Abstract
The pool of microbes inhabiting our body is known as “microbiota” and their collective genomes as “microbiome”. The colon is the most densely populated organ in the human body, although other parts, such as the skin, vaginal mucosa, or respiratory tract, also harbour specific microbiota. This microbial community regulates some important metabolic and physiological functions of the host, and drives the maturation of the immune system in early life, contributing to its homeostasis during life. Alterations of the intestinal microbiota can occur by changes in composition (dysbiosis), function, or microbiota-host interactions and they can be directly correlated with several diseases. The only disease in which a clear causal role of a dysbiotic microbiota has been demonstrated is the case of Clostridium difficile infections. Nonetheless, alterations in composition and function of the microbiota have been associated with several gastrointestinal diseases (inflammatory bowel disease, colorectal cancer, or irritable bowel syndrome), as well as extra-intestinal pathologies, such as those affecting the liver, or the respiratory tract (e.g., allergy, bronchial asthma, and cystic fibrosis), among others. Species of Bifidobacterium genus are the normal inhabitants of a healthy human gut and alterations in number and composition of their populations is one of the most frequent features present in these diseases. The use of probiotics, including bifidobacteria strains, in preventive medicine to maintain a healthy intestinal function is well documented. Probiotics are also proposed as therapeutic agents for gastrointestinal disorders and other pathologies. The World Gastroenterology Organization recently published potential clinical applications for several probiotic formulations, in which species of lactobacilli are predominant. This review is focused on probiotic preparations containing Bifidobacterium strains, alone or in combination with other bacteria, which have been tested in human clinical studies. In spite of extensive literature on and research into this topic, the degree of scientific evidence of the effectiveness of probiotics is still insufficient in most cases. More effort need to be made to design and conduct accurate human studies demonstrating the efficacy of probiotics in the prevention, alleviation, or treatment of different pathologies.
Keywords: Intestinal microbiota, Bifidobacterium, Probiotics, Dysbiosis, Inflammatory bowel disease, Irritable bowel syndrome, Colorectal cancer, Liver disease, Respiratory disease, Functional foods
Core tip: In this review we focus on how bifidobacteria can contribute to maintain a proper health status through their interactions with gut microbiota and the host. We present several gastrointestinal and extra-intestinal pathologies associated with imbalances in the microbiota composition and function, including bifidobacteria-associated dysbiosis. We review up-to-date scientific evidence sustaining the use of probiotic bifidobacteria to prevent, or treat, several disorders, and we include a list of specific Bifidobacterium strains that have been tested in human clinical studies.
INTRODUCTION
About 100 trillion (1014) microbes inhabit the human gut, which represents 10 fold the number of eukaryotic cells in the body and contributes 1.5-2 kg of total body weight[1]. The number and complexity of these microbial populations gradually increase from the stomach to the colon, where microorganisms reach levels of up to 1011 cells per gram of intestinal content[2]. Although the colon is the more densely populated organ, microorganisms are also normal inhabitants of other parts of the body, such as the skin, vagina, throat and the upper respiratory tract[3]. This pool of microbes is known as “microbiota” and the ensemble of their genes is named “microbiome”.
Microbiota composition
The recent advent of next generation sequencing techniques has greatly contributed to demonstrate that the human body harbours more than 1000 phylotypes at species-level, but most intestinal bacteria belong to just a few phyla. In adults, Bacteroidetes and Firmicutes usually dominate the intestinal microbiota, whereas Actinobacteria, Proteobacteria and Verrucomicrobia are in considerably minor proportion (Figure 1). Methanogenic archaea (represented by Methanobrevibacter smithii), eukaryotes (mainly yeast) and viruses (mainly bacteriophages) are also components of this microbiota[3,4]. A recent work identified three enterotypes in the human gut microbiome differing in species and functional characteristics[5]. In spite of a consistency in the global composition, the intestinal microbiota seems to be highly variable among individuals at species-level phylotypes; usually Faecalibacterium prausnitzii (F. prausnitzii), Roseburia intestinalis, Bacteroides uniformis, and species of bifidobacteria and lactobacilli are present in most people[6]. The microbial colonization of the gut begins in infants immediately after birth. Facultative anaerobes, such as enterobacteria, enterococci and lactobacilli are the first colonizers (Figure 1). Anaerobic microorganisms, including Bifidobacterium, Bacteroides and Clostridium establish gradually, and contribute to a progressive decrease of the facultative anaerobes to strict anaerobes ratio in time[7]. At about 3 years of age, the gut microbiota reaches a composition and diversity similar to adults and remains more or less stable over time in adulthood. New changes appear in the senescence, the microbiota of elderly people differing from the core microbiota and diversity levels of younger adults[8,9].
Figure 1.
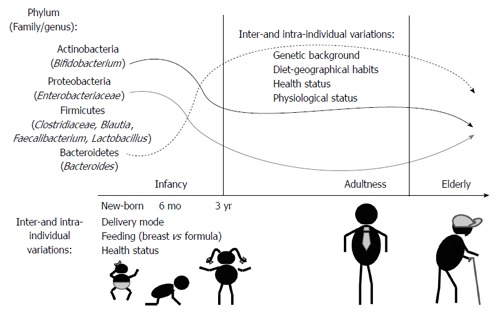
Evolution of some representatives of the intestinal microbiota accordingly to age.
