Abstract
Purpose
To provide a method for the optimal selection of sampling points for myocardial T1 mapping, and to evaluate how this selection affects the precision.
Theory
The Cramér-Rao lower bound on the variance of the unbiased estimator was derived for the sampling of the longitudinal magnetization curve, as a function of T1, signal-to-noise ratio (SNR) and noise mean. The bound was then minimized numerically over a search space of possible sampling points to find the optimal selection of sampling points.
Methods
Numerical simulations were carried out for a saturation recovery based T1 mapping sequence, comparing the proposed point selection method to a uniform distribution of sampling points along the recovery curve for various T1 ranges of interest, as well as number of sampling points. Phantom imaging was performed to replicate the scenarios in numerical simulations. Invivo imaging for myocardial T1 mapping was also performed in healthy subjects.
Results
Numerical simulations show that the precision can be improved by 13-25% by selecting the sampling points according to the target T1 values of interest. Results of the phantom imaging were not significantly different than the theoretical predictions for different sampling strategies, SNR and number of sampling points. In-vivo imaging showed precision can be improved in myocardial T1 mapping by using the proposed point selection method as predicted by theory.
Conclusion
The framework presented can be used to select the sampling points in order to improve the precision without penalties on accuracy or scan time.
Keywords: Cramér-Rao bound, precision, sampling points, parameter estimation, Fisher information, myocardial T1 mapping, tissue characterization
Introduction
Quantitative mapping of various MR parameters, such as longitudinal (T1) or transverse (T2) relaxation times may provide diagnostic information beyond conventional contrast-weighted images (1). These quantitative techniques have found a range of applications in neurological (2,3), oncologic (4,5) and cardiac (6,7) applications. In particular, recently, myocardial T1 mapping (8-12) has been shown to provide a technique for assessment of interstitial diffuse fibrosis (13). Furthermore, if myocardial T1 maps are acquired both prior to and following contrast injection, they can further be used to measure the extracellular volume fraction (14), which has shown utility for detection of diffuse myocardial fibrosis (6).
Quantitative mapping approaches rely on acquiring multiple images of the same anatomy with different contrast weightings, achieved by varying one or more imaging parameter(s). These parameters may include flip angles (15), echo times (16) and/or inversion/saturation times (17). Then a closed-form expression, which characterizes how the magnetization evolves, as a function of the unknown parameters of interest and the known parameter(s) that is being varied, is voxel-wise fitted to these series of images. Clearly, the number of images in this series has to be greater than or equal to the number of unknown parameters of interest. Thus, multiple images are acquired in the series with different contrasts, which correspond to different sampling points on the magnetization curve that is parameterized by the unknown quantities.
To achieve a sufficient level of precision in the estimation, a high number of images are acquired with different choices of sampling points (18), typically with a uniform distribution along the curve (10,19), resulting in a long acquisition time. However, there is a degree of freedom on the selection of the location and the number of the sampling points on the parametric curve, which is controlled by the sequence designer. Furthermore, in myocardial parameter mapping, the respiratory motion has to be compensated for. In myocardial T1 mapping, this is commonly achieved by performing breath-holds, typically ranging from 11 to 17 heartbeats (8,10,11,20). Thus it is critical to improve the precision of the sequence with this constraint on the total scan time.
The precision of the T1 estimation process can be characterized by the Cramér-Rao Bound, which (CRB) provides a lower bound on the variance of an unbiased estimator of a deterministic parameter (21). Furthermore it characterizes when an estimator that achieves this lower bound exists. CRB has been used in a number of MRI applications, including diffusion kurtosis imaging (22,23) and parameter mapping (24-26). By considering a parametric model involving the gradient vector directions, as well as the diffusion and kurtosis tensors, an estimation theoretic approach was used to optimize the direction and b-value of the diffusion weighting gradients (23). It was shown that the optimal selection was different than the conventionally used set of standard gradient settings, and this selection lead to improvements in precision. In parameter mapping, an approach for determining experimental parameters were proposed in (24) for rapid simultaneous T1 and T2 measurement. This CRB-based technique was used in (25) for characterizing optimal sampling times to determine the T2 relaxation time. The optimal scheme yielded a distribution concentrated at certain points related to the T2 value, unlike the traditional uniform distribution utilized in other studies. Most similar to our current work is that of (26), which derived the CRB for T1 mapping. This was then used to characterize the selection strategies for certain ordered sampling schemes, which lead to tractable analytical expressions.
Because the search space in (26) was limited to these ordered sampling schemes, the resulting sampling schemes were not similar to that of (25) in T2 mapping, which exhibited concentrations at certain areas of the parametric curve. Furthermore, only numerical simulations were used in (26), with no validation for phantom or in-vivo data.
In this work, we sought to use an estimation theoretic framework for bounding the precision of a given myocardial T1 mapping sequence in the presence of noise. We extend the bounding approach of (26) to accommodate a wide range of T1 values of interest, by providing a Bayesian lower bound on the precision of the T1 estimator, defined as the standard deviation of error for a given noise level. We then propose to determine the optimal selection of sampling points by numerically minimizing this bound. To test the theory in practice, we choose a saturation-based T1 mapping sequence, and minimize the corresponding closed form expression for the lower bound over a large search space of sampling points to select an improved distribution of sampling points for a given range of T1 values of interest. Numerical experiments, phantom and in-vivo imaging are performed to evaluate the proposed sampling point strategy versus a uniform distribution of sampling points along the curve.
Theory
Cramér-Rao Lower Bound for Parameter Mapping
For completeness, we describe the signal model and derive the CRB for T1 mapping, which is similar to the derivation of (26). In myocardial T1 mapping, at each sampled image voxel, we observe
| [1] |
for k ∈ {1, ..., K} representing images with different contrast weightings along the longitudinal magnetization curve. nk is measurement noise. Furthermore,
| [2] |
is the magnetization at xk on the magnetization curve, parameterized by a, the signal-to-noise ratio of the image with fully recovered magnetization (with appropriate normalization for the noise variable nk); b, which captures the saturation/inversion efficiency, the effects of imaging pulses on the magnetization recovery (10) and the mean of noise in the magnitude images (27), and T1, the longitudinal relaxation time.
For the K observations, we let Y = {y1, y2, ..., yK}. Based on the noise distribution, the probability density function p(Y|a,b,T1) can be determined. This in turn can be used to find the Fisher information matrix (21), whose info (i,j)th entry is given by:
| [3] |
where, for ease of notation, θ1 = a, θ2 = b, θ3 = T1. Then the CRB states, if p(Y|a,b,T1) satisfies certain regularity conditions (21), the estimator of T1, denoted by , has variance bounded by
| [4] |
Since the Rician noise in the magnitude images is well-approximated as Gaussian noise with sufficient baseline SNR (27), and since the maximum-likelihood estimate for Gaussian noise corresponds to the least-squares estimator that is commonly used in the context of MR parameter estimation (2,8), we restrict ourselves to a Gaussian noise model. Without loss of generality, we consider unit-variance zero-mean Gaussian noise, since the non-zero mean can be captured using the parameters, a and b in the T1 mapping model. In this setting,
| [5] |
where C is a constant. We also note that the maximum-likelihood estimator is unbiased and attains this lower bound for this log-likelihood function (21).
Optimization of Sampling Point Selection
The model described by Equations [1] and [5] leads to:
| [6] |
Thus the Fisher information matrix for the T1 mapping model of [6] is
| [7] |
The goal of the optimization procedure is to minimize the variance of the estimator of T1. From Equation [4], , where
| [8] |
We minimize as a surrogate for minimizing . While this is a closed form expression in terms of {xk} given the values of [a, b, T1] that we are interested in, the analytical minimization of the objective function is not tractable. Thus we propose to solve
| [9] |
numerically over a grid of values for {xk} for the desired values of a, b, T1. Furthermore, in Appendix A, we show that
| [10] |
Thus the values of a, b only scales the lower bound itself, and the minimization over the grid of values for {xk} is independent of a, b, and only depends on the T1 value of interest.
