Abstract
Lipid heterogeneities, such as lipid rafts, are widely considered to be important for the sequestering of membrane proteins in plasma membranes, thereby influencing membrane protein functionality. However, the underlying mechanisms of such sequestration processes remain elusive, in part, due to the small size and often transient nature of these functional membrane heterogeneities in cellular membranes. To overcome these challenges, here we report the sequestration behavior of urokinase receptor (uPAR), a glycosylphosphatidylinositol-anchored protein, in a planar model membrane platform with raft-mimicking lipid mixtures of well-defined compositions using a powerful optical imaging platform consisting of confocal spectroscopy XY-scans, photon counting histogram, and fluorescence correlation spectroscopy analyses. This methodology provides parallel information about receptor sequestration, oligomerization state, and lateral mobility with single molecule sensitivity. Most notably, our experiments demonstrate that moderate changes in uPAR sequestration are not only associated with modifications in uPAR dimerization levels, but may also be linked to ligand-mediated allosteric changes of these membrane receptors. Our data show that these modifications in uPAR sequestration can be induced by exposure to specific ligands (urokinase plasminogen activator, vitronectin), but not via adjustment of the cholesterol level in the planar model membrane system. Good agreement of our key findings with published results on cell membranes confirms the validity of our model membrane approach. We hypothesize that the observed mechanism of receptor translocation in the presence of raft-mimicking lipid mixtures is also applicable to other glycosylphosphatidylinositol-anchored proteins.
Introduction
The heterogeneous distribution of lipids in the plasma membrane is increasingly recognized as an important regulatory factor that influences membrane protein distribution and functionality (1,2). Lipid rafts, which are enriched in cholesterol (CHOL) and sphingolipids, represent one prominent type of lipid heterogeneity (3,4). Raft microdomains have been associated with multiple cellular activities, including signaling (5), pathogenesis (6), and regulation of cell adhesion, cell morphology, and angiogenesis (7). Their functional importance has been largely linked to the ability to sequester membrane proteins of different raft affinity (8). Membrane protein raft affinity has been in part attributed to several molecular motifs, such as protein acylation and glycosylphosphatidylinositol (GPI) anchors (3).
Raft-mediated receptor clustering and associated change in receptor function represent potentially important roles for raft domains in cellular membranes (9). For example, Paladino et al. (10) reported that oligomerization of apically sorted GPI-anchored proteins (GPI-AP) during their transport to the plasma membrane of Madin-Darby canine kidney (MDCK) cells is associated with lipid rafts. These authors observed that CHOL depletion not only impaired raft affinity and apical sorting, but also protein oligomerization of GPI-APs in the Golgi. Similarly, Kusumi and co-workers (11) attributed the stabilization of GPI-AP homodimers in the plasma membrane of CHO-K1 cells to raft-lipid interactions forming GPI-AP homodimer rafts. Their interpretation was largely based on the observation that GPI-anchored GFP showed longer dimer lifetimes than GFP counterparts with a transmembrane (TM) anchor. Ligand-induced receptor oligomerization plays a key role in multiple TM signaling processes (12). Lipid rafts are considered to be important in the redistribution of membrane proteins in response to ligand-induced changes in the membrane protein oligomerization state. For example, raft partitioning of urokinase plasminogen activator receptor (uPAR) in human embryonic kidney 293 cells was reported to be linked to alterations of the receptor dimerization state upon ligand binding (13). Here, urokinase plasminogen activator (uPA) binding was found to reduce dimerization and raft partitioning of this GPI-anchored membrane receptor, whereas enhanced uPAR dimerization and raft partitioning was observed upon vitronectin (VN) addition. Of importance, ligand-mediated alterations in receptor dimerization and raft partitioning are not only limited to GPI-APs. A similar mechanism was recently proposed for G-protein-coupled receptors (GPCRs) on the basis of computational results (14). Despite these supporting results, there is still uncertainty about the functional relationship between raft domains and receptor oligomerization and function. This uncertainty is not only due to a lack of knowledge about the role of potential ligand-induced allosteric changes of receptor conformation on raft affinity, but should also be attributed to the small size and transient nature of such membrane domains in the plasma membrane (15,16). As a consequence, optical visualization of stable raft domains in cell membranes typically requires the use of cross-linking agents, such as cross-linking antibodies or GM1-cholera toxin B. Not surprisingly, Lingwood and Simons (17) recently pointed out parallels to Heisenberg’s uncertainty principle: by introducing a cross-linker, the observer influences the object of study. Another potential problem related to the characterization of raft domains and raft-associated molecular processes in cells is that traditional biochemical techniques, such as analysis of detergent-resistant membrane fractions and CHOL depletion, are prone to artifacts (18,19).
Therefore, model membrane studies have emerged as a complementary platform, in which properties of raft-like domains can be studied in well-defined membrane environments. Raft-mimicking model membrane mixtures may result in stable lipid-lipid phase separations into stable CHOL-rich liquid-ordered (lo) and CHOL-deficient liquid-disordered (ld) domains, which depending on composition may range from submicron to several microns in size (20,21). Previously, the model membrane approach was successfully employed to explore the distribution of membrane proteins in the absence and presence of cross-linking agents in lipid mixtures with coexisting lo and ld domains (22–25). One particular strength of model membrane experiments lies in the ability to explore the sequestration of membrane proteins in the presence of stable, optically visible domains in the absence of artificial cross-linking agents. Recently, our group built on this concept and developed an experimental platform that allows the analysis of membrane protein sequestration and oligomerization in planar polymer-tethered lipid bilayers with single molecule sensitivity. Such membrane architectures are well suited for the investigation of membrane proteins in well-defined lipid environments (26–28). By applying this powerful experimental platform, we showed that native ligands alter integrin sequestration but not oligomerization in raft-mimicking lipid mixtures (29). In another study, this experimental strategy was pursued to demonstrate that bilayer asymmetry (i.e., presence of lo-ld phase separations exclusively in the top leaflet of the bilayer versus bilayer-spanning lo-ld phase separations) influences integrin sequestration in such lipid mixtures as well (30). Most notably, in the latter work, αvβ3 and α5β1 integrins (in the absence of ligands) were found to have a preference for the lo phase in the asymmetric bilayer, but exhibit an ld phase affinity in their symmetric counterparts.
In the current work, we employ a comparable experimental model membrane strategy to investigate the fascinating functional relationship between raft domains and receptor oligomerization, and the effect of native ligands on this relationship. Specifically, the sequestration and oligomerization behavior of uPAR is examined in a planar polymer-tethered lipid bilayer containing coexisting bilayer-spanning lo and ld phase separations (Fig. 1). This GPI-AP, which consists of three domains (numbered from the N-terminus, domains D1, D2, and D3), represents a key component of the urokinase plasminogen activation system (31,32). Our experiments show that uPAR has a pronounced affinity for lo lipid regions and exhibits a substantial dimer population. Addition of uPA not only causes the efficient suppression of uPAR dimers, but also results in reduced lo affinity. In contrast, addition of VN promotes uPAR dimerization and causes enhanced affinity for CHOL-enriched lipid domains. Complementary experiments on binary lipid-CHOL mixtures furthermore demonstrate that uPAR dimerization levels are largely independent of CHOL content in the bilayer. Our results are significant because they agree with previous findings about the role of uPA and VN in uPAR dimerization and raft affinity in plasma membranes, thus validating our model membrane approach (13). Moreover, the current work provides valuable insight into the roles of native ligands and lipid composition in receptor sequestration in biological membranes.
Figure 1.
