Abstract
Nucleotide selection is essential for fidelity control in gene replication and transcription. Recent work on T7 RNA polymerase suggested that a small posttranslocation free energy bias stabilizes Tyr639 in the active site to aid nucleotide selection. However, it was not clear exactly how Tyr639 assists the selection. Here we report a molecular-dynamics simulation study revealing atomistic detail of this critical selectivity. The study shows first that Tyr639 blocks the active site at posttranslocation by marginally stacking to the end basepair of the DNA-RNA hybrid. The study then demonstrates that at the nucleotide preinsertion state, a cognate RNA nucleotide does not affect the local Tyr639 stabilization, whereas a noncognate nucleotide substantially stabilizes Tyr639 so that Tyr639 keeps blocking the active site. As a result, further nucleotide insertion into the active site, which requires moving Tyr639 out of the site, would be hindered for the noncognate nucleotide, but not for the cognate nucleotide. In particular, we note that water molecules assist the ribose recognition in the RNA nucleotide preinsertion, and help Tyr639 stacking to the end basepair in the case of a DNA nucleotide. It was also seen that a base-mismatched nucleotide at preinsertion directly grabs Tyr639 for the active site stabilization. We also find that in a mutant polymerase Y639F the strong stabilization of residue 639 in the active site cannot establish upon the DNA nucleotide preinsertion. The finding explains the reduced differentiation between ribo- and deoxyribonucleotides that has been recorded experimentally for the mutant polymerase.
Introduction
Gene replication and transcription are fundamental processes in molecular biology. The essential enzymes that direct the processes are DNA and RNA polymerases (1). The polymerase copies information from template DNA/RNA to a newly synthesized polynucleic strand, largely based on Watson-Crick (WC) basepairing between the template nucleotide (nt) and the incoming nucleoside triphosphate (NTP). The basepairing is fully established as the incoming NTP inserts into the active site of the polymerase, followed by chemical addition of the nucleoside monophosphate moiety to the synthesizing strand along with pyrophosphate ion dissociation. In the absence of the polymerase, polymerization is slow and error-prone, due to a large activation barrier for chemical reaction and a small free energy differentiation between cognate (correct) and noncognate (incorrect) nucleotide incorporation in free solution (2). The polymerase enzyme tremendously accelerates the polymerization and improves fidelity by recruiting the nucleotide into a dehydrated or partially dehydrated environment (3). Nucleotide selection is accordingly performed upon the entry of the nucleotide, and continues until the end of the chemical addition. Next, proofreading or editing can also be employed in some of the polymerases, so that errors leaked from previous selection steps can be corrected to improve fidelity of the polymerization (4–7).
Modeling and computational studies in recent years on polymerases have provided fruitful insights into molecular mechanisms of the elongation (8–15). In particular, molecular dynamics (MD) simulations are able to zoom into atomistic detail of the polymerases at various stages of the elongation (16–20). Nevertheless, there have been only a few studies that focused on fidelity control of the polymerases (8,9,12,14), and even fewer on their nucleotide selectivity (14). In this study, we use a prototypical polymerase from bacterial phage T7 to probe its nucleotide selectivity as a basic way of fidelity control, employing the MD simulation.
T7 RNAP polymerase (RNAP) is a single-subunit transcription enzyme, with a handlike structure that is also representative of a group of DNA polymerases for gene replication (21). The finger subdomain of the enzyme switches from open to closed to recruit an incoming NTP from a preinsertion binding site (22), which is relatively accessible to the bulk solution, into the active site that is more buried inside the palm subdomain. Experiments showed that T7 RNAP achieves an error rate of ∼10−4 in elongation, with no proofreading activities detected (23). Hence, the fidelity control of this system seems to fully rely on nucleotide selection during a multistep incorporation process (24): upon NTP preinsertion; from preinsertion to insertion; upon NTP full insertion; and during chemical addition or catalysis. In this study, we focus on the selectivity of T7 RNAP starting from NTP preinsertion to insertion. The process is highly coordinated by a critical amino-acid residue Tyr639, which we illustrate below.
T7 RNAP is comparatively well studied from kinetic ensemble measurements (24) to studies at the single molecule level, with both high-resolution structural characterization and single-molecule dynamic measurements (22,25–27). In our previous modeling work on T7 RNAP, it was suggested that a small posttranslocation free energy bias aids the nucleotide selection of this polymerase (28). Basically, the polymerase works as a Brownian ratchet (29) that oscillates freely between a pretranslocated and a posttranslocated state before NTP binding, presumably with no free energy bias in the translocation. NTP binding and insertion then inhibit the backward movements of the polymerase, and consequently act as a pawl to support forward elongation. Interestingly, however, a small free energy bias toward the posttranslocated state was detected from single-molecule experiments (26,27). Kinetic modeling combined with structural analyses indicated that a residue Tyr639 is stabilized by the small free energy bias at the active site upon posttranslocation and is not displaced until full nucleotide insertion (22). Hence, the residue can closely assist nucleotide selection upon nucleotide preinsertion as well as during the nucleotide insertion (28).
The RNA polymerase is specialized in synthesizing RNA rather than DNA strands, and previous biochemical study on T7 RNAP indicated that Tyr639 is essential in differentiating a ribo-nucleotide (rNTP) from a deoxy-ribonucleotide (dNTP) (30). It is remarkable in that study that a mutant polymerase Y639F works simultaneously as an RNA polymerase and as a DNA polymerase: The mutant gains significantly enhanced activities in the dNTP incorporation, compared to the wild-type (WT) polymerase, while it maintains the WT level of activity in the rNTP incorporation. Indeed, the residue at the position of Y639 has been found to be highly conserved as either tyrosine or phenylalanine across several species of DNA and RNA polymerases (31,32). The homologous tyrosine or phenylalanine at this position in some of the Pol-I family DNA polymerases has been found to be critical for distinguishing deoxy- and dideoxy-ribonucleotides (33).
In T7 RNAP, it is highly likely that Tyr639 participates in the nucleotide selection during the nucleotide insertion. Tyr639 is located at the C-terminal of an O-helix structure on the finger domain, which maintains an open configuration from the posttranslocation to the nucleotide binding or preinsertion state. Tyr639 occupies the active site during this stage, occluding nucleotide insertion (22,25). The preinsertion site is located near the active site, so that Tyr639 is spatially available to associate closely with the preinserted NTP. A previous structural study suggested that Tyr639 distinguishes ribose from deoxy-ribose at preinsertion via the coordinated bond interaction, which is mediated by a magnesium ion located between the Tyr639 hydroxyl group and 2′-OH of the rNTP ribose (22). Nevertheless, details regarding the structural dynamics were lacking to determine the differentiation mechanism. Indeed, to allow the nucleotide insertion, Tyr639 has to move out of the active site during the insertion process. At the same time, the O-helix along with the finger domain rotates from open to closed to assist the nucleotide insertion into the active site (25). Hence, the selectivity of Tyr639 can be manifested from the nucleotide preinsertion to the insertion.
