Abstract
Aberrant methylation of CpG dinucleotides is a commonly observed epigenetic modification in human cancer. Thus, detection of aberrant gene promoter methylation as a tool for diagnosis of tumors or as a prognostic marker has been widely described for many types of cancers, including nonsmall cell lung cancer (NSCLC). Emerging evidence indicates that CDH13 is a candidate tumor suppressor in several types of human tumors, including NSCLC. However, the correlation between CDH13 hypermethylation and clinicopathological characteristics of NSCLC remains unclear. In the current study, we conducted a systematic review and meta-analysis to quantitatively evaluate the effects of CDH13 hypermethylation on the incidence of NSCLC and clinicopathological characteristics. Final analysis of 803 NSCLC patients from eleven eligible studies was performed. CDH13 hypermethylation was observed to be significantly higher in NSCLC than in normal lung tissue, with the pooled odds ratio (OR) from seven studies including 448 NSCLC and 345 normal lung tissue (OR, 7.85; 95% confidence interval, 5.12–12.03; P<0.00001). CDH13 hypermethylation was also associated with pathological types. The pooled OR was obtained from four studies, including 111 squamous cell carcinoma and 106 adenocarcinoma (OR, 0.35; 95% confidence interval, 0.19–0.66; P=0.001), which indicated that CDH13 hypermethylation plays a more important role in the pathogenesis of adenocarcinoma. NSCLC with CDH13 hypermethylation was found more frequently in poorly differentiated NSCLC patients. NSCLC patients with CDH13 hypermethylation had a lower survival rate than those without CDH13 hypermethylation. In addition, CDH13 mRNA high expression was found to correlate with better overall survival for all NSCLC patients followed for 20 years (hazard ratio, 0.81; P=0.0056). Interestingly, CDH13 mRNA overexpression was found to correlate with better overall survival only in adenocarcinoma patients (hazard ratio, 0.42; P=9.6e–09), not in squamous cell carcinoma patients (hazard ratio, 0.93; P=0.59). The results of this meta-analysis suggest that CDH13 hypermethylation is associated with an increased risk and worse survival in NSCLC. CDH13 hypermethylation and mRNA expression play an important role in carcinogenesis, progression, and development, as well as clinical outcomes.
Keywords: prognosis, methylation, lung cancer, tumor suppressor gene, meta-analysis, odds ratio, hazard ratio
Introduction
Epigenetic modification of gene expression plays an important role in carcinogenesis. Aberrant methylation of CpG dinucleotides is a commonly observed epigenetic modification in human cancer.1–3 Loss of function in cancer suppressor genes may hinder cancer cell growth inhibition, which leads to malignant transcription and translation during replication of DNA. Thus, analysis of specific gene promoter methylation as a tool for diagnosis of tumors and its use as a prognostic marker have been widely used for many different cancers, including nonsmall cell lung cancer (NSCLC).4 A number of genes including the cyclin-dependent kinase inhibitor (p16), the tumor suppressor gene cystic fibrosis transmembrane conductance regulator, the DNA repair gene MGMT, Ras association domain family protein 1A, and Kelch-like ECH-associating protein 1, and so on are demonstratively methylated across NSCLC.5–8 Cadherins, which function as membrane receptors mediating outside-in signals, activating small GTPases and the β-catenin/Wnt pathway, and resulting in dynamic cytoskeleton reorganization and changes in the phenotype, are important determinants of tumor progression, serving as a suppressors of invasion and metastasis in many contexts.9,10 Cadherin 13 (CDH13), also known as T-cadherin or H-cadherin (heart), is a unique member of the cadherin superfamily, as it lacks the transmembrane and cytoplasmic domains and is anchored to the cell membrane through the glycosylphosphatidylinisotol (GPI) anchor.11–13 Although signaling partners and adapter proteins for CDH13 remain to be elucidated, it is well known that CDH13 is involved in low-density lipoproteins, hormone-like effects on Ca2+-mobilization and increased cell migration, phenotype changes, insulin-dependent signaling, eNOS activation, and angiogenesis.14–16 Its precise function has been intensively studied in several tumors, with upregulation of inducing cell cycle arrest, apoptosis, and inhibition of angiogenesis.17–22 Lack of protein expression of CDH13 by promoter methylation (hypermethylation) and/or gene deletion has been found to play an important role in lung alveolar differentiation regulation and epithelial tumorigenesis.23–27 However, its roles in NSCLC and clinical significance have not been thoroughly investigated. In this study, we review and update the published clinical investigations regarding the effect of CDH13 on patients with NSCLC.
Material and methods
Search strategy and selection criteria
We searched PubMed, Embase and ISI Web of Knowledge to identify studies from May 1, 1998, to March 1, 2014, using the search terms “lung,” “cancer or tumor or neoplasm or carcinoma,” “methylation,” and “CDH13 or H-cadherin or T-cadherin or cadherin 13.” We also manually searched the reference lists of the retrieved articles and reviews for additional articles.
Although our search did not have language limits initially, for the full-text reading and final evaluation, we only performed a review of the studies published in the English language. After exclusion of irrelevant and/or redundant publications from the different databases, the remaining papers were evaluated in the full-text version for inclusion and exclusion criteria, and for relevant articles in the reference lists. All searched data were retrieved. Authors’ bibliographies and references from selected studies were also searched for other relevant studies. The most complete study was chosen to avoid duplication if the same patient populations were reported in several publications.