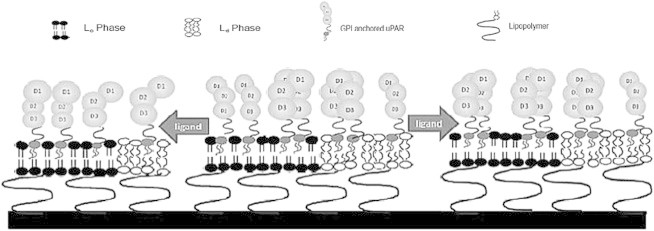
Schematic of a polymer-tethered lipid bilayer with functionally incorporated uPAR. This GPI-AP is characterized by a monomer-dimer equilibrium. The role of specific ligands (uPA and VN) in uPAR dimerization and sequestering is investigated in a raft-mimicking lipid mixture forming coexisting lo and ld domains.
Materials and Methods
Materials
The phospholipids 1,2-dioleoyl-sn-glycro-3-phosphocholine (DOPC) and 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), as well as CHOL were obtained from Avanti Polar Lipids (Alabaster, AL). The lipopolymer 1,2-dioctadecyl-sn-glycero-3-N-poly(2-methyl-2-oxazoline) 50 (diC18M50) was synthesized as reported previously (33). The dye-labeled phospholipids N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)-1,2-dihexadec-anoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt (NBD-DHPE) and N-(6-tetramethylrhodamine-thiocarbamoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (TRITC-DHPE) were purchased from Invitrogen (Carlsbad, CA). Purified recombinant Homo sapiens uPAR with C-terminal MYC/DDK tag and Anti-DDK monoclonal antibody were purchased from OriGene Technologies (Rockville, MD). Anti-uPAR (N-terminal) antibody, was purchased from Proteintech (Chicago, IL). Recombinant Human uPA and human purified VN were obtained from Millipore (Billerica, MA). Rhodamine-6-G applied as standard fluorescent markers was purchased from Sigma-Aldrich (St. Louis, MO). To facilitate optical imaging experiments, antibodies were labeled using Alexa 555, 488, or 594 labeling kits (Invitrogen). The surfactant n-octyl-β-glycopyranoside (OG) was purchased from Fisher BioReagents (Fairlawn, NJ). Chloroform (HPLC grade; Fisher Scientific, Pittsburgh, PA) was employed as spreading solvent of lipid monolayers at the air-water interface. Milli-Q water (pH 5.5, 18 MU-cm resistivity) applied as a subphase material in the Langmuir-Blodgett trough was taken from a Milli-Q system (Millipore). Glass coverslips used as solid support of polymer tethered lipid bilayer systems were baked and cleaned as previously described (27,34).
Construction of polymer-tethered phospholipid bilayers
Polymer-tethered phospholipid bilayers were constructed via subsequent Langmuir-Blodgett (LB) and Langmuir-Schaefer (LS) monolayer transfers as previously described (35). Before each monolayer transfer, chloroform solutions of lipopolymer/lipid and lipid mixtures were spread at the air-water interface of an LB trough with dipper (Labcon, Darlington, UK). Each monolayer was compressed to a film pressure of 30 mN/m and equilibrated for ∼40 min. LB transfers were accomplished via the dipper of the LB trough. LS transfers were pursued by gently pushing the coverglass with the LB monolayer though the LS monolayer at the air-water interface and by placing the LB/LS bilayer substrate onto a depression slide in the Milli-Q subphase. Next, all solid-supported LB/LS bilayers were transferred to a petri dish and Milli-Q was replaced with phosphate buffered saline (PBS) (Fisher Scientific, 10× concentrations, diluted in Milli-Q) for protein reconstitution. Specifically, two types of bilayer systems were constructed, a binary DOPC-CHOL mixed bilayer of different CHOL content (0, 5, 15, 25, and 35 mol % CHOL) and a ternary DOPC/DPPC/CHOL mixed bilayer with equal amounts of DOPC/DPPC and 28, 33 mol % CHOL, separately. To lift up the bilayer from the underlying glass substrate, both types of bilayer systems contain 5 mol % of the lipopolymer diC18M50 in their LB monolayer.
Reconstitution of uPAR into bilayers and labeling with Alexa555-tagged antibodies
The reconstitution of uPAR into the planar polymer-tethered lipid bilayer was conducted by adapting the modified Rigaud technique applied to integrins previously (29,30). Specifically, 1.3 × 10−11 mol uPAR and 250 μM OG were added together to the preassembled bilayer system and allowed to incubate for 2 h. This surfactant concentration in the presence of the bilayer sample corresponds to ∼0.002 cmc. Next, OG was removed from the bilayer via a single layer of SM-2 Bio-Beads (Bio-Rad, Hercules, CA) (incubation time: 10 min) followed by Bio-Bead removal and extensive rinsing with PBS. To allow the fluorescence detection of functionally reconstituted uPAR, 2 × 10−11 mol of Alexa-555 labeled anti-DDK monoclonal antibody (MAb) was added to the uPAR-containing bilayer sample and allowed to incubate for 2 h. Before imaging, excess (unbound) MAbs were removed by thorough rinsing with PBS. Epifluorescence (EPI) micrographs were acquired, which confirmed the homogeneous distribution of MAb-labeled uPAR in DOPC and DOPC + 35 mol % CHOL bilayers (Fig. S1 in the Supporting Material). To explore the role of native ligands on uPAR dimerization and sequestration, uPA or VN were added with an equimolar ratio relative to uPAR and kept overnight to allow specific binding to the bilayer-incorporated uPAR. Again, before imaging, excess unbound ligands were removed by rinsing with PBS. Imaging data were acquired from the same bilayer substrate to compare differences before and after ligand binding. To verify specific MAb-receptor binding, control experiments were conducted, where Alexa-555 labeled anti-DDK MAbs were added to the bilayer system without uPAR. This control assay was also applied in the presence of uPA or VN to exclude the possibility of non-specific binding of ligands. As described in more detail in the Supporting Material section, separate control experiments on different concentrations of TRITC-DHPE and uPAR (labeled with Alexa-555 labeled anti-DDK MAbs) (Fig. S2) showed that the functional reconstitution of uPAR in the bilayer was achieved with 86 ± 6% efficiency and that antibody-mediated uPAR cross-linking is rather insignificant.
Microscopy techniques
A Confocor 2 microscopy system (Carl Zeiss, Jena, Germany) equipped with an Axiovert 200M (Carl Zeiss, Oberkochen, Germany), a Zeiss C-Apochromat objective (water immersion, 40 × NA = 1.2), and a Zeiss AxioCam MRm monochrome digital camera was employed to investigate uPAR dimerization levels and sequestration in polymer-tethered lipid bilayer samples. As reported previously, the microscopy system allows EPI microscopy, confocal fluorescence intensity detection, fluorescence correlation spectroscopy (FCS), and photon counting histogram analysis (PCH) (29,30). EPI analysis was conducted to confirm the integrity of the polymer-tethered lipid bilayer and the existence of coexisting lo and ld lipid phases in ternary raft-mimicking lipid mixtures. Confocal spectroscopy XY (CS-XY) scans of bilayer regions were pursued via the piezo stage of the microscope (maximum scan size: 10 μm × 10 μm; step size: 0.5 μm), thus obtaining parallel information about the distributions of Alexa 555 MAb-tagged uPAR and NBD-DHPE in the same bilayer regions. Here, Alexa 555 detection was facilitated using a 1.0-mW HeNe laser (543 nm) with a 560–615-nm emission filter (Alexa 555 channel), whereas NBD analysis was achieved using the 488 nm line of a 30-mW Argon laser with 505–530 nm emission filter (NBD channel). To correct for background, control experiments were performed through the Alexa 555 channel, in which the bilayer contained NBD-DHPE, but lacked Alexa 555 MAb-tagged uPAR.