Methods
All imaging was performed on a 1.5T Philips Achieva (Philips Healthcare, Best, The Netherlands) system. Phantom imaging was performed with a body coil, and in-vivo imaging was performed with a 32-channel cardiac phased-array receiver coil. For this HIPAA-compliant study, the imaging protocol was approved by our institutional review board, and written informed consent was obtained from all participants.
Saturation and inversion recovery are the most commonly employed techniques for sampling of the longitudinal magnetization recovery. Currently, there are no myocardial T1 mapping sequences that sample an unperturbed inversion recovery (IR) curve in a single breath-hold, though there are Look-Locker based variants (8,9). Hence, we concentrate on the saturation recovery (SR) curve. We choose a SR-based T1 mapping sequence, SASHA (10), for validation in this study. This sequence is electrocardiogram-triggered, and applies a saturation pulse at every heart beat, followed by imaging after a recovery time of xk. The first heart beat is acquired without magnetization preparation, corresponding to an infinite SR time. Since different SR times can be selected for each heart beat in this sequence, this increases the degrees of freedom for the selection of the optimal sampling points.
Numerical Simulations
Simulation Setup
We note from Equation [8], depends directly on 1/a2, i.e. the lower bound on the variance of the estimator scales inversely with the square of the SNR. Thus the choice of a does not affect the evaluation of the function, only changing the scaling. Hence a = 40 was arbitrarily chosen as the baseline SNR for the simulations. b = 0.9 was used for the simulations, selected from the typical experimental range of b values for SASHA between 0.9 and 1.1 (10). A trigger delay of 780 ms at 60 bpm and an acquisition window of 190 ms were used. Including the duration of the saturation pulse and the duration of the read-outs to the k-space center (with the assumption of a linear profile order), the allowable saturation times ranged between Tmin = 140 ms and Tmax = 760 ms.
Experiments
Two sets of experiments were performed. In Numerical Experiment A, the bound was evaluated for various T1 values of interest for K = 11 sampling points: i) 1250 ms (myocardium pre-contrast), ii) 450 ms (myocardium post-contrast), iii) 950 – 1250 ms (pre-contrast T1 range), iv) 400 – 600 ms (post-contrast T1 range), v) 450 & 1250 ms (myocardium pre- and post-contrast). In Numerical Experiment B, the bound was evaluated for various K values of {5, 7, 9, 11, 13, 15} for T1 values of interest from 950 to 1250 ms.
Additional experiments, the effects of changing Tmax, and allowing for multiple sampling of the point at infinity were performed, and are included in Appendix B.
Numerical Optimization
The K sampling points {xk} were selected numerically as follows: The search space was generated as all the possible saturation times from Tmin to Tmax with a step size of 10 ms, and a saturation time at infinity, corresponding to sampling without any magnetization preparation. Every possible ordered samplings of length K, allowing repetitions, was generated from the search space. The point at infinity was only allowed to be included once in any sampling, since any imaging pulse performed during acquisition disturbs the magnetization at this point. Thus, to sample the same point at infinity requires multiple rest periods for the longitudinal magnetization to re-grow, which decreases efficiency, especially considering the constraints on scan duration with the breath-hold acquisition. For every ordered sampling, was evaluated for the given [a, b, T1].
For the T1 values in a continuous range, a Bayesian CRB variant was used (28), where
| [11] |
was evaluated instead. The integral was performed numerically with a step size of 5 ms, while making sure the (nearly)-uniform distribution on T1 was 0 at the boundary points as required in (28). For the bi-modal T1 value of optimization (v), the procedure was evaluated with both equal weights and weights proportional to 1/T12 for the variances associated with the two T1 values.
The CRB on precision, defined as the square root of and denoted by CRBprec, was evaluated for the points selected by the optimization procedure, and compared to a uniform distribution of sampling points, where K – 1 sampling points are uniformly spread in the range Tmin to Tmax, and a point at infinity as in (10).
Phantom Imaging
Imaging Setup
To characterize the effect of the choice of sampling points on the precision of the T1 estimates, phantom imaging was performed using 14 NiCl2 doped agarose vials (29), whose T1 and T2 values spanned the ranges of values found in the blood and myocardium pre- and post- contrast. A single-shot steady-state free precession (SSFP) sequence with the following parameters was used: 2D single-slice, FOV = 210×170 mm2, in-plane resolution = 1.9×2.5 mm2, slice-thickness = 8 mm, TR/TE = 2.7 ms/1.35 ms, flip angle = 70°, 10 ramp-up pulses, acquisition window = 190 ms, linear k-space ordering. All scans were repeated 5 times to average out random variations.
Experiments
The first set of experiments compared different sets of sampling points for K = 11. The sets of sampling points tested were the ones chosen from the numerical simulations for T1 values of interest varying from 950 to 1250 ms, from 400 to 600 ms, and 450 and 1250 ms. For comparison, acquisitions with saturation times uniformly distributed in the range Tmin to Tmax, plus a point at infinity (referred to as “uniform”) were performed. Each scan was acquired with number of signal averages (NSA) = 5 for sufficient baseline SNR.
The second set of experiments evaluated the precision of the uniform and proposed point selection techniques for T1 values of interest varying from 950 to 1250 ms, for different number of sampling points, K = {5, 7, 9, 11, 13, 15}. NSA = 5 was used for these scans as well.
The third set of experiments was performed to validate the inverse linear dependence of precision to baseline SNR. The sequence was designed to last for K = 11 heartbeats, and the scans were acquired with NSA = {1, 3, 5, 10}. The uniform point selection and the proposed point selection for T1 values of interest varying from 950 to 1250 ms were utilized.
In-vivo Imaging
Myocardial T1 maps were acquired in 5 healthy adult subjects (4 women, 23.4 ± 3.3 years) without a contrast injection. Images were acquired in the short-axis of the heart. The SASHA sequence was used with a single-shot acquisition and an SSFP readout with the following parameters: 2D single-slice, FOV = 300×300 mm2, in-plane resolution = 1.9×2.5 mm2, slice-thickness = 8 mm, TR/TE = 3.1 ms/1.55 ms, flip angle = 70°, 10 ramp-up pulses, SENSE rate = 2, acquisition window = 190 ms, linear k-space ordering. The sequence duration was 11 heartbeats, and it was performed during breath-hold. Acquisitions were performed using a uniformly distributed point selection and the proposed selection of points for T1 values between 950 and 1250 ms. For each point selection strategy, the acquisition was repeated 5 times to average out random variations.
Image Analysis
T1 estimation was performed offline using MATLAB (v7.6, MathWorks, Natick, MA) using the 3-point model (10) of Equation [2]. Any point acquired without a saturation preparation was assumed to be sampled at an infinite (∞) saturation time. Multiple points acquired at the same point on the magnetization recovery curve were used individually for fitting, instead of averaging prior to fitting.
For T1 measurements, a region-of-interest (ROI) analysis was performed for both phantom and in-vivo imaging. The mean value and standard deviation in the ROI was recorded for each acquisition. The estimated T1 value, T1est, is reported as an average ± standard deviation of the mean values in the ROI (over the 5 acquisitions for each sampling strategy), which is a surrogate for accuracy and the inter-scan reproducibility. Under the assumption that the T1 values are homogenous in the small ROI (11), the precision, prec(T1est), is reported as the average ± standard deviation of the spatial standard deviation of the T1 values in the ROI (over 5 acquisitions for each sampling strategy).
For phantom imaging, circular ROIs were drawn across each of the 14 vials, starting from the center of the vial and containing ~300 voxels. The ratios of the mean prec(T1est) for the two sampling strategies were then compared to the ratios predicted by the theoretical model for different K and T1 values in the union of the ranges of optimization using paired t-test. For the SNR experiment, the correlation between prec(T1est) for different NSAs were compared to 1/NSA2 using Pearson's linear correlation coefficient for both sampling strategies. For in-vivo imaging, ROIs were manually drawn, by two independent experienced readers, in the left ventricle (LV) myocardium and the blood pool. T1est and prec(T1est) were calculated for both strategies.