Initial nucleotide selection can start immediately upon the nucleotide entry to the binding or preinsertion site, as the noncognate nucleotide is rejected quickly back to solution with a large unbinding or off-rate. Selection can then happen during the process of nucleotide insertion, as the noncognate nucleotide is inhibited from inserting into the active site with a low rate of the insertion. Further nucleotide selection can happen upon the full insertion of the nucleotide, and through catalysis. However, because Tyr639 moves away from the active site upon the full nucleotide insertion, we consider only the earlier two steps of nucleotide selection as relevant to the Tyr639 selectivity.
First, due to the significant role of Tyr639 in differentiating rNTP and dNTP (30), it is natural to wonder how exactly the differentiation arises, in contrast to the mutant Y639F that exhibits the diminished differentiation. Second, we questioned whether Tyr639 also distinguishes the base-mismatched nucleotides from the matched ones, a capability that is rendered only when the nucleotide base becomes distinguishable according to the template nt. We then wanted to find out whether Tyr639 participates in both the nucleotide preinsertion rejection and insertion inhibition, as mentioned above for the two early steps of the nucleotide selection.
In this work, we implemented atomistic MD simulations to probe the detailed mechanisms of nucleotide selection assisted by Tyr639 in T7 RNAP elongation. The focus is on how the polymerase detects the nucleotide species upon preinsertion, before the full nucleotide insertion and chemical addition. As a control, we first examined how Tyr639 is stabilized at the posttranslocated state right before NTP binds/preinserts. We then systematically studied whether, and how, Tyr639 senses the cognate and noncognate NTPs upon preinsertion. Multiple species of nucleotides binding individually to the preinsertion site were simulated, revealing common characteristics that differentiate the correct species from the incorrect ones. We present analyses of local energetic contributions to the stabilization of Tyr639 at the active site upon different nucleotide preinsertion. Comparative analyses on the mutant polymerase Y639F are also provided. A mechanism for Tyr639 assisting the early nucleotide selection in T7 RNAP elongation is consequently proposed.
Methods
The MD simulations were performed using the GROMACS-4.6.5 software package (34,35). The AMBER99sb force field with PARMBSC0 nucleic-acid parameters was used (36–39). The ATP/GTP parameters were obtained from Meagher et al. (40). The parameters of dATP, UTP, and CTP were obtained by comparing the monophosphate in the AMBER force field with that of triphosphate in the ATP/GTP. The high-resolution structures of the T7 RNAP elongation complex (protein, nucleic acids, or NAs, with or without the NTP substrate) were captured by crystallization studies in four conformational states: posttranslocated, preinsertion, substrate-insertion, and product (with PDB:1MSW, PDB:1S0V, PDB:1S76, and PDB:1S77, respectively) (22,25,41), from among which we intensively simulated the former two structures in this work. In particular, preinsertion of ATP, GTP, dATP, UTP, and CTP were simulated in the WT polymerase, whereas preinsertion of ATP and dATP were simulated in the mutant polymerase Y639F. Missing residues in one conformational state of the polymerase complex were rebuilt by using the corresponding parts from another state(s). The crystal waters within 10 Å of ATP molecule and two Mg2+ ions in the active site were retained.
The RNAP complex was solvated with explicit TIP3P water (42) in a cubic box and the minimum distance from the protein to the wall was 10 Å. To neutralize the system and make the salt concentration 0.1 M, 115 Na+ ions and 81 Cl− ions were added. The entire simulation system contained ∼134,000 atoms. The van der Waals (vdW) and short-range electrostatic interactions used a cutoff at 10 Å. Long-range electrostatic interactions were treated by the particle-mesh Ewald method (43,44). The MD simulations were run at 1 bar and 310 K using the Parrinello-Rahman barostat (45,46) and the velocity rescaling thermostat (47), respectively. The LINCS algorithm was used to constrain all the chemical bonds (48). The time-step was 2 fs and the neighbor list was updated every five steps. The solvated system was minimized with the steepest-descent algorithm, followed by 100 ps of MD simulation within the canonical ensemble and 200 ps of simulation within the NPT ensemble. Position restraints on the heavy atoms of the protein and NA chains were imposed at the beginning of the simulation. After the constrained simulation, unconstrained MD simulations were carried out for 100 ns, for each preinsertion or posttranslocation elongation complex, within the NPT ensemble.
To examine local associations among key structural elements around the active site (residue 639, the end basepair from the DNA-RNA hybrid, the template DNA nt for the incoming nucleotide, and the incoming/preinsertion nucleotide), we calculated atomistic pairwise nonbonded interactions, including the Coulomb electrostatic potential and the Lennard-Jones vdW potential (49,50), between each two structural elements. In this study, we assume local entropic and solvation variations are not significant for the different nucleotide preinsertion cases. Hence, nonbonded potential energies can be dominant in determining how those structural elements interact locally to each other, and the relative local stabilization on Tyr639.
Results
Below we present results from systematic MD simulations of the T7 RNAP elongation complex from the posttranslocation to the nucleotide preinsertion state. We first examined the posttranslocation complex without nucleotide bound, and then focused on the preinsertion complexes bound with different species of nucleotides. Individual simulations were conducted for 100 ns each and then analyzed. In particular, we analyzed how Tyr639 responds to cognate and noncognate nucleotides at preinsertion. Further, we comparatively studied the mutant Y639F, focusing on its differentiation between ribo- and deoxy-ribonucleotides.
Tyr639 is stabilized marginally by the end basepair of the DNA-RNA hybrid at posttranslocation
It had been suggested that the polymerase works as a Brownian ratchet (29,51,52), i.e., translocating back and forth without any free energy bias. The polymerase is then prevented from moving backward when an incoming NTP binds and inserts into the active site. Nevertheless, single-molecule experiments on T7 RNAP had detected a slight free energy bias toward the posttranslocated state (26), directly after the translocation but before the NTP binding. Our previous modeling work suggested that the slight free energy bias, without essentially changing the overall Brownian ratchet mechanism, would facilitate nucleotide selection by stabilizing Tyr639 in the active site (28). The stabilization free energy between the in and out configurations of Tyr639 at posttranslocation was estimated to be ∼−2 kBT, by numerical fitting to the single-molecule experimental data (26,27). In this work, we first probed what accounts for the local stabilization of Tyr639 in the active site.
