Abstract
Background and Objective
Genes encoding RNA-binding proteins, including FUS and TDP43, play a central role in different neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Recently, a mutation located in the nuclear export signal (NES) of the FUS gene has been reported to cause an autosomal dominant form of familial Essential tremor.
Material and Methods
We sequenced the exons coding the NES domains of five RNA-binding proteins (TARDBP, hnRNPA2B1, hnRNPA1, TAF15 and EWSR1) that have been previously implicated in neurodegeneration in a series of 257 essential tremor (ET) cases and 376 healthy controls. We genotyped 404 additional ET subjects and 510 healthy controls to assess the frequency of the EWSR1 p.R471C substitution.
Results
We identified a rare EWSR1 p.R471C substitution, which is highly conserved, in a single subject with familial ET. The pathogenicity of this substitution remains equivocal, as DNA samples from relatives were not available and the genotyping of 404 additional ET subjects did not reveal any further carriers. No other variants were observed with significant allele frequency differences compared to controls in the NES coding regions.
Conclusions
The present study demonstrates that the NES domains of RNA-binding proteins are highly conserved. The role of the EWSR1 p.R471C substitution needs to be further evaluated in future studies.
Introduction
Essential tremor (ET) is the most common movement disorder of the elderly and is characterized by a postural or motion tremor [1]. Recently, exome sequencing in a large pedigree with an autosomal dominant form of familial ET proposed a rare mutation in the nuclear exporting signal region (NES; p.Q290X) of Fused in Sarcoma gene (FUS; OMIM*137070) as pathogenic [2]–[4]. FUS is a RNA-binding protein carrying a canonical RNA recognition motif (RRM), an NES and a putative prion-like domain (Figure 1).
Figure 1. Nuclear export signal (NES) prediction of candidate proteins based on the NetNES 1.1 prediction tool [16].
NN = neural network algorithm; HMM = hidden Markov Model algorithm; NES score = combination of NN and HMM algorithms; QGSY-rich = glutamine, glycine, serine, tyrosine rich region; G-rich = glycine rich region; RRM = RNA recognition motif; RGG = Arg-Gly-Gly rich domain; Zn = zinc finger domain; The ? denotes that the NES predicted location does not surpass the NetNES established threshold. The thin black line denotes the prion-like domain location and the thick black line represents the highest score core region according to the Alberti algorithm [20].
Several RNA-binding proteins harboring these domains have been implicated in the development of different neurodegenerative diseases. For example, mutations in TAR DNA binding protein (TARDBP; OMIM*605078) and FUS cause familial amyotrophic lateral sclerosis (ALS) [5]–[10]. Moreover, mutations in heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1; OMIM*164017) and A2/B1 (hnRNPA2B1; OMIM*600124) have been described in families with multisystem proteinopathy and ALS [11], TATA box-binding protein-associated factor 2N (TAF15; OMIM*601574) and Ewing sarcoma breakpoint region 1 (EWRS1; OMIM*133450) have been implicated both in ALS and frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTDL-U) [12]–[14].
Our previous sequencing studies of the entire coding region of FUS gene in ET did not identify any additional pathogenic variants within the NES domain [3], [4]. In the present study we have sequenced the predicted NES locations of these additional RNA-binding proteins in a cohort of ET subjects and controls, in order to identify novel mutations in those regions that may be responsible for disease.
Materials and Methods
All individuals gave their written informed consent and the study was approved by the Mayo Clinic Institutional Review Board, Jacksonville, Florida and the respective local Ethical Committees.
The Mayo Clinic ET series includes 257 patients, 151 individuals with familial ET and 106 sporadic ET subjects (Table 1). A diagnosis of ET was established according to the Consensus Statement of the Movement Disorder Society on Tremor [15] by an experienced neurologist specialized on movement disorders (JAvG, RJU and ZKW). A series of 376 healthy subjects from Mayo Clinic, Jacksonville, was sequenced to establish the minor allele frequency (MAF) of identified mutations in a control population. All participants in the study are unrelated, non-Hispanic Caucasians recruited at Mayo Clinic, Jacksonville. In order to replicate the results of our study, we further genotyped two additional ET series and matched healthy controls to establish the frequency of the EWSR1 p.R471C substitution which is located in a highly conserved region of the protein (Figure 2). We genotyped 291 Canadian ET patients and 328 matched healthy subjects and 113 Spanish ET patients and 182 matched healthy subjects (Table 1).