Microbiota function
Gut microbiota provides nutrients and energy for the host through the fermentation of non-digestible dietary components in the large intestine. The main products of the substrate fermentation in the gut are short chain fatty acids (SCFA), which interact with the intestinal microbiota and host cells. The microbiota regulates, via different mechanisms, some important physiological functions of the host, such as those related to energy expenditure, satiety and glucose homeostasis[10,11]. It can also act as a barrier against the establishment of foodborne pathogens. Remarkably, the intestinal microbiota is also in contact with the second largest (after the brain) neural pool of cells in the body, as well as with the largest group of immune cells in our organism[12]. Therefore, the microbiota drives the maturation of the immune system in infancy and contributes to the maintenance of its homeostasis during life[13,14]. Moreover, the possible influence of the microbiota in the development of the nervous system and in cognitive function currently constitutes a hot target for biomedical research[15].
Factors affecting microbiota
Some external and internal factors of the host can influence the composition and metabolic activity of the intestinal microbiota. Diet strongly affects human health, partly through its interaction with intestinal microbiota[16]. Distinctive features have been clearly evidenced recently across different geographic locations, which can be partly explained by differing diets[17]. Functional immaturity of the immune system and intestinal epithelium can influence the aberrant intestinal colonization pattern occurring in preterm neonates[18]. The type of feeding in early infancy (breast-fed vs formula fed) also seems to condition the microbiota establishment, as well as the maturation of the immune system. Medication, especially chronic medication, can exert a strong impact on intestinal microbiota[19]. A misbalance of this intestinal microbial community can act as a source of infection, or chronic inflammation, and can be involved, as well, in gastrointestinal diseases and other extra-intestinal disorders.
MICROBIOTA AND GASTROINTESTINAL PATHOLOGY
As the gut microbiota has a well-established role in host homeostasis, several highly prevalent gastrointestinal diseases have been associated with shifts, or imbalances, in microbiota composition (dysbiosis) and function, as well as in microbiota-host interactions[20,21]. The term dysbiosis is unhelpful if used only to describe a change in the microbiota, which is assumed to be deleterious to the host. In some instances, changed microbiota may be an appropriate response to a change in the host, or may represent an epiphenomenon without pathophysiological implications[22]. Therefore, as the entire functional complexity of the gut microbiota is incompletely defined, with just a determination of composition by taxonomic assignment[6], a functional analysis may provide information about metabolic and immunologic functions, as well as microbiota-host interactions[23].
Clostridium difficile infection
The only disease process in which a dysbiotic microbiota plays an undoubtedly proven role is in the case of Clostridium difficile infection (CDI). Treatment with antibiotics transiently alters the microbiota composition, providing the niche in which this pathogen can expand[20]. The restoration of normal healthy microbiota, by faecal microbiota transplantation (FMT) is an effective therapy to treat CDI[24]. FMT consists of the engraftment of microbiota from a healthy donor(s) into a recipient, which results in the restoration of the normal gut microbial community structure, with the aim of recovering metabolic and immunologic balance[25]. FMT introduces a complete, stable, and durable community of gut microbiota[26], unlike probiotics, that temporarily alter the metabolic, or immunologic activity, of the native gut microbiota[27]. The mechanism(s) that facilitates this microbiota normalization, as well as the occurrence of putative long-term side effects, still has to be determined. However, FMT represents an emerging therapy for disease states related to dysbiosis, and is a current, recommended treatment for relapsing and non-responding CDI[28].
Inflammatory bowel diseases
There is increasing evidence of the pathogenic implication of the host microbiota in inflammatory bowel disease (IBD), as several taxonomic and functional changes, as well as imbalances in the host-microbiota cross-talk, have been described[29]. This may be understood as a bidirectional relationship between altered immune function (mucosal barrier, innate bacterial killing, or immune regulation) and altered bacterial community (its features, functions, or metabolites)[30]. A deregulated immune response against commensal gut bacteria, in which local tolerance mechanisms towards commensal microbes seem to be impaired, may contribute to the onset or perpetuation of IBD[31]. Studies of faecal and gut mucosal-associated microbiota have demonstrated quantitative and qualitative changes in composition and function associated with IBD, with a shift towards an inflammatory-promoting microbiome[32]. There is a decreased complexity in composition, with a loss of normal anaerobic bacteria[33], a temporary composition instability during clinical remission[34], and a dysbiosis towards selected microorganisms, with over- and under-representation of certain species[35]. An excessive abundance of Desulfovibrio species has been described in ulcerative colitis (UC), which has pathogenic potential due to its ability to generate sulfides[36]. In addition, an increase in microbial genes involved in the metabolism of cysteine (sulphur-containing amino acid) and sulphate transport systems have been reported[37]. UC and Crohn′s disease (CD) present a low abundance of F. prausnitzii, which has known anti-inflammatory properties[38], and this is associated with a higher risk of postoperative recurrence of ileal disease[29], and with a concomitant increase in the abundance of Escherichia coli (E. coli)[39]. Adherent-invasive E. coli strains are specifically associated with the CD ileal phenotype[40]. In addition, a decrease of SCFA levels, and of genes related to SCFA metabolism, is described in CD[41]. Clinical and basic evidence suggests that dysbiosis has a key role in the initiation and progression of chronic inflammation in the pouch reservoir[29]. A notable increase in Proteobacteria (E. coli and other enterobacteria), with a marked decrease in Bacteroides and F. prausnitzii, has been described[42].
