, where a dimer to monomer transition occurs upon DSS1 binding43. In avian DT40 cells, a transition from a slower diffusion species (thus larger macromolecular assemblies) of BRCA2 to one capable of faster diffusion (thus smaller entities) is observed upon DNA damage44. Dimer to monomer transitions have previously been proposed as key regulatory mechanisms for a number of DNA damage signaling and repair proteins, such as ATM or ATR45. However, in contrast to ATM or ATR, BRCA2 dimers are active in terms of ssDNA binding and RAD51 filament formation. Whether or not BRCA2 monomers can promote filament formation remains to be determined.
Figure 6. A proposed mode of action for BRCA2 in RAD51 filament nucleation.
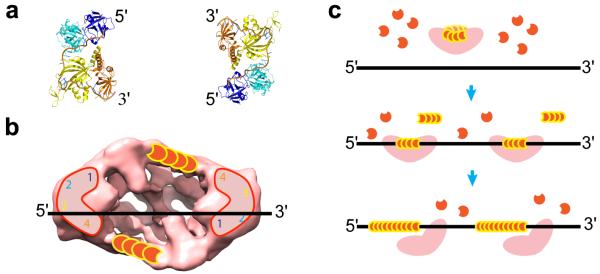
(a) Crystal structure of RPA bound to 30 nt ssDNA, showing a compact configuration and bending of the ssDNA into a U-shape. DNA binding domains of BRCA2 could adapt similar conformations. The polarity of the ssDNA is indicated. The two RPA molecules, related by 2-fold symmetry, could represent the DNA binding domains in the BRCA2 dimer as indicated below. (b) The DNA binding domains (1,2,3,4) of BRCA2 are depicted in similar conformations as those shown for RPA in (a), such that ssDNA could simultaneously bind to domains 3-4 (OB2-OB3) at the 5′ end (left hand side) of one BRCA2 monomer while domains 1-2 (alpha-helical domain and OB1) at the 3′ end (right hand side) of the second monomer. Two sets of RAD51 molecules bind the BRCA2 dimer in opposing directions. Only one set can be productive in ssDNA binding. (c) Model for filament formation and elongation using multiple BRCA2-RAD51 nucleation sites with BRCA2 acting as a molecular chaperone for RAD51.
How might ssDNA, which has a defined polarity, bind across BRCA2? The BRCA2 DNA binding region consists of four domains including an α-helical domain and three OB folds (denoted domains 1-2-3-4; Fig. 6), with OB2 and OB3 having a higher affinity to ssDNA than α-helcial-OB129. This region binds ssDNA of 12-20 nucleotides weakly and 36 nucleotides tightly, suggesting that it resembles the ssDNA binding protein RPA. In support of the analogy between the BRCA2 and RPA DNA binding domains, a heterologous fusion protein containing BRC repeats fused to the large subunit of RPA can at least partially complement BRCA2 mutant cells46. RPA uses two of its five OB folds for the weak 8 nucleotides binding mode and four OB folds for tight 30 nucleotide binding29,47. Crystallographic analysis of RPA in complex with a 30 mer revealed that the four ssDNA binding OB folds, which are arranged in a compact quaternary structure, cause the ssDNA to take a U-turn48 (Fig. 6a). The four DNA binding domains of BRCA2 may be arranged in a similar compact fashion to those in RPA and only two out of the four domains could be utilized for ssDNA binding (Fig. 6a). Thus, irrespective of its relative polarity, ssDNA could bind to two of the four DNA binding domains in one BRCA2 monomer (for example, Fig. 6a and b, domains 3-4 (OB2-OB3), left hand side monomer). Due to the symmetry imposed by the dimer, two different domains (domains 1-2: helical domain and OB1) in the other BRCA2 monomer would bind to ssDNA with the same polarity (Fig. 6a and b, right hand side monomer), thus allowing the correct polarity for ssDNA binding in both monomers. Our observation that BRCA2 requires 40 nt ssDNA for weak binding, and >70 nt for maximal binding, supports the notion that the two DNA binding domains in each monomer act in concert.