Results
Numerical Simulations
Point selection for different T1 values of interest
The 11 sampling points selected by the proposed method for the first 4 different cases of T1 values of interest, and the associated CRBprec are depicted in Table 1 for SNR = 40, along with the corresponding CRBprec for a uniform distribution of sampling points. The proposed choice of sampling points yields a tri-modal distribution: 1 point at ∞, 3-4 points at Tmin and 6-7 points close to the T1 value(s) of interest. The proposed selection leads to a reduction in CRBprec in all cases (13%, 26%, 24%, 12.1% reduction for T1 values of interest of 450 ms, 1250 ms, 950 – 1250 ms, 400 – 600 ms respectively) compared to the uniform selection.
Table 1.
Results of the proposed optimization procedure for sampling point selection for various T1 values of interest and 11 sampling points. The saturation recovery times are chosen between Tmin = 140 ms and Tmax = 760 ms (and possibly one point at ∞, representing an acquisition with no preparation). The associated Cramér-Rao lower bound (CRB) on the standard deviation (std) of the T1 estimator (CRBprec) when using the proposed strategy and a strategy selecting uniformly separated points are also depicted. The results indicate that the optimization procedure favors selection of multiple points at the same locations, with multiple points at Tmin, as well as multiple points close to the T1 value of interest (or the closest to it if these values are greater than Tmax). In all cases, the optimized procedure has a lower CRB associated with the variance and standard deviation of the T1 estimator, when compared to a uniform distribution of sampling points.
| T1 values of interest (ms) | Sampling Point Selection Strategy | Saturation Recovery Sampling Points (ms) | CRBprec (ms) |
|---|---|---|---|
| 1250 | proposed | (140, 140, 140, 140, 760, 760, 760, 760, 760, 760, ∞) | 82.6 |
| uniformly distributed | (140, 209, 278, 347, 416, 485, 554, 623, 692, 760, ∞) | 111.3 | |
| 450 | proposed | (140, 140, 140, 480, 480, 480, 480, 480, 480, 480, ∞) | 33.9 |
| uniformly distributed | (140, 209, 278, 347, 416, 485, 554, 623, 692, 760, ∞) | 39.1 | |
| 950 - 1250 | proposed | (140, 140, 140, 140, 760, 760, 760, 760, 760, 760, ∞) | 71.8 |
| uniformly distributed | (140, 209, 278, 347, 416, 485, 554, 623, 692, 760, ∞) | 94.0 | |
| 400 - 600 | proposed | (140, 140, 140, 510, 510, 510, 510, 510, 510, 510, ∞) | 36.9 |
| uniformly distributed | (140, 209, 278, 347, 416, 485, 554, 623, 692, 760, ∞) | 42.0 |
For the fifth case of T1 values, the optimization with equal weights yielded 4 points at 140 ms, 6 at 760 ms and 1 at ∞. In this case, CRBprec associated with the T1 = 450 ms estimation was 42.3 ms, whereas CRBprec for T1 = 1250 ms was of 82.6 ms, where the former is worse than for uniform selection. Note that for equal weights, the variance of the higher T1 value dominates the optimization procedure, yielding the same choice of points as for just T1 = 1250 ms. When weights of 1/T12 are used to capture the percentage variation in estimation, the optimal selection becomes 4 points at 140 ms, 4 at 570 ms, 2 at 760 ms and 1 at ∞, resulting in reductions in CRBprec of 5% and 18% (for T1 of 450 and 1250 ms respectively) compared to uniform selection. This latter selection of points is used in subsequent imaging for these T1 values of interest.
Effect of changing the number of sampling points
Table 2 depicts the results of the proposed point selection strategy and CRBprec for both point selection strategies for T1 values of interest of 950-1250 ms for various numbers of sampling points, K. The trend for the optimization strategy is to select one point at ∞, floor(K/2)-1 or floor(K/2) points at Tmin, and floor(K/2) or floor(K/2)+1 points at Tmax (since T1 > Tmax). The reduction in CRBprec compared to uniform sampling is 18.7%, 24.4%, 25.6%, 25.8%, 25.7%, 24.9% for K = 5, 7, 9, 11, 13, 15 points respectively.
Table 2.
The variation of the selected points for different number of sampling points, K, as well as the associated CRB on the standard deviation of the T1 estimator (CRBprec) when using the proposed strategy and a strategy selecting uniformly separated points. The range of T1 values of interest was from 950 ms to 1250 ms, with Tmin = 140 ms and Tmax = 760 ms. The proposed selection procedure again favors a bi-modal distribution of points among Tmin and Tmax, which results in an improvement over the CRB for variance and std of the T1 estimator with respect to a uniform distribution of selection points for all K.
| # sampling points, K | Point Selection Strategy | Saturation Recovery Sampling Points (ms) | CRBprec (ms) |
|---|---|---|---|
| 5 | proposed | (140, 140, 760, 760, ∞) | 117.7 |
| uniform | (140, 347, 554, 760, ∞) | 144.8 | |
| 7 | proposed | (140, 140, 760, 760, 760, 760, ∞) | 98.6 |
| uniform | (140, 264, 388, 512, 636, 760, ∞) | 130.4 | |
| 9 | proposed | (140, 140, 140, 760, 760, 760, 760, 760, ∞) | 88.8 |
| uniform | (140, 229, 318, 407, 496, 585, 674, 760, ∞) | 119.4 | |
| 11 | proposed | (140, 140, 140, 140, 760, 760, 760, 760, 760, 760, ∞) | 82.6 |
| uniform | (140, 209, 278, 347, 416, 485, 554, 623, 692, 760, ∞) | 111.3 | |
| 13 | proposed | (140, 140, 140, 140, 140, 760, 760, 760, 760, 760, 760, 760, ∞) | 78.2 |
| uniform | (140, 196, 252, 308, 364, 420, 476, 532, 588, 644, 700, 760, ∞) | 105.2 | |
| 15 | proposed | (140, 140, 140, 140, 140, 760, 760, 760, 760, 760, 760, 760, 760, 760, ∞) | 74.8 |
| uniform | (140, 188, 236, 284, 332, 380, 428, 476, 524, 572, 620, 668, 716, 760, ∞) | 99.6 |
Phantom Imaging
Precision for different sampling point selections
Table 3 depicts the results of the phantom measurements over all 14 vials with different T1 values for various sampling strategies with NSA = 5 for each acquisition and K = 11. The estimated T1 values for different sampling strategies for each vial are in good agreement with theory, even though there are variations between the 5 acquisitions due to noise (reflected as the standard deviation in the T1est). The ratio of prec(T1est) for each proposed sampling strategy and that of the uniform sampling strategy is reported as “precision (prec) with respect to (wrt) uniform.” The points optimized for T1 values from 950 to 1250 ms show consistent improvement (as a ratio < 1) in this range with respect to the uniform strategy. Similarly, the points optimized for T1 values from 400 to 600 ms show consistent improvement for smaller T1 values < 1150 ms. The points optimized for two T1 values of 450 and 1250 ms show improvement in standard deviation of the estimator for all T1 values. Theoretical prediction for the ratios of prec(T1est) with respect to that of the uniform strategy is also reported for all selection of points. These are not statistically different than the experimental ratios (P = 0.26, P = 0.09, P = 0.05 for T1 values of 950 – 1250 ms, 400 – 600 ms, 450 & 1250 ms respectively) for vials with T1 values in the union of the ranges of optimization.
Table 3.