We performed a ∼100-ns equilibration simulation of the T7 RNAP complex in the posttranslocated state. The root-mean-square deviation (RMSD) values of the protein are shown in Fig. 1 a. In the posttranslocated state, the side chain of Tyr639 remains in the active site. Because tyrosine contains an aromatic side ring, it is able to stack with the nucleotide base. We show in particular the nonbonded interaction between Tyr639 and the end basepair (bp) of the RNA-DNA hybrid (at 3′-end of the synthesizing RNA strand). It is measured at −2.0 ± 1.2 kcal/mol (see Fig. 1 b), with contributions from both the electrostatic (0.2 ± 0.6 kcal/mol) and vdW interactions (−2.2 ± 1.0 kcal/mol). The average Tyr639-end bp association ∼−3 kBT comes primarily from the vdW attraction, while the electrostatic interaction accounts mainly for the energy fluctuation and slight destabilization. Because the association would be abolished when Tyr639 moves out of the active site, the energetics likely contributes well to the in and out free energy stabilization ∼−2 kBT, as estimated from the previous kinetic modeling (28).
Figure 1.
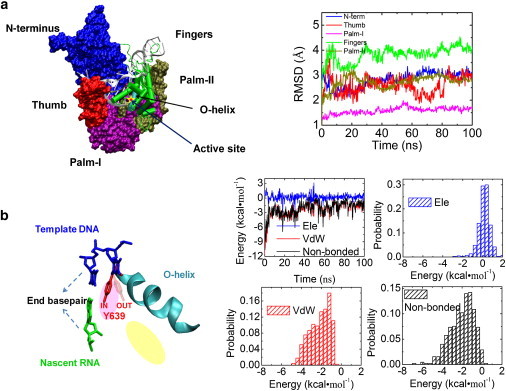
MD simulation on T7 RNAP at the posttranslocated state. (a) (Left) A structure of T7 RNAP elongation complex. (Silver) Nucleic acids. Each protein subdomain is colored differently. (Blue) N-terminus (residues 1∼325); (red) thumb (326∼411); (purple) palm-I (412∼553); (green) fingers (554∼784); (brown) palm-II (785∼883). (Right) The RMSD values of the protein subdomains with corresponding colors. The active site (yellow star) for the nucleotide addition is located on the palm-I subdomain. The O-helix (cyan) on the fingers subdomain close to the active site is also indicated. (b) (Left) The stacking of Tyr639 (red) to the end bp of the DNA-RNA hybrid (blue and green) in cartoon representation. Tyr639 locates on the C-terminal of the O-helix, with its side chain inserted in the active site (pink oval) and close to the preinsertion site (yellow oval). (Right) The interaction energy between Tyr639 and the end bp. The nonbonded interaction energies (black) between Tyr639 and the end bp, along with the electrostatic (ele) (blue) and vdW (red) contributions, are shown in time series (from the 100-ns simulation) and histograms (measured −2.0 ± 1.2 kcal/mol for nonbonded, 0.2 ± 0.6 kcal/mol ele, and −2.2 ± 1.0 kcal/mol vdW).
Preinsertion of a cognate rATP does not enhance the marginal stabilization of Tyr639 in the active site
We then studied how Tyr639 reacts differently to bindings of different nucleotides at the preinsertion site. The molecular views of the active site upon preinsertion of different NTPs are shown in Fig. 2 (RMSD values of the simulation systems are provided in Fig. S1 in the Supporting Material). The views from both the crystal structure (22) and the equilibrated structure of the preinsertion complex at the end of the 100-ns simulation are shown (Fig. 2, a and b). In the crystal structure, the template DNA nt to pair with the incoming NTP is a thymine (dT), so the ribo-adenine (ATP, rATP, or rA) is the cognate or correct NTP. Likewise, a ribo-guanine (GTP, rGTP, or rG), a ribo-uracil (UTP, rUTP, or rU), and a ribo-cytosine (CTP, rCTP, or rC), are regarded as noncognate or incorrect due to their mismatched bases, whereas a deoxyribo-adenine (dATP or dA) is regarded as noncognate or incorrect due to its sugar deficiency. The structures bound with the noncognate NTPs were obtained by replacing rA in the crystal structure with the noncognate NTPs, and each complex structure was then equilibrated for 100 ns (Fig. 2, c–f).
Figure 2.
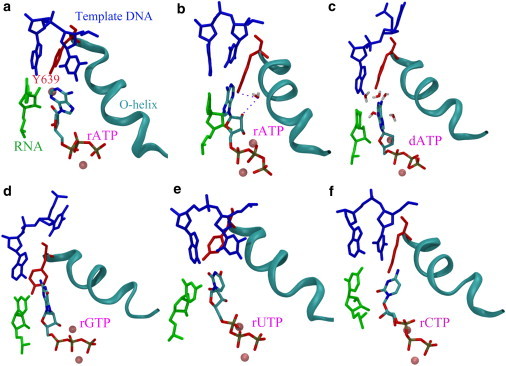
Molecular views of the active site in the preinsertion complex with different nucleotides bound. (a) From the crystal structure complex (PDB:1S0V), in which rA (the cognate NTP) is bound. The other views are obtained from the equilibrated structures at end of the ∼100 ns simulations, bound with rA (b), dA (c), rG (d), rU (e), and rC (f). The preinserted NTP is colored according to the atom type. (blue) Template DNA; (green) 3′-end of RNA; (red) Tyr639; and (cyan) O-helix (the same as in Fig. 1). Two magnesium ions (pink) are also shown.
In the rA preinsertion, we noted that the template DNA nt (dT) stayed close to the active site, that is, close to its adjacent upstream DNA nt, close to Tyr639, and close to the preinserted rA. One important feature we wanted to investigate is whether the WC basepairing can establish between the template dT and the cognate rA at preinsertion. In the crystal structure, the preinserted rA is located close to the template nt to allow some specific interaction, but the basepairing was not yet identified (9). In the simulation, two hydrogen bonds (HBs) of the standard WC basepairing between the template dT and the preinserted rA (N6H60…:O4 and N3H3…:N1) were able to form stably after ∼50 ns (see Fig. 3 a), and persisted to the end of the simulation. The structural, energetic, and dynamical properties of the dT-rA association are demonstrated in Fig. 3. The energetics is described through the nonbonded interaction between the template dT and preinserted rA, sampled from the 100-ns simulation (∼−2.4 ± 3.8 kcal/mol, see Fig. 3 b). The energetics between the template dT and the preinserted noncognate NTP are also shown (Fig. 3 b), and will be addressed later.
Figure 3.