Data analysis
The characterization of uPAR dimerization levels and sequestration in raft-mimicking lipid mixtures was achieved by adapting analysis procedures recently reported for integrins in comparable membrane systems (29). In short, FCS autocorrelation analysis was conducted to determine the average brightness and lateral mobility of Alexa 555-labeled MAbs in solution and bound to bilayer-reconstituted uPAR. In the latter case, the confocal spot was kept at a fixed bilayer position and the photon counts through the Alexa 555 channel were collected over 50 s. In these experiments, the correct confocal position was identified by maximizing the photon-count rate of probe molecule of interest. In light of the uncertainties about the geometry of the confocal spot (knowledge about confocal shape is required for accurate acquisition of diffusion coefficients via FCS), diffusion coefficients of bilayer-incorporated probe molecules were determined by using a TRITC-DHPE standard, for which the FCS diffusion time could be compared to a diffusion coefficient determined using wide-field single molecule fluorescence microscopy (30). Raw data of photon counts at a fixed bilayer position over time were also used for PCH analysis. The PCH method was used to acquire the average brightness and number of uPAR monomers (ε, Navg) and dimers (εdimer(=2ε), Navgdimer) in binary DOPC-CHOL mixtures and lo and ld lipid phases of ternary DOPC/DPPC/CHOL mixtures thereby adapting methods reported previously (29,36). The dimerization level can be quantified in terms of the mole fraction of dimers, xdimer. The PCH method was previously tested using fluorescent dyes and CdSe/ZnS quantum dots in solution and bound to lipids in a planar lipid bilayer (29). The surface functionalization of such quantum dots and their specific linkage to lipid bilayers has been described elsewhere (37). As described in more detail in the Supporting Material section, particle brightness and sample-specific background are first determined in separate control experiments to allow an accurate determination of xdimer using the PCH method. With the sample-specific brightness and background values being determined experimentally, the numbers of monomers and dimers are the only variable parameters in the PCH model fit analysis. The impact of changing xdimer values on the shape of PCH curves is illustrated in Fig. S3.
In addition to the described FCS and PCH characterization, confocal XY scans (CS-XY) were applied to determine the NBD-DHPE and Alexa 555 MAb-tagged uPAR distributions in the same bilayer regions of different composition bilayers. Each raw data set was corrected for NBD-DHPE bleed through the Alexa 555 channel, which is ∼6% of the total background. Nonspecifically bound Alexa-555 MAbs represent another potential source of background. This background contribution was determined in separate control CS-XY scan experiments on bilayer samples, in which comparable amounts of Alexa 555-tagged MAbs were added to uPAR-free bilayers of identical lipid composition (compared to uPAR-containing bilayers) and, following an incubation time of 2 h, unbound antibodies were removed by rinsing with PBS. With this additional background information in place, the overall signal/background (through the Alexa 555 channel) for Alexa 555 MAb-tagged uPAR in the bilayer in the presence of NBD-DHPE was identified to be ∼4:1. As reported before, the receptor distribution in the bilayer in the presence of coexisting lo and ld lipid phases can be quantified in terms of the parameter Eraft, which is defined as
| (1) |
where Ilo and Ild describe the background-corrected average signal intensities of the membrane protein probe molecule in the lo and ld phases, respectively (29). To guarantee reproducibility, all Eraft values provided are based on at least three different bilayer samples with at least five spots (12 × 12 μm2) being analyzed from each bilayer. The statistical significance of all presented data (lateral diffusion coefficients, xdimer, Eraft, uPAR cleavage assay) was verified by comparing independent data sets using T- and/or ANOVA-tests.
Results and Discussion
VN and uPA influence uPAR dimerization levels qualitatively differently
Protein dimerization level is considered as an important factor determining raft association of membrane proteins in cellular membranes, thereby influencing crucial cellular processes, such as membrane protein sorting and raft-mediated signaling. However, the characterization of this important functional relationship remains challenging, due to the small size and often transient nature of raft domains in cellular membranes. To overcome these experimental challenges, we therefore explored the relationship between the receptor dimerization level and raft affinity using the GPI-AP uPAR reconstituted into a polymer-tethered lipid bilayer of well-defined lipid compositions. Specifically, uPAR (labeled with Alexa-555 labeled anti-DDK MAb) was initially incorporated into a polymer-tethered DOPC bilayer (inner monolayer also contains 5 mol % diC18M50) and its dimerization behavior was analyzed using the PCH method following the procedures described in the Materials and Methods section. Specifically, PCH analysis of bilayer-reconstituted uPAR was conducted in the absence of any ligands, as well as in the presence of uPA and VN, respectively. Fig. 2 illustrates PCH data + fitting curves (left) and resulting xdimer values (right). Typically, uPAR was visualized using an Alexa-555-tagged anti-DDK (FLAG) MAb, which binds to the DDK tag near the C-terminal region, close to the GPI anchor, of the genetically modified uPAR (domain D3 in Fig. 1). As illustrated in Fig. 2, in the absence of any ligands, the obtained value of xdimer = 0.42 suggests substantial populations of both uPAR monomers and dimers. Remarkably, uPA addition almost completely suppresses uPAR dimer formation. In contrast, in the presence of VN (without uPA), receptor dimerization levels increase relative to the ligand-free case. The data in Fig. 2 are significant because they illustrate that uPAR dimerization critically depends on the type of ligand exposure. To confirm our PCH data, we next analyzed the lateral mobility of uPAR in a polymer-tethered DOPC bilayer for the same types of ligand exposure conditions using FCS autocorrelation analysis. FCS autocorrelation curves from these experiments are provided in Fig. S4. This method was pursued because the lateral diffusion of membrane proteins in a planar solid-supported model membrane, as employed herein, represents an alternative parameter that sensitively reflects changes in receptor oligomerization state in the membrane (38). Indeed, the observed trend in the identified diffusion coefficients (t-test: p < 0.01) of D = 1.37 ± 0.26 μm2/s (no ligand), D = 1.65 ± 0.52 μm2/s (uPA added), and D = 1.02 ± 0.34 μm2/s (VN added) coincides with the findings from the PCH analysis in Fig. 2. Each listed diffusion coefficient is based on 40 FCS autocorrelation analysis readings from 10 different bilayer samples (provided error values represent corresponding standard deviations). VN addition leads to the lowest lateral mobility, which can be rationalized in terms of the highest level of uPAR dimerization. In contrast, presence of uPA results in the highest uPAR lateral mobility associated with the lowest level of uPAR dimerization. As expected, in the absence of ligands, intermediate uPAR dimerization levels lead to intermediate uPAR diffusion coefficients. Interestingly, the lateral mobility of uPAR + uPA is almost indistinguishable with the corresponding value of TRITC-DHPE of D = 1.68 ± 0.20 μm2/s obtained in a comparable lipid composition, thus suggesting predominant uPAR monomer formation in the presence of uPA (30). Notably, the described PCH and FCS data are in very good qualitative agreement with corresponding results of uPAR behavior in human embryonic kidney 293 cells reported previously (13). In these cellular studies, VN addition also resulted in enhanced uPAR dimer formation, whereas uPA and uPA-PAI-I addition, respectively, caused the breakup of uPAR dimers into monomers. It should be pointed out that previous reports about uPAR dimerization levels without ligands have been somewhat inconsistent ranging from predominantly monomeric to almost completely dimerized states (39,40). Currently, the underlying molecular mechanisms of uPAR dimerization remain unclear. However, the structures of soluble form uPAR (suPAR)-VN suPAR-VN and suPAR-uPA complexes have been resolved (41). These studies show that both uPA and VN interact with the D1 and D2 domains of uPAR, albeit at opposite sides of the receptor. Interestingly, ligand-induced changes in the receptor oligomerization state have also been reported on several other membrane proteins including integral and lipid-anchored proteins (42–44). In these cases, contact between ligands and receptor is often associated with the formation of connecting structures, such as dimerization loops (43) or disulfide bonds (45), which promote receptor dimerization/oligomerization. Dimerization levels of GPI-anchored proteins may also be influenced by multiple ligands (44).