Results of the phantom imaging over all 14 vials with different T1 values for various sampling strategies, where each acquisition was repeated 5 times and K = 11. The ratio of the standard deviation of the T1 estimator for each proposed sampling strategy and that of the uniform sampling strategy is reported as “precision (prec) with respect to (wrt) uniform.” The points from numerical experiment #3, #4 and #5 show improvement on the standard deviation of the uniform sampling strategy for T1 values > 600 ms, T1 values < 1150 ms and all T1 values, respectively. The theoretical prediction corresponding to the ratios of standard deviations are also reported for each sampling strategy, which are in good agreement with the experimental ratios (P = 0.26, P = 0.09, P = 0.05 for the point selections of different T1 values of interest respectively) for vials with T1 values in the union of the ranges of optimization.
| uniformly distributed points | points optimized for T = 950 - 1250 ms | po i n t s o pt i mized for T1 = 400 - 600 ms | po i nts o pt imized for T1 = 450 & 1250 ms | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vial | (T1est (ms) | prec (T1est) (ms) | T1est (ms) | prec (T1est) (ms) | prec wrt. uniform | theory prec wrt. uniform | T1est (ms) | prec T1est (ms) | prec wrt. uniform | theory prec wrt. uniform | T1est (ms) | prec (T1est) (ms) | prec wrt. uniform | theory prec wrt. uniform |
| 1 | 1457 ± 7.7 | 69.2 ± 6.3 | 1456 ± 7.4 | 48.1 ± 5.2 | 0.70 | 0.71 | 1432 ± 12.7 | 72.0 ± 5.2 | 1.04 | 1.01 | 1450 ± 26.5 | 48.8 ± 4.4 | 0.71 | 0.81 |
| 2 | 1144 ± 14.5 | 55.7 ± 6.7 | 1130 ± 7.1 | 41.0 ± 2.5 | 0.74 | 0.76 | 1127 ± 6.8 | 54.1 ± 2.7 | 0.97 | 0.98 | 1125 ± 12.9 | 44.7 ± 2.3 | 0.80 | 0.82 |
| 3 | 1151 ± 11.5 | 53.2 ± 3.8 | 1155 ± 8.6 | 43.1 ± 1.4 | 0.81 | 0.76 | 1146 ± 12.2 | 54.0 ± 6.7 | 1.02 | 0.98 | 1147 ± 20.3 | 45.3 ± 1.9 | 0.85 | 0.82 |
| 4 | 274 ± 5.0 | 15.3 ± 1.2 | 275 ± 6.8 | 23.2 ± 2.5 | 1.51 | 1.71 | 271 ± 7.7 | 14.7 ± 0.8 | 0.96 | 0.99 | 269 ± 4.5 | 15.0 ± 0.9 | 0.98 | 1.17 |
| 5 | 429 ± 6.6 | 17.3 ± 1.1 | 431 ± 2.3 | 18.8 ± 1.5 | 1.09 | 1.11 | 426 ± 6.8 | 14.1 ± 1.0 | 0.81 | 0.87 | 426 ± 5.9 | 15.2 ± 1.0 | 0.88 | 0.96 |
| 6 | 729 ± 10.3 | 31.3 ± 1.2 | 724 ± 2.1 | 26.1 ± 3.3 | 0.83 | 0.86 | 724 ± 4.6 | 29.0 ± 2.6 | 0.93 | 0.91 | 727 ± 13.9 | 27.7 ± 1.6 | 0.88 | 0.87 |
| 7 | 595 ± 6.6 | 20.0 ± 1.6 | 599 ± 6.0 | 18.8 ± 1.6 | 0.94 | 0.94 | 590 ± 11.3 | 17.8 ± 1.9 | 0.89 | 0.89 | 588 ± 11.8 | 16.6 ± 1.2 | 0.83 | 0.89 |
| 8 | 597 ± 5.0 | 25.9 ± 0.9 | 597 ± 5.0 | 23.3 ± 3.0 | 0.90 | 0.94 | 592 ± 5.3 | 22.8 ± 1.3 | 0.88 | 0.89 | 594 ± 11.8 | 23.1 ± 0.3 | 0.89 | 0.89 |
| 9 | 980 ± 11.2 | 34.6 ± 1.2 | 981 ± 10.4 | 25.2 ± 0.6 | 0.73 | 0.78 | 971 ± 8.5 | 29.0 ± 2.5 | 0.84 | 0.96 | 983 ± 13.5 | 24.2 ± 1.0 | 0.70 | 0.84 |
| 10 | 823 ± 13.3 | 29.8 ± 2.1 | 822 ± 7.7 | 24.2 ± 2.0 | 0.81 | 0.83 | 816 ± 8.3 | 29.0 ± 2.4 | 0.97 | 0.93 | 816 ± 15.0 | 22.1 ± 1.3 | 0.74 | 0.86 |
| 11 | 1148 ± 18.4 | 52.6 ± 6.4 | 1144 ± 8.3 | 37.8 ± 1.5 | 0.72 | 0.76 | 1129 ± 15.5 | 45.9 ± 2.4 | 0.87 | 0.98 | 1151 ± 18.7 | 42.0 ± 2.8 | 0.80 | 0.82 |
| 12 | 341 ± 4.1 | 19.4 ± 1.7 | 342 ± 5.8 | 22.6 ± 2.4 | 1.17 | 1.29 | 337 ± 7.0 | 16.3 ± 1.0 | 0.84 | 0.89 | 335 ± 8.7 | 17.3 ± 0.3 | 0.89 | 1.01 |
| 13 | 963 ± 13.8 | 49.8 ± 3.4 | 962 ± 6.2 | 35.4 ± 2.2 | 0.71 | 0.79 | 956 ± 3.7 | 47.1 ± 5.4 | 0.95 | 0.95 | 964 ± 8.6 | 37.9 ± 4.1 | 0.76 | 0.83 |
| 14 | 1130 ± 10.6 | 55.8 ± 5.6 | 1137 ± 10.5 | 45.0 ± 3.9 | 0.81 | 0.76 | 1121 ± 4.5 | 52.9 ± 2.2 | 0.95 | 0.98 | 1139 ± 15.8 | 45.0 ± 1.7 | 0.81 | 0.82 |
Precision versus number of sampling points
Table 4 shows phantom results evaluating how prec(T1est) varies with number of sampling points, K. The ratio of prec(T1est) between the proposed and uniform point selections are depicted for K = {5, 7, 9, 11, 13, 15} for the vials, whose T1 values are between 950 and 1250 ms. Corresponding ratios predicted by theory for these vials are also reported. The experimental and theoretical ratios are in good agreement across all vials considered (P = 0.97).
Table 4.
The results of phantom imaging for various values of number of sampling points, K, depicting the precision values, the ratio of the standard deviation of the proposed point selection for T1 values ranging from 950 ms to 1250 ms and that of the uniformly distributed point selection, as well as the ratio predicted by theory. The ratios are in good agreement across all vials considered (P = 0.97).
