The identification of activating mutations in the receptor tyrosine kinase c-KIT in acute myelogenous leukemia (AML) coupled with the development of potent c-KIT inhibitors provides an approach for potentially improving survival in a disease where outcomes remain unsatisfactory1. However, despite the in vitro sensitivity of c-KIT mutant cell lines to c-KIT inhibitors, primary resistance in patients remains a significant concern. Direct interactions between leukemia cells and bone marrow stromal cells in the hematopoietic microenvironment (HM) as well as soluble factors secreted by stromal cells can support leukemic cell proliferation and survival in the presence of cytotoxic and molecularly targeted therapies2,3. Accordingly, defining the mechanisms by which the microenvironment contributes to leukemia growth and therapy resistance is essential for ensuring the success of c-KIT inhibitors, and other new therapies, as they advance from the laboratory to the clinic. This is particularly relevant for c-KIT mutated AML as a phase II clinical trial (NCT01830361) is currently underway to assess the efficacy of adding the c-KIT inhibitor midostaurin (PKC412) to standard therapy in patients with newly diagnosed c-KIT or FLT3-ITD mutated t(8;21) AML. In this study, we demonstrate that ex vivo modeling of components of the hematopoietic microenvironment provides valuable insights into mechanisms of primary therapy resistance in c-KIT dependent AML.
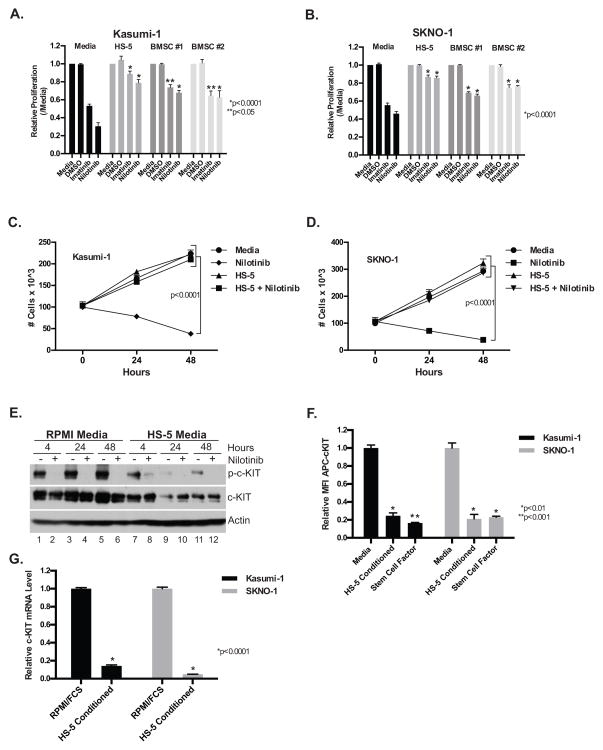
In agreement with previous studies4,5, the c-KIT inhibitors imatinib and nilotinib decreased the proliferation of the c-KIT mutant AML cell lines Kasumi-1 and SKNO-1 relative to DMSO treated controls in regular media consisting of RPMI and 20% FBS (Figures 1A & B). However, the effect of c-KIT inhibition on proliferation was significantly attenuated when the cell lines were cultured in RPMI/FBS 20% that had been conditioned for 48–72 hours with either i) the human bone marrow stromal cell line HS-5, or ii) primary human bone marrow stromal cells from two independent donors (Figures 1A & B). Consistent with the proliferation assay, nilotinib decreased the cell number of both cell lines in the regular, but not HS-5 conditioned media when cultured over a 48 hour time-course (Figure 1C & D). Furthermore, HS-5 conditioned media significantly diminished nilotinib-induced apoptosis as determined by annexin-V/7-AAD staining and flow cytometry, caspase 3/7 activity, and morphology (Supplementary Figures S1A–C). In agreement with the primary effect of conditioned media being on a reduction in apoptosis, conditioned media also showed no significant effect on cell cycle progression as analyzed by DAPI staining (Supplementary Figure S1D).