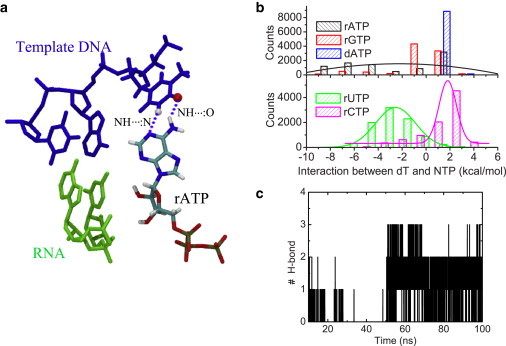
The structural, energetic, and dynamical features of associations between the template dT and the preinserted NTP. (a) Structural view of the WC basepairing between dT and rA; (b) nonbonded interactions between dT and the preinserted NTP (rATP, dATP, rGTP, rUTP, and rCTP); and (c) number of hydrogen bonds (N6H60…:O4, and N3H3…:N1 formed between dT and rA) during the 100-ns simulation of the rA preinsertion complex.
In particular, we found that the nonbonded association between Tyr639 and the end bp of the DNA-RNA hybrid is ∼−2.6 ± 1.3 kcal/mol upon the rA preinsertion, similar to that in the posttranslocation state. We further measured the interactions between Tyr639 and the preinserted rA to be ∼−2.9 ± 2.7 kcal/mol.
In the simulation, we did not observe hydrogen bonding interaction between Y639-OH and 2′-OH of rA ribose, although in the end of the simulation the two groups approached closely to each other (see Fig. S2). Neither did we observe that the magnesium ion was kept in between Y639-OH and 2′-OH of rA (22). In the crystal structure of the preinsertion complex, one magnesium is located in between Y639-OH and 2′-OH of rA, with distances ∼2.2 Å and 2.6 Å to the respective oxygen (22). In our simulation without any constraint, the magnesium ion soon drifted away from its initial position and moved toward the phosphate or sugar ring of NTP (see Fig. 2 b). In a controlled simulation with a constraint applied to the magnesium ion at its initial position for ∼3 ns, the ion still drifted away from Y639-OH once the constraint was removed.
Nevertheless, we found that Y639-OH recognizes 2′-OH of the cognate rNTP through water-mediated interaction. There were usually ∼2–4 water molecules approaching within ∼3.5 Å of the respective -OH groups (see Fig. S2). Interestingly, one could identify that some water molecules transiently (∼5% of simulation time) bridge in between Y639-OH and 2′-OH of rA, by forming HBs with respective -OH groups (see Fig. 2 b and see Movie S1 in the Supporting Material). All movies were made using VMD (53). Inasmuch as the dwell time in the preinsertion state reaches up to milliseconds due to the slow transition rate (∼220 s−1) toward the insertion state (24), the transient water can greatly assist Tyr639 to differentiate between ribo- and deoxy-ribonucleotides.
Preinsertion of a noncognate dATP strengthens Tyr639 stacking to the end bp of the DNA-RNA hybrid
In the case of the dA preinsertion, however, the template dT moves away from the active site, keeping far from its upstream DNA nt and Tyr639. The bp stabilization of Tyr639 strengthens substantially to ∼−9.2 ± 1.2 kcal/mol, ∼7 kcal/mol stronger than that in the rA preinsertion. The large stabilization is contributed dominantly by stacking or the vdW association between Tyr639 and the DNA nt in the end bp (∼−8 kcal/mol; see Table S1 in the Supporting Material).
The nonbonded association between Tyr639 and dA at preinsertion is ∼−3.9 ± 2.3 kcal/mol, slightly (∼1 kcal/mol) more stabilized than that between Tyr639 and rA. Interestingly, although the interaction is not very strong, we note that the Tyr639 side-ring pairs with the dA base, as being facilitated by water. There are approximately five water molecules staying in between the Tyr639 hydroxyl group (Y639-OH) and the dA base (see Fig. 2 c and Movie S2), assisting Tyr639-dA association and enhancing the stacking of the Tyr639 side-ring to the DNA-RNA end bp. Upon constant collisions from the water molecules, one HB was able to form in between Tyr639-OH and N1 on the dA base (see Fig. S3). In other cases of the rNTP preinsertion, no such kind of stabilized water molecules were observed at that location.
Consequently, the interaction between the template dT and dA is weak (∼−0.7 ± 0.1 kcal/mol). In general, the preinserted nucleotide seems to be located fairly close to either the template nt or Tyr639, so that a cognate nucleotide can basepair with the template nt whereas the noncognate nucleotide associates closely with Tyr639. In the case of the dA preinsertion, even when the incoming base appears correct according to the template dT, the WC basepairing cannot form for dT-dA. This can be explained by the above observation: Tyr639-dA seems to mimic basepairing through water-mediated interactions between the side ring of Tyr639 and the base of dA, therefore, dA can no longer basepair with the template dT.
The mismatched rNTP at preinsertion grabs Tyr639 strongly in the active site
In the rG preinsertion, the template dT also moves away from the active site, staying far from its upstream DNA nt and Tyr639 (see Fig. 2 d). Similar to the dA preinsertion, the Tyr639-end bp association strengthens to ∼−6.0 ± 3.1 kcal/mol, or ∼3 kcal/mol enhanced compared to that in the rA preinsertion. The stabilization is also contributed mainly by the vdW interaction (see Table S1). Significantly different from the dA preinsertion, the interaction between rG and Tyr639 is strong at ∼−7.7 ± 4.2 kcal/mol, or ∼5 kcal/mol stronger than that in the rA preinsertion. A large contribution to the strong Tyr639-rG association comes from the electrostatic interaction (see Table S2).
In the rC and rU preinsertion, however, the template dT stays close to the active site. Tyr639 drifts away from the end bp, and moves close to rC or rU (see Fig. 2, e and f). In contrast to the dA or rG preinsertion, we observed a fairly small Tyr639-end bp association ∼−1.7 ± 0.7 kcal/mol (rU) or ∼−3.6 ± 2.9 kcal/mol (rC), around or even less than that in the rA preinsertion. In the simulation, the single-ring base in rU or rC indeed created more space around the active site to allow the template dT to stay close and partially stacks to its upstream nt, thus causing Tyr639 to shift away from the end bp. On the other hand, the association between Tyr639 and rU or rC at preinsertion was significantly strong as ∼−8.3 ± 1.1 kcal/mol or ∼−9.8 ± 5.1 kcal/mol.
In Fig. 4, we report the nonbonded interactions between Tyr639 and the end bp and that between Tyr639 and the preinserted NTP from MD simulations of different NTPs at preinsertion (rA, dA, rG, rC, and rU, sampled between10 ns and 100 ns; similar results obtained between 40 ns and 100 ns are shown in Fig. S4). To summarize, both the Tyr639 association with the end bp and that with the preinserted NTP are comparatively weak in the presence of a cognate rA (∼−5.5 kcal/mol is the average sum of the two types of interactions). However, in the case of a noncognate NTP at preinsertion, either the Tyr639-end bp stacking strengthens significantly (as for dA), or the Tyr639-NTP interaction becomes very strong (for the mismatched rNTP), or both are strong (as for rG). Overall, the average sums of the two types of interactions for the noncognate NTPs are ∼−13 kcal/mol for the dA preinsertion, ∼−14 kcal/mol for rG, ∼−10 kcal/mol for rU, and ∼−13 kcal/mol for rC. We also calculated interactions between Tyr639 and the reminder of the preinsertion complex (see Fig. S5). We found that except for the above two types of signature interactions (shown in Fig. 4), no other interaction components change significantly or demonstrate substantially different characteristics between the cognate and noncognate NTP preinsertion.