| Vial # | Evaluation Metric | K = 5 | K = 7 | K = 9 | K = 11 | K = 13 | K = 15 |
|---|---|---|---|---|---|---|---|
| 2 | uniform - prec(T1est) (ms) | 70 | 59 | 58 | 56 | 50 | 47 |
| proposed - prec (T1est) (ms) | 53 | 45 | 44 | 41 | 39 | 35 | |
| std ratio proposed vs. uniform | 0.76 | 0.77 | 0.73 | 0.73 | 0.80 | 0.76 | |
| theoretical ratio | 0.83 | 0.77 | 0.76 | 0.76 | 0.76 | 0.77 | |
| 3 | uniform - prec(T1est) (ms) | 62 | 56 | 55 | 53 | 50 | 47 |
| proposed - prec(T1est) (ms) | 53 | 45 | 44 | 43 | 40 | 37 | |
| std ratio proposed vs. uniform | 0.86 | 0.81 | 0.79 | 0.81 | 0.79 | 0.77 | |
| theoretical ratio | 0.82 | 0.76 | 0.75 | 0.75 | 0.76 | 0.77 | |
| 9 | uniform - prec(T1est) (ms) | 43 | 36 | 35 | 34 | 34 | 30 |
| proposed - prec(T1est) (ms) | 35 | 28 | 28 | 25 | 24 | 25 | |
| std ratio proposed vs. uniform | 0.81 | 0.78 | 0.81 | 0.73 | 0.70 | 0.85 | |
| theoretical ratio | 0.85 | 0.79 | 0.78 | 0.79 | 0.79 | 0.80 | |
| 11 | uniform - prec(T1est) (ms) | 59 | 54 | 52 | 53 | 45 | 45 |
| proposed - prec(T1est) (ms) | 49 | 39 | 41 | 37 | 35 | 34 | |
| std ratio proposed vs. uniform | 0.84 | 0.73 | 0.80 | 0.71 | 0.79 | 0.76 | |
| theoretical ratio | 0.82 | 0.76 | 0.75 | 0.75 | 0.76 | 0.76 | |
| 13 | uniform - prec(T1est) (ms) | 53 | 49 | 48 | 49 | 44 | 40 |
| proposed - prec(T1est) (ms) | 45 | 41 | 37 | 35 | 34 | 33 | |
| std ratio proposed vs. uniform | 0.85 | 0.84 | 0.78 | 0.71 | 0.78 | 0.82 | |
| theoretical ratio | 0.85 | 0.79 | 0.79 | 0.79 | 0.80 | 0.80 | |
| 14 | uniform - prec(T1est) (ms) | 63 | 62 | 58 | 56 | 50 | 48 |
| proposed - prec(T1est) (ms) | 53 | 44 | 45 | 45 | 39 | 36 | |
| std ratio proposed vs. uniform | 0.84 | 0.70 | 0.79 | 0.81 | 0.77 | 0.75 | |
| theoretical ratio | 0.82 | 0.77 | 0.76 | 0.76 | 0.76 | 0.77 |
Precision versus baseline SNR
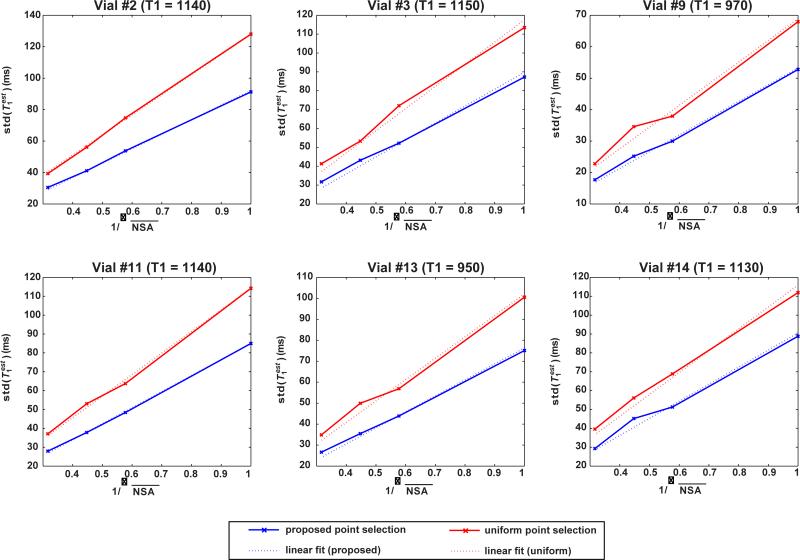
Figure 1 depicts prec(T1est) with changing NSA (equivalently √SNR) for all vials with T1 values between 950 and 1250 ms and for both point selection strategies. The best linear fits (y = ax) are also shown (dashed lines) as a function of changing NSA. As predicted by Equation [8], these are in good agreement with the experimentally measured prec(T1est) in all cases, with Pearson correlation coefficients >0.99 for all vials and for both point selection strategies. The baseline SNR was 29.2 ± 2.3 for the case of full longitudinal recovery (i.e. with SR time of ∞) image with NSA = 1.
Figure 1.
Standard deviation of the T1 estimator as a function of varying NSA (or equivalently √SNR) for the proposed point selection for T1 values ranging from 950 to 1250 ms and the uniformly distributed point selection. The experimental results are in good agreement with the best linear fit of the form y = ax (dashed lines) with Pearson coefficients > 0.99 for all cases.
In-vivo Imaging
Table 5 depicts the results of in-vivo T1 measurements for the five healthy subjects using the proposed and uniform sampling strategies. The reduction in prec(T1est) as measured by reader 1 were 13.9% and 20.1% for the myocardium and blood respectively, with the proposed approach compared to the uniform selection strategy. The corresponding reductions measured by reader 2 were 20.2% and 26.6% for the myocardium and blood respectively. Overall, there was a 17.0% and 23.5% reduction in the prec(T1est) in the myocardium and blood respectively using the proposed approach compared to the uniform selection.
Table 5.
The results of in-vivo imaging on five healthy subjects using the proposed and uniform sampling strategies, where each acquisition was repeated 5 times, as measured by two independent readers. T1est is reported as the mean ± std of the spatial average T1 values in the ROI across 5 scans, as a surrogate for accuracy and inter-scan reproducibility. prec(T1est) is reported as the mean ± std of the spatial std of the T1 values in the ROI across 5 scans, as a surrogate for the precision within the scan. Std wrt. uniform is the ratio of the mean values of prec(T1est) using the proposed and uniform point selection, characterizing the percentage gain in precision. prec(T1est) in the myocardium and blood was reduced by 17.0% and 23.5% respectively using the proposed approach.
| uniform point selection | proposed point selection | |||||||
|---|---|---|---|---|---|---|---|---|
| subject | heart rate (bpm) | anatomy | T1est (ms) | prec (T1est) (ms) | T1est (ms) | prec (T1est) (ms) | std wrt.uniform | |
| Reader #1 | 1 | 65 | myocardium | 1163 ± 24.2 | 104.9 ± 15.6 | 1155 ± 33.3 | 103.7 ± 11.3 | 0.99 |
| blood | 1926 ± 17.2 | 162.5 ± 15.3 | 1897 ± 26.8 | 114.7 ± 10.2 | 0.71 | |||
| 2 | 75 | myocardium | 1350 ± 19.6 | 177.9 ± 11.5 | 1348 ± 30.0 | 103.7 ± 11.3 | 0.89 | |
| blood | 1751 ± 46.4 | 187.6 ± 30.3 | 1766 ± 59.6 | 149.0 ± 5.8 | 0.79 | |||
| 3 | 70 | myocardium | 1242 ± 40.7 | 153.4 ± 21.1 | 1255 ± 15.6 | 124.5 ± 14.5 | 0.81 | |
| blood | 1799 ± 9.5 | 189.0 ± 20.7 | 1811.2 ± 36.5 | 155.0 ± 12.0 | 0.82 | |||
| 4 | 68 | myocardium | 1200 ± 33.9 | 110.9 ± 11.5 | 1193 ± 13.6 | 93.7 ± 9.9 | 0.84 | |
| blood | 1733 ± 44.2 | 140.8 ± 14.8 | 1754 ± 16.1 | 128.1 ± 17.8 | 0.91 | |||
| 5 | 58 | myocardium | 1135 ± 77.9 | 105.1 ± 18.9 | 1185 ± 16.5 | 81.8 ± 10.3 | 0.78 | |
| blood | 1757 ± 52.1 | 150.2 ± 15.1 | 1751 ± 15.1 | 116.8 ± 9.6 | 0.78 | |||
| Reader #2 | 1 | 65 | myocardium | 1188 ± 15.6 | 115.7 ± 16.2 | 1194 ± 15.3 | 91.7 ± 11.0 | 0.79 |
| blood | 1925 ± 25.1 | 166.7 ± 16.5 | 1903 ± 19.2 | 111.8 ± 7.7 | 0.67 | |||
| 2 | 75 | myocardium | 1314 ± 25.4 | 174.4 ± 28.6 | 1307 ± 21.5 | 150.1 ± 21.2 | 0.86 | |
| blood | 1772 ± 24.1 | 205.0 ± 32.3 | 1755 ± 20.8 | 148.2 ± 7.2 | 0.72 | |||
| 3 | 70 | myocardium | 1204 ± 70.2 | 117.0 ± 16.7 | 1218 ± 37.8 | 83.0 ± 13.5 | 0.71 | |
| blood | 1787 ± 37.3 | 179.7 ± 26.5 | 1809 ± 35.4 | 138.1 ± 14.9 | 0.77 | |||
| 4 | 68 | myocardium | 1213 ± 49.5 | 107.3 ± 16.7 | 1207 ± 31.7 | 85.0 ± 5.3 | 0.79 | |
| blood | 1755 ± 34.7 | 161.4 ± 16.7 | 1780 ± 19.2 | 131.4 ± 12.8 | 0.81 | |||
| 5 | 58 | myocardium | 1168 ± 54.3 | 95.3 ± 6.8 | 1187 ± 14.8 | 76.6 ± 5.0 | 0.80 | |
| blood | 1772 ± 47.8 | 164.7 ± 18.9 | 1761 ± 20.8 | 114.2 ± 10.4 | 0.69 | |||
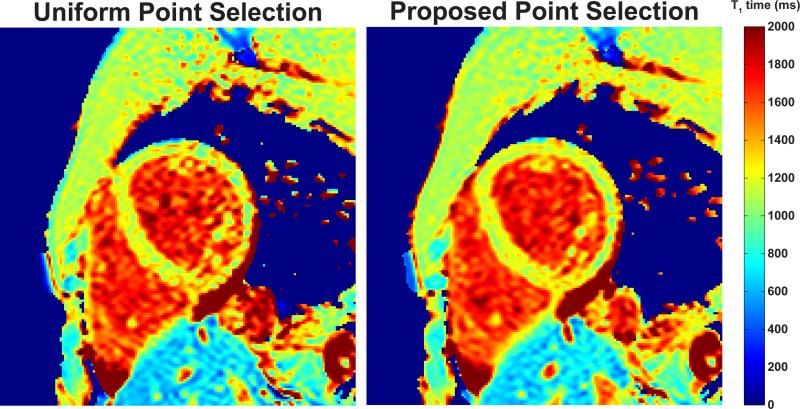
Figure 2 shows example myocardial T1 maps from a healthy subject (subject #5), acquired using the proposed and uniform point selection strategies at a heart rate of 60 bpm. Homogeneity of the T1 maps is visibly improved in the blood pools, where the T1 value is higher compared to the myocardium. prec(T1est) in the myocardium averaged over the 5 acquisitions showed 22% and 20% improvement with the proposed selection as measured by readers 1 and 2 respectively. Similarly, prec(T1est) for the blood pool showed 22% and 31% improvement as measured by readers 1 and 2 respectively. In comparison, the theory predicts reductions of 24.6% and 30.3% for CRBprec of the myocardium and blood respectively.