The criteria that an eligible study had to meet were as follows: CDH13 hypermethylation evaluated in the primary NSCLC tissues, studies revealed the relationship between CDH13 hypermethylation and NSCLC clinicopathological parameters and prognosis, CDH13 hypermethylation examined by polymerase chain reaction, and studies provided sufficient information to estimate hazard ratio (HR) about overall survival (OS) and 95% confidence intervals (CIs). The exclusion criteria included the following: letters, reviews, case reports, conference abstracts, editorials, and expert opinion; all publications regarding in vitro/ex vivo studies, cell lines, and human xenografts were also excluded.
Data extraction and methodological assessment
Two authors (RX, CY) independently reviewed and extracted data from eligible studies. Disagreements were resolved by discussion and consensus. Two authors (FZ, DL) reviewed all of the articles that fit inclusion and exclusion criteria. The following information was recorded for each study: first author name, year of publication, sample source, number of cases, clinicopathological parameters, cancer tumor node metastasis stage, methylation detection method, methylation rate and/or expression, and follow-up. Data for study characteristics and clinical responses were summarized and turned into a table. The heterogeneity of the investigation was evaluated to determine whether the data of the various studies could be analyzed for a meta-analysis.
For the methodological evaluation of the studies, three investigators (DL, CY, and FZ) read through the publications independently and assessed and scored them according to Reporting Recommendations for Tumor Marker Prognostic Studies guidelines and the European Lung Cancer Working Party quality scale.28,29 The three readers provided the quality scores, compared them, and then reached a consensus value for each item.
Patient survival analysis
An online database30 was used to assess relevance of CDH13 mRNA expression to relapse-free survival. The database was established using gene expression data and survival information from 1,405 NSCLC patients downloaded from Gene Expression Omnibus. Briefly, CDH13 was entered into the database (http://kmplot.com/analysis/index.php?p=service&cancer=lung) to obtain Kaplan-Meier survival plots in which the number at risk is indicated below the main plot. HR and 95% CIs and logrank P were calculated and displayed on the Web page.
Statistical analysis
Analysis was conducted using the STATA 12.0 (Stata Corporation) and Review Manager 5.2 (Cochrane Collaboration). The pooled frequency of CDH13 hypermethylation and 95% CIs were estimated. The frequency of CDH13 hypermethylation was compared in different tumor characteristics. Heterogeneity among studies was evaluated with Cochran’s Q test31 and the I2 statistic.32,33 When heterogeneity was not an issue (I2 values <50%), a fixed-effect model was used to calculate parameters. If there was substantial heterogeneity (I2 values ≥ 50%), a random-effects model was used to pool data and to attempt to identify potential sources of heterogeneity based on subgroup analyses. The pooled OR was estimated for the association between CDH13 hypermethylation and clinicopathological features. P-values two-tailed less than 0.05 were considered statistically significant.
Publication bias was assessed by using a method reported by Egger and colleagues.34 We also explored reasons for statistical heterogeneity using meta-regression, subgroup analysis, and sensitivity analysis. The analysis of meta-regression and publication bias was performed by using STATA version 10.0.
Results
Identification of relevant studies
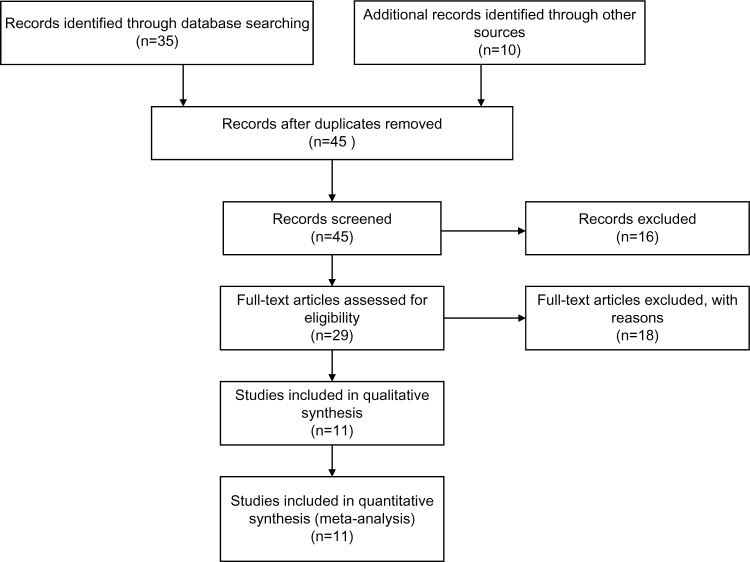
Forty-five publications were identified by the search method, as described earlier. Thirty-four of those were excluded because they were laboratory studies, nonoriginal articles (reviews), or studies irrelevant to the current analysis. Eventually, there were eleven studies included in final meta-analysis, as shown in Figure 1.
Figure 1.
Flow chart of study selection.
Study characteristics
Eleven studies published from 2001 to 2012 were eligible for meta-analysis. A total of 803 NSCLC patients from China, Japan, South Korea, Italy, Serbia, and the United States were enrolled. There were five studies from Western countries, in which CDH13 hypermethylation rates were from 15.6%∼65.6%, with an average of 44.0%. There were six studies from Asian countries, in which CDH13 hypermethylation rates were from 26.7%∼53.6%, with an average of 39.3%. There are no big differences in CDH13 hypermethylation rates between Western and Asian patients. Their basic characteristics are summarized in Table 1.
Table 1.