Colorectal cancer
There is a strong genetic component in the development of colorectal adenomas and colorectal cancer (CRC), but there are also environmental factors linked to this disease[43]. Considering the continuous exposure of the colonic mucosa to the microbiota and its metabolites, this microbial community has been proposed as contributing to carcinogenesis. However, the mechanisms of this association remain unknown, although there are several ways in which an altered microbiota may promote CRC[44]. Chronic intestinal inflammation has been associated with the development of CRC and it can result from an aberrant ratio of protective (tolerogenic) to aggressive (pro-inflammatory, pro-tumorigenic) microbiota[45]. In this regard, Bacteroides fragilis (B. fragilis) and Streptococcus bovis (S. bovis) could be linked to the development of CRC by activating immune cells to release pro-mitogenic and pro-angiogenic cytokines, mostly Interleukin-17 (IL-17)[46]. There are also some biological activities of the intestinal microbiota that are presumed to generate metabolites involved in CRC carcinogenesis: secondary bile salt transformations, desulfuration of bile acids, production of hydrogen sulfide, production of aglycones from inactivated harmful compounds, bacterial β-glucuronidases, production of aromatic amines by azoreductases and nitroreductases, generation of acetaldehyde, and generation of reactive oxygen species[47].
Several changes in microbiota composition are described in CRC. An increased diversity in Clostridium leptum (C. leptum) and Clostridium coccoides is present, while temporary instability and decreased complexity in composition are linked to colitis-associated CRC[48,49]. The microenvironment within colorectal neoplastic lesions is significantly different from normal intestine, thus it can promote the accumulation of additional mutations and epigenetic changes. Many bacterial species were found to be enriched in colorectal tumour samples and adjacent tissue: B. fragilis, Bacteroides vulgatus, Bifidobacterium longum, Clostridium butyricum, Mitsuokella multiacida, E. coli, Enterococcus faecalis, and S. bovis[45].
Functional gastrointestinal disorders
Functional gastrointestinal disorders are defined and categorized based upon clusters of chronic, or recurrent symptoms that can be attributed to the gastrointestinal tract in the absence of any discernible organic abnormality[50]. An increasing body of evidence supports the physio-pathological role of microbiota in irritable bowel syndrome (IBS), although the clinical relevance of many findings still remains unclear[51]. Clinical studies show that up to 20% of IBS is preceded by an enteric infection, which produces a profound alteration of the host microbiota[52]. The precise mechanisms by which this alteration determines the persistency of IBS symptoms after the acute episode are not fully elucidated; a genetic susceptibility, an abnormal mucosal barrier integrity, variations in SCFA production, and an increase in mucosal entero-endocrine cells may contribute[53,54]. A meta-analysis review showed that small intestinal bacterial overgrowth (SIBO) was present between 4% and up to 54% in IBS[55], suggesting that microbiota alterations may play a role in a subset of patients. However, the large differences between the studies, methodological problems, such as the lack of standardization, poor sensitivity and specificity of breath tests, as well as the questioned cut-off value of cultured duodenum/jejunum aspirates (> 105 CFU/mL), make the importance of SIBO in IBS unclear[56]. Interventional studies showing the positive effects of treatments directed at gut microbiota also support the role of microbial alterations in IBD[57,58]. Indeed, several alterations in microbiota composition have been described, although the heterogeneity of IBS, as well as methodological variations, has resulted in contradictory reports. Nonetheless, emerging data support the existence of dysbiosis in a subset of IBS[59]: a decreased complexity in composition[60], temporary instability[61], and changes in the mucosa-associated microbiota, with an increase in Bacteroides and Clostridia and a reduction in Bifidobacterium[62].
There are several functional changes that may contribute to the pathophysiology of IBS. An altered fermentation process, with an increase in faecal SCFA and their producing bacteria, has been described; the highest levels being related to the severest symptoms[63]. The alteration of intestinal barrier function, with increased gut permeability, has a major role in IBS[64]; nevertheless, its relationship with altered microbiota remains to be fully understood. Modulation of enteric sensorimotor function, through alterations in bile acid metabolism by microbiota, has been described in several in vitro studies[59]. An increase in primary bile acids in faeces, associated with the decrease in C. leptum (able to transform primary into secondary bile acid), was correlated with the stool consistency in diarrhoea predominant IBS[65]. There is a growing appreciation for the hypothesis that IBS may be a condition of low-grade inflammation without strong tissue damage, but enough to alter the sensorimotor function and produce symptoms[66]. However, there are few studies assessing a direct link between alterations in gut microbiota and low-grade inflammation or immune activation in the gut. Finally, data from animal and human studies have demonstrated the effects on the brain-gut axis at peripheral and central levels, with an impact on the enteric nervous system, brain chemistry and behaviour[67]; although caution must be exercised when in speculating on the implications of these findings[68].
MICROBIOTA AND EXTRA-INTESTINAL PATHOLOGY
It has been denoted that aberrancies in intestinal microbiota may be involved in intestinal diseases. Moreover, there is increasing evidence that dysbiosis in this microbial community could be related with some extra-intestinal pathologies, such as allergies, obesity and metabolic syndrome, rheumatic disease, and degenerative processes, among others[69-71].