Figure 2.
T1 maps from a healthy subject (subject #5), acquired using the proposed and uniform point selection strategies. The T1 map generated using the proposed point selection is more homogenous, as visible in the blood pool. prec(T1est) averaged over the 5 acquisitions were 105.1 ± 18.9 ms and 81.8 ± 10.3 ms for the myocardium as measured by reader 1, and 95.3 ± 6.8 ms and 76.6 ± 5.0 ms as measured by reader 2 for uniform and proposed selection strategies respectively. prec(T1est) for the blood pool were 150.2 ± 15.1 ms and 116.8 ± 9.6 ms as measured by reader 1, and 164.7 ± 18.9 ms and 114.2 ± 10.4 ms as observed by reader 2 for uniform and proposed selection strategies respectively.
Discussion
In this study, we used an estimation theoretic framework for evaluating the precision of myocardial T1 mapping techniques to devise a sampling point selection strategy. This was achieved by minimizing the CRB on the variance of the estimators over the search space of possible sampling points. Subsequently, we evaluated this point selection strategy in a saturation-recovery based T1 mapping sequence, and compared it to a uniform distribution of sampling points along the SR curve. The improvements in precision in phantom and in-vivo imaging matched those from the numerical simulations derived from the theory.
The proposed optimized selection procedure yielded sampling points that are concentrated in 3 areas of the T1 relaxation curve: one at a low saturation (or inversion) recovery time (Tmin), one with a saturation time near (or due to sequence limitations, Tmax, the closest possible value to) the T1 value of interest, and the third point at ∞. The multiplicity of the points at these locations varies with the total number of points, as well as the sequence being simulated (e.g. taking multiple points at ∞ may not be feasible in cardiac MR). In all cases, these lead to an improved precision with respect to a uniform distribution of the points along the T1 relaxation curve. Thus, from a theoretical perspective it is better to improve the SNR at specific well-chosen points along the magnetization recovery curve by sampling multiple times at these locations than to distribute the sampling points along the curve. We note that this choice of points tends to be different than another existing hypothesis that points on the curve with higher magnitude derivatives with respect to time are more influential on the final estimate (30).
The CRB for T1 mapping was also derived previously in (26). This was then used to select a new set of sampling points, however the search space was only limited to ordered (linear, quadratic, cubic) sampling schemes due to the analytical tractability of these expressions. Thus, the tri-modal distribution resulting from our numerical optimization scheme with a more extensive search space, was not observed in (26). We note that the tri-modal distribution of optimal points presented here is similar to the bi-modal distribution observed in T2 mapping where a 2-parameter model was utilized (25). Apart from the differences in search spaces, which lead to substantially different selection of optimal sampling points, there are other differences between our work and (26). In particular, only numerical simulations were provided in (26), while we have also provided phantom and in-vivo data to validate the precision gains of our point selection approach directly. Furthermore, we have also evaluated our selection strategy using a Bayesian CRB to accommodate a wide range of T1 values of interest.
From both the theoretical and experimental perspectives it is observed that the standard deviation of the estimator increases with increasing T1 values, which is reflected in the choice of sampling points when a range of T1 values are of interest. This is also reflected in the corresponding gains of the optimized point selection versus uniformly distributed point selection, with more substantial gains in the precision for higher T1 values. Conversely, for shorter T1 values, both the percentage and the absolute reduction in the standard deviation of the estimator were smaller when using the optimized point selection.
Optimization of the sampling points for one range of T1 values does not guarantee a systematic improvement in precision across all T1 values. This is consistent with previous studies in parameter mapping (25,26). This was further validated in phantom imaging, where the sampling points optimized for T1 values from 950 to 1250 ms, did worse than uniform distribution in terms of precision for low T1 values < 400 ms. Optimizing for a larger spread of values also requires careful consideration as to how the final variance should be weighted. For instance, equal weights for T1 values of interest of 450 and 1250 ms yielded the same distribution as just optimizing for 1250 ms, since the variance of the estimator is dominated by the larger T1 value. However, by switching to weights scaling as 1/T12, thus characterizing the weighted error as a percentage of the T1 values, an optimal point selection that systematically outperformed the uniform sampling point distribution in numerical and phantom experiments was obtained.
The point selection procedure is not dependent on the SNR of the fully-recovered image, a or saturation/inversion efficiency, b, as shown in Appendix A. However, the standard deviation of the T1 estimator scales inverse-linearly with a, and with b. Thus, the percentage gain in precision between optimal point selection and uniform point distribution is constant for varying SNR, but the numerical difference becomes small for high values of a. Hence, for high SNR sequences, the gains from the proposed point selection may not be noticeable. Since b also inverse-linearly scales the standard deviation, the dynamic range acquired by doing an inversion recovery T1 mapping doubles the precision with respect to SR, assuming accurate flip angles and ignoring effects of imaging pulses until the acquisition of central k-space.
The effects of increasing K versus improving the SNR are different based on the constraints on what points on the curve can be sampled with a given sequence. In a practical myocardial T1 mapping scenario, where an infinite SR time can be imaged only once, going from K = 5 to 15 lead to an improvement in the standard deviation of the T1 estimator that was worse than the corresponding improvement achieved by increasing the SNR by √3. However, in an ideal scenario, where any point can be sampled multiple times (in Appendix B), going from K = 5 to 10 was better for improving the precision than increasing the SNR by √2. Thus, based on the constraints on the sampling points and the sequence, the designer can perform the numerical simulations based on the theory and conclude whether to improve the SNR or increase K for improved precision in estimation.