Table 1. Demographic data of Discovery and Replication Samples.
| Discovery Sample | Replication Samples | Whole Sample | ||||||||||||||
| USA | Replication Sample 1 (Canada) | Replication Sample 2 (Spain) | ||||||||||||||
| Fam ET | Spo ET | All ET | Cont | Fam ET | Spo ET | All ET | Cont | Fam ET | Spo ET | All ET | Cont | Fam ET | Spo ET | All ET | Cont | |
| (n = 151) | (n = 106) | (n = 257) | (n = 376) | (n = 113) | (n = 178) | (n = 291) | (n = 328) | (n = 39) | (n = 74) | (n = 113) | (n = 182) | (n = 303) | (n = 358) | (n = 661) | (n = 886) | |
| Age*(SD) | 74.28(11.66) | 75.77(12.02) | 74.90(11.83) | 67.12(12.26) | 71.76(15.08) | 74.91(12.93) | 73.65(13.92) | 73.26(12.37) | 60.00(15.10) | 65.96(14.55) | 63.88(15.01) | 68.92(10.43) | 71.54(14.20) | 73.30(13.56) | 72.48(13.89) | 69.63(12.25) |
| Age*range | 35–96 | 46–98 | 35–98 | 29–88 | 32–99 | 23–97 | 23–99 | 27–99 | 19–88 | 22–91 | 19–91 | 12–110 | 19–99 | 22–98 | 19–99 | 12–110 |
| AAO§(SD) | 47.04(19.91) | 56.10(18.73) | 50.66(19.95) | NA | 52.10(19.06) | 56.33(16.39) | 54.74(17.56) | NA | 49.77(18.83) | 55.85(20.87) | 53.86(20.43) | NA | 49.21(19.61) | 56.16(18.12) | 53.00(19.12 | NA |
| AAO§range | 5–84 | 6–88 | 5–88 | NA | 15–87 | 12–87 | 12–87 | NA | 14–78 | 10–89 | 10–89 | NA | 5–87 | 6–89 | 5–89 | NA |
| Female,% | 54.31 | 51.89 | 53.31 | 55.32 | 58.41 | 55.62 | 56.70 | 69.21 | 53.85 | 50.00 | 51.33 | 54.95 | 55.78 | 53.35 | 54.46 | 60.38 |
Fam ET = familial essential tremor; Spo ET = sporadic essential tremor; Cont = Healthy controls; y = years; AAO = age at onset; SD = standard deviation; NA = data not applicable.
*Age was not available for 57 subjects (6 familial, 17 sporadic cases and 34 controls) from Replication Sample 1 and for 11 subjects (1 familial, 3 sporadic cases and 7 controls) from Replication Sample 2.
AAO was not available for 22 subjects (10 familial and 12 sporadic cases) from the Discovery Sample, for 27 subjects (14 familial and 13 sporadic cases) from the Replication Sample 1 and for 6 subjects (4 familial and 2 sporadic cases) from Replication Sample 2.
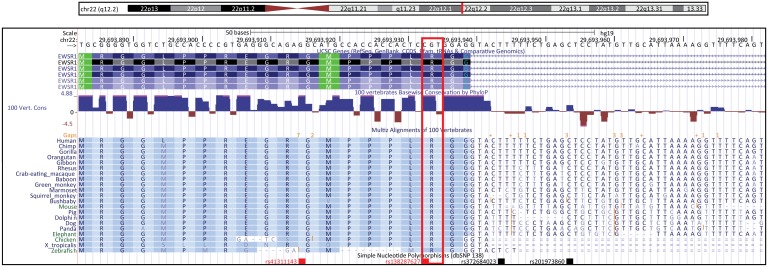
Figure 2. Conservation of EWSR1 p.R471 amino acid.
Species alignment of EWSR1 p.R471 amino acid showing its highly preservation across evolution and the exact location of p.R471C substitution (rs138287627). Picture extracted from the UCSC Genome Browser (http://genome.ucsc.edu/cgi-bin/hgGateway).