Liver diseases
Modifications in intestinal microbiota have been associated with the physiopathology of non-alcoholic fatty liver disease (NAFLD), alcoholic liver disease, and total parenteral nutrition-intestinal failure liver disease, as well as in the precipitation of infectious and non-infectious complications of liver cirrhosis[72]. Among the potential contributors of the microbiota to complications of liver cirrhosis is the presence of SIBO[73], and a dysfunctional mucosal barrier which can contribute to bacterial (or endotoxin) translocation to the portal circulation[74]. The pivotal role of gut microbiota in the pathogenesis of hepatic encephalopathy is supported by interventional studies showing the positive effects of treatments directed at gut microbiota in primary prophylaxis[75]. Gut microbiota is also involved in the pathogenesis of NAFLD and its progression to non-alcoholic steatohepatitis. In obese patients, the accumulation of triglyceride-derived metabolites in the liver is due to an increased release of fatty acids from dysfunctional and insulin-resistant adipocytes. A specific microbial taxonomic and functional profile has been associated with obesity[76] and obesity-related liver disease[77], thus suggesting a link between microbiota and liver pathology[78].
Respiratory diseases
The relationship between intestinal microbiota and chronic respiratory diseases is poorly understood. However, a few works have been able to establish a connection between an aberrant microbiota, allergic diseases and bronchial asthma. Several epidemiological studies have shown a clear inverse link between early exposure to microorganisms and the incidence of asthma, referring to this phenomenon as the “hygiene hypothesis”[79]. Also, the increased use of antibiotics and changes in dietary patterns in developed countries, may lead to alterations in the composition of the intestinal microbiota and a higher risk of bronchial asthma[80-82]. Thus, a reduction of Bacteroidetes, lactobacilli and bifidobacteria has been associated with an asthma phenotype[83]. Recent research, focused on the study of airway microbiota and their potential consequences in asthmatic patients, shows that biodiversity is much higher in asthmatic patients with bronchial hyper-reactivity compared with healthy subjects[84].
The results of some studies on other chronic respiratory diseases suggest that pulmonary inflammation and intestinal inflammation are somehow related to each other, and both contribute to the progression of chronic lung disease. This relationship is more evident in some diseases such as cystic fibrosis (CF), in which the lung and intestinal habitats are interconnected through a lung- intestine axis. Evidence suggests that in CF patients the lung disease determines the vital prognosis[85]. CF is an autosomal recessive genetic disease caused by mutations in the gene coding for the regulatory transmembrane conductance regulator (CFTR), which results in progressive lung disease, pancreatic insufficiency, and deficiencies in growth and nutrition. CFTR functions as a chloride ion channel that controls the transportation of water and ions across the apical membrane of epithelial cells. There are different genetic mutations, responsible for different degrees of severity, the DF508 mutation being associated with the most serious symptoms, and one of the most common ones. In this sense, the results of a recent study have shown that the composition of the intestinal microbiota in patients with CF is significantly different depending on the genetic variation[86]; patients with the DF508 mutation have a marked intestinal dysbiosis, in which harmful species, such as E. coli and Eubacterium biforme, are abundant, whereas potentially beneficial ones, such as F. prausnitzii and Bifidobacterium spp., are decreased[86]. In general, these patients, regardless of their genetic background, display a permanent dysbiosis, motivated in part by the characteristics of the disease (thick mucus secretion and pancreatic insufficiency) and the effect related to the aggressive antibiotic therapy[87]. Moreover, it is important to try to correlate the alterations of the airway microbiota with alterations in intestinal microbiota in CF patients. Some authors suggest a link between nutrition and the development of microbial communities in the respiratory tract[88]. These authors note how dietary changes give rise to alterations in the microbiota of the respiratory tract, suggesting that nutritional factors and the pattern of intestinal colonization are determining microbial growth in the respiratory airways, and giving us the opportunity to evaluate the response of CF patients to probiotic and prebiotic interventions[88].
Oral administration of prebiotics or probiotics, or a combination of both, can lead to a change in the composition of the intestinal microbiota. This can directly influence the composition of the microbial communities of the airway by the release of bacterial products or metabolites that reach the lung. These mechanisms could theoretically lead to the restoration of a microbiota that promotes a healthy status, thus having a therapeutic effect on chronic diseases[89]. It is also important to note that probiotic treatment may modulate immune response in the lung; in particular gut microbial stimulation can enhance T regulatory response in the airway, which emphasizes the potential role of probiotics in the pulmonary inflammatory response[89]. However, the mechanisms underlying these specific effects of probiotics are far from being completely understood. Therefore, it will be of key importance to know if airway microbiota is derived from, or linked to, intestinal microbiota. This would mean that a modulation of the intestinal microbiota could have a positive effect on lung disease. Considering this, characterization, modification and the possible functional consequences of airway microbiota are emerging fields that can constitute a new way of acting on chronic lung diseases.