Figure 4.
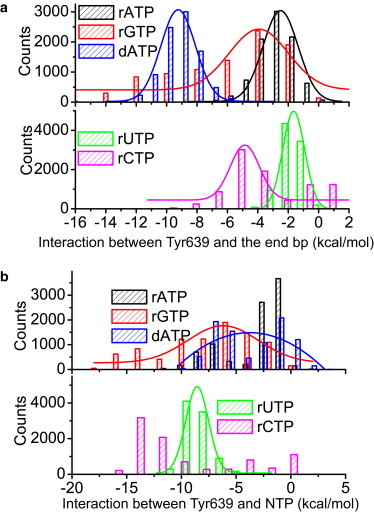
The local interactions that stabilize Tyr639 in the active site of the preinsertion complex with different nucleotides bound. The histograms are sampled from 10 to 100 ns in each individual simulation. (a) The interactions between Tyr639 and the end bp of the DNA-RNA hybrid. (b) The interactions between Tyr639 and the preinserted nucleotide, rATP, dATP, rGTP, rUTP, and rCTP.
In terms of the HB association between the preinsertion NTP and the template dT, we found that no HB could be sustained between the template dT and the preinserted rG or rC. In the preinsertion of rU, two HBs that were not the standard WC basepairing type formed between rU and dT (see Fig. S6 for more details). According to the nonbonded interaction between the template dT the preinserted NTP (see Fig. 3 b), rA-dT and rU-dT are similarly stabilized at ∼−2.4 ± 3.8 kcal/mol and ∼−2.4 ± 1.7 kcal/mol, respectively, with both the electrostatic and vdW interactions contributed toward the stabilization (see Table S3). On the other hand, the template dT interaction with rG and rC is either weak or slightly repulsive (∼−0.4 ± 0.4 kcal/mol and ∼1.1 ± 1.8 kcal/mol).
Note that in the simulation of the rNTP preinsertion complex, one of the magnesium ions left its initial position next to the Tyr639 hydroxyl and moved close to the β-phosphate in the preinserted rNTP. In the dA preinsertion complex, the magnesium ion moved close to the sugar of dA.
The rNTP and dNTP preinsertions are not quite distinguishable in the mutant polymerase Y639F
We further studied the mutant polymerase Y639F for a comparison. We examined the association between Phe639 and the DNA-RNA hybrid end bp and found that the association is not quite distinguishable between the rA and dA preinsertion: In the rA preinsertion, Phe639 was not stabilized by the end bp (∼0.1 ± 0.4 kcal/mol); in the dA preinsertion, the end bp stabilization on Phe639 is also weak (∼−1.5 ± 1.5 kcal/mol). In both cases, the template dT stays close to the active site, so that Phe639 stacking to the end bp is interfered by the template nt (see Fig. 5). In contrast, in the WT, the template dT stays away from the active site in the dA preinsertion, and the end-bp association with Tyr639 clearly differentiates the rA and dA preinsertion (∼−2.6 kcal/mol for rA, and ∼−9.2 kcal/mol for dA).
Figure 5.
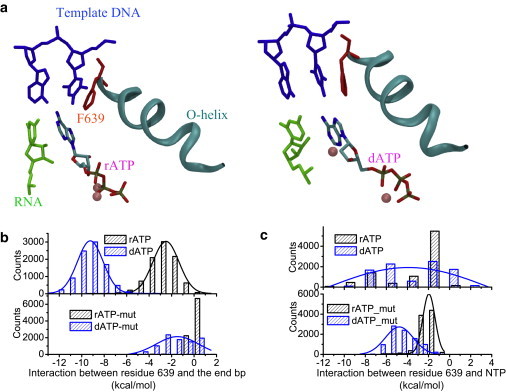
Differentiation between ribo- and deoxy-ribonucleotides in the Y639F mutant polymerase. (a) The structural details showing the preinsertion configurations of rA and dA in the mutant. (b) The nonbonded interaction energies between residue 639 and the end bp of the DNA-RNA hybrid, and (c) the nonbonded interaction energies between the residue 639 and the preinserted NTP (rA or dA), in both the wild-type polymerase (top row) and the mutant Y639F (bottom row). The wild-type data are the same as that in Fig. 4, shown here for an easy comparison.
In terms of the Phe639 association with the preinserted nucleotide, one sees that the association with rA is ∼−2.1 ± 0.7 kcal/mol (−2.9 ± 2.7 kcal/mol in the WT), whereas that with dA is ∼−4.6 ± 1.2 kcal/mol (−3.9 ± 3.0 kcal/mol in the WT). Hence, the differentiation through residue 639-NTP association is not significant and does not change much between the mutant and the WT (see Fig. 5 c).
Indeed, the only difference between the residues tyrosine and phenylalanine is that the former has a hydroxyl group (-OH) on the side ring, whereas the latter has instead a hydrogen atom at the corresponding position. Interestingly, we noted that the nonbonded interaction between the tyrosine hydroxyl group (Y639-OH) and rA is ∼1.8 ± 2.5 kcal/mol, and that between Y639-OH and dA is ∼−0.2 ± 1.9 kcal/mol. In the mutant, the nonbonded interaction between the corresponding-site hydrogen atom (F639-H) and rA, and that between F639-H and dA, are identical at ∼−3.1 kcal/mol (see Table S4). That is, the hydroxyl group from Tyr639 can energetically differentiate rA and dA, whereas Phe639 lacks the capability.
In the WT, Tyr639 is indeed the nearest residue to the 2′-OH group on the rA sugar ring. The slight repelling of rA by Y639-OH seems to help rA to position correctly to form the WC basepairing with the template dT. Importantly, transient water molecule that bridges Y639-OH and 2′-OH of rA can help Tyr639 to recognize ribonucleotide, which does not happen for Phe639 in Y639F.
We noted that most of time only one HB was able to form between the template nt and the preinserted rA or dA in the mutant Y639F simulations (see Fig. S6 c). In the case of the rA preinsertion, the WC basepairing appeared toward the end of the simulation, yet to be stabilized. In the case of the dA preinsertion, the HB interaction disappeared at the end of the simulation.
As mentioned above for the WT, we observed several water molecules staying between Y639-OH and the preinserted dA. No stabilized water molecules were found in between F639 and the preinserted dA in the mutant. On the other hand, we observed an overall increased hydration level around the mutant active site (55∼60 water molecules), compared to that in the WT (45 ∼50 water molecules), for both the preinsertion of rA and dA (see Fig. S7). The increased level of the hydration in the active site further suggests a lowered differentiation capability between the rA and dA substrates in the mutant polymerase.