Recombination is initiated at long 3′-ssDNA tails that form by DSB resection. These tails serve as targets for multiple BRCA2-mediated nucleation events merging to form a single RAD51-ssDNA filament that will initiate homologous pairing. Our analysis of the BRCA2-RAD51-ssDNA complex indicates that BRCA2 is located at the 3′-end of each nucleating RAD51 filament, supporting a model in which RAD51 filaments grow in the 3′-5′ direction, consistent with the polarity of RAD51-mediated strand exchange40,41. One possibility is that BRCA2 ‘caps’ one end of the filament (the 3′-end) and blocks dissociation, while allowing RAD51 to self-polymerize and grow at the other end. RAD51 polymerization in the 3′-5′ direction may lead to a situation in which several nucleotides at the 3′-terminus itself will not be bound by RAD51. This is unlikely to be a problem, however, as a free end is not required for pairing50,51, and it has been shown that RAD51 can promote the initial ssDNA-dsDNA pairing interactions at both 3′- and 5′-extended ssDNA tails52,53. Once pairing is initiated, however, only an invading 3′-terminus can act as a primer for DNA repair synthesis.
Interestingly, the Brh2 protein preferentially binds at single and double-stranded junctions in DNA and stimulates RAD51 filament formation at a 3′ ssDNA overhang54. On the other hand, BRCA2 fails to exhibit a preference for binding junctions over ssDNA20,21. Given that BRCA2 caps the 3′ end of the RAD51 filament, which grows 3′-5′, these observations suggest that some differences exist between BRCA2 and Brh2, but whether these differences reflect mechanistic diversities between the two proteins or are due to different experimental conditions is presently unclear.
In contrast to RAD51, the E. coli RecA binds ssDNA and promotes strand exchange with a 5′-3′ polarity55-57. This difference might be due to the presence of an additional N-terminal domain in RAD51, whereas RecA has a C-terminal addition, since these domains are positioned on opposing sides of the ATPase core relative to ssDNA (Supplemental Fig. 6a). In RAD51, filament formation involves a β-strand connecting the N-terminal domain with the ATPase domain, and this is stabilized and ordered upon binding to the ATPase domain of the adjacent monomer. This interaction surface is therefore enhanced and stabilized at the growing end of the filament towards the N-terminal, or 5′ direction (Supplemental Fig. 6b). In RecA, however, there is a loop involved in subunit interactions that is only ordered in the filament. This loop is located towards the 3′ direction, such that subunit addition in this direction would stabilize the loop and subsequent interactions (Fig. 6b). Due to the distinct domain configurations and the locations of the loop regions, RAD51 and RecA filaments thus grow in opposite directions (Fig. 6b and c).
In summary, the results presented here indicate a model in which a BRCA2 dimer binds to two sets of RAD51 molecules. Upon ssDNA binding, only one set of RAD51 molecules can bind ssDNA due to the inherent polarity of the bound ssDNA. The RAD51 filament then grows in the 3′-5′ direction, governed by its molecular architecture, leaving BRCA2 to cap the 3′-end of the filament (Fig. 6c). Given that only very small amounts of BRCA2 are required for RAD51 filament nucleation, we suggest that BRCA2 dissociates once the nascent filament undergoes extension, and can subsequently be utilized once more to initiate the formation of another filament. In so doing, BRCA2 acts as a molecular chaperone for filament assembly.
Online Methods
Proteins
C-terminally GFP-Flag tagged BRCA2 protein was purified using a modified method to that described previously20. In brief, BRCA2 was purified from 20 liters of HeLa cells carrying the BAC-BRCA2FLAP vector. Following cell collection and lysis, BRCA2 was affinity purified using M2-Flag agarose (Sigma) and GFP-Trap A agarose (Chromotek). In some preparations the order of the Flag and GFP affinity steps were reversed without any obvious differences. The final protein (200 μl at 2.5 μg/ml) was stored in small aliquots at −80°C. RAD51 was purified as described58. For the initial EM analyses, to provide a more monodisperse preparation of BRCA2, the purified protein was subjected to an additional gel filtration step through Superose 6 10/300 GL (GE Healthcare).