Basic characteristics of the included studies
| Study | Country | Patients | Methods | Primary aim | Methylation site | CDH13 expression |
|---|---|---|---|---|---|---|
| Kontic et al44 | Serbia | 65 | MSP | Determine the frequency of CDH13 hypermethylation in NSCLC | Promoter, CpG islands | – |
| Zhang et al55 | People’s Republic of China | 78 | MSP | Determine the methylation status of multiple genes in NSCLC | Promoter, CpG islands | – |
| Kubo et al56 | Japan | 100 | MSP | Determine the methylation status of five tumor suppressor in NSCLC | Promoter, CpG islands | – |
| Feng et al57 | United States | 49 | DNA methylation (MethyLight) analysis | Determine the clinical significance of seven tumor suppressors in NSCLC | Promoter, CpG islands | – |
| Brock et al23 | United States | 79 | MSP | Determine the association between gene methylation and recurrence in NSCLC | Promoter, CpG islands | – |
| Wang et al58 | People’s Republic of China | 28 | DNA microarray-coupled polymerase chain reaction | Determine 15 genes’ methylation status in NSCLC | Promoter, CpG islands | – |
| Kim et al50 | South Korea | 88 | MSP | Determine the role of CDH13 in pathogenesis of NSCLC | Promoter, CpG islands | – |
| Hsu et al59 | People’s Republic of China | 63 | MSP | Determine methylation patterns of six tumor suppressor gene in NSCLC | Promoter, CpG islands | – |
| Suzuki et al54 | Japan | 150 | MSP | The methylation profile of nine genes for NSCLC were analyzed and correlated with clinical data | Promoter, CpG islands | – |
| Ulivi et al60 | Italy | 61 | MSP | Detection of promoter methylation of p16INK4A and CDH13 genes in NSCLC | Promoter, CpG islands | – |
| Toyooka et al26 | United States | 42 | MSP | Analyze DNA methylation status of CDH13 gene in NSCLC patients | Promoter, CpG islands | + |
Abbreviations: NSCLC, nonsmall cell lung cancer; MSP, methylation-specific polymerase chain reaction.
The correlation of CDH13 hypermethylation with clinicopathological features
The inactivation of CDH13 through hypermethylation in NSCLC
The loss of CDH13 mRNA and/or protein expression was strongly correlated with the promoter hypermethylation in several types of cancer including NSCLC.25,26,35–43 CDH13 hypermethylation was significantly higher in NSCLC than normal lung tissue. The pooled OR from seven studies including 448 NSCLCs and 345 normal lung tissues is shown in Figure 2 (OR, 7.85; 95% CI, 5.12–12.03; P<0.00001), indicating that CDH13 inactivation through hypermethylation plays an important role in the carcinogenesis of NSCLC.
Figure 2.
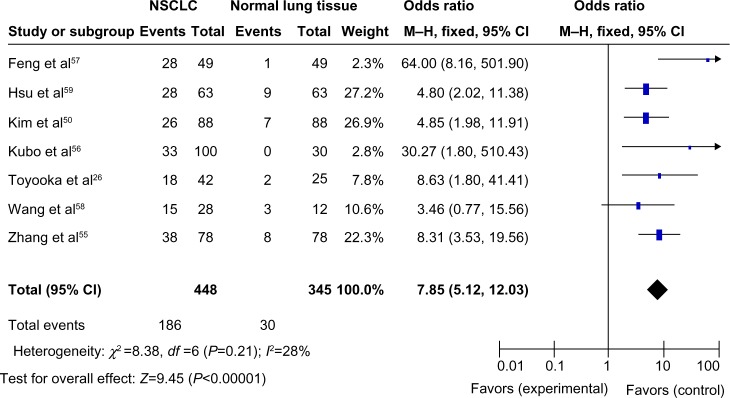
The pooled odds ratio from seven studies including 448 NSCLCs and 345 normal lung tissues.
Notes: Odds ratio, 7.85; 95% CI, 5.12–12.03; P<0.00001.
Abbreviations: NSCLC, nonsmall cell lung cancer; CI, confidence interval; M–H, Mantel–Haenszel.
Relationship between the frequency of CDH13 hypermethylation and pathological types
Histology was associated with CDH13 hypermethylation, which was observed more frequently in patients with adenocarcinoma.44 The pooled OR from 4 studies, including 111 of squamous cell carcinoma and 106 of adenocarcinoma, is shown in Figure 3 (OR, 0.35; 95% CI, 0.19–0.66; P=0.001), indicating that CDH13 hypermethylation plays more important role in the pathogenesis of adenocarcinoma.
Figure 3.
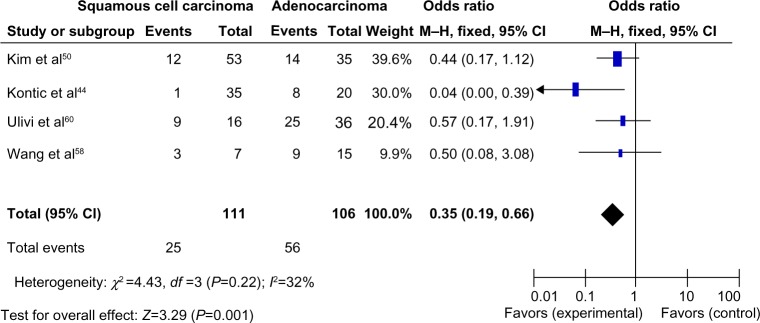
Pooled odds ratio from four studies, including 111 squamous cell carcinomas and 106 adenocarcinomas.
Notes: Odds ratio, 0.35; 95% CI, 0.19–0.66; P=0.001, which indicated that CDH13 hypermethylation plays a more important role in the pathogenesis of adenocarcinoma.
Abbreviations: CI, confidence interval; M–H, Mantel–Haenszel.