Discussion
In this work, we studied in detail how residue Tyr639 senses the nucleotide at preinsertion so as to selectively recruit or gate the nucleotide during its insertion in T7 RNAP. Inasmuch as a successful nucleotide insertion serves for a pawl in the Brownian ratcheting of the polymerase, the selective nucleotide gating allows for selective ratcheting.
Tyr639 had been determined in early experiments as being critical for differentiating ribo- versus deoxy-ribonucleotides (30). Our previous modeling work suggested that Tyr639 is stabilized in the active site at posttranslocation, so that it can assist nucleotide selection for the elongation (28). Accordingly, we examined first how Tyr639 is stabilized at posttranslocation. It is interesting to note that Tyr639 is marginally stabilized by stacking to the end bp of the RNA-DNA hybrid. The stabilizing energetic seems to contribute toward the small posttranslocation free energy bias detected from single-molecule experiments (26), as suggested from the previous modeling (28).
Then we intensively investigated T7 RNAP preinsertion complexes bound with different nucleotide species. In the WT polymerase, when a cognate NTP binds, the marginal stabilization of Tyr639 in the active site is not much affected. When a noncognate species of NTP preinserts, however, we found that either the association of Tyr639 with the end bp is enhanced, or a strong interaction between the preinserted NTP and Tyr639 is built. In Fig. 6 a, we illustrate schematically these signature stabilization effects. Remarkably, we see that the end bp stabilization is largely enhanced in the dA preinsertion, whereas the base mismatched rNTPs directly grabs Tyr639 to enhance the local stabilization.
Figure 6.
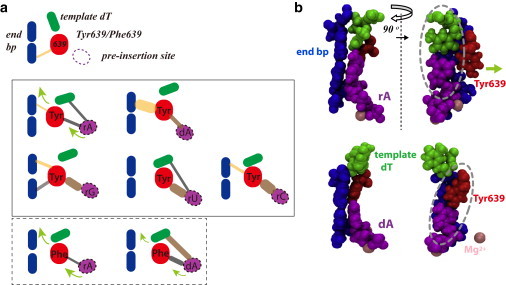
The Tyr639-assisted nucleotide selection mechanism in T7 RNAP elongation. (a) Schematics show how Tyr639 senses the cognate and noncognate NTPs at preinsertion to allow selective gating during NTP insertion. The four key elements are shown in different colors: (red) residue 639; (blue) end bp of the DNA-RNA hybrid; (magenta) preinsertion NTP; and (green) template DNA nt. Lines linking the elements indicate the nonbonded associations within each pair: (light orange) vdW-dominated; (brown) electrostatic-dominated; (gray) combined; (round head) attraction, i.e., no significant repulsion detected; (thin) weak interaction; (thick) strong interaction. Note that very weak interaction with a strength <1.2 kcal/mol is not shown. The first row shows the posttranslocation configuration. The window (solid line) in the middle shows the preinsertion configurations in the WT, with the cognate rA or noncognate dA/rG/rU/rC bound at the preinsertion site. The window (dashed line) on the bottom shows the preinsertion configurations in the mutant Y639F, with rA and dA bound, respectively. (Light green arrows) NTP insertion as residue 639 moves away. (b) Front and side atomistic views of the key elements captured at the end of the simulations for the respective preinsertions of rA and dA in the WT (see Movie S3 and Movie S4 and additional views of other NTP preinsertion configurations in Fig. S9 of the Supporting Material). The atoms are shown in vdW spheres, colored for Tyr639 (red), the end bp (blue), the template dT (green), and the preinserted rA or dA (magenta). In addition, two magnesium ions are also shown (pink). (Light green arrow) indicating the direction Tyr639 moves out of the active site. (Two gray ovals) dT-rA basepairing and Tyr639-dA association in respective preinsertions of rA and dA.
From these observations, one can infer that it is relatively easy for Tyr639 to move out of the active site to allow the insertion of a cognate NTP, because Tyr639 is neither held tightly by the preinserted NTP nor by the end bp of the DNA-RNA hybrid. Upon Tyr639 moving away, the cognate NTP inserts into the active site so that its WC basepairing with the template is further stabilized; with the proper basepairing, stacking, and local alignments, chemical addition then follows. On the other hand, for the noncognate NTP, the insertion process would be difficult because Tyr639 is stabilized there so as to block the active site. As a result, the noncognate NTP cannot move easily into the active site; it is then highly likely that the noncognate NTP will be rejected back to the solution after it is kept long in the preinsertion site. We present this as the selective gating mechanism of Tyr639 in the T7 RNAP nucleotide selection.
Although the nucleotide detection varies in detail for different nucleotide preinsertions, the detection is essentially led by the template nt and coordinated by Tyr639, as if Tyr639 competes with the template nt for the end bp or the nucleotide association. For example, in the case of the cognate rA, the template dT stays close to the active site and interferes with the Tyr639 for local association (see Movie S3). Before the WC basepairing of dT-rA, the Tyr639-end bp or Tyr639-rA association appeared relatively strong in the simulation, whereas the association was substantially reduced once the dT-rA basepairing established (see Fig. S8). In the case of the dA preinsertion, the template dT moves away and the dT-dA basepairing could not form due to the Tyr639 intervention: The Tyr639-dA pair association somehow mimics the standard basepairing, so that the Tyr639 side ring stacks nicely with the upstream DNA nt in the end bp (see Movie S4). A further comparison of the rA and dA preinsertion is provided in Fig. 6 b (in both front and side views of the active site).
We note that the detection of rA or dA at preinsertion is facilitated by water: The transient water molecule assists recognition of rA by the Tyr639 hydroxyl (see Movie S1), and the localized water molecules help the Tyr639-dA pair association (see Movie S2). In the mismatched rG preinsertion, dT simply moves away so that Tyr639 associates well with both the end bp and rG. In the case of the mismatched rU or rC preinsertion, dT does not move away but partially stacks with the upstream nt in the end bp, thus Tyr639 cannot stack well with the end bp; yet Tyr639 can grab tightly on rU or rC, conquering the intervening from dT. Additional views of rG/rU/rC preinsertion are provided in Fig. S9.
In the mutant polymerase Y639F, in contrast, the template nt is able to localize closely to the active site in both the rA and dA preinsertion. Phe639 seems less capable of stacking well with the end bp than Tyr639 of a higher polarity. Consequently, the preinsertion of dA does not trigger strong stacking of Phe639 to the end bp. There is also no stabilized water molecule in between Phe639 and the dA base to assist the pair association as there is for Tyr639-dA. Lack of the hydroxyl group on the side ring, Phe639, cannot differentiate well between rA and dA, either energetically or through the water-mediated HB interaction, as done in the WT. Furthermore, Phe639 seems to allow more water molecules into the active site than that in the WT case, lowering further the substrate differentiation capability.