Figure 2.
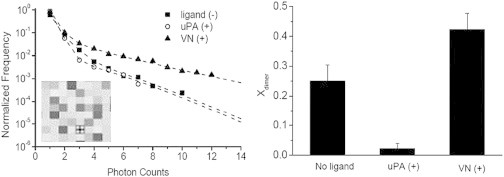
PCH data and fitting curves (left) and determined fraction of dimers, xdimer, of uPAR, uPAR + uPA, and uPAR + VN (right) in a polymer tethered DOPC bilayer. Typical CS-XY scan data of uPAR in such a bilayer environment, which lack any large-scale phase separations, are provided as an inset together with the PCH data (Box size 4 × 4 μm2). The xdimer data (ANOVA test: p < 0.01) suggest qualitatively different behavior of uPA and VN on uPAR dimerization levels. Note that, due to sample-specific differences in brightness and background, PCH curves of different xdimer may look similar (detailed description of uPAR dimerization analysis in the Supporting Material section). Each xdimer histogram bar is based on 30 individual PCH readings from 6 bilayers. Error bars represent corresponding standard deviations. ANOVA test for each presented xdimer value was conducted with 3 data sets of 10 PCH readings per data set.
As reported by earlier coimmunoprecipitation experiments exhibiting reduction of uPAR-uPAR interactions with increasing uPAR cleavage (13), the observed uPA-mediated suppression of uPAR dimerization may be caused, at least in part, by uPA-induced cleavage of the D1 domain of the uPAR receptor. N-terminal sequencing of cleaved and uncleaved uPAR from monocyte-like cells previously showed that uPA causes the removal of the D1 domain under physiological conditions by cleaving two specific sites at the D1-D2 linking region (46–48). Remarkably, soluble uPAR, which lacks the GPI anchor, was found to be less prone to uPA-mediated cleavage of the D1 domain than uPAR with the GPI anchor (49). To probe the proteolytic activity of uPA on the urokinase receptor in the lipid bilayer, we conducted a set of experiments using an alternate antibody for uPAR, one that interacts with the N-terminus at the D1 domain. For these experiments, the N-terminal antibody was conjugated to Alexa 555 and the fluorescence intensity was monitored through the Alexa-555 channel of the Confocor FCS system in solution, above the uPAR-containing bilayer, over time. This fluorescence-based assay was chosen because of its unparalleled single molecule sensitivity, which is warranted in light of the very low protein concentrations (∼1.0 × 10−11 mol) in the bilayer sample. Traditional methods, such as Western blotting, were found to be not sensitive enough. Fig. 3 illustrates results from these experiments. Specifically, uPAR tagged with anti-uPAR MAb (targets N-terminus, domain D1) was investigated under three different conditions 1), uPAR + uPA; 2), uPAR (without ligands); and 3), uPAR + VN. Fig. 3 shows that an increase of the fluorescence intensity in solution over time can only be observed in the case of anti-uPAR (N-terminal) antibody + uPA. To further understand these results, uPAR was also labeled with the anti-DDK MAb that binds to the DDK tag between D3 domain and GPI anchor, uPA was added and the intensity in solution above the bilayer was monitored. In this circumstance, no increase in fluorescence intensity was observed (data not shown). Together, these two results support a mechanism in which uPA induces a cleavage within the extracellular part of the urokinase receptor. The lack of fluorescence intensity increase in the presence of Alexa-555 labeled anti-uPAR (N-terminal) antibody + VN demonstrates, furthermore, the different influence of uPA and VN on uPAR cleavage, which is consistent with corresponding cellular studies (31,46,48). Unfortunately, the N-terminal anti-uPAR antibody showed significant non-specific adsorption on the bilayer surface, which prevented a more quantitative analysis of cleaved and uncleaved uPAR in the bilayer using PCH analysis (data not shown). Nevertheless, the findings in Figs. 2 and 3 are significant because they illustrate a tantalizing relationship between ligand-dependent dimerization and cleavage of uPAR, which is in good agreement with previous results on this receptor in a plasma membrane environment (46,47). Another notable result is that uPA-initiated cleavage of bilayer-reconstituted uPAR can be accomplished without the presence of adjuvant proteins, cofactors, or even CHOL, because these experiments were conducted on model DOPC bilayers in PBS solution. Our results in Fig. 3 are plausible in light of earlier reports regarding uPA-mediated uPAR cleavage in plasma membranes, which identified the linking region between D1 and D2 as the cleavage site for uPA (48–50). Specifically, uPA-induced cleavage was determined to occur between Arg83 and Ala84 and between Arg89 and Ser90, in the D1-D2 linking region of the uPAR receptor (49).
Figure 3.
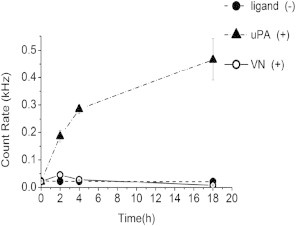
Time evolution of confocal photon count rate in solution, above the planar polymer-tethered lipid bilayer with incorporated uPAR. Experiments were conducted on uPAR (without ligands), uPAR + uPA, and uPAR + VN. The statistically significant (ANOVA test: p < 0.05) increase of fluorescence count rate in the presence of uPA supports a mechanism of uPA-mediated cleavage of the D1 domain of uPAR. Fluorescence intensity data were collected from 5 bilayer samples and 10 readings per bilayer were recorded at each individual time spot (standard deviations indicated by error bars). ANOVA test was conducted with 5 data sets of 10 fluorescence readings per data set.
CHOL content does not affect uPAR dimerization
GPI-APs are well known to be apically sorted in several polarized epithelial cells (51,52). Initially, it has been proposed that the GPI anchor acts as an apical sorting motif because GPI-APs are associated with lipid rafts (3,51). Later, this biophysical sorting concept was questioned by the finding that GPI-APs are sorted to both the apical and basolateral membrane regions of Fisher rat thyroid and MDCK cells (53,54). Interestingly, Paladino et al. previously reported that only apical GPI-APs are able to oligomerize and that the presence of CHOL is a key requirement for GPI-AP oligomerization to occur (10,55). These authors therefore proposed oligomerization of GPI-APs as a possible apical sorting mechanism, in which enhanced oligomerization of GPI-AP enhances their affinity for lipid rafts. An important question related to such a proposed mechanism is whether receptor oligomerization is dependent on CHOL content in the membrane. Currently, the picture about the relationship between membrane protein clustering and CHOL content is rather complex, as several contributing factors should be considered including protein localization and protein species. For example, Ras oligomerization shows distinct CHOL-dependencies in different parts of the cell membrane (56). On the other hand, depletion of plasma membrane CHOL was found to affect the micro-clustering of several GPI-APs, such as GH-DAF and FR-GPI (57). Considering this complexity, our model membrane system provides an attractive experimental platform to explore the influence of CHOL on protein oligomerization states.