To form the BRCA2-RAD51 complex, BRCA2 was partially purified using GFP-Trap agarose and incubated with excess RAD51 (0.5 μM) for 30 min at 4°C. The BRCA2-RAD51 complex was then purified away from the remaining RAD51 by binding to M2-Flag agarose, extensively washed and then eluted using 3x FLAG peptide.
DNAs
Linear φX174 single-stranded DNA was prepared by annealing a 22 nt oligonucleotide (5′-TAAAACTCTGCAGGTTGGATAC-3′) to φX174 ssDNA followed by treatment with PstI. The linearized ssDNA was then gel purified. The ssDNA oligonucleotides, ranging from 20 – 74 nucleotides in length, were all 5′-derivatives of the 74 mer (written 5′ - 3′):
GCGCTACCAGTGATCACCAATGGATTGCTAGGACATCTTTGCCCACCTGCTACCCCGGGTTCGAAATCGATAAG
The 100-mer was:
GGGCGAATTGGGCCCGACGTCGCATGCTCCTCTAGACTCGAGGAATTCGGTACCCCGGGTTCGAAATCGATAAGCTTACAGTCTCCATTTAAAGGACAAG
Two related gapped duplex DNAs were prepared. gDNA1 had two 50 mer oligos annealed to the 5′- and 3′-ends of a 200 mer, resulting in a gap of 100 nucleotides. gDNA2 was similar except that the complementary oligos were 65 nt in length, resulting in a 70 nucleotide ssDNA gap. The 200 mer had the following sequence (5′-3′):
GAGTTTTATCGCTTCCATGACGCAGAAGTTAACACTTTCGGATATTTCTGATGAGTCGAAAAATTATCTTGATAAAGCAGGAATTACTACTGCTTGTTTACGAATTAAATCGAAGTGGACTGCTGGCGGAAAATGAGAAAATTCGACCTATCCTTGCGCAGCTCGAGAAGCTCTTACTTTGCGACCTTTCGCCATCAACT
The 5′-biotinylated ssDNA substrate for gold labeling was generated by PCR amplification using a single primer (5′-biotin-TAAAACTCTGCAGGTTGGATACGCCAATC-3′) and φX174 RFI DNA as template. The PCR product, 5′-biotinylated linear ssDNA, was gel purified and 5′-end labeled with 5 nm gold particles coupled to streptavidin.
BRCA2-ssDNA binding assays
Reactions (10 μl) contained 5′-32P-end-labeled ssDNA oligonucleotides (1 nM) and BRCA2 (1 nM) in binding buffer (20 mM HEPES pH 7.5, 2 mM ATP, 2 mM MgCl2, 1 mM dithiothreitol). Protein was added and mixed as indicated. After 10 min at 20°C, complexes were fixed by the addition of 0.25% glutaraldehyde and incubation at 20°C for 15 minutes. Protein-DNA complexes were analyzed by electrophoresis through 1% agarose gels run in TAE buffer at 4 V/cm for 2 h, dried onto filter paper and visualized by autoradiography.
Electron microscopic analysis of BRCA2-RAD51-ssDNA complexes
Unless indicated otherwise, RAD51-ssDNA complexes were prepared by incubating RAD51 (150 nM) with linear φX174 ssDNA (0.2 nM) at room temperature in binding buffer. For the analysis of BRCA2-RAD51 complexes, BRCA2 (0.1 nM) was incubated with RAD51 (150 nM) for 1 min at room temperature in binding buffer prior to the addition of ssDNA (0.2 nM). Incubation was then continued for a further 3 min, before the protein-DNA complexes were applied to glow-discharged carbon-coated grids. After 2 min, the grids were negatively stained with 2% uranyl acetate, and images obtained using a Gatan Orius 3x4K CCD camera on a FEI Tecnai G2 Spirit T12 Twin Transmission Electron Microscope.