INTESTINAL BIFIDOBACTERIA
The genus Bifidobacterium belongs to the phylum Actinobacteria and comprises over 45 species/subspecies of high G + C (guanine and cytosine) content in their genomes; they are gram-positive, polymorphic rod-shaped bacteria and normal inhabitants of the gastro-intestinal tract of humans and animals[90]. In the human gut the most commonly found species of Bifidobacterium genus include B. adolescentis, B. angulatum, B. bifidum, B. breve, B. catenulatum, B. dentium, B. longum, B. pseudocatenulatum, and B. pseudolongum[91], whereas B. animalis subsp. lactis is the species most often included in functional foods and food supplements[92]. Bifidobacteria is a dominant microbial group in healthy breastfed babies and, during an adult′s life the levels remain relatively stable, tending to decrease in the senescence[93]. During recent years this genus has been extensively studied due to both its important role within the human intestinal microbiota and the extensive use of certain Bifidobacterium strains in probiotic products.
As has been highlighted in the previous sections, intestinal microbiota dysbiosis has been described in different diseases. Given the attention traditionally paid to the genus Bifidobacterium, aberrancies in specific bifidobacterial microbiota, such as decreased numbers, or atypical species composition, have been identified in some of them (Figure 2); these include atopic disease[94,95], IBS[96,97], IBD[98-101], CRC[102], or celiac disease[103,104]. Additionally, bifidobacteria dysbiosis has been reported to precede obesity[105]. It is important to stress that no definitive proof is available on the relationship between reduced bifidobacterial levels, or altered species composition, and disease. However, an aberrant bifidobacterial number or composition is perhaps the most frequently observed intestinal microbiota alteration, being present in many different diseases. This fact suggests an important role for the bifidobacteria population in the intestinal homeostasis. Therefore, on one hand, the bifidobacteria fingerprint could be used as a potential biomarker to understand the intestinal status pointing to a putative dysbiosis. On the other hand, increasing bifidobacterial levels in the gastrointestinal tract could be considered a target to prevent and/or alleviate microbiota-associated diseases. In this regard, several health-promoting effects have been attributed to some specific strains of this genus. The use of functional foods towards the oral delivery of beneficial bifidobacteria (probiotics), alone, or in combination with substrates (prebiotics) that promote the growth of beneficial microbes in the gut, are the basis of dietetic intervention strategies to amend or attenuate intestinal dysbiosis.
Figure 2.
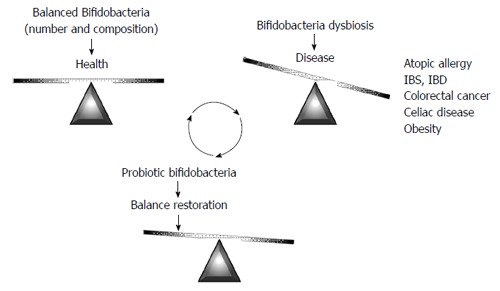
Bifidobacterial dysbiosis and its relationship with diseases: A target for probiotic intervention. IBD: Inflammatory bowel disease; IBS: Irritable bowel syndrome.
Probiotics were defined in 2001 by a group of experts, joined by the World Health Organization and the Food Agriculture Organization of the United Nations, as “live microorganisms, which when administered in adequate amounts confer a health benefit on the host”[106]. However, interest in the use of living bacteria as therapeutic agents started at the beginning of the 1900s with the observations of the Nobel Prize winner Ellie Metchnikoff, and the pediatrician Henry Tissier, both working at the Pasteur Institute. Indeed, Tissier detected that bacteria with “bifid” shape, which were abundant in the faeces of healthy children, were absent in those suffering from diarrhoea; thus he proposed the use of (bifido)-bacteria to restore a healthy microbiota[107]. Afterwards, the use of pro-life (probiotic) microorganisms did not receive much attention in the scientific community in the West. However, around 1930 Dr. Minoru Shirota, working at the Medicine School of Kyoto, cultured a bacterium strain (Lactobacillus casei strain Shirota) that become the first probiotic commercialized in 1935 by the Japanese company Yakut[108]. The market for foods containing probiotics, including bifidobacteria, is growing at a high rate in recent years. Research on this topic is also receiving a lot of attention, since the use of probiotics in preventive medicine to keep a healthy intestinal function is well documented, and probiotics are also proposed as therapeutic agents for gastrointestinal disorders and other pathologies[27].
A search in the Cochrane Library (http://onlinelibrary.wiley.com/cochranelibrary/search) for research into “probiotics” in human health shows several systematic reviews about the use of these microorganisms for prevention, or treatment, of several diseases (Table 1)[109-127]. The overall picture of these studies indicates that the degree of scientific evidence of the effectiveness of probiotics on several pathologies, mainly gut-associated diseases, is insufficient, or the data were not enough to arrive at a conclusion. In most reviews, the selection of human intervention trials with probiotics was a miscellaneous of studies, without defined criteria: probiotic strains were not completely defined (at strain level, dose, vehicle of delivery, duration of the intervention, etc.); the placebo group was not accurate; often probiotics were used in combination with other therapies (e.g., antibiotics); the human target population was not clearly delimited (age, physio-pathological state of the disease, etc.); among others. This lack of definition could mask any beneficial findings obtained with probiotics, and it could hinder a real progress in obtaining effective products. Therefore, more attention should be paid to design, conduct and accurate reporting human studies on probiotics.
Table 1.