Specifically, we systematically varied the CHOL content in binary DOPC-CHOL mixtures from 0 to 35 mol % CHOL and investigated the uPAR dimerization levels using PCH analysis. Experiments on such binary mixtures were chosen because they lack large-scale lipid-lipid phase separations. Representative PCH-data and model fits for uPAR in binary DOPC-CHOL mixtures are shown in Fig. S5. As Fig. 4 shows, uPAR dimerization levels (expressed in terms of xdimer) obtained from the PCH analysis are slightly lower in the absence of CHOL, but remain largely unchanged over the whole range of CHOL molar concentrations investigated. Of importance, the qualitatively different impact of uPA and VN on uPAR dimerization observed in a polymer-tethered DOPC bilayer (Fig. 2) remains conserved in different binary DOPC-CHOL mixtures (Fig. 4). Together these data imply that the impact of the GPI anchor and its interaction with the local model membrane environment do not play a substantial role in the uPAR dimerization process. Notably, these model membrane findings are in good agreement with corresponding results on uPAR in cell membranes, which showed that lowering of membrane CHOL does not alter uPAR dimerization and that raftophobic TM uPAR chimera dimerize with similar efficiency than their GPI-anchored counterparts (13). Furthermore, dimerization was also observed on suPAR, which lack the GPI-anchor (39). In other words, the good qualitative agreement between model and cell membrane data not only corroborates our model membrane approach, but also illustrates that the local membrane environment (i.e., composition, lipid packing density, etc.) does not directly influence uPAR dimerization levels regardless of the complexity of the membrane composition.
Figure 4.
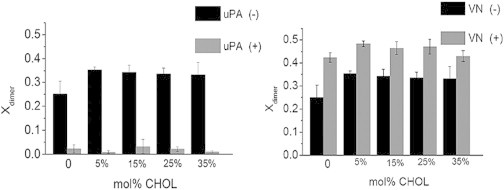
Fraction of dimers, xdimer, of uPAR, uPAR + uPA, and uPAR + VN in different binary DOPC-CHOL mixtures. The statistically significant xdimer data (ANOVA test: p < 0.01) show that uPA suppresses uPAR dimerization (left), whereas VN has the opposite effect (right). In both cases, changes in CHOL molar concentration have no notable impact on the dimerization levels of uPAR. Presented xdimer values are based on a total of 60 individual PCH readings from 6 bilayers. Error bars represent corresponding standard deviations. ANOVA test was conducted with 3 data sets of 10 PCH readings per data set.
Influence of native ligands on uPAR sequestration and dimerization in ternary raft-mimicking lipid mixtures
To explore the mechanisms that drive GPI-AP association with lipid rafts, we next explored the sequestering and oligomerization behavior of uPAR in a polymer-tethered lipid bilayer in the presence of coexisting lo and ld domains. As described in the Materials and Methods section, in this model membrane, raft-mimicking lipid-lipid phase separations are achieved by using ternary DOPC/DPPC/CHOL lipid mixtures of 33/33/33 and 36/36/28, which are characterized by equimolar DOPC-DPPC concentrations and different CHOL content of 28 and 33 mol % CHOL. The bottom leaflet (LB monolayer) of the bilayer systems also contains 5 mol % diC18M50 to lift up the bilayer from the underlying glass substrate. In each imaging experiment, ∼10−3 mol % uPAR are incorporated into the bilayer system (procedure of uPAR incorporation into the lipid bilayer described in the Materials and Methods section). To conduct fluorescence detection experiments, the lo-ld lipid-lipid phase separation was visualized using NBD-DHPE (prefers lo), whereas uPAR labeling was typically accomplished using Alexa-555 tagged anti-DDK MAbs.
Fig. 5 presents results of the uPAR sequestration in ternary DOPC/DPPC/CHOL lipid mixtures acquired through the NBD (lipid distribution) and Alexa-555 channels (uPAR distribution) of the confocal fluorescence detection system. As shown in the representative CS-XY scans of the 33/33/33 DOPC/DPPC/CHOL lipid mixture (A–H), uPAR sequestration was investigated without ligands (A, C, E, and G) and after either addition of uPA or VN (B and D (uPA); F and H (VN)). The CS-XY data in Fig. 5 show that the lo-ld lipid-lipid phase separations are well reflected by the corresponding uPAR distributions regardless of the presence or absence of uPA or VN. Moreover, comparison of NBD and Alexa-555 channel data in Fig. 5, A–H, shows that uPAR (without ligands), uPAR + uPA, and uPAR + VN all have an affinity for lo lipid regions of the bilayer. This lo phase affinity is expected because uPAR is a GPI-AP, a typical raft marker. Notably, the raftophilic behavior of GPI-APs is not only well documented by experiments on cellular membranes (58), but also confirmed in previous model membrane studies with coexisting lo-ld lipid phase separations (59,60). Fig. 5 I shows Eraft data of uPAR distribution (without ligands, as well as with uPA or VN) in the 33/33/33 DOPC/DPPC/CHOL lipid mixture. The Eraft parameter has been recently introduced to quantify the sequestering behavior of integrins in a comparable membrane system by finding the normalized preference of a protein or lipid for lo domains versus ld domains (29). As already qualitatively shown in the CS-XY scans of uPAR distribution in Fig. 5, C, D, G, and H, the Eraft data in Fig. 5 I are all positive, thus indicating the preferred affinity for the lo region of the bilayer sample. More importantly, Fig. 5 I also shows that uPA and VN influences the uPAR sequestration within coexisting lo and ld lipid phase regions in a qualitatively different way. Addition of uPA causes the reduction of Eraft, whereas VN leads to a measurable increase in this parameter relative to the ligand-free case.
Figure 5.
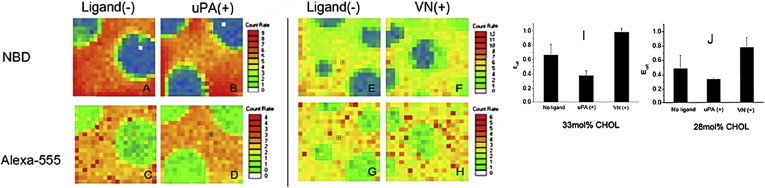
Representative CS-XY scans (raw data) of uPAR distribution in a polymer-tethered lipid bilayer with coexisting lo and ld domains (33 mol % CHOL, left) before (A, C, E, and G) and upon addition of the ligands uPA (B and D) and VN (F and H). The lo and ld lipid phase separation is visualized through the NBD channel (top row: A, B, E, and F) and the corresponding uPAR distribution is monitored by the Alexa-555 channel (bottom row: C, D, G, and H). Note that the presented CS-XY scans have different scaling. Fig. 5, I and J, represent Eraft values (t-test: p < 0.01) of uPAR (without ligands), uPAR + uPA, and uPAR + VN in ternary mixtures with 33 mol % CHOL (I) and 28 mol % CHOL (J) as determined using CS-XY analysis in a polymer-tethered lipid bilayer with coexisting lo and ld domains. Box size 10 × 10μm2. Eraft data for each ligand exposure state in Fig. 5, I and J, are based on 4 bilayers (5 readings per bilayer). Error bars represent corresponding standard deviations. t-test was conducted with two data sets consisting of 10 Eraft readings per data set.