The role of CDH13 hypermethylation in NSCLC progression
We analyzed 363 NSCLC patients pooled from 4 studies to assess whether the aberrant CDH13 hypermethylation in NSCLC was associated with advanced stage. As shown in Figure 4, aberrant CDH13 hypermethylation was not significantly higher in advanced NSCLC (stage III and IV) than in early-stage NSCLC (stage I and II; OR, 0.93; 95% CI, 0.57–1.53; P=0.79). However, as shown in Figure 5, aberrant CDH13 hypermethylation was significantly higher in poorly differentiated NSCLC than in moderately and highly differentiated NSCLC (OR, 3.61; 95% CI, 1.76–7.39; P=0.0004). These results suggest that CDH13 hypermethylation may play an important role in NSCLC progression and development.
Figure 4.
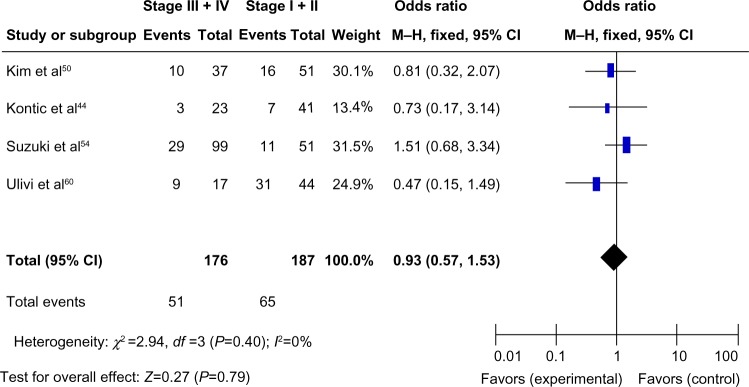
Three hundred and sixty-three nonsmall cell lung cancer patients were pooled from four studies to assess whether the aberrant CDH13 hypermethylation in nonsmall cell lung cancer was associated with advanced stage.
Notes: Aberrant CDH13 hypermethylation was not significantly higher in advanced nonsmall cell lung cancer (III and IV) than that in early-staged nonsmall cell lung cancer (I and II). Odds ratio, 0.93; 95% CI, 0.57–1.53; P=0.79.
Abbreviations: CI, confidence interval; M–H, Mantel–Haenszel.
Figure 5.
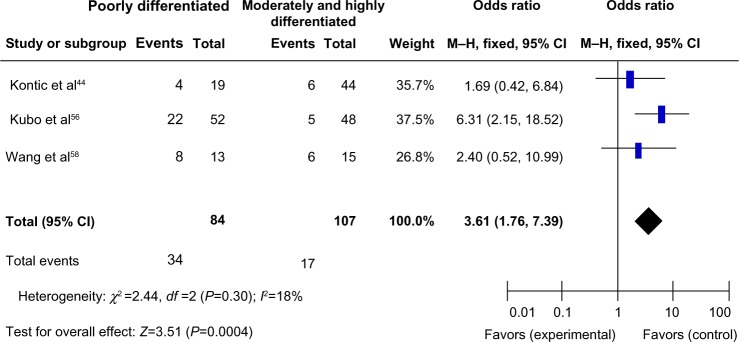
Aberrant CDH13 hypermethylation was significantly higher in poorly differentiated nonsmall cell lung cancer than that in moderately and highly differentiated nonsmall cell lung cancer.
Notes: Odds ratio, 3.61; 95% CI, 1.76–7.39; P=0.0004.
Abbreviations: CI, confidence interval; M–H, Mantel–Haenszel.
CDH13 hypermethylation as a prognostic factor for NSCLC
There were four studies that estimated the relationship between CDH13 hypermethylation and OS in NSCLC. The pooled HR for OS showed that CDH13 hypermethylation was associated with worse survival in NSCLC patients, as shown in Figure 6 (HR, 3.21; 95% CI, 1.41–7.31; P=0.005).
Figure 6.
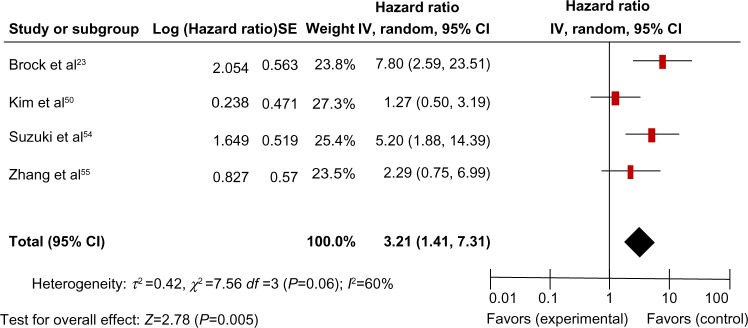
The four studies included investigated the relationship between overall survival and CDH13 hypermethylation.
Notes: The pooled hazard ratio for overall survival showed that CDH13 hypermethylation was associated with worse survival in nonsmall cell lung cancer. Hazard ratio, 3.21; 95% CI, 1.41–7.31; P=0.005.
Abbreviations: SE, standard error; CI, confidence interval.
Sensitivity analyses and publication bias
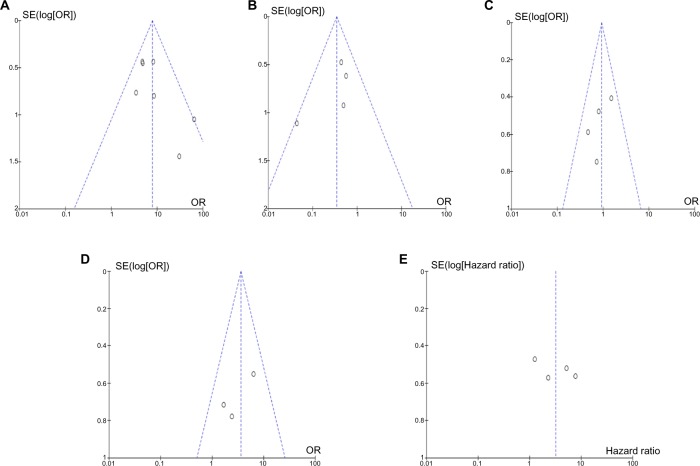
A sensitivity analysis, in which one study was removed at a time, was conducted to assess the result stability. The pooled ORs and HRs were not significantly changed, indicating the stability of our analyses. The funnel plots were largely symmetric (Figure 7), suggesting there were no publication biases in the meta-analysis of CDH13 hypermethylation and clinicopathological features.