To analyze the effect of BRCA2 on RAD51 filament nucleation, RAD51 (40 nM), in the presence or absence of BRCA2 (0.2 nM), was incubated for 1 min at room temperature in binding buffer prior to the addition of ssDNA (0.2 nM) and incubation was continued for 1 min. Next, additional RAD51 (160 nM) was added and the protein-DNA complexes were applied to glow-discharged carbon-coated grids. After 1 min, the grids were stained with uranyl acetate and images obtained. Nucleation sites were quantified from randomly collected areas of each grid (204 for RAD51 and 149 for BRCA2-RAD51).
Immunoaffinity gold labeling of BRCA2 was carried out using a monoclonal Flag antibody (Sigma F3165) conjugated to 20 nm gold particles using the InnovaCoat conjugation kit (Innova Bioscience). First, BRCA2 (0.2 nM) was incubated with 0.1 μg/ml antibody-conjugated gold particles for 15 min at room temperature in binding buffer without dithiothreitol. RAD51 (150 nM) was then added for a further 1 min followed by the addition of 0.2 nM ssDNA, and incubation was continued for a further 1 min. Samples were then analyzed by electron microscopy. Images were recorded at a magnification of ×67,000.
To determine the polarity of RAD51 filament formation, BRCA2 (0.2 nM) and RAD51 (150 nM) were premixed in binding buffer without dithiothreitol, and after 1 min 5′-gold-labeled ssDNA was added. Incubation was continued for a further 1 min, and protein-DNA complexes were adsorbed on carbon-coated grids.
Structural analysis of BRCA2 and BRCA2-RAD51 complexes
Protein samples were adsorbed onto glow-discharged carbon-coated copper Quantifoil R2/2 grids or TAAB 300-mesh continuous carbon grids for 2 minutes at room temperature. The grids were washed twice with water and negatively stained with 2% uranyl acetate. Images were collected on a Philips CM200 series FEG (field emission gun) transmission electron microscope (TEM), equipped with a TVIPS 4k × 4k pixel CCD camera at an operational voltage of 200 kV, at 50,000× magnification with an underfocus of 1.5-3 μm and an exposure of 40 e−Å−2. Raw micrographs were coarsened by a factor of 2, yielding a pixel size of 3.52 Å pixel−1. A total of 8873 and 6877 individual particles for BRCA2 and BRCA2-RAD51 complexes, respectively, were manually picked using the EMAN2 boxer program. We selected all particles that are not too big (i.e. aggregates) or too small (i.e. potential degradation products) and these particles exhibited dimensions of 120 – 250 Å for BRCA2 samples and slightly larger for BRCA2-RAD51 samples.
Image processing
Images were processed using IMAGIC31 and aligned using the brute-force ‘timalign’ program from the Tigris package (http://tigris.sourceforge.net). Initial reference-free image classification by multivariate statistical analysis (MSA) and multi-reference alignment (MRA) methods generated a set of class averages. Three orthogonal class averages based on their size and shapes were selected for angular assignment using the IMAGIC C1 start-up procedure. These were then used to assign angles to the rest of the class averages and to generate a preliminary 3D map. After several iterative rounds of MRA, MSA-based classification, and angle assignment by angular reconstitution, clear 2-fold symmetry was apparent in both the class averages and the 3D reconstructions (Supplemental Figs. 3 and 5), hence subsequent refinements were carried out with C2 symmetry imposed. Two-fold symmetry had also been apparent in the initial eigen-images from the centered dataset of particles (Supplemental Figs. 3 and 5). Further to confirm that BRCA2 indeed exists predominantly as dimers, we segmented the dimer reconstructions into monomers and subjected the whole dataset for competitive alignments to both dimer and monomer models. 80% and 85% of the BRCA2 and BRCA2-RAD51 datasets respectively aligned to the dimer models, reinforcing that BRCA2 and BRCA2-RAD51 complexes exist as dimers. Finally, a further refinement step involving projection matching was carried out. The final 3D reconstructions incorporated the vast majority of the datasets, 568 out of the 578 class averages for the BRCA2 dataset and 553 out of 578 class averages for the BRCA2-RAD51 dataset and represent a broad range of angular distributions (Supplemental Fig. 4). To avoid any potential model bias, the BRCA2 and BRCA2-RAD51 datasets were processed independently. The reprojections of the 3D model are in excellent agreement with the corresponding class averages and the reconstructions have a resolution of approx. 19-20 Å, respectively, determined by the 0.5 Fourier shell correlation (FSC) criterion by applying a soft spherical mask of 0.7 radius (Supplemental Fig. 4).