Fig. 5 J shows the results of the corresponding Eraft analysis of uPAR sequestration obtained from the ternary 36/36/28 DOPC/DPPC/CHOL lipid mixture. Comparison of Figs. 5, I and J, reveals several interesting results. Most notably, the overall footprint of the Eraft data is very similar for ternary lipid mixtures with 28 and 33 mol % CHOL. In both cases, Eraft values obtained for uPAR, uPAR + uPA, and uPAR +VN are all positive, thereby following the order in terms of magnitude of: uPAR + uPA < uPAR < uPAR + VN. Even absolute Eraft values of both ternary lipid mixtures are comparable for a given ligand exposure state. The data in Fig. 5 J are important because they confirm the distinct impact of uPA and VN on uPAR sequestration behavior shown in Fig. 5 I. The results in Fig. 5, I and J, which indicate significant translocations of uPAR either out of (uPA) or into (VN) raft-mimicking lo phase regions of the bilayer, are intriguing in light of the observed impact of uPA and VN on uPAR dimerization in DOPC and DOPC-CHOL mixtures (Figs. 2 and 4). Together these data suggest a link between ligand-mediated regulation of uPAR dimerization and sequestration in the raft-mimicking lipid mixture.
To explore the possibility of such a link, we next pursued domain-specific PCH analysis of the laterally mobile uPAR receptors in the polymer-tethered lipid bilayer with coexisting lo and ld lipid regions. Again, ternary raft-mimicking lipid mixtures were investigated, which contain an equimolar ratio of DOPC and DPPC, as well as CHOL molar concentrations of 28 and 33 mol %, respectively. Representative PCH-data and model fits for uPAR in ternary DOPC/DPPC/CHOL mixtures are provided in Fig. S6. Fig. 6 presents the xdimer data from these PCH experiments. In excellent agreement with our findings in DOPC and DOPC-CHOL mixtures (Figs. 2 and 4), Fig. 6 shows again that uPA and VN influence the uPAR dimerization level in a qualitatively different manner. Although uPAR dimerization is substantially reduced by uPA, it is measurably enhanced in the presence of VN. These results agree well with a previous cellular study, which also shows that uPA reduces uPAR dimerization and raft partitioning, whereas uPAR dimerization and raft partitioning increase upon VN addition (13). Interestingly, Fig. 6 also shows that there is no measurable difference between xdimer values in lo and ld regions for a corresponding ligand exposure state (uPAR, uPAR + uPA, uPAR + VN) regardless of the CHOL molar concentration used. Although this result is consistent with our results on binary DOPC-CHOL mixtures in that xdimer values are largely independent of CHOL molar concentration, it does not support a mechanism, in which receptor sequestration is entirely regulated by the receptor dimerization level. Instead, the Eraft and xdimer data in Figs. 5 and 6, respectively, suggest that other contributing factors may be involved in the regulation of uPAR sequestration in coexisting lo-ld domains.
Figure 6.
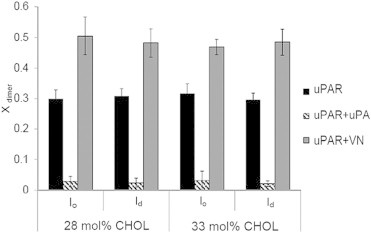
Influence of uPA and VN on xdimer of uPAR in a polymer-tethered lipid bilayer with coexisting lo and ld domains containing 28 mol % (left) and 33 mol % CHOL (right). In agreement with findings on binary phospholipid-CHOL mixtures (Fig. 4), both ligands alter xdimer (ANOVA test: p < 0.01) in a qualitatively different manner. No measurable difference in uPAR dimerization is observed in lo versus ld domains regardless of the absence or presence of ligands. Furthermore, in the ternary systems (as in the binary systems) the CHOL level does not influence the dimerization level in either the presence or absence of ligands. Determined xdimer values are based on 30 individual PCH readings from 6 bilayers. Error bars show corresponding standard deviations. ANOVA test was conducted with 3 data sets of 10 PCH readings per data set.
One potential factor of uPAR sequestration regulation could be linked to ligand-specific allosteric changes of membrane receptors affecting receptor-bilayer interactions. For example, using αvβ3 and α5β1 integrins in corresponding model membrane systems, we showed that addition of native ligands VN (αvβ3) and fibronectin (α5β1) leads to substantial integrin translocations from ld to lo phase regions of the bilayer, which can be attributed to ligand-induced allosteric changes of these membrane receptors (29). Because such allosteric changes are known to not only affect the extracellular part of integrins, but also their TM and cytosolic regions (61,62), the observed ligand-mediated integrin translocations to lo regions could be rationalized in terms of different hydrophobic matching conditions. Similarly, the affinity of the multispan TM protein perfringolysin O for ordered lipid environments has been interpreted in terms of hydrophobic matching arguments (63). In the case of GPI-APs, such as uPAR, relevant allosteric changes affecting receptor sequestration should be limited to the protein’s extracellular region. Indeed, ligand-induced conformational changes of the extracellular region of uPAR have been reported previously (31,63). Interestingly, the weak dependence of CHOL concentration on uPAR dimerization and sequestration also suggests that the role of the GPI anchor in receptor sequestration changes is rather limited. Although, the observed lo-phase preference of uPAR with or without ligands depicted in Fig. 5 supports the notion that the GPI-anchor represents the intrinsic molecular motif that determines the raft affinity of GPI-APs.
In light of our findings, we should point out that experiments on ternary raft-mimicking model membrane mixtures consisting of monounsaturated lipids, saturated lipids, and CHOL may be limited in their ability to test the hypothesis that the dimerization level of GPI-APs can be seen as a membrane protein sorting principle in the presence of raft domains in cellular membranes. The main reason for caution is that, unlike their counterparts in cellular membranes, lo and ld domains in model ternary lipid mixtures are characterized by surprisingly similar lipid compositions (i.e., significant amounts of monounsaturated lipids and CHOL can be found in both lo and ld domains) (64,65). Consequently, differences in lipid packing between both lo and ld regions, a parameter affecting protein sequestration, is expected to be less significant (relative to raft and nonraft regions in cellular membranes). In other words, our experiments cannot completely exclude the possibility that changes in receptor dimerization represent a key mechanism that regulates sequestration of GPI-APs in cellular membranes.
Conclusion
The current study shows that raft-mimicking model membrane mixtures in combination with single molecule-sensitive detection techniques represent an attractive tool to explore the influence of native ligands on sequestration and dimerization in biological membranes. A particular strength of such a model membrane approach is that raft-associated processes of membrane proteins can be investigated without artificial cross-linking agents, which may impair experimental findings. Specifically, our experimental strategy was applied to explore the influence of native ligands (uPA, VN) on dimerization level and sequestering behavior of uPAR, a GPI-anchored membrane protein. Most notably, our work shows that VN and uPA not only have a distinct influence on uPAR dimerization levels in the bilayer, but also impact uPAR sequestration in the presence of coexisting lo and ld domains in a ligand-specific way. This finding is in good agreement with previous results about the role of ligands in uPAR dimerization and sequestration in cellular membranes, which validates our model membrane approach (13,66). The described model membrane experiments also show that variations of CHOL content do not have a comparable influence on uPAR dimerization. The latter finding suggests that the GPI anchor and the local lipid environment, regardless of complexity, are less significant in the regulation of uPAR dimerization than the extracellular region of this GPI-AP. The current study illustrates that model membrane experiments can play an important role in supporting cell membrane experiments by confirming membrane-associated molecular processes under less complex conditions. At the same time, our PCH experiments of uPAR in ternary raft-mimicking lipid mixtures also exhibit that the model membrane strategy may be limited in its ability to make accurate quantitative predictions about the sequestration of receptor monomers and dimers in complex plasma membrane environments. Despite these limitations, this study is significant because it illustrates the potentially important role of native ligands in the regulation of membrane protein sequestration in the presence of lipid rafts. In that sense, our results may contribute to a better understanding about the differential effects of ligands in the assembly of protein signaling complexes in such functional domains.