The NetNES prediction server (http://www.cbs.dtu.dk/services/NetNES/) [16] was used to identify potential NES signals in the candidate genes. This online tool works with amino acid sequences and combines both neural networks (NN) and hidden Markov models (hMM) in its prediction algorithm. The integration of both models allows us to combine the superior observed specificity of the hMM with the observed superior sensitivity of the NN [16]. Once the NES amino acid signals were identified (Figure 1), we performed bidirectional sequencing of the exons of each gene coding for these specific regions (see Table S1 for specific primers).
Variants were numbered according to standard nomenclature based on RefSeq mRNA and peptide accession numbers for each gene (TARDBP: NM_007375.3, NP_031401.1; hnRNPA1: NM_002136.2, NP_002127.1; hnRNPA2B1: NM_031243.2, NP_112533.1; TAF15: NM_139215.2, NP_631961.1; EWSR1: NM_005243.3, NP_005234.1). PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/index.shtml) and SIFT (http://sift.bii.a-star.edu.sg/) algorithms were used to assess the impact of the amino acid substitutions on the protein structure. The virtual effect of intronic variants on splicing was assessed using the Human Splicing Finder (HSF) algorithm (http://www.umd.be/SSF/) [17]. Allelic association analysis and Bonferroni correction for multiple testing were performed with PLINK v.1.07 software (http://pngu.mgh.harvard.edu/purcell/plink/) [18].
Results
Our sequencing analysis of 257 patients with ET did not identify any novel variants in the predicted NES coding regions of the candidate genes. We identified two previously described missense variants in the EWSR1 gene (rs41311143, p.G465S and rs138287627, p.R471C; Table 2), which are predicted to be probably damaging and benign by the Polyphen-2 algorithm, respectively. The minor allele frequency (MAF) of the EWSR1 p.G465S variant was the same in ET cases and controls (MAF 1.5%) whereas the p.R471C substitution was observed in only one patient with autosomal dominant familial ET. Unfortunately, DNA from the rest of the family members was not available to asses for co-segregation of this variant with disease. The genotyping of 404 additional ET patients and 510 healthy controls did not reveal any additional EWSR1 p.R471C carriers.
Table 2. Genotype counts of variants identified in predicted NES regions of candidate genes from the Discovery Sample.
| ET Patients (n = 257) | Controls (n = 376) | ||||||||||||||||
| Gene | Location | rs number | Mutation | AA change | Major | Het | Minor | MAF | Major | Het | Minor | MAF | EVS MAF | 1 kG MAF | P value a | PolyPhen-2 | SIFT score |
| EWSR1 | Int 10 | rs41309649 | c.1046–17C>G | Intronic | 250 | 7 | 0 | 0.014 | 366 | 8 | 0 | 0.010 | 0.01 | 0.01 | 0.63 | NA | NA |
| EWSR1 | Int 11 | rs3761426 | c.1164+37T>G | Intronic | 186 | 63 | 8 | 0.154 | 263 | 90 | 21 | 0.120 | 0.12 | 0.15 | 0.29 | NA | NA |
| EWSR1 | Ex 13 | rs41311143 | c.1393G>A | p.G465S | 250 | 7 | 0 | 0.014 | 358 | 11 | 0 | 0.015 | 0.01 | 0.01 | 0.85 | Probably damaging (0.96) | Tolerated (0.19) |
| EWSR1 | Ex 13 | rs138287627 | c.1411C>T | p.R471C | 256 | 1 | 0 | 0.002 | 369 | 0 | 0 | 0 | 7.68×10−5 | NA | 0.23 | Benign (0.013) | Tolerated (0.07) |
| EWSR1 | Int 13 | rs3747142 | c.1417+51A>G | Intronic | 184 | 68 | 5 | 0.152 | 258 | 103 | 8 | 0.137 | 0.12 | 0.14 | 0.65 | NA | NA |
| EWSR1 | Int 13 | NA | c.1417+68_1417+71delGATT | Intronic | 252 | 3 | 0 | 0.006 | 361 | 8 | 0 | 0.011 | NA | NA | 0.36 | NA | NA |