Figure 1. Bone marrow stromal cell conditioned media attenuates the therapeutic effect of c-KIT inhibitors and down-regulates c-KIT expression.
Kasumi-1 (A) and SKNO-1 (B) leukemia cells were treated with c-KIT inhibitors (1 μM) for 48 hours in either regular media, HS-5 bone marrow stromal cell line conditioned media, or conditioned media from primary bone marrow stromal cells from two independent donors (Lonza and Stem Cell Technologies) and assessed with the CellTiter-Glo viability assay (Promega). The data are normalized to the untreated controls and are the mean +/− SEM from three independent experiments. In (A), p ≥ 0.0001 for nilotinib in regular media when compared with all other conditioned medias. For imatinib, p ≥ 0.0001 for HS-5 conditioned media and p ≥ 0.05 for both primary bone marrow stromal cell conditioned medias. In (B), p ≥ 0.0001 for nilotinib and imatinib in regular media when compared with all other conditioned medias. Kasumi-1 (C) and SKNO-1 (D) cells were treated with c-KIT inhibitors (1 μM) in either regular or HS-5 conditioned media and cell number determined by trypan blue exclusion at 0, 24, and 48 hours. The data are the mean +/− SEM from three independent experiments. For both Kasumi-1 and SKNO-1 p<0.0001 when comparing nilotinib treated and untreated cells in regular media at 24 and 48 hours. (E) Western blot analysis of whole cell lysates prepared from Kasumi-1 cells grown in either regular (lanes 1–6) or HS-5 conditioned media (lanes 7–12) in the presence or absence of nilotinib. Blots were probed with antibodies specific to c-KIT (Cell Signaling Technology), phospho-c-KIT (Tyr719) (Cell Signaling Technology), and β-actin (Sigma). This is a representative blot from two independent experiments. (F) Cell surface expression of c-KIT in either regular media, HS-5 conditioned media, or regular media supplemented with SCF 20 ng/mL was determined by flow cytometry with an APC-conjugated c-KIT antibody (eBiosciences). The data are normalized to the regular media control and are the mean +/− SEM from three independent experiments. (G) Quantitative RT-PCR analysis of total RNA isolated from Kasumi-1 and SKNO-1 cells after culturing for 48 hours in either regular or HS-5 conditioned media. c-KIT mRNA levels were normalized to GAPDH. Reactions were performed in triplicate and the data are the mean +/− SEM from three independent experiments.
To confirm that the effects of the drugs on apoptosis correlated with c-KIT inhibition, western blots for phospho-c-KIT and total c-KIT were performed on Kasumi-1 cells that had been treated with nilotinib for 4, 24, or 48 hours in either regular or HS-5 conditioned media (Figure 1E). Nilotinib treatment in regular media diminished c-KIT phosphorylation relative to total c-KIT protein levels at all time-points, consistent with a rapid and sustained c-KIT inhibition. In contrast, while nilotinib still inhibited phosphorylation of c-KIT in HS-5 conditioned media, the level of total c-KIT protein was also significantly diminished relative to regular media in the absence of nilotinib (Figure 1E). In agreement, the level of c-KIT cell surface expression, as assessed with an APC-conjugated c-KIT antibody and flow cytometry, was reduced in both the Kasumi-1 and SKNO-1 cell lines in HS-5 conditioned media (Figure 1F). The presence of stem cell factor (SCF), the ligand for c-KIT in HS-5 conditioned media, likely contributes to c-KIT down-regulation as c-KIT is rapidly internalized and degraded following ligand binding6 (Figure 1F). However, in addition to the effects of HS-5 conditioned media on c-KIT protein stability, Kasumi-1 and SKNO-1 cells cultured in HS-5 conditioned media also exhibited significantly decreased c-KIT mRNA levels (Figure 1G). Together, these data suggest that the cause of c-KIT down-regulation in HS-5 conditioned media is likely multi-factorial and occurs at both a transcriptional and post-translational level.