Reviews obtained from the search into “probiotics”
| Probiotics | Author's conclusions | Ref. |
| Treating acute infectious diarrhea | Beneficial effects | [109] |
| Prevention of pediatric antibiotic associated diarrhea | Evidence of a protective effect | [110] |
| Treating persistent diarrhea in children | Limited evidence | [111] |
| Treatment of Clostridium difficile (C. difficile)-associated colitis in adults | Insufficient evidence | [112] |
| Prevention of C. difficile-associated diarrhea in adults and children | Moderate quality evidence | [113] |
| Prevention of necrotizing enterocolitis in preterm infants | Prevents severe necrotizing enterocolitis | [114] |
| Induction of remission in ulcerative colitis | Limited evidence | [115] |
| Treatment and prevention of pouchitis after ileal pouch-anal anastomosis for chronic ulcerative colitis | Probiotic VSL#3 was more effective than placebo | [116] |
| Maintenance of remission in ulcerative colitis | Insufficient evidence | [117] |
| Maintenance of remission in Crohn's disease | No evidence | [118] |
| Induction of remission in Crohn's disease | Insufficient evidence | [119] |
| Prevention of post-operative recurrence of Crohn's disease | Insufficient randomized trials | [120] |
| Non-alcoholic fatty liver disease and/or steatohepatitis | Lack of randomized clinical trials | [121] |
| Patients with hepatic encephalopathy | Need of efficacy demonstration | [122] |
| Treatment of bacterial vaginosis | No sufficient evidence | [123] |
| Preventing preterm labour | Insufficient data | [124] |
| For prevention of allergic disease and food hypersensitivity (in infants) | Insufficient evidence | [125] |
| Treating eczema | No effective treatment | [126] |
| Preventing acute upper respiratory tract infections | Limited evidence | [127] |
Reviews obtained from the search into “probiotics” in the Cochrane Library (http://onlinelibrary.wiley.com/cochranelibrary/search) of the Cochrane Database of Systematic Reviews journal.
Bifidobacteria as probiotic in gastrointestinal pathology
In spite of the above mentioned limitations, the World Gastroenterology Organization (WGO) has recently published potential clinical applications for several probiotic formulations, in which species of Lactobacillus play a predominant role[128]. In the current review, we have compiled a series of human intervention studies, showing beneficial effects in gastrointestinal health, which have been made with bifidobacterial strains (Table 2)[129-154]. It is worth noting that only a few Bifidobacterium strains, such as the case of B. animalis subsp. lactis HN019 and B. infantis 35624, have been studied alone, and not in combination with other strains, which limits the conclusions obtained regarding the specific effects of bifidobacteria strains. The studies carried out with the strain 35624 show that it was effective in reducing the pro-inflammatory state of patients with IBD[142,143], as well as with UC[144]; besides, the immune modulation effect of B. infantis 35624 was extended to systemic level since it was effective in reducing inflammatory biomarkers in patients with non-gastrointestinal inflammatory processes, such as chronic fatigue syndrome and psoriasis[144]. Most bifidobacteria strains were tested in combination with other probiotics (lactobacilli and propionibacteria) and/or food-starter bacteria (Streptococcus thermophilus, Lactobacillus delbrueckii subsp. bulgaricus and Lactococcus lactis) (Table 2); thus, the reported beneficial effects cannot be exclusively assigned to a specific bifidobacterial strain. Indeed, the two strains of B. breve included in the table were tested together with some of these bacteria[139,141], or even with prebiotics such as galacto-oligosaccharides[140]. In the case of B. animalis subsp. lactis strains, they were often used as bio-ingredients of yogurt-like fermented milks; thus the additional beneficial effect of this food itself cannot be obviated. Nevertheless, most human studies carried out with this species, mainly with the strains Bb12, DN-173010 and HN019, showed that they are effective in reducing IBS symptoms helping to improve the well-being of the individuals[131,132,134-137]. Different bifidobacteria species (B. animalis subsp. lactis, B. bifidum, or B. infantis), mostly combined with St. thermophilus and/or Lb. acidophilus, have been shown to be effective in the prevention and treatment of antibiotic associated diarrhea[129,130], as well as in the prevention of necrotizing enterocolitis in children[138,145,146]. Finally, the inflammatory conditions of different IBD types (Crohn′s disease, UC and pouchitis) were reduced, to variable extents, after the use of bifidobacteria-containing probiotic products. Particularly the product VSL#3, which includes three bifidobacteria in combination with five lactic acid bacteria, seems to be efficient in the reduction of symptoms, or maintaining remission of IBD in children and adults[147-154].
Table 2.