Acknowledgments
This work was supported in part by the National Science Foundation (grant MCB-0920134), the Indiana University-Purdue University Indianapolis Nanoscale Imaging Center, and the Integrated Nanosystems Development Institute.
Supporting Material
References
- 1.Edidin M. Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends Cell Biol. 2001;11:492–496. doi: 10.1016/s0962-8924(01)02139-0. [DOI] [PubMed] [Google Scholar]
- 2.Kusumi A., Suzuki K.G.N., Fujiwara T.K. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem. Sci. 2011;36:604–615. doi: 10.1016/j.tibs.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Simons K., Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 4.Brown D.A., London E. Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J. Biol. Chem. 2000;275:17221–17224. doi: 10.1074/jbc.R000005200. [DOI] [PubMed] [Google Scholar]
- 5.Holowka D., Gosse J.A., Baird B. Lipid segregation and IgE receptor signaling: a decade of progress. BBA- Mol. Cell Res. 2005;1746:252–259. doi: 10.1016/j.bbamcr.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 6.Pelkmans L. Secrets of caveolae-and lipid raft-mediated endocytosis revealed by mammalian viruses. BBA- Mol. Cell Res. 2005;1746:295–304. doi: 10.1016/j.bbamcr.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 7.Carman C.V., Springer T.A. Integrin avidity regulation: are changes in affinity and conformation underemphasized? Curr. Opin. Cell Biol. 2003;15:547–556. doi: 10.1016/j.ceb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 8.Brown D.A., London E. Functions of lipid rafts in biological membranes. Annu. Rev. Cell Dev. Biol. 1998;14:111–136. doi: 10.1146/annurev.cellbio.14.1.111. [DOI] [PubMed] [Google Scholar]
- 9.Seddon A.M., Curnow P., Booth P.J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta. Biomembr. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Paladino S., Sarnataro D., Zurzolo C. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J. Cell Biol. 2004;167:699–709. doi: 10.1083/jcb.200407094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki K.G.N., Kasai R.S., Kusumi A. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat. Chem. Biol. 2012;8:774–783. doi: 10.1038/nchembio.1028. [DOI] [PubMed] [Google Scholar]
- 12.Lemmon M.A., Schlessinger J. Regulation of signal transduction and signal diversity by receptor oligomerization. Trends Biochem. Sci. 1994;19:459–463. doi: 10.1016/0968-0004(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham O., Andolfo A., Sidenius N. Dimerization controls the lipid raft partitioning of uPAR/CD87 and regulates its biological functions. EMBO J. 2003;22:5994–6003. doi: 10.1093/emboj/cdg588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fallahi-Sichani M., Linderman J.J. Lipid raft-mediated regulation of G-protein coupled receptor signaling by ligands which influence receptor dimerization: a computational study. PLoS ONE. 2009;4:e6604. doi: 10.1371/journal.pone.0006604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simons K., Gerl M.J. Revitalizing membrane rafts: new tools and insights. Nat. Rev. Mol. Cell Biol. 2010;11:688–699. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 16.Owen D.M., Magenau A., Gaus K. The lipid raft hypothesis revisited—new insights on raft composition and function from super-resolution fluorescence microscopy. BioEssays. 2012;34:739–747. doi: 10.1002/bies.201200044. [DOI] [PubMed] [Google Scholar]
- 17.Lingwood D., Simons K. Lipid rafts as a membrane-organizing principle. Science. 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 18.Ganguly S., Chattopadhyay A. Cholesterol depletion mimics the effect of cytoskeletal destabilization on membrane dynamics of the serotonin1A receptor: a zFCS study. Biophys. J. 2010;99:1397–1407. doi: 10.1016/j.bpj.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lichtenberg D., Goñi F.M., Heerklotz H. Detergent-resistant membranes should not be identified with membrane rafts. Trends Biochem. Sci. 2005;30:430–436. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 20.Veatch S.L., Keller S.L. Organization in lipid membranes containing cholesterol. Phys. Rev. Lett. 2002;89:268101. doi: 10.1103/PhysRevLett.89.268101. [DOI] [PubMed] [Google Scholar]
- 21.Feigenson G.W. Phase diagrams and lipid domains in multicomponent lipid bilayer mixtures. Biochim. Biophys. Acta. 2009;1788:47–52. doi: 10.1016/j.bbamem.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumgart T., Hammond A.T., Webb W.W. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc. Natl. Acad. Sci. USA. 2007;104:3165–3170. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahya N., Brown D.A., Schwille P. Raft partitioning and dynamic behavior of human placental alkaline phosphatase in giant unilamellar vesicles. Biochemistry. 2005;44:7479–7489. doi: 10.1021/bi047429d. [DOI] [PubMed] [Google Scholar]
- 24.Sengupta P., Hammond A., Baird B. Structural determinants for partitioning of lipids and proteins between coexisting fluid phases in giant plasma membrane vesicles. Biochim. Biophys. Acta. Biomembr. 2008;1778:20–32. doi: 10.1016/j.bbamem.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milhiet P.-E., Gubellini F., Lévy D. High-resolution AFM of membrane proteins directly incorporated at high density in planar lipid bilayer. Biophys. J. 2006;91:3268–3275. doi: 10.1529/biophysj.106.087791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner M.L., Tamm L.K. Tethered polymer-supported planar lipid bilayers for reconstitution of integral membrane proteins: silane-polyethyleneglycol-lipid as a cushion and covalent linker. Biophys. J. 2000;79:1400–1414. doi: 10.1016/S0006-3495(00)76392-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deverall M.A., Gindl E., Naumann C.A. Membrane lateral mobility obstructed by polymer-tethered lipids studied at the single molecule level. Biophys. J. 2005;88:1875–1886. doi: 10.1529/biophysj.104.050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purrucker O., Gönnenwein S., Tanaka M. Polymer-tethered membranes as quantitative models for the study of integrin-mediated cell adhesion. Soft Matter. 2007;3:333–336. doi: 10.1039/b612069e. [DOI] [PubMed] [Google Scholar]
- 29.Siegel A.P., Kimble-Hill A., Naumann C.A. Native ligands change integrin sequestering but not oligomerization in raft-mimicking lipid mixtures. Biophys. J. 2011;101:1642–1650. doi: 10.1016/j.bpj.2011.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hussain N.F., Siegel A.P., Naumann C.A. Bilayer asymmetry influences integrin sequestering in raft-mimicking lipid mixtures. Biophys. J. 2013;104:2212–2221. doi: 10.1016/j.bpj.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith H.W., Marshall C.J. Regulation of cell signalling by uPAR. Nat. Rev. Mol. Cell Biol. 2010;11:23–36. doi: 10.1038/nrm2821. [DOI] [PubMed] [Google Scholar]
- 32.Blasi F., Carmeliet P. uPAR: a versatile signalling orchestrator. Nat. Rev. Mol. Cell Biol. 2002;3:932–943. doi: 10.1038/nrm977. [DOI] [PubMed] [Google Scholar]
- 33.Lüdtke K., Jordan R., Naumann C.A. Lipopolymers from new 2-substituted-2-oxazolines for artificial cell membrane constructs. Macromol. Biosci. 2005;5:384–393. doi: 10.1002/mabi.200500004. [DOI] [PubMed] [Google Scholar]