Figure 7.
The funnel plots were largely symmetric, suggesting there were no publication biases in the meta-analysis of CDH13 hypermethylation and clinicopathological features.
Notes: (A) The funnel plot from seven studies comparing nonsmall cell lung cancer and normal lung tissue. (B) The funnel plot from four studies comparing CDH13 hypermethylation between squamous cell carcinoma and adenocarcinoma. (C) The funnel plot from four studies in determining CDH13 hypermethylation in differently staged nonsmall cell lung cancer. (D) The funnel plot from three studies determining CDH13 hypermethylation in different differentiated nonsmall cell lung cancer. (E) The funnel plot from four studies determining the relationship between CDH13 hypermethylation and overall survival in nonsmall cell lung cancer.
Abbreviations: OR, odds ratio; SE, standard error.
Effect of CDH13 mRNA expression on prognosis of NSCLC
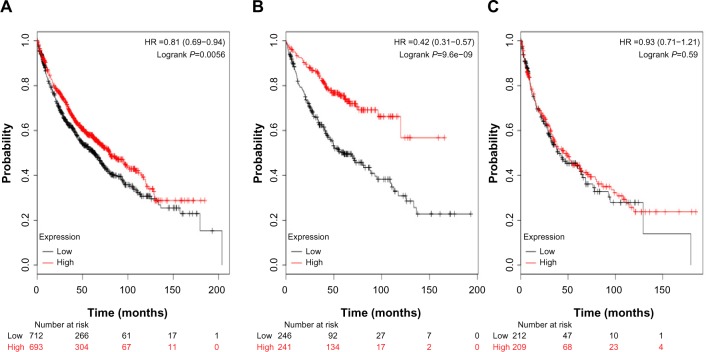
The clinical relevance of CDH13 was further corroborated in a patient survival analysis, using an online database containing the expression of 22,277 genes and 20-year survival information of 1,809 patients.30 The database has recently been updated to include survival information of 1,405 NSCLC patients (http://www.kmplot.com/analysis/). CDH13 mRNA high expression was found to correlate with better OS for all NSCLC patients followed for 20 years (Figure 8A; HR, 0.81; P=0.0056). In addition, CDH13 mRNA high expression was found to correlate with better relapse OS only in adenocarcinoma patients (Figure 8B; HR, 0.42; P=9.6e–09), but not squamous cell carcinoma patients (Figure 8C; HR, 0.93; P=0.59).
Figure 8.
The clinical relevance of CDH13 was corroborated in a patient survival analysis using an online database containing the expression of 22,277 genes and 20-year survival information of 1,405 patients.
Notes: CDH13 mRNA high expression was found to correlate with better overall survival for all nonsmall cell lung cancer patients followed for 20 years. (A) HR, 0.81; P=0.0056. In addition, CDH13 mRNA high expression was found to correlate with better overall survival only in adenocarcinoma patients. (B) HR, 0.42; P=9.6e–09, but not patients with squamous cell carcinoma. (C) HR, 0.93; P=0.59.
Abbreviation: HR, hazard ratio.
Discussion
CDH13 has been reported to encode T-cadherin, an adiponectin receptor discovered after adipoR1 and adipoR2.45 Inactivation of CDH13 by promoter hypermethylation plays an important role during normal development and in tumorigenesis in several types of tumors, including NSCLC.37,44,46–51 To date, there have been some studies describing the precise expression, prognostic effect, and methylation status of CDH13 in NSCLC; however, the roles of inactivation of CDH13 in NSCLC and clinical significance have not been thoroughly investigated. We conducted the meta-analysis to determine the correlation between CDH13 hypermethylation and clinicopathological characteristics in NSCLC. Analysis of the pooled data showed that NSCLC had a higher hypermethylation than normal lung tissue, CDH13 hypermethylation plays a more important role in the pathogenesis of adenocarcinoma, and aberrant CDH13 hypermethylation was not significantly higher in advanced NSCLC (stage III and IV) than in early-staged NSCLC (stage I and II). However, aberrant CDH13 hypermethylation was significantly higher in poorly differentiated NSCLC than in moderately and highly differentiated NSCLC. In addition, NSCLC patients with CDH13 hypermethylation had a lower survival rate than those without CDH13 hypermethylation. The results from the current study demonstrated that the hypermethylation rate of the CDH13 gene promoter in NSCLC was significantly higher than that in the normal lung tissues, indicating that CDH13 promoter hypermethylation was common in NSCLC. Because changes in CDH13 promoter hypermethylation are reversible, drug treatment through demethylation may be useful to delay carcinogenesis and progression and improve prognosis. This approach may bring new direction and hope for cancer treatment through gene-targeted therapy.