Structural validation
3-D reconstructions were validated using versions of the tilt-pair and tilt-series validation methods59-61. The particle sets used in this analysis each constituted a tilt series ranging between nominal tilts of 0° and 40°, in steps of 5°. Each particle was independently assigned Euler angles using projection matching based on 3-D reconstructions. The particle with a nominal tilt of 0° was used as a reference to calculate all the other relative tilt transformations. The measured tilts angles lie within an average of ~2° of the expected tilt, confirming the overall correctness of the reconstructions.
BRCA2 and BRCA2-RAD51 antibody labeling
BRCA2-antibody complexes were prepared by incubating BRCA2 with 10-30 nM of each antibody for 30 min at 4°C. For BRCA2FLAP we used anti-Flag M2-HRP antibody (Sigma-Aldrich) and the anti-BRCA2 OP95 mouse antibody (Merck Millipore, Ab-1). The anti-RAD51 antibody 14B4 (Abcam ab213) was used for BRCA2-RAD51 complexes.
Guided by the distinct Y shape of the antibody, individual BRCA2 particles with antibody bound were selected from micrographs. We used projection matching to assign angles using our BRCA2 3D reconstruction. To avoid mis-assignment, the antibody was masked out during the angular assignment procedure. With multiple differently orientated particles, the 3D position of the antibody relative to the BRCA2 3D reconstruction could then be obtained by visual triangulation (Figs. 1 and 2).
Prior to EM analysis, the binding efficiency of each antibody towards BRCA2 or BRCA2-RAD51 was determined by immunoprecipitation. To do this, antibody (5 μg) was incubated with 2 ml of BRCA2 lysate or 10 ng purified BRCA2-RAD51 complex in 2 ml of IP buffer for 2 hours with constant mixing at 4°C. Dynabeads Protein G (50 μl; Life Technologies) was then added and incubation was continued for another 2 hours to capture antibody-bound BRCA2. The beads were washed five times with 1 ml of IP buffer and resuspended in SDS-PAGE gel loading buffer and analyzed by SDS-PAGE. Western blots were performed to detect captured BRCA2.
Scanning Transmission Electron Microscopy (STEM) analysis
STEM was performed at the Brookhaven National Laboratory (BNL) STEM facility, as described on http://www.bnl.gov/biology/stem/. Further details are described62. The program PCMass was used for mass measurements with tobacco mosaic virus as an internal control.
BRCA2-DNA complex
To prepare BRCA2-DNA complexes for visualization by electron microscopy, purified BRCA2 was incubated with gapped duplex DNA (100 nM) for 30 min at 37°C in a low salt buffer (50 mM NaCl). Binding of BRCA2 to this substrate was confirmed by gel-shift assays (Supplemental Fig. 1). Images with higher contrast were required to visualize the DNA strands, hence BRCA2-DNA images were collected on a FEI Tecnai F20 TEM equipped with a Falcon II 4k × 4k direct electron detector. A magnification of 50,000×, an underfocus of 4 μm and exposure of 20 e−Å−2, were used for image acquisition, yielding a pixel size of 2.05 Å. Images were band-pass filtered to 60-270 Å to improve visibility of the DNA strands.
3D electron density analysis and fitting
The UCSF Chimera package 35 and PyMol were used in all structural analyses and visualization, volume segmentation and fitting. They were also used to produce representations of 3D maps and fittings.
Supplementary Material
Acknowledgements
We thank R. Carzaniga and L. Collinson at the London Research Institute Electron Microscope facility for advice and input, and our colleagues for their comments. We thank Innova Biosicence for providing InnovaCoat conjugation kit. This work was supported by the UK Medical Research Council and the Wellcome Trust (XZ), and by Cancer Research UK, the European Research Council, the Breast Cancer Campaign, Swiss Bridge and the Louis-Jeantet foundation (SCW). LM was supported in part by an European Molecular Biology Organization Fellowship. JS was supported in part by Marie Curie Intra-European Fellowship.