Some Bifidobacterium strains tested in human intervention studies showing positive effects on gastrointestinal functions
| Bifido species | Bifido strain | Other species | Reported effect | Ref. |
| B. animalis subs. lactis | Bb12 | + S. thermophilus | Antibiotic associated diarrhoea–children | [129] |
| No | Prevent infection in child care centres | [130] | ||
| + L. rhamnosus (GG and Lc705), P. freudenreichii ssp. shermani JS | Alleviate symptoms of IBS | [131] | ||
| DN-173010 | + S. thermophilus, L. delbureckii subsp. bulgaricus, L. lactis (= fermented milk) | Improve symptoms of IBS | [132,133] | |
| Improve GI well-being in women with minor digestive symptoms | [134,135] | |||
| Affect brain activity (emotion and sensation) in healthy women | [136] | |||
| HN019 | No | Improve functional GI symptoms in adults | [137] | |
| B. bifidum | NCDO 1453 | + L. acidophilus NCDO 1748 | Prevent NEC in very low birth weight preterm infants | [138] |
| B. breve | Bb99 | + L. rhamnosus (GG and Lc705), P. freudenreichii ssp. shermani JS | Alleviate symptoms of IBS | [139] |
| Yakult | + prebiotic GOS | Improve clinical conditions of patients with UC | [140] | |
| + L. casei Shirota | Improve symptoms and decrease H2 production in lactose-intolerant patients | [141] | ||
| B. infantis | 35624 | No | Alleviate symptoms of IBS | [142] |
| No | Relive symptoms of IBS in woman | [143] | ||
| No | Reduce systemic pro-inflammatory biomarkers in GI and no-GI conditions | [144] | ||
| Infloran® (B. infantis) | Unknown | + L. acidophilus | Prevent NEC in very low birth weight preterm infants | [145] |
| ABC Dophilus (B. infantis, B. bifidus) | Unknown | + S. thermophilus | Prevent NEC in very low birth weight neonates | [146] |
| VSL#3® (B. breve, B. infantis, B. longum) | Unknown | + S. thermophilus, L. acidophilus, L. plantarum, L. paracasei, L. delbrueckii subsp. bulgaricus | Remission of UC in children | [147] |
| Ameliorate symptoms in children with IBS | [148] | |||
| Reduce symptoms in mild to moderate UC patients | [149,150] | |||
| Maintain remission in recurrent or refractory pouchitis | [151,152] | |||
| Reduce the pouchitis activity index in IPAA patients | [153] | |||
| Reduce the incidence of AAD in risk hospital inpatients | [154] |
AAD: Antibiotic associated diarrhoea; GI: Gastrointestinal; IBS: Irritable bowel syndrome; IPAA: Ileal pouch anal anastomosis; NEC: Necrotizing enterocolitis; UC: Ulcerative colitis; S. thermophilus: Streptococcus thermophilus; L. rhamnosus: Lactobacillus rhamnosus; L. delbureckii: Lactobacillus delbrueckii; L. lactis: Lactococcus lactis; L. acidophilus: Lactobacillus acidophilus; L. casei: Lactobacillus casei; L. plantarum: Lactobacillus plantarum; L. paracasei: Lactobacillus paracasei; P. freudenreichii: Propionibacterium freudenreichii; B. animalis: Bifidobacterium animalis; B. bifidum: Bifidobacterium bifidum; B. breve: Bifidobacterium breve; B. infantis: Bifidobacterium infantis; B. longum: Bifidobacterium longum.
Bifidobacteria as probiotic in respiratory pathology
During the last few years, it has been shown that a few probiotic strains have a potential role in reducing the symptoms of asthma and other allergic respiratory troubles. For instance, a hydrolyzed formula with B. breve and a galacto/fructo-oligosaccharide mixture (prebiotics) was able to prevent asthma-like symptoms in infants with atopic dermatitis[155]. However, one of the more promising applications of probiotic bacteria in respiratory diseases is the use of some Lactobacillus and Bifidobacterium strains in the treatment of allergic rhinitis. In a double-blind, randomized, cross-over study, involving 31 adults with allergic rhinitis to grass pollen, Perrin and colleagues demonstrated that short-term consumption of Lactobacillus paracasei NCC2461 reduced subjective nasal pruritus, whilst not affecting nasal congestion[156]. Regarding Bifidobacterium strains, oral administration of B. animalis subsp. lactis NCC2818 mitigates immune parameters and allergic symptoms in adult subjects suffering from allergic rhinitis during seasonal exposure[157]. Also, the strain B. longum BB536 has shown promising applications in relieving symptoms of cedar pollinosis[158,159]. Furthermore, a combination of probiotics, including one strain of L. acidophilus and two strains of B. animalis subsp. lactis, was shown to be associated with changes in faecal microbiota composition and was able to prevent the pollen-induced infiltration of eosinophils into the nasal mucosa in birch pollen allergy, and indicated a trend for reduced nasal symptoms[160].
CONCLUSION
The human gut is colonised by a myriad of microorganisms commonly referred to as intestinal microbiota. This complex and dynamic bacterial community plays an important role in human health. Alterations, or dysbioses, in microbiota composition and function have been related to different intestinal and extra-intestinal diseases. Among human gut microbiota members the genus Bifidobacterium has attracted lot of scientific interest. Alterations in intestinal bifidobacteria levels, or species composition, are often present in cases of intestinal microbiota dysbiosis. Indeed, deviations in intestinal bifidobacteria have been observed in different diseases, including allergies, IBD, IBS or CRC. Therefore, modulating the intestinal bifidobacteria population has often been considered a target for dietary interventions, providing the rational for the use of microorganisms of the genus Bifidobacterium as probiotics. Different strains have been assessed as probiotics for different diseases, with different results obtained depending on both the strain and the disease tested. Nevertheless, some Bifidobacterium strains have shown very promising results, improving IBD, IBS, diarrhoea, and allergy symptoms.