| hnRNPA1 | Int 1 | rs2071391 | c.16–57G>A | Intronic | 186 | 59 | 7 | 0.145 | 265 | 103 | 8 | 0.137 | NA | 0.24 | 0.51 | NA | NA |
| hnRNPA2B1 | Int 2 | NA | c.42+17T>C | Intronic | 256 | 1 | 0 | 0.002 | 375 | 0 | 0 | 0 | NA | NA | 0.23 | NA | NA |
| hnRNPA2B1 | Int 3 | rs41275982 | c.153+4T>C | Intronic | 242 | 15 | 0 | 0.029 | 355 | 19 | 1 | 0.025 | 0.03 | 0.03 | 0.90 | NA | NA |
| TAF15 | Int 12 | rs4251774 | c.1006+34A>G | Intronic | 247 | 10 | 0 | 0.019 | 373 | 3 | 0 | 0.004 | 0.01 | 0.01 | 0.01 | NA | NA |
| TARDBP | Ex 6 | rs147795017 | c.1122T>C | p.Y374Y | 255 | 1 | 0 | 0.002 | 365 | 0 | 0 | 0 | 7.7×10−5 | NA | 0.23 | NA | Tolerated (1) |
Genes are sorted in alphabetical order. ET = essential tremor; AA = Amino acid; EVS = Exome Variant Server; 1 kG = 1000 Genomes; MAF = minor allele frequency; Ex = exon; Int = intron; Het = count of heterozygous carriers; NA = not applicable; SIFT = Sorting Tolerant From Intolerant algorithm.
Uncorrected p-value calculated with PLINK case-control association analysis.
Across the candidate genes we observed a known synonymous variant in the TARDBP gene (rs147795017, p.Y374Y; Table 2) and eight intronic variants, two of which were novel (hnRNPA2B1 c.42+17T>C and EWSR1 c.1417+68_1417+71delGATT; Table 2). The hnRNPA2B1 c.42+17T>C mutation was present in a single sporadic case and was absent in a series of 376 healthy controls. The HSF algorithm predicts this mutation to change the exon 2 splicing site including 15 intronic nucleotides between exons two and three, which would cause an in-frame insertion of five amino acids (VLCQQ), but RNA from the mutation carrier was not available for examination. The EWSR1 c.1417+68_1417+71delGATT variant was present in three ET subjects, two familial ET cases and one sporadic subject (MAF = 0.006), but was also present in 8 out of 376 healthy controls (MAF = 0.011) excluding a possible role in ET pathology. The frequency of TAF15 c.1006+34A>G intronic variant was significantly different between cases and controls (p = 0.007), however this level of significance would not remain after Bonferroni correction for multiple testing.
Discussion
The involvement of mutated RNA-binding proteins in several neurodegenerative disorders suggests that this family of proteins may be relevant across heterogeneous disease phenotypes. The identification of a nonsense mutation in the NES domain of the FUS protein (p.Q290X) in a large kindred with autosomal dominant ET has raised interest in the role of these genes in this common movement disorder. In the present study we screened the predicted NES regions of other RNA-binding proteins that have been associated with neurodegeneration but did not identify any novel variants related to the ET phenotype.
FUS mutations have been proposed to be involved both in ALS [5], [7] and in ET [2]. However, the described mutations in both diseases are located in different domains of the protein. While the ALS mutations affect the RGG domain [7], the mutation causing ET results in a premature stop codon located in the NES region of the protein [2]. Additionally, functional analyses have shown that the pathogenic effect of the ET-specific FUS mutation, whose mRNA is degraded by the nonsense-mediated decay (NMD) pathway, differ from those of the ALS mutations, whose mRNAs do not undergo this kind of degradation [2]. This fact suggests that the affected domain of the protein and type of mutation plays a critical role in determining the disease phenotype developed by mutation carriers.