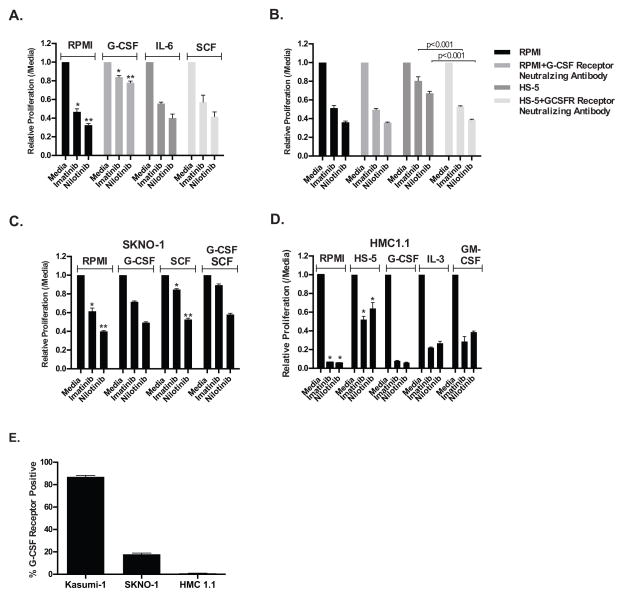
In an attempt to identify the soluble factor(s) mediating rescue of c-KIT inhibition we next examined a variety of cytokines secreted by HS-5 cells that are known to stimulate hematopoietic cells. We initially tested the effect of G-CSF, IL-6, and SCF on Kasumi-1 proliferation in the presence of c-KIT inhibitors. Figure 2A shows that G-CSF alone, but not IL-6 or SCF, could mimic the effects seen with HS-5 conditioned media. Furthermore, the concentration of G-CSF in the minimal amount of HS-5 conditioned media that affords rescue (240 pg/mL G-CSF) correlates well with the results of a G-CSF titration in the presence of nilotinib (EC50=140 pg/mL; Supplementary Figures S2A–C). Although this G-CSF concentration is greater than the average level of 25 pg/mL measured in healthy, normal adults7 it is well below levels measured in patients immediately following myeloablative therapy (699 pg/mL)8 or in patients with documented infections (731.8 pg/mL)9 and thus may be clinically relevant. Further illustrating this dynamic regulation of cytokine production, we also found that treatment of the low cytokine secreting HS-27A human bone marrow stromal cell line with non-toxic doses of cytarabine and daunorubicin stimulated the release of sufficient G-CSF to partially rescue the effects of c-KIT inhibition on Kasumi-1 cells (Supplementary Figure S3C, D & E). Finally, a neutralizing antibody for the G-CSF receptor partially restored sensitivity of Kasumi-1 cells to c- KIT inhibitors in the presence of HS-5 conditioned media (Figure 2B). The lack of complete rescue may be due to incomplete neutralization of the receptor by the antibody or the ability of soluble factors other than G-CSF to provide partial rescue.
Figure 2. Cytokines attenuate the therapeutic effect of c-KIT inhibitors. (A).
The proliferation of Kasumi-1 leukemia cells was assessed with the CellTiter-Glo viability assay following treatment with the specified c-KIT inhibitor (1 μM) for 48 hours in regular media supplemented with 10 ng/mL G-CSF, SCF, or IL-6. *=p<0.05 (imatinib) and **=p<0.01 (nilotinib) when comparing regular and G-CSF supplemented media. (B) The proliferation of Kasumi-1 cells was assessed with the CellTiter-Glo viability assay following treatment with the specified c-KIT inhibitor (1 μM) for 48 hours in either regular media, regular media supplemented with a G-CSF receptor neutralizing antibody (1 μg/mL; R&D Systems), 0.05% HS-5 conditioned media, or 0.05% HS-5 conditioned media supplemented with a G-CSF receptor neutralizing antibody. The data are normalized to the untreated controls and are the mean +/− SEM from three independent experiments. For both imatinib and nilotinib p ≥ 0.001 when comparing HS-5 conditioned media against all other conditions. The SKNO-1 (C) and HMC1.1 (D) c-KIT dependent leukemia cells lines were treated with a c-KIT inhibitor (1 μM for SKNO-1 and 200 nM for HMC1.1) for 48 hours in regular media supplemented with 10 ng/mL G-CSF, SCF, GM-CSF, or IL-3 and assessed with the CellTiter-Glo viability assay. The data are normalized to the untreated controls and are the mean +/− SEM from three independent experiments. In (C), *=p ≥ 0.001 (imatinib) and **=p<0.05 (nilotinib) when comparing regular media and SCF supplemented media. In (D), *=p ≥ 0.0001 for both imatinib and nilotinib when comparing regular and HS-5 conditioned media. (E) Kasumi-1, SKNO-1, and HMC1.1 cells were stained with a PE conjugated anti-G-CSF receptor antibody (Biolegend) and assessed by flow cytometry. The data are the mean +/− SEM from three independent experiments.