Epigenetic alterations, particularly aberrant DNA methylation, one of the best-characterized epigenetic modifications, contribute to tumor initiation and progression.2,3 CDH13 is thought to affect cellular function and behavior largely through its signaling properties.22 CDH13 reexpression in most cancer cell lines inhibits cell proliferation and invasiveness, increases susceptibility to apoptosis, and reduces tumor growth in vivo models.36,52,53 Therefore, CDH13 can be considered a tumor suppressor, and its inactivation could contribute to tumor progression and poor prognosis. Although only four studies evaluated the relationship between overall survival and CDH13 hypermethylation in NSCLC, they showed very similar results.23,50,54,55 According to this meta-analysis, we may consider that CDH13 hypermethylation in NSCLC tends to indicate a poor prognosis. We further determined the clinical relevance of CDH13 mRNA expression in a patient survival analysis, using an online data-base containing the expression of 22,277 genes and 20-year survival information of 1,405 patients.30 As expected, CDH13 mRNA high expression was found to correlate with OS for all NSCLC patients followed for 20 years. Interestingly, CDH13 mRNA high expression was found to correlate with better OS only in adenocarcinoma patients, but not squamous cell carcinoma patients.
Consistent results were shown in sensitivity analyses, and no evidence of publication bias was found. This study has several potential limitations. First, the possibility of information and selection biases and unidentified confounders could not be completely excluded because all of the included studies were observational. Second, the searching strategy was restricted to articles published in English. Articles with potentially high-quality data that were published in other languages were not included because of anticipated difficulties in obtaining accurate medical translation. Hence, caution should be taken when our findings are interpreted among the general population.
In conclusion, our meta-analysis showed CDH13 may play an important role in NSCLC initiation and progression. In addition, CDH13 hypermethylation is associated with an increased risk and worse survival in NSCLC. Further large-scale studies, especially multicenter and well-matched cohort research, will provide more insight into the role of CDH13 in the prognosis and clinical implementation of NSCLC patients.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ghavifekr Fakhr M, Farshdousti Hagh M, Shanehbandi D, Baradaran B. DNA methylation pattern as important epigenetic criterion in cancer. Genet Res Int. 2013;2013:317569. doi: 10.1155/2013/317569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delpu Y, Cordelier P, Cho WC, Torrisani J. DNA methylation and cancer diagnosis. Int J Mol Sci. 2013;14(7):15029–15058. doi: 10.3390/ijms140715029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma X, Wang YW, Zhang MQ, Gazdar AF. DNA methylation data analysis and its application to cancer research. Epigenomics. 2013;5(3):301–316. doi: 10.2217/epi.13.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleischhacker M, Dietrich D, Liebenberg V, Field JK, Schmidt B. The role of DNA methylation as biomarkers in the clinical management of lung cancer. Expert Rev Respir Med. 2013;7(4):363–383. doi: 10.1586/17476348.2013.814397. [DOI] [PubMed] [Google Scholar]
- 5.Guo D, Wu B, Yan J, Li X, Sun H, Zhou D. A possible gene silencing mechanism: hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells. Biochem Biophys Res Commun. 2012;428(1):80–85. doi: 10.1016/j.bbrc.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Kurakawa E, Shimamoto T, Utsumi K, Hirano T, Kato H, Ohyashiki K. Hypermethylation of p16(INK4a) and p15(INK4b) genes in non-small cell lung cancer. Int J Oncol. 2001;19(2):277–281. [PubMed] [Google Scholar]
- 7.Gao T, Wang S, He B, et al. The association of RAS association domain family Protein1A (RASSF1A) methylation states and bladder cancer risk: a systematic review and meta-analysis. PLoS ONE. 2012;7(11):e48300. doi: 10.1371/journal.pone.0048300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Son JW, Kim YJ, Cho HM, et al. Promoter hypermethylation of the CFTR gene and clinical/pathological features associated with non-small cell lung cancer. Respirology. 2011;16(8):1203–1209. doi: 10.1111/j.1440-1843.2011.01994.x. [DOI] [PubMed] [Google Scholar]
- 9.Berx G, van Roy F. Involvement of members of the cadherin superfamily in cancer. Cold Spring Harb Perspect Biol. 2009;1(6):a003129. doi: 10.1101/cshperspect.a003129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27(55):6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angst BD, Marcozzi C, Magee AI. The cadherin superfamily: diversity in form and function. J Cell Sci. 2001;114(Pt 4):629–641. doi: 10.1242/jcs.114.4.629. [DOI] [PubMed] [Google Scholar]
- 12.Takeuchi T, Ohtsuki Y. Recent progress in T-cadherin (CDH13, H-cadherin) research. Histol Histopathol. 2001;16(4):1287–1293. doi: 10.14670/HH-16.1287. [DOI] [PubMed] [Google Scholar]
- 13.Philippova M, Joshi MB, Kyriakakis E, Pfaff D, Erne P, Resink TJ. A guide and guard: the many faces of T-cadherin. Cell Signal. 2009;21(7):1035–1044. doi: 10.1016/j.cellsig.2009.01.035. [DOI] [PubMed] [Google Scholar]