The general estimation theoretic framework for evaluation of variance lower bounds can be used for a wide range of sequences and applications (22-26). Since the differences predicted by the theory for different sampling point selection schemes matches experimental values well, the bounds themselves may be useful in comparing multiple sequences in terms of their precision theoretically in a quantitative manner. Alternatively this can also be studied with Monte-Carlo simulations (31), although this does not lead to closed form expressions, and thus does not allow for a computationally tractable method of optimizing point selection.
In this study, we have arbitrarily chosen a SR approach utilized in myocardial T1 mapping for validation. This sequence has certain limitations, most notably the need to apply the saturation pulse and imaging during the same R-R interval. This limits the maximum possible saturation time to Tmax, which decreases as the heart rate increases. Thus, for higher heart rates (or larger acquisition windows), the optimal selection at Tmax will shift to smaller values, reducing the precision of the sequence for the same number of sampling points. Furthermore, the results in Appendix B indicate that the precision can be improved if Tmax is higher than the T1 value of interest. Thus, precision may be improved by applying the preparation pulse(s) in one R-R interval and imaging in the consequent one, which was explored in other works (32,33).
The accuracy of the T1 estimation using SASHA sequence with respect to a gold standard inversion recovery spin echo sequence using phantom measurements were not evaluated, as this has already been reported (10), and further studied elsewhere (12,34). Our focus is on the choice of sampling points and how this can reduce the variance of the estimator, i.e. the precision, as evaluated by signal homogeneity in an ROI where T1 is expected to be homogenous. We do note that the CRB theory presented here is for unbiased estimators, which is an assumption satisfied by the SASHA sequence with sufficient SNR and the 3-point T1 fitting of Equation [2] (10). The theory of CRB can be extended to biased estimators, provided that the bias term can be characterized in terms of the parameters that are being estimated (28). An interesting implication of the CRB for biased estimators is that a biased estimator does not necessarily have worse precision than an unbiased estimator, and this extension may be appropriate for alternative myocardial T1 mapping sequences based on Look-Locker experiments (8,9,35).
We have based the probabilistic model in our CRB derivation on least squares estimation, the most commonly used T1 curve fitting procedure (8-12). This has one-to-one correspondence with a Gaussian noise model in the images. However, the noise in the magnitude images is Rician. Thus, there is an underlying assumption that the noise in the magnitude images can be well-approximated by Gaussian noise, which is true with sufficient SNR (27). However, this assumption may not hold for images acquired close to the saturation pulse in the SASHA sequence. This, in turn, may translate to an apparent bias in the T1 estimates if sufficiently many of the acquired images have low SNR. For instance, such cases may happen when the heart-rate of the subject is high, thus Tmax is low, which was discussed in (10). In our case, this was observed in phantom experiments for vials with high T1 values, when sampling points were selected for low T1 values, but were used to image vials with high T1 values. Thus, the unbiased nature of SASHA should be considered, when using the optimal selection procedure for one range of T1 values, but imaging another higher range of T1 values.
Our study has several limitations. We have provided a general theory for evaluating the variance of estimators in myocardial parameter mapping, but we have only evaluated this approach in saturation-recovery based myocardial T1 mapping, the choice of which was arbitrary. Further studies for confirming the theoretical results for other parameter mapping techniques are warranted. The in-vivo data presented was acquired using breath-hold, but respiratory drift may be present among subsequent contrast-weighted images (corresponding to different sampling points along the SR curve), which can corrupt the estimated T1 maps and their homogeneity. In the in-vivo data, in-flow and off-resonance effects (36) may also increase signal inhomogeneity, affecting the final reported mean and precision of the T1 values.
Conclusion
We have used an estimation theoretic framework to evaluating the precision of a T1 mapping technique in the presence of noise, and to choose the location and number of sampling points on the parametric magnetization curve to achieve a given level of precision. The point selection can be optimized for different values of interest of a given parameter to achieve higher levels of precision in estimation without increasing the scan time.
Acknowledgements
The project described was supported by NIH R01EB008743-01A2, NIH K99HL111410-01.
Appendix A
Equation [8] states
| [A1] |
We let
| [A2] |
Thus, the numerator of [A1] becomes
| [A3] |
since with a simple change of indices. Similarly, the denominator of [A1] is given by
| [A4] |
since , and after change of indices.
From these expressions we have
| [A5] |
Since γk and βk are functions of {xk} and T1 only, this shows is inversely proportional to a2b2. In particular,
| [A6] |
Appendix B
Two additional numerical simulations of theoretical interest were performed. First, for the T1 value of 1250 ms with K = 11, Tmax was set to 1300 ms, which is higher than the trigger delay time, and the optimization procedure was repeated. Second, to compare the effects of increasing SNR versus increasing K under the assumption that any sampling point can be selected multiple times, we simulated the sequence with K = 5 and K = 10 for T1 values of interest varying from 950 to 1250 ms, allowing for multiple sampling of all points, including the point at infinity.
For the set of experiments, where Tmax = 1300 ms, the proposed selection of points yielded (140, 140, 140, 1060, 1060, 1060, 1060, 1060, 1060, 1060, ∞), whereas the uniform distribution resulted in (140, 269, 398, 527, 656, 785, 914, 1043, 1172, 1300, ∞). These lead to CRBprec of 77.2 ms vs. 89.1 ms respectively (13.4% reduction). The distribution of points using the proposed method shows the trend of the optimization procedure to select 1 point at ∞, 3 points at Tmin, and the remaining 7 points at 1060 ms.
For the second set of experiments, where any sampling point can be selected multiple times, the optimization for the sequence with K = 5 yielded (140, 1265, 1265, ∞, ∞) for a CRBprec of 86.7 ms, whereas K = 10 yielded (140, 140, 1220, 1220, 1220, 1220, 1220, ∞, ∞, ∞) for a CRBprec of 60.1 ms. We note that the point selection still favors choosing points in a concentrated region, favoring multiplicities of the points with higher signal content. It is also worth noting that going from 5 to 10 points yields an improvement of 31% in the variance, which is better than acquiring the K = 5 points twice for double the SNR (29%), although the difference is not large.