- 34.Naumann C., Prucker O., Frank C.W. The polymer-supported phospholipid bilayer: tethering as a new approach to substrate-membrane stabilization. Biomacromolecules. 2002;3:27–35. doi: 10.1021/bm0100211. [DOI] [PubMed] [Google Scholar]
- 35.Garg S., Rühe J., Naumann C.A. Domain registration in raft-mimicking lipid mixtures studied using polymer-tethered lipid bilayers. Biophys. J. 2007;92:1263–1270. doi: 10.1529/biophysj.106.091082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y., Müller J.D., Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys. J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murcia M.J., Minner D.E., Naumann C.A. Design of quantum dot-conjugated lipids for long-term, high-speed tracking experiments on cell surfaces. J. Am. Chem. Soc. 2008;130:15054–15062. doi: 10.1021/ja803325b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziemba B.P., Falke J.J. Lateral diffusion of peripheral membrane proteins on supported lipid bilayers is controlled by the additive frictional drags of (1) bound lipids and (2) protein domains penetrating into the bilayer hydrocarbon core. Chem. Phys. Lipids. 2013;172-173:67–77. doi: 10.1016/j.chemphyslip.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shliom O., Huang M., Higazi A.A. Novel interactions between urokinase and its receptor. J. Biol. Chem. 2000;275:24304–24312. doi: 10.1074/jbc.M002024200. [DOI] [PubMed] [Google Scholar]
- 40.Hellriegel C., Caiolfa V.R., Zamai M. Number and brightness image analysis reveals ATF-induced dimerization kinetics of uPAR in the cell membrane. FASEB J. 2011;25:2883–2897. doi: 10.1096/fj.11-181537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huai Q., Zhou A., Huang M. Crystal structures of two human vitronectin, urokinase and urokinase receptor complexes. Nat. Struct. Mol. Biol. 2008;15:422–423. doi: 10.1038/nsmb.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- 43.Mellroth P., Karlsson J., Steiner H. Ligand-induced dimerization of Drosophila peptidoglycan recognition proteins in vitro. Proc. Natl. Acad. Sci. USA. 2005;102:6455–6460. doi: 10.1073/pnas.0407559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arthur J.F., Shen Y., Gardiner E.E. Ligand binding rapidly induces disulfide-dependent dimerization of glycoprotein VI on the platelet plasma membrane. J. Biol. Chem. 2007;282:30434–30441. doi: 10.1074/jbc.M701330200. [DOI] [PubMed] [Google Scholar]
- 45.Montuori N., Carriero M.V., Ragno P. The cleavage of the urokinase receptor regulates its multiple functions. J. Biol. Chem. 2002;277:46932–46939. doi: 10.1074/jbc.M207494200. [DOI] [PubMed] [Google Scholar]
- 46.Sidenius N., Sier C.F.M., Blasi F. Shedding and cleavage of the urokinase receptor (uPAR): identification and characterisation of uPAR fragments in vitro and in vivo. FEBS Lett. 2000;475:52–56. doi: 10.1016/s0014-5793(00)01624-0. [DOI] [PubMed] [Google Scholar]
- 47.Høyer-Hansen G., Rønne E., Danø K. Urokinase plasminogen activator cleaves its cell surface receptor releasing the ligand-binding domain. J. Biol. Chem. 1992;267:18224–18229. [PubMed] [Google Scholar]
- 48.Høyer-Hansen G., Pessara U., Behrendt N. Urokinase-catalysed cleavage of the urokinase receptor requires an intact glycolipid anchor. Biochem. J. 2001;358:673–679. doi: 10.1042/0264-6021:3580673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plesner T., Behrendt N., Ploug M. Structure, function and expression on blood and bone marrow cells of the urokinase-type plasminogen activator receptor, uPAR. Stem Cells. 1997;15:398–408. doi: 10.1002/stem.150398. [DOI] [PubMed] [Google Scholar]
- 50.Lisanti M.P., Caras I.W., Rodriguez-Boulan E. A glycophospholipid membrane anchor acts as an apical targeting signal in polarized epithelial cells. J. Cell Biol. 1989;109:2145–2156. doi: 10.1083/jcb.109.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benting J.H., Rietveld A.G., Simons K. N-Glycans mediate the apical sorting of a GPI-anchored, raft-associated protein in Madin-Darby canine kidney cells. J. Cell Biol. 1999;146:313–320. doi: 10.1083/jcb.146.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown D.A., Rose J.K. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 53.Lipardi C., Nitsch L., Zurzolo C. Detergent-insoluble GPI-anchored proteins are apically sorted in fischer rat thyroid cells, but interference with cholesterol or sphingolipids differentially affects detergent insolubility and apical sorting. Mol. Biol. Cell. 2000;11:531–542. doi: 10.1091/mbc.11.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paladino S., Lebreton S., Zurzolo C. Different GPI-attachment signals affect the oligomerization of GPI-anchored proteins and their apical sorting. J. Cell Sci. 2008;121:4001–4007. doi: 10.1242/jcs.036038. [DOI] [PubMed] [Google Scholar]
- 55.Prior I.A., Muncke C., Hancock J.F. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J. Cell Biol. 2003;160:165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedrichson T., Kurzchalia T.V. Microdomains of GPI-anchored proteins in living cells revealed by cross-linking. Nature. 1998;394:802–805. doi: 10.1038/29570. [DOI] [PubMed] [Google Scholar]
- 57.Harder T., Scheiffele P., Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J. Cell Biol. 1998;141:929–942. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jacobson K., Dietrich C. Looking at lipid rafts? Trends Cell Biol. 1999;9:87–91. doi: 10.1016/s0962-8924(98)01495-0. [DOI] [PubMed] [Google Scholar]
- 59.Lingwood D., Ries J., Simons K. Plasma membranes are poised for activation of raft phase coalescence at physiological temperature. Proc. Natl. Acad. Sci. USA. 2008;105:10005–10010. doi: 10.1073/pnas.0804374105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 61.Takada Y., Ye X., Simon S. The integrins. Genome Biol. 2007;8:215.1–215.9. doi: 10.1186/gb-2007-8-5-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin Q., London E. Altering hydrophobic sequence lengths shows that hydrophobic mismatch controls affinity for ordered lipid domains (rafts) in the multitransmembrane strand protein perfringolysin O. J. Biol. Chem. 2013;288:1340–1352. doi: 10.1074/jbc.M112.415596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huai Q., Mazar A.P., Huang M. Structure of human urokinase plasminogen activator in complex with its receptor. Science. 2006;311:656–659. doi: 10.1126/science.1121143. [DOI] [PubMed] [Google Scholar]
- 64.Li L., Wang H., Cheng J.-X. Quantitative coherent anti-Stokes Raman scattering imaging of lipid distribution in coexisting domains. Biophys. J. 2005;89:3480–3490. doi: 10.1529/biophysj.105.065607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bunge A., Müller P., Huster D. Characterization of the ternary mixture of sphingomyelin, POPC, and cholesterol: support for an inhomogeneous lipid distribution at high temperatures. Biophys. J. 2008;94:2680–2690. doi: 10.1529/biophysj.107.112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Caiolfa V.R., Zamai M., Sidenius N. Monomer dimer dynamics and distribution of GPI-anchored uPAR are determined by cell surface protein assemblies. J. Cell Biol. 2007;179:1067–1082. doi: 10.1083/jcb.200702151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