Additionally, we wondered whether Tyr639 assists nucleotide selection by also contributing to the rejection of the noncognate NTPs back to solution from the preinsertion site. To achieve the rejection, the off-rate (koff) of the noncognate NTP should be higher than that of the cognate NTP (kwoff > kroff) at the preinsertion site. Because one assumes that the binding or on-rates of different NTPs are the same, the noncognate NTP should be less stabilized at the preinsertion site than the cognate NTP. From the Results, it is seen that the nonbonded interactions between Tyr639 and mismatched nucleotides are indeed much more stabilizing than those between Tyr639 and the cognate rA, hence, Tyr639 seems unable to destabilize a noncognate NTP relative to the cognate NTP. Nevertheless, one can identify a slight vdW destabilization (∼1 kcal/mol) of dA relative to rA at the preinsertion site, contributed by Tyr639 (see Fig. S10). Other than that, one cannot infer any selectivity of Tyr639 against the noncognate nucleotide binding at the preinsertion site. Hence, the selectivity of Tyr639 is achieved mainly from nucleotide preinsertion into the insertion, rather than at the preinsertion.
Conclusion
In this work, we implemented intensive atomistic simulations to investigate how nucleotide selection is assisted by a critical residue Tyr639 in T7 RNAP transcription. Our study demonstrates that Tyr639 recognizes the nucleotide species early at preinsertion to recruit the cognate species into the active site for chemical addition. Interestingly, the study reveals signature local interactions that distinguish the cognate nucleotides from the noncognate ones at preinsertion and modulate the stability of Tyr639 in the active site. The signature interactions come partly from the Tyr639 stacking to the end bp of the DNA-RNA hybrid at the 3′-end of the synthesizing RNA, and partly from the direct association between Tyr639 and the nucleotide at preinsertion.
Indeed, Tyr639 seems to compete with the template nt for these local interactions. In the case of a cognate NTP at preinsertion, the Watson-Crick basepairing was detected between the template nt and the cognate NTP during the simulation. The template nt stays close to the active site and prevents further stabilization of Tyr639 by these local interactions. As a result, Tyr639 can be easily removed from the active site to allow a proper nucleotide insertion. In the case of a noncognate NTP at preinsertion, however, the template nt cannot stay close to the active site or grab the preinserted NTP. Tyr639 is thus stabilized by the full signature interactions or at least part of them, and keeps blocking the active site to prevent the noncognate NTP insertion. As a result, the noncognate NTPs are likely rejected back into the solution by overall destabilization at the preinsertion site.
In particular, when a DNA nucleotide preinserts into the WT T7 RNAP, a nice pair association forms between dNTP and Tyr639 under the mediation of localized water molecules, and the Tyr639 side ring stacks strongly to the end bp. The template nt shifts away, and Tyr639 keeps blocking the active site so that dNTP is prevented from insertion. In the mutant polymerase Y639F, however, the template nt stays close to the active site in both the rNTP and dNTP preinsertion. Phe639 lacks the hydroxyl group on the side ring. It does not associate well with the preinserted NTP nor energetically differentiate the dNTP and rNTP. The low polarity of Phe639 also makes it less capable of stacking to the end bp than Tyr639. Hence, Phe639 does not distinguish well between the ribo- and deoxy-ribonucleotide at preinsertion. In addition, hydration of the active site increases in the Y639F mutant and further blurs the substrate differentiation. The findings explain well the previous experimental discovery that the mutant Y639F works as both an RNA and a DNA polymerase.
With equilibrium atomistic MD simulations totaling up to 1 μm, we show how Tyr639 differentiates cognate and noncognate nucleotides at preinsertion. Nevertheless, the corresponding nucleotide selection happens from the nucleotide preinsertion to the insertion, which is slow at a millisecond timescale. Hence, it is not yet possible to simulate the full process of the nucleotide insertion or selection. Certain sampling and simulation techniques, however, allow calculating the activation barrier differences for the nucleotide insertion between the cognate and noncognate nucleotides, as well as the binding free energy differences between the cognate and noncognate nucleotides at the preinsertion site (unpublished work), although the calculation does not necessarily reveal individual residue contributions.
From this study, one is able to see that Tyr639 is unlikely to be involved in the initial nucleotide selection; that is, to destabilize the noncognate nucleotide relative to the cognate one at the preinsertion site. Nevertheless, other local protein-NTP interactions at the preinsertion site may help to reject the noncognate nucleotides, so that the polymerase can conduct an efficient nucleotide selection or initial screening immediately upon the nucleotide binding. After that, the nucleotide selectivity assisted by Tyr639 from preinsertion to insertion serves for the next, but is likely a major selection checkpoint. It is notable that this simulation work, based on high-resolution structural studies, also links to mutagenesis and single-molecule experiments. With a perspective built for full elongation kinetics of the polymerase under the Brownian ratchet mechanism, the study reveals, with unprecedented molecular detail, information about one critical residue in the mechanism of transcription fidelity control.
Acknowledgments
Thanks are due to Dr. Yibing Shan for comments on the manuscript.