References
- 1.Bottomley PA, Hardy CJ, Argersinger RE, Allen-Moore G. A review of 1H nuclear magnetic resonance relaxation in pathology: are T1 and T2 diagnostic? Med Phys. 1987;14(1):1–37. doi: 10.1118/1.596111. [DOI] [PubMed] [Google Scholar]
- 2.Warntjes JB, Leinhard OD, West J, Lundberg P. Rapid magnetic resonance quantification on the brain: Optimization for clinical usage. Magn Reson Med. 2008;60(2):320–329. doi: 10.1002/mrm.21635. [DOI] [PubMed] [Google Scholar]
- 3.Alexander AL, Hurley SA, Samsonov AA, Adluru N, Hosseinbor AP, Mossahebi P, Tromp do PM, Zakszewski E, Field AS. Characterization of cerebral white matter properties using quantitative magnetic resonance imaging stains. Brain Connect. 2011;1(6):423–446. doi: 10.1089/brain.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith SR, Roberts N, Percy DF, Edwards RH. Detection of bone marrow abnormalities in patients with Hodgkin's disease by T1 mapping of MR images of lumbar vertebral bone marrow. Br J Cancer. 1992;65(2):246–251. doi: 10.1038/bjc.1992.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hattingen E, Jurcoane A, Daneshvar K, Pilatus U, Mittelbronn M, Steinbach JP, Bahr O. Quantitative T2 mapping of recurrent glioblastoma under bevacizumab improves monitoring for non-enhancing tumor progression and predicts overall survival. Neuro Oncol. 2013;15(10):1395–1404. doi: 10.1093/neuonc/not105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mewton N, Liu CY, Croisille P, Bluemke D, Lima JA. Assessment of myocardial fibrosis with cardiovascular magnetic resonance. J Am Coll Cardiol. 2011;57(8):891–903. doi: 10.1016/j.jacc.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marwick TH, Schwaiger M. The future of cardiovascular imaging in the diagnosis and management of heart failure, part 1: tasks and tools. Circ Cardiovasc Imaging. 2008;1(1):58–69. doi: 10.1161/CIRCIMAGING.108.792408. [DOI] [PubMed] [Google Scholar]
- 8.Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn Reson Med. 2004;52(1):141–146. doi: 10.1002/mrm.20110. [DOI] [PubMed] [Google Scholar]
- 9.Piechnik SK, Ferreira VM, Dall'Armellina E, Cochlin LE, Greiser A, Neubauer S, Robson MD. Shortened Modified Look-Locker Inversion recovery (ShMOLLI) for clinical myocardial T1-mapping at 1.5 and 3 T within a 9 heartbeat breathhold. J Cardiovasc Magn Reson. 2010;12:69. doi: 10.1186/1532-429X-12-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chow K, Flewitt JA, Green JD, Pagano JJ, Friedrich MG, Thompson RB. Saturation recovery single-shot acquisition (SASHA) for myocardial T mapping. Magn Reson Med. 2013 doi: 10.1002/mrm.24878. doi: 10.1002/mrm.24878. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.Weingartner S, Akcakaya M, Basha T, Kissinger KV, Goddu B, Berg S, Manning WJ, Nezafat R. Combined saturation/inversion recovery sequences for improved evaluation of scar and diffuse fibrosis in patients with arrhythmia or heart rate variability. Magn Reson Med. 2013 doi: 10.1002/mrm.24761. doi: 10.1002/mrm.24761. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 12.Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J Cardiovasc Magn Reson. 2014;16(1):2. doi: 10.1186/1532-429X-16-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz-Menger J, Friedrich MG. Magnetic resonance imaging in patients with cardiomyopathies: when and why. Herz. 2000;25(4):384–391. doi: 10.1007/s000590050030. [DOI] [PubMed] [Google Scholar]
- 14.Arheden H, Saeed M, Higgins CB, Gao DW, Bremerich J, Wyttenbach R, Dae MW, Wendland MF. Measurement of the distribution volume of gadopentetate dimeglumine at echo-planar MR imaging to quantify myocardial infarction: comparison with 99mTc-DTPA autoradiography in rats. Radiology. 1999;211(3):698–708. doi: 10.1148/radiology.211.3.r99jn41698. [DOI] [PubMed] [Google Scholar]
- 15.Deoni SC, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn Reson Med. 2003;49(3):515–526. doi: 10.1002/mrm.10407. [DOI] [PubMed] [Google Scholar]
- 16.Liney GP, Knowles AJ, Manton DJ, Turnbull LW, Blackband SJ, Horsman A. Comparison of conventional single echo and multi-echo sequences with a fast spin-echo sequence for quantitative T2 mapping: application to the prostate. J Magn Reson Imaging. 1996;6(4):603–607. doi: 10.1002/jmri.1880060408. [DOI] [PubMed] [Google Scholar]
- 17.Look DC, Locker DR. Time Saving in Measurement of NMR and EPR Relaxation Times. Review of Scientific Instruments. 1970;41(2):250–251. [Google Scholar]
- 18.Steinhoff S, Zaitsev M, Zilles K, Shah NJ. Fast T(1) mapping with volume coverage. Magn Reson Med. 2001;46(1):131–140. doi: 10.1002/mrm.1168. [DOI] [PubMed] [Google Scholar]
- 19.Clare S, Jezzard P. Rapid T(1) mapping using multislice echo planar imaging. Magn Reson Med. 2001;45(4):630–634. doi: 10.1002/mrm.1085. [DOI] [PubMed] [Google Scholar]
- 20.Kellman P, Arai AE, Xue H. T1 and extracellular volume mapping in the heart: estimation of error maps and the influence of noise on precision. J Cardiovasc Magn Reson. 2013;15:56. doi: 10.1186/1532-429X-15-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kay S. Fundamentals of Statistical Signal Processing, Volume I: Estimation Theory. Prentice Hall; 1993. [Google Scholar]
- 22.Brihuega-Moreno O, Heese FP, Hall LD. Optimization of diffusion measurements using Cramer-Rao lower bound theory and its application to articular cartilage. Magn Reson Med. 2003;50(5):1069–1076. doi: 10.1002/mrm.10628. [DOI] [PubMed] [Google Scholar]
- 23.Poot DH, den Dekker AJ, Achten E, Verhoye M, Sijbers J. Optimal experimental design for diffusion kurtosis imaging. IEEE Trans Med Imaging. 2010;29(3):819–829. doi: 10.1109/TMI.2009.2037915. [DOI] [PubMed] [Google Scholar]
- 24.Sezginer A, Kleinberg RL, Fukuhara M, Latour LL. Very rapid simultaneous measurement of nuclear magnetic resonance spin-lattice relaxation time and spin-spin relaxation time. J Magn Res. 1991;92:504–527. [Google Scholar]
- 25.Jones JA, Hodgkinson P, Barker AL, Hore PJ. Optimal sampling strategies for the measurement of spin–spin relaxation times. J Magn Res - Series B. 1996;113:25–34. [Google Scholar]
- 26.Zhang Y, Yeung HN, O'Donnell M, Carson PL. Determination of sample time for T1 measurement. J Magn Reson Imaging. 1998;8(3):675–681. doi: 10.1002/jmri.1880080324. [DOI] [PubMed] [Google Scholar]
- 27.Gudbjartsson H, Patz S. The Rician distribution of noisy MRI data. Magn Reson Med. 1995;34(6):910–914. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill RD, Levit BY. Applications of the van Trees inequality: a Bayesian Cramér-Rao bound. Bernoulli. 1995;1(1-2):59–79. [Google Scholar]
- 29.Kraft KA, Fatouros PP, Clarke GD, Kishore PR. An MRI phantom material for quantitative relaxometry. Magn Reson Med. 1987;5(6):555–562. doi: 10.1002/mrm.1910050606. [DOI] [PubMed] [Google Scholar]
- 30.Xue H, Greiser A, Zuehlsdorff S, Jolly MP, Guehring J, Arai AE, Kellman P. Phase-sensitive inversion recovery for myocardial T1 mapping with motion correction and parametric fitting. Magn Reson Med. 2013;69(5):1408–1420. doi: 10.1002/mrm.24385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellman P, Xue H, Hansen MS. Comparison of myocardial T1-mapping protocols: accuracy and precision 2013 May Salt Lake City. Proceedings of the 21st Scientific Meeting of ISMRM. :1394. [Google Scholar]
- 32.Weingartner S, Akçakaya M, Kissinger KV, Manning WJ, Nezafat R. Heart-rate independent non-contrast myocardial T1 mapping. 2013 May Salt Lake City. Proceedings of the 21st Scientific Meeting of ISMRM. :1358. [Google Scholar]
- 33.Slavin GS, Stanisby JA. True T1 mapping with SMART1Map: A comparison with MOLLI. 2013 May Salt Lake City. Proceedings of the 21st Scientific Meeting of ISMRM. :1416. [Google Scholar]
- 34.Roujol S, Weingartner S, Foppa M, Chow K, Kawaji K, Kissinger KV, Goddu B, Berg S, Kellman P, Manning WJ, Thompson RB, Nezafat R. Accuracy and reproducibility of four T1 mapping sequences: A head-to-head comparison of MOLLI, ShMOLLI, SASHA, and SAPPHIRE. J Cardiovasc Magn Reson. 2014;16(Suppl 1):O26. doi: 10.1148/radiol.14140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robson MD, Piechnik SK, Tunnicliffe EM, Neubauer S. T measurements in the human myocardium: The effects of magnetization transfer on the SASHA and MOLLI sequences. Magn Reson Med. 2013 doi: 10.1002/mrm.24867. doi: 10.1002/mrm.24867. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 36.Kellman P, Herzka DA, Arai AE, Hansen MS. Influence of Off-resonance in myocardial T1-mapping using SSFP based MOLLI method. J Cardiovasc Magn Reson. 2013;15:63. doi: 10.1186/1532-429X-15-63. [DOI] [PMC free article] [PubMed] [Google Scholar]