Having demonstrated a clear role for G-CSF in modulating the response of Kasumi-1 cells to c- KIT inhibition, we next examined the effect of G-CSF and other cytokines on the sensitivity of other c-KIT mutant leukemia cell lines to c-KIT inhibition. Surprisingly, SCF provided a more substantial rescue of the effects of c-KIT inhibition than G-CSF for the SKNO-1 cell line (Figure 2C). Similarly, for HMC1.110, a mast cell leukemia cell line containing an activating c-KIT mutation, G-CSF did not rescue the effect of c-KIT inhibition, whereas IL-3 and GM-CSF both provided modest rescue (Figure 2D). To investigate whether the ability of G-CSF to rescue proliferation was correlated with receptor expression, the level of the G-CSF receptor expression on the leukemia cell surface was determined by flow cytometry. As seen in Figure 2E, over 80% of Kasumi-1 cells exhibited cell surface expression of the G-CSF receptor compared to <20% of SKNO-1 and HMC1.1 cells. In agreement, it has been previously shown that Kasumi-1 cells express significantly more G-CSF receptor mRNA than SKNO-1 cells11. These data suggest that the cell surface expression of the G-CSF receptor is correlated with the capacity of the cytokine to reverse sensitivity to c-KIT inhibition in Kasumi-1 cells and its failure to do so in SKNO-1 and HMC1.1 cells. Furthermore, the data suggest that directly targeting individual cytokines to restore c-KIT inhibitor sensitivity, as demonstrated in Figure 2B, may not be a viable therapeutic option unless the cytokine sensitivity is profiled for each c-KIT dependent leukemia.
These results also have additional implications for understanding leukemia development, maintenance, and therapy. In fitting with the two-step model of AML development12, it is hypothesized that the t(8;21) (AML1/ETO) or inv(16) translocation impairs hematopoietic differentiation and cooperates with an activating c-KIT mutation to confer a proliferative advantage and full leukemic transformation13. Murine models have elegantly supported this model as activating c-KIT mutations cooperate with the full length AML1/ETO translocation to induce leukemia in the mouse14. However, the requirements for leukemia maintenance may be different than those required for leukemia development. Consistent with this, when c-KIT mutant leukemia cell lines are cultured with stromal conditioned media, c-KIT signaling appears to be dispensable for leukemia cell maintenance as i) the c-KIT leukemia cell lines are relatively resistant to c-KIT inhibitors in stromal conditioned media despite adequate c-KIT inhibition as seen by western blot (Figure 1E), and ii) c-KIT dependent leukemia cells proliferate in stromal conditioned media despite extensive down-regulation of c-KIT (Figure 1E, F, & G). These data suggest that mutant c-KIT signaling may not be required for leukemia maintenance in the presence of leukemia stem cell niche-derived pro-survival cytokines. This has implications for both understanding leukemia development and raises concerns about the efficacy of targeting mutant c-KIT in AML, although when combined with conventional cytotoxic therapy there may be an additive or synergistic effect that would render it a more effective therapeutic strategy.
Supplementary Material
Acknowledgments
We thank Drs. Stegmaier and Butterfield for kindly providing the SKNO-1 and HMC-1.1 cells lines, respectively. We also thank Maria Suarez and Natasha Rossi for administrative assistance and members of the Williams laboratory for helpful discussions and suggestions. PMG is supported by NIH Grant 5K08CA154782-02. DAW is supported by 5R01DK062757, 5R01CA113969-08, and the William Lawrence & Blanche Hughes Foundation.