- 14.Philippova M, Joshi MB, Pfaff D, et al. T-cadherin attenuates insulin-dependent signalling, eNOS activation, and angiogenesis in vascular endothelial cells. Cardiovasc Res. 2012;93(3):498–507. doi: 10.1093/cvr/cvs004. [DOI] [PubMed] [Google Scholar]
- 15.Rubina K, Kalinina N, Potekhina A, et al. T-cadherin suppresses angiogenesis in vivo by inhibiting migration of endothelial cells. Angiogenesis. 2007;10(3):183–195. doi: 10.1007/s10456-007-9072-2. [DOI] [PubMed] [Google Scholar]
- 16.Rubina K, Talovskaya E, Cherenkov V, et al. LDL induces intracellular signalling and cell migration via atypical LDL-binding protein T-cadherin. Mol Cell Biochem. 2005;273(1–2):33–41. doi: 10.1007/s11010-005-0250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosserhoff AK, Ellmann L, Quast AS, Eberle J, Boyle GM, Kuphal S. Loss of T-cadherin (CDH-13) regulates AKT signaling and desensitizes cells to apoptosis in melanoma. Mol Carcinog. 2014;53(8):635–647. doi: 10.1002/mc.22018. [DOI] [PubMed] [Google Scholar]
- 18.Ghosh S, Joshi MB, Ivanov D, et al. Use of multicellular tumor spheroids to dissect endothelial cell-tumor cell interactions: a role for T-cadherin in tumor angiogenesis. FEBS Lett. 2007;581(23):4523–4528. doi: 10.1016/j.febslet.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 19.Wang XD, Wang BE, Soriano R, et al. Expression profiling of the mouse prostate after castration and hormone replacement: implication of H-cadherin in prostate tumorigenesis. Differentiation. 2007;75(3):219–234. doi: 10.1111/j.1432-0436.2006.00135.x. [DOI] [PubMed] [Google Scholar]
- 20.Riou P, Saffroy R, Chenailler C, et al. Expression of T-cadherin in tumor cells influences invasive potential of human hepatocellular carcinoma. FASEB J. 2006;20(13):2291–2301. doi: 10.1096/fj.06-6085com. [DOI] [PubMed] [Google Scholar]
- 21.Duan XS, Lu J, Ge ZH, Xing EH, Lu HT, Sun LX. Effects of T- cadherin expression on B16F10 melanoma cells. Oncol Lett. 2013;5(4):1205–1210. doi: 10.3892/ol.2013.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreeva AV, Kutuzov MA. Cadherin 13 in cancer. Genes Chromosomes Cancer. 2010;49(9):775–790. doi: 10.1002/gcc.20787. [DOI] [PubMed] [Google Scholar]
- 23.Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358(11):1118–1128. doi: 10.1056/NEJMoa0706550. [DOI] [PubMed] [Google Scholar]
- 24.Zhong Y, Delgado Y, Gomez J, Lee SW, Perez-Soler R. Loss of H-cadherin protein expression in human non-small cell lung cancer is associated with tumorigenicity. Clin Cancer Res. 2001;7(6):1683–1687. [PubMed] [Google Scholar]
- 25.Sato M, Mori Y, Sakurada A, Fujimura S, Horii A. The H-cadherin (CDH13) gene is inactivated in human lung cancer. Hum Genet. 1998;103(1):96–101. doi: 10.1007/s004390050790. [DOI] [PubMed] [Google Scholar]
- 26.Toyooka KO, Toyooka S, Virmani AK, et al. Loss of expression and aberrant methylation of the CDH13 (H-cadherin) gene in breast and lung carcinomas. Cancer Res. 2001;61(11):4556–4560. [PubMed] [Google Scholar]
- 27.Jin M, Kawakami K, Fukui Y, et al. Different histological types of non-small cell lung cancer have distinct folate and DNA methylation levels. Cancer Sci. 2009;100(12):2325–2330. doi: 10.1111/j.1349-7006.2009.01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics Reporting recommendations for tumor marker prognostic studies (REMARK) J Natl Cancer Inst. 2005;97(16):1180–1184. doi: 10.1093/jnci/dji237. [DOI] [PubMed] [Google Scholar]
- 29.Steels E, Paesmans M, Berghmans T, et al. Role of p53 as a prognostic factor for survival in lung cancer: a systematic review of the literature with a meta-analysis. Eur Respir J. 2001;18(4):705–719. doi: 10.1183/09031936.01.00062201. [DOI] [PubMed] [Google Scholar]
- 30.Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 31.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 32.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med. 1996;15(12):1237–1248. doi: 10.1002/(SICI)1097-0258(19960630)15:12<1237::AID-SIM301>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren JZ, Huo JR. 5-aza-2′-deoxycytidine-induced inhibition of CDH13 expression and its inhibitory effect on methylation status in human colon cancer cells in vitro and on growth of xenograft in nude mice. Zhonghua Zhong Liu Za Zhi. 2012;34(1):6–10. [PubMed] [Google Scholar]
- 36.Ren JZ, Huo JR. Correlation between T-cadherin gene expression and aberrant methylation of T-cadherin promoter in human colon carcinoma cells. Med Oncol. 2012;29(2):915–918. doi: 10.1007/s12032-011-9836-9. [DOI] [PubMed] [Google Scholar]
- 37.Jin Z, Cheng Y, Olaru A, et al. Promoter hypermethylation of CDH13 is a common, early event in human esophageal adenocarcinogenesis and correlates with clinical risk factors. Int J Cancer. 2008;123(10):2331–2336. doi: 10.1002/ijc.23804. [DOI] [PubMed] [Google Scholar]
- 38.Qian ZR, Sano T, Yoshimoto K, et al. Tumor-specific downregulation and methylation of the CDH13 (H-cadherin) and CDH1 (E-cadherin) genes correlate with aggressiveness of human pituitary adenomas. Mod Pathol. 2007;20(12):1269–1277. doi: 10.1038/modpathol.3800965. [DOI] [PubMed] [Google Scholar]