This work is supported by National Science Foundation of China grant No. 11275022.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supporting Material
References
- 1.Buc H., Strick T., editors. RNA Polymerase as Molecular Motors. The Royal Society of Chemistry; Cambridge, United Kingdom: 2009. [Google Scholar]
- 2.Johnson K.A. Conformational coupling in DNA polymerase fidelity. Annu. Rev. Biochem. 1993;62:685–713. doi: 10.1146/annurev.bi.62.070193.003345. [DOI] [PubMed] [Google Scholar]
- 3.Petruska J., Sowers L.C., Goodman M.F. Comparison of nucleotide interactions in water, proteins, and vacuum: model for DNA polymerase fidelity. Proc. Natl. Acad. Sci. USA. 1986;83:1559–1562. doi: 10.1073/pnas.83.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hopfield J.J. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. USA. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 6.Loeb L.A., Kunkel T.A. Fidelity of DNA synthesis. Annu. Rev. Biochem. 1982;51:429–457. doi: 10.1146/annurev.bi.51.070182.002241. [DOI] [PubMed] [Google Scholar]
- 7.Sydow J.F., Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr. Opin. Struct. Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 8.Voliotis M., Cohen N., Liverpool T.B. Backtracking and proofreading in DNA transcription. Phys. Rev. Lett. 2009;102:258101. doi: 10.1103/PhysRevLett.102.258101. [DOI] [PubMed] [Google Scholar]
- 9.Yamada Y.R., Peskin C.S. A look-ahead model for the elongation dynamics of transcription. Biophys. J. 2009;96:3015–3031. doi: 10.1016/j.bpj.2008.12.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maoiléidigh D.O., Tadigotla V.R., Ruckenstein A.E. A unified model of transcription elongation: what have we learned from single-molecule experiments? Biophys. J. 2011;100:1157–1166. doi: 10.1016/j.bpj.2010.12.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golosov A.A., Warren J.J., Karplus M. The mechanism of the translocation step in DNA replication by DNA polymerase I: a computer simulation analysis. Structure. 2010;18:83–93. doi: 10.1016/j.str.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moustafa I.M., Shen H., Cameron C.E. Molecular dynamics simulations of viral RNA polymerases link conserved and correlated motions of functional elements to fidelity. J. Mol. Biol. 2011;410:159–181. doi: 10.1016/j.jmb.2011.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang B., Predeus A.V., Feig M. Energetic and structural details of the trigger-loop closing transition in RNA polymerase II. Biophys. J. 2013;105:767–775. doi: 10.1016/j.bpj.2013.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang X., Wang D., Levitt M. RNA polymerase II trigger loop residues stabilize and position the incoming nucleotide triphosphate in transcription. Proc. Natl. Acad. Sci. USA. 2010;107:15745–15750. doi: 10.1073/pnas.1009898107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pardo-Avila F., Da L.-T., Huang X. Theoretical investigations on elucidating fundamental mechanisms of catalysis and dynamics involved in transcription by RNA polymerase. J. Theor. Comput. Chem. 2013;12:1341005. [Google Scholar]
- 16.Feig M., Burton Z.F. RNA polymerase II with open and closed trigger loops: active site dynamics and nucleic acid translocation. Biophys. J. 2010;99:2577–2586. doi: 10.1016/j.bpj.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kireeva M.L., Opron K., Burton Z.F. Molecular dynamics and mutational analysis of the catalytic and translocation cycle of RNA polymerase. BMC Biophys. 2012 doi: 10.1186/2046-1682-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Da L.T., Wang D., Huang X. Dynamics of pyrophosphate ion release and its coupled trigger loop motion from closed to open state in RNA polymerase II. J. Am. Chem. Soc. 2012;134:2399–2406. doi: 10.1021/ja210656k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Da L.-T., Pardo Avila F., Huang X. A two-state model for the dynamics of the pyrophosphate ion release in bacterial RNA polymerase. PLOS Comput. Biol. 2013;9:e1003020. doi: 10.1371/journal.pcbi.1003020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva D.A., Weiss D.R., Huang X. Millisecond dynamics of RNA polymerase II translocation at atomic resolution. Proc. Natl. Acad. Sci. USA. 2014;111:7665–7670. doi: 10.1073/pnas.1315751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joyce C.M., Steitz T.A. Function and structure relationships in DNA polymerases. Annu. Rev. Biochem. 1994;63:777–822. doi: 10.1146/annurev.bi.63.070194.004021. [DOI] [PubMed] [Google Scholar]
- 22.Temiakov D., Patlan V., Vassylyev D.G. Structural basis for substrate selection by t7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 23.Huang J., Brieba L.G., Sousa R. Misincorporation by wild-type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry. 2000;39:11571–11580. doi: 10.1021/bi000579d. [DOI] [PubMed] [Google Scholar]
- 24.Anand V.S., Patel S.S. Transient state kinetics of transcription elongation by T7 RNA polymerase. J. Biol. Chem. 2006;281:35677–35685. doi: 10.1074/jbc.M608180200. [DOI] [PubMed] [Google Scholar]
- 25.Yin Y.W., Steitz T.A. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 26.Thomen P., Lopez P.J., Heslot F. Unraveling the mechanism of RNA-polymerase forward motion by using mechanical force. Phys. Rev. Lett. 2005;94:128102. doi: 10.1103/PhysRevLett.94.128102. [DOI] [PubMed] [Google Scholar]
- 27.Thomen P., Lopez P.J., Heslot F. T7 RNA polymerase studied by force measurements varying cofactor concentration. Biophys. J. 2008;95:2423–2433. doi: 10.1529/biophysj.107.125096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu J., Oster G. A small posttranslocation energy bias aids nucleotide selection in T7 RNA polymerase transcription. Biophys. J. 2012;102:532–541. doi: 10.1016/j.bpj.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H., Oster G. Ratchets, power strokes, and molecular motors. Appl. Phys., A Mater. Sci. Process. 2002;75:315–323. [Google Scholar]
- 30.Sousa R., Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joyce C.M. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc. Natl. Acad. Sci. USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delarue M., Poch O., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- 33.Tabor S., Richardson C.C. A single residue in DNA polymerases of the Escherichia coli DNA polymerase I family is critical for distinguishing between deoxy- and dideoxyribonucleotides. Proc. Natl. Acad. Sci. USA. 1995;92:6339–6343. doi: 10.1073/pnas.92.14.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 35.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 36.Guy A.T., Piggot T.J., Khalid S. Single-stranded DNA within nanopores: conformational dynamics and implications for sequencing; a molecular dynamics simulation study. Biophys. J. 2012;103:1028–1036. doi: 10.1016/j.bpj.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hornak V., Abel R., Simmerling C. Comparison of multiple AMBER force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joung I.S., Cheatham T.E., 3rd Determination of alkali and halide monovalent ion parameters for use in explicitly solvated biomolecular simulations. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joung I.S., Cheatham T.E., 3rd Molecular dynamics simulations of the dynamic and energetic properties of alkali and halide ions using water-model-specific ion parameters. J. Phys. Chem. B. 2009;113:13279–13290. doi: 10.1021/jp902584c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meagher K.L., Redman L.T., Carlson H.A. Development of polyphosphate parameters for use with the AMBER force field. J. Comput. Chem. 2003;24:1016–1025. doi: 10.1002/jcc.10262. [DOI] [PubMed] [Google Scholar]
- 41.Yin Y.W., Steitz T.A. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 42.Price D.J., Brooks C.L., 3rd A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 43.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 44.Essmann U., Perera L., Pedersen L.G. a smooth particle mesh Ewald method. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 45.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52:7182–7190. [Google Scholar]
- 46.Nosé S., Klein M.L. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983;50:1055–1076. [Google Scholar]
- 47.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 48.Hess B., Bekker H., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- 49.Leach A.R. 2nd Ed. Prentice-Hall; Upper Saddle River, NJ: 2001. Molecular Modeling: Principles and Applications. [Google Scholar]
- 50.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 51.Bar-Nahum G., Epshtein V., Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 52.Guo Q., Sousa R. Translocation by T7 RNA polymerase: a sensitively poised Brownian ratchet. J. Mol. Biol. 2006;358:241–254. doi: 10.1016/j.jmb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 53.Humphrey W., Dalke W., Schulten K. VMD - Visual Molecular Dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