Footnotes
Accession Codes: The reconstructions of BRCA2 (EMD-12805) and BRCA2-RAD51 (EMD-12806) have been deposited in the Electron Microscopy Data Bank.
References
- 1.Stratton MR, Rahman N. The emerging landscape of breast cancer susceptibility. Nat. Genet. 2008;40:17–22. doi: 10.1038/ng.2007.53. [DOI] [PubMed] [Google Scholar]
- 2.Wooster R, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 3.King MC, Marks JH, Mandell JB, Grp NYBCS. Breast and ovarian cancer risks due to inherited mutations in BRCA1 and BRCA2. Science. 2003;302:643–646. doi: 10.1126/science.1088759. [DOI] [PubMed] [Google Scholar]
- 4.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 5.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell. 2002;108:171–182. doi: 10.1016/s0092-8674(02)00615-3. [DOI] [PubMed] [Google Scholar]
- 7.Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001;7:263–272. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 9.Lomonosov M, Anand S, Sangrithi M, Davies R, Venkitaraman AR. Stabilization of stalled DNA replication forks by the BRCA2 breast cancer susceptibility protein. Genes and Dev. 2003;17:3017–3022. doi: 10.1101/gad.279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlacher K, et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan S-SF, et al. BRCA2 is required for ionizing radiation-induced assembly of RAD51 complex in vivo. Cancer Res. 1999;59:3547–3551. [PubMed] [Google Scholar]
- 12.Tarsounas M, Davies D, West SC. BRCA2-dependent and independent formation of RAD51 nuclear foci. Oncogene. 2003;22:1115–1123. doi: 10.1038/sj.onc.1206263. [DOI] [PubMed] [Google Scholar]
- 13.Bork P, Blomberg N, Nilges M. Internal repeats in the BRCA2 protein sequence. Nat. Genet. 1996;13:22–23. doi: 10.1038/ng0596-22. [DOI] [PubMed] [Google Scholar]
- 14.Bignell G, Micklem G, Stratton MR, Ashworth A, Wooster R. The BRC repeats are conserved in mammalian BRCA2 proteins. Human Mol. Genet. 1997;6:53–58. doi: 10.1093/hmg/6.1.53. [DOI] [PubMed] [Google Scholar]
- 15.Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 16.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat. Struct. Mol. Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 17.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA-binding specificity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 19.Tal A, Arbel-Goren R, Stavans J. Cancer-associated mutations in BRC domains of BRCA2 affect homologous recombination induced by RAD51. J. Mol. Biol. 2009;393:1007–1012. doi: 10.1016/j.jmb.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 20.Thorslund T, et al. The breast cancer tumour suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–83. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SA, Roques C, Magwood AC, Masson JY, Baker MD. Recovery of deficient homologous recombination in BRCA2-depleted mouse cells by wild-type RAD51 expression. DNA Repair. 2009;8:170–181. doi: 10.1016/j.dnarep.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Buisson R, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat. Struct. Mol. Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dray E, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat. Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sy SMH, Huen MSY, Zhu YY, Chen JJ. PALB2 regulates recombinational repair through chromatin association and oligomerization. J. Biol. Chem. 2009;284:18302–18310. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorslund T, Esashi F, West SC. Interactions between human BRCA2 protein and the meiosis-specific recombinase DMC1. EMBO J. 2007;26:2915–2922. doi: 10.1038/sj.emboj.7601739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang HJ, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–1848. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 30.van Heel M, et al. Single-particle electron cryo-microscopy: towards atomic resolution. Q. Revs. Biophys. 2000;33:307–369. doi: 10.1017/s0033583500003644. [DOI] [PubMed] [Google Scholar]