- 39.Ogama Y, Ouchida M, Yoshino T, et al. Prevalent hyper-methylation of the CDH13 gene promoter in malignant B cell lymphomas. Int J Oncol. 2004;25(3):685–691. [PubMed] [Google Scholar]
- 40.Roman-Gomez J, Castillejo JA, Jimenez A, et al. Cadherin-13, a mediator of calcium-dependent cell-cell adhesion, is silenced by methylation in chronic myeloid leukemia and correlates with pretreatment risk profile and cytogenetic response to interferon alfa. J Clin Oncol. 2003;21(8):1472–1479. doi: 10.1200/JCO.2003.08.166. [DOI] [PubMed] [Google Scholar]
- 41.Toyooka S, Toyooka KO, Harada K, et al. Aberrant methylation of the CDH13 (H-cadherin) promoter region in colorectal cancers and adenomas. Cancer Res. 2002;62(12):3382–3386. [PubMed] [Google Scholar]
- 42.Sun D, Zhang Z, Van N, Huang G, Ernberg I, Hu L. Aberrant methylation of CDH13 gene in nasopharyngeal carcinoma could serve as a potential diagnostic biomarker. Oral Oncol. 2007;43(1):82–87. doi: 10.1016/j.oraloncology.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 43.Sakai M, Hibi K, Koshikawa K, et al. Frequent promoter methylation and gene silencing of CDH13 in pancreatic cancer. Cancer Sci. 2004;95(7):588–591. doi: 10.1111/j.1349-7006.2004.tb02491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kontic M, Stojsic J, Jovanovic D, et al. Aberrant promoter methylation of CDH13 and MGMT genes is associated with clinicopathologic characteristics of primary non-small-cell lung carcinoma. Clin Lung Cancer. 2012;13(4):297–303. doi: 10.1016/j.cllc.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hug C, Wang J, Ahmad NS, Bogan JS, Tsao TS, Lodish HF. T- cadherin is a receptor for hexameric and high-molecular-weight forms of Acrp30/adiponectin. Proc Natl Acad Sci U S A. 2004;101(28):10308–10313. doi: 10.1073/pnas.0403382101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abudukadeer A, Bakry R, Goebel G, et al. Clinical Relevance of CDH1 and CDH13 DNA-Methylation in Serum of Cervical Cancer Patients. Int J Mol Sci. 2012;13(7):8353–8363. doi: 10.3390/ijms13078353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin YL, Sun G, Liu XQ, Li WP, Ma JG. Clinical significance of CDH13 promoter methylation in serum samples from patients with bladder transitional cell carcinoma. J Int Med Res. 2011;39(1):179–186. doi: 10.1177/147323001103900119. [DOI] [PubMed] [Google Scholar]
- 48.Missaoui N, Hmissa S, Trabelsi A, et al. Promoter hypermethylation of CDH13, DAPK1 and TWIST1 genes in precancerous and cancerous lesions of the uterine cervix. Pathol Res Pract. 2011;207(1):37–42. doi: 10.1016/j.prp.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Suehiro Y, Okada T, Okada T, et al. Aneuploidy predicts outcome in patients with endometrial carcinoma and is related to lack of CDH13 hypermethylation. Clin Cancer Res. 2008;14(11):3354–3361. doi: 10.1158/1078-0432.CCR-07-4609. [DOI] [PubMed] [Google Scholar]
- 50.Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, Park JY. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer. 2007;110(12):2785–2792. doi: 10.1002/cncr.23113. [DOI] [PubMed] [Google Scholar]
- 51.Hibi K, Kodera Y, Ito K, Akiyama S, Nakao A. Methylation pattern of CDH13 gene in digestive tract cancers. Br J Cancer. 2004;91(6):1139–1142. doi: 10.1038/sj.bjc.6602095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ellmann L, Joshi MB, Resink TJ, Bosserhoff AK, Kuphal S. BRN2 is a transcriptional repressor of CDH13 (T-cadherin) in melanoma cells. Lab Invest. 2012;92(12):1788–1800. doi: 10.1038/labinvest.2012.140. [DOI] [PubMed] [Google Scholar]
- 53.Kuphal S, Martyn AC, Pedley J, et al. H-cadherin expression reduces invasion of malignant melanoma. Pigment Cell Melanoma Res. 2009;22(3):296–306. doi: 10.1111/j.1755-148X.2009.00568.x. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki M, Shigematsu H, Iizasa T, et al. Exclusive mutation in epidermal growth factor receptor gene, HER-2, and KRAS, and synchronous methylation of nonsmall cell lung cancer. Cancer. 2006;106(10):2200–2207. doi: 10.1002/cncr.21853. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303(1):21–28. doi: 10.1016/j.canlet.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 56.Kubo T, Yamamoto H, Ichimura K, et al. DNA methylation in small lung adenocarcinoma with bronchioloalveolar carcinoma components. Lung Cancer. 2009;65(3):328–332. doi: 10.1016/j.lungcan.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 57.Feng Q, Hawes SE, Stern JE, et al. DNA methylation in tumor and matched normal tissues from non-small cell lung cancer patients. Cancer Epidemiol Biomarkers Prev. 2008;17(3):645–654. doi: 10.1158/1055-9965.EPI-07-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Zhang D, Zheng W, Luo J, Bai Y, Lu Z. Multiple gene methylation of nonsmall cell lung cancers evaluated with 3-dimensional microarray. Cancer. 2008;112(6):1325–1336. doi: 10.1002/cncr.23312. [DOI] [PubMed] [Google Scholar]
- 59.Hsu HS, Chen TP, Hung CH, et al. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer. 2007;110(9):2019–2026. doi: 10.1002/cncr.23001. [DOI] [PubMed] [Google Scholar]
- 60.Ulivi P, Zoli W, Calistri D, et al. p16INK4A and CDH13 hypermethylation in tumor and serum of non-small cell lung cancer patients. J Cell Physiol. 2006;206(3):611–615. doi: 10.1002/jcp.20503. [DOI] [PubMed] [Google Scholar]