Supplementary information is available at Leukemia’s website.
Footnotes
The authors declare no competing financial interests.
References
- 1.Pollard JA, Alonzo TA, Gerbing RB, Ho PA, Zeng R, Ravindranath Y, et al. Prevalence and prognostic significance of KIT mutations in pediatric patients with core binding factor AML enrolled on serial pediatric cooperative trials for de novo AML. Blood. 2010;115:2372–9. doi: 10.1182/blood-2009-09-241075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tabe Y, Konopleva M. Advances in understanding the leukaemia microenvironment. Br J Haematol. 2014;164:767–78. doi: 10.1111/bjh.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weisberg E, Barrett R, Liu Q, Stone R, Gray N, Griffin JD. FLT3 inhibition and mechanisms of drug resistance in mutant FLT3-positive AML. Drug Resist Updat. 2009;12:81–9. doi: 10.1016/j.drup.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beghini A, Bellini M, Magnani I, Colapietro P, Cairoli R, Morra E, et al. STI 571 inhibition effect on KITAsn822Lys-mediated signal transduction cascade. Exp Hematol. 2005;33:682–8. doi: 10.1016/j.exphem.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Mpakou VE, Kontsioti F, Papageorgiou S, Spathis A, Kottaridi C, Girkas K, et al. Dasatinib inhibits proliferation and induces apoptosis in the KASUMI-1 cell line bearing the t(8;21)(q22;q22) and the N822K c-kit mutation. Leuk Res. 2013;37:175–82. doi: 10.1016/j.leukres.2012.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Lennartsson J, Rönnstrand L. Stem Cell Factor Receptor/c-Kit: From Basic Science to Clinical Implications. Physiol Rev. 2012;92:1619–49. doi: 10.1152/physrev.00046.2011. [DOI] [PubMed] [Google Scholar]
- 7.Watari K, Asano S, Shirafuji N, Kodo H, Ozawa K, Takaku F, et al. Serum granulocyte colony-stimulating factor levels in healthy volunteers and patients with various disorders as estimated by enzyme immunoassay. Blood. 1989;73:117–22. [PubMed] [Google Scholar]
- 8.Cairo MS, Suen Y, Sender L, Gillan ER, Ho W, Plunkett JM, et al. Circulating granulocyte colony-stimulating factor (G-CSF) levels after allogeneic and autologous bone marrow transplantation: endogenous G-CSF production correlates with myeloid engraftment. Blood. 1992;79:1869–73. [PubMed] [Google Scholar]
- 9.Kawakami M, Tsutsumi H, Kumakawa T, Abe H, Hirai M, Kurosawa S, et al. Levels of serum granulocyte colony-stimulating factor in patients with infections. Blood. 1990;76:1962–4. [PubMed] [Google Scholar]
- 10.Butterfield JH, Weiler DA, Hunt LW, Wynn SR, Roche PC. Purification of tryptase from a human mast cell line. J Leukoc Biol. 1990;47:409–19. doi: 10.1002/jlb.47.5.409. [DOI] [PubMed] [Google Scholar]
- 11.Watari K, Ozawa K, Tajika K, Tojo A, Tani K, Kamachi S, et al. Production of human granulocyte colony stimulating factor by various kinds of stromal cells in vitro detected by enzyme immunoassay and in situ hybridization. Stem Cells. 1994;12:416–23. doi: 10.1002/stem.5530120409. [DOI] [PubMed] [Google Scholar]
- 12.Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–13. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y-Y, Zhou G-B, Yin T, Chen B, Shi J-Y, Liang W-X, et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: implication in stepwise leukemogenesis and response to Gleevec. Proc Natl Acad Sci USA. 2005;102:1104–9. doi: 10.1073/pnas.0408831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y-Y, Zhao L-J, Wu C-F, Liu P, Shi L, Liang Y, et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci USA. 2011;108:2450–5. doi: 10.1073/pnas.1019625108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.