A recent study has shown that ∼1% of human protein-coding genes contain a potential prion-like domain and of this 1%, there is a 12-fold enrichment from proteins containing also a canonical RRM [19]. Thus, ∼11.7% of human protein-coding genes containing a candidate prion domain also harbors an RRM. The high percentage of RRM proteins among prion-like candidates suggests that human RNA-binding proteins could have a greater trend towards aggregation and therefore play a role in neurodegeneration [19]. Therefore, although we sequenced only the NES domain of these candidate genes, further investigation of other domains such as the RRM may be warranted. However, whether ET is a neurodegenerative disorder remains controversial and there is no clear evidence of protein aggregation in the disease pathology. Thus, further understanding of the genetic determinants underlying ET risk will hopefully clarify the pathogenic processes and provide a clearer picture of disease etiology.
Although we could not establish the cosegregation of the EWSR1 p.R471C substitution in the index family due to the lack of DNA samples from affected relatives, nor its presence in other ET populations, the EWSR1 p.R471C substitution is a candidate variant that needs to be further screened in future ET studies.
Supporting Information
Sequencing primers.
(DOCX)
Acknowledgments
We wish to thank the patients and families who participated in the study.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported in part by a Morris K. Udall Parkinson's Disease Research Center of Excellence (NINDS P50 #NS072187) and NINDS R01 NS078086, by the Spanish Ministry of Economy and Competitivity grants SAF2006-10126 (2006-2009) and SAF2010-22329-C02-01 (2011-2013) to PP and by the UTE project FIMA to PP and SOC and by the Canada Research Chair program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Puschmann A, Wszolek ZK (2011) Diagnosis and treatment of common forms of tremor. Semin Neurol 31: 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Merner ND, Girard SL, Catoire H, Bourassa CV, Belzil VV, et al. (2012) Exome sequencing identifies FUS mutations as a cause of essential tremor. Am J Hum Genet 91: 313–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Labbe C, Soto-Ortolaza AI, Rayaprolu S, Harriott AM, Strongosky AJ, et al. (2013) Investigating the role of FUS exonic variants in essential tremor. Parkinsonism Relat Disord 19: 755–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ortega-Cubero S, Lorenzo-Betancor O, Lorenzo E, Alonso E, Coria F, et al.. (2013) Fused in Sarcoma (FUS) gene mutations are not a frequent cause of essential tremor in Europeans. Neurobiol Aging 34: 2441 e2449–2441 e2411. [DOI] [PubMed]
- 5. Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, et al. (2009) Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, et al. (2006) Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133. [DOI] [PubMed] [Google Scholar]
- 7. Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, et al. (2009) Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208. [DOI] [PubMed] [Google Scholar]
- 8. Da Cruz S, Cleveland DW (2011) Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol 21: 904–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen-Plotkin AS, Lee VM, Trojanowski JQ (2010) TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol 6: 211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, et al. (2006) TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 351: 602–611. [DOI] [PubMed] [Google Scholar]
- 11. Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, et al. (2013) Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Couthouis J, Hart MP, Shorter J, DeJesus-Hernandez M, Erion R, et al. (2011) A yeast functional screen predicts new candidate ALS disease genes. Proc Natl Acad Sci U S A 108: 20881–20890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Neumann M, Bentmann E, Dormann D, Jawaid A, DeJesus-Hernandez M, et al. (2011) FET proteins TAF15 and EWS are selective markers that distinguish FTLD with FUS pathology from amyotrophic lateral sclerosis with FUS mutations. Brain 134: 2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ticozzi N, Vance C, Leclerc AL, Keagle P, Glass JD, et al. (2011) Mutational analysis reveals the FUS homolog TAF15 as a candidate gene for familial amyotrophic lateral sclerosis. Am J Med Genet B Neuropsychiatr Genet 156B: 285–290. [DOI] [PubMed] [Google Scholar]
- 15. Deuschl G, Bain P, Brin M (1998) Consensus statement of the Movement Disorder Society on Tremor. Ad Hoc Scientific Committee. Mov Disord 13 Suppl 3: 2–23. [DOI] [PubMed] [Google Scholar]
- 16. la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, et al. (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17: 527–536. [DOI] [PubMed] [Google Scholar]
- 17. Desmet FO, Hamroun D, Lalande M, Collod-Beroud G, Claustres M, et al. (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. King OD, Gitler AD, Shorter J (2012) The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 1462: 61–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Alberti S, Halfmann R, King O, Kapila A, Lindquist S (2009) A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 137: 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequencing primers.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.