- 31.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J. Struct. Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 32.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Sauerwald A, et al. Structure of active dimeric human telomerase. Nat. Struct. Mol. Biol. 2013;20 doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong AKC, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene BRCA2. J. Biol. Chem. 1997;272:31941–31944. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 35.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Benson FE, Stasiak A, West SC. Purification and characterisation of the human RAD51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baumann P, Benson FE, West SC. Human RAD51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 38.Shivji MKK, et al. The BRC repeats of human BRCA2 differentially regulate RAD51 binding on single- versus double-stranded DNA to stimulate strand exchange. Proc. Natl. Acad. Sci. U. S. A. 2009;106:13254–13259. doi: 10.1073/pnas.0906208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ho Y, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- 40.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 41.Baumann P, West SC. The human RAD51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hilario J, Amitani I, Baskin RJ, Kowalczykowski SC. Direct imaging of human RAD51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. U. S. A. 2009;106:361–368. doi: 10.1073/pnas.0811965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou QW, et al. Dss1 interaction with Brh2 as a regulatory mechanism for recombinational repair. Mol. Cell. Biol. 2007;27:2512–2526. doi: 10.1128/MCB.01907-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jeyasekharana AD, et al. DNA damage regulates the mobility of BRCA2 within the nucleoplasm of living cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21937–21942. doi: 10.1073/pnas.1009577107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 46.Saeki H, et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bochkarev A, Bochkareva E. From RPA to BRCA2: lessons from single-stranded DNA binding by the OB-fold. Curr. Opin. Struct. Biol. 2004;14:36–42. doi: 10.1016/j.sbi.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 48.Fan J, Pavletich NP. Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev. 2012;26:2337–2347. doi: 10.1101/gad.194787.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van der Heijden T, et al. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucl. Acids Res. 2007;35:5646–5657. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bianchi M, Das Gupta C, Radding CM. Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell. 1983;34:931–939. doi: 10.1016/0092-8674(83)90550-0. [DOI] [PubMed] [Google Scholar]
- 51.Bortner C, Griffith J. Three-stranded paranemic joints: Architecture, topological constraints and movement. J. Mol. Biol. 1990;215:623–634. doi: 10.1016/S0022-2836(05)80173-1. [DOI] [PubMed] [Google Scholar]
- 52.McIlwraith MJ, et al. Reconstitution of the strand invasion step of double-strand break repair using human RAD51, RAD52 and RPA proteins. J. Mol. Biol. 2000;304:151–164. doi: 10.1006/jmbi.2000.4180. [DOI] [PubMed] [Google Scholar]
- 53.Namsaraev EA, Berg P. Rad51 uses one mechanism to drive DNA strand exchange in both directions. J. Biol. Chem. 2000;275:3970–3976. doi: 10.1074/jbc.275.6.3970. [DOI] [PubMed] [Google Scholar]
- 54.Yang H, Li Q, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 55.West SC, Cassuto E, Howard-Flanders P. Heteroduplex formation by RecA protein: Polarity of strand exchange. Proc. Natl. Acad. Sci. U.S.A. 1981;78:6149–6153. doi: 10.1073/pnas.78.10.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kahn R, Cunningham RP, DasGupta C, Radding CM. Polarity of heteroduplex formation promoted by Escherichia coli RecA protein. Proc. Natl. Acad. Sci. U.S.A. 1981;78:4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cox MM, Lehman IR. Directionality and polarity in RecA protein-promoted branch migration. Proc. Natl. Acad. Sci. U.S.A. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baumann P, Benson FE, Hajibagheri N, West SC. Purification of human RAD51 protein by selective spermidine precipitation. Mut. Res. DNA Repair. 1997;384:65–72. doi: 10.1016/s0921-8777(97)00028-1. [DOI] [PubMed] [Google Scholar]
- 59.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. Journal of molecular biology. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 60.Henderson R, et al. Tilt-pair analysis of images from a range of different specimens in single-particle electron cryomicroscopy. Journal of molecular biology. 2011;413:1028–46. doi: 10.1016/j.jmb.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sauerwald A, et al. Structure of active dimeric human telomerase. Nat Struct Mol Biol. 2013;20:454–60. doi: 10.1038/nsmb.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wall JS, Hainfeld JF, Simon MN. Scanning transmission electron microscopy of nuclear structures. Methods Cell Biol. 1998;53:139–64. doi: 10.1016/s0091-679x(08)60878-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.