Footnotes
Supported by The Spanish Ministry of Economy and Competitiveness and by FEDER European Union funds, projects No. AGL2010-14952, No. AGL2010-16525 and No. AGL2012-33278
P- Reviewer: Ortiz LT, Zhang J S- Editor: Gou SX L- Editor: A E- Editor: Wang CH
References
- 1.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 2.Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim BS, Jeon YS, Chun J. Current status and future promise of the human microbiome. Pediatr Gastroenterol Hepatol Nutr. 2013;16:71–79. doi: 10.5223/pghn.2013.16.2.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arboleya S, Solís G, Fernández N, de los Reyes-Gavilán CG, Gueimonde M. Facultative to strict anaerobes ratio in the preterm infant microbiota: a target for intervention? Gut Microbes. 2012;3:583–588. doi: 10.4161/gmic.21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA. 2011;108 Suppl 1:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salazar N, López P, Valdés L, Margolles A, Suárez A, Patterson AM, Cuervo A, de los Reyes-Gavilán CG, Ruas-Madiedo P, González S, et al. Microbial targets for the development of functional foods accordingly with nutritional and immune parameters altered in the elderly. J Am Coll Nutr. 2013;32:399–406. doi: 10.1080/07315724.2013.827047. [DOI] [PubMed] [Google Scholar]
- 10.Cani PD, Everard A, Duparc T. Gut microbiota, enteroendocrine functions and metabolism. Curr Opin Pharmacol. 2013;13:935–940. doi: 10.1016/j.coph.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 11.Reigstad CS, Kashyap PC. Beyond phylotyping: understanding the impact of gut microbiota on host biology. Neurogastroenterol Motil. 2013;25:358–372. doi: 10.1111/nmo.12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 13.Sjögren YM, Tomicic S, Lundberg A, Böttcher MF, Björkstén B, Sverremark-Ekström E, Jenmalm MC. Influence of early gut microbiota on the maturation of childhood mucosal and systemic immune responses. Clin Exp Allergy. 2009;39:1842–1851. doi: 10.1111/j.1365-2222.2009.03326.x. [DOI] [PubMed] [Google Scholar]
- 14.Martin R, Nauta AJ, Ben Amor K, Knippels LM, Knol J, Garssen J. Early life: gut microbiota and immune development in infancy. Benef Microbes. 2010;1:367–382. doi: 10.3920/BM2010.0027. [DOI] [PubMed] [Google Scholar]
- 15.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334:105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arboleya S, Ang L, Margolles A, Yiyuan L, Dongya Z, Liang X, Solís G, Fernández N, de Los Reyes-Gavilán CG, Gueimonde M. Deep 16S rRNA metagenomics and quantitative PCR analyses of the premature infant fecal microbiota. Anaerobe. 2012;18:378–380. doi: 10.1016/j.anaerobe.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 19.Pérez-Cobas AE, Artacho A, Knecht H, Ferrús ML, Friedrichs A, Ott SJ, Moya A, Latorre A, Gosalbes MJ. Differential effects of antibiotic therapy on the structure and function of human gut microbiota. PLoS One. 2013;8:e80201. doi: 10.1371/journal.pone.0080201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aziz Q, Doré J, Emmanuel A, Guarner F, Quigley EM. Gut microbiota and gastrointestinal health: current concepts and future directions. Neurogastroenterol Motil. 2013;25:4–15. doi: 10.1111/nmo.12046. [DOI] [PubMed] [Google Scholar]
- 21.Wu GD, Lewis JD. Analysis of the human gut microbiome and association with disease. Clin Gastroenterol Hepatol. 2013;11:774–777. doi: 10.1016/j.cgh.2013.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shanahan F, Quigley EM. Manipulation of the microbiota for treatment of IBS and IBD-challenges and controversies. Gastroenterology. 2014;146:1554–1563. doi: 10.1053/j.gastro.2014.01.050. [DOI] [PubMed] [Google Scholar]
- 23.Fraher MH, O’Toole PW, Quigley EM. Techniques used to characterize the gut microbiota: a guide for the clinician. Nat Rev Gastroenterol Hepatol. 2012;9:312–322. doi: 10.1038/nrgastro.2012.44. [DOI] [PubMed] [Google Scholar]
- 24.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM, Visser CE, Kuijper EJ, Bartelsman JF, Tijssen JG, et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med. 2013;368:407–415. doi: 10.1056/NEJMoa1205037. [DOI] [PubMed] [Google Scholar]
- 25.Borody TJ, Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9:88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- 26.Smits LP, Bouter KE, de Vos WM, Borody TJ, Nieuwdorp M. Therapeutic potential of fecal microbiota transplantation. Gastroenterology. 2013;145:946–953. doi: 10.1053/j.gastro.2013.08.058. [DOI] [PubMed] [Google Scholar]
- 27.Sanders ME, Guarner F, Guerrant R, Holt PR, Quigley EM, Sartor RB, Sherman PM, Mayer EA. An update on the use and investigation of probiotics in health and disease. Gut. 2013;62:787–796. doi: 10.1136/gutjnl-2012-302504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Borody TJ, Brandt LJ, Paramsothy S. Therapeutic faecal microbiota transplantation: current status and future developments. Curr Opin Gastroenterol. 2014;30:97–105. doi: 10.1097/MOG.0000000000000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manichanh C, Borruel N, Casellas F, Guarner F. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol. 2012;9:599–608. doi: 10.1038/nrgastro.2012.152. [DOI] [PubMed] [Google Scholar]
- 30.Knights D, Lassen KG, Xavier RJ. Advances in inflammatory bowel disease pathogenesis: linking host genetics and the microbiome. Gut. 2013;62:1505–1510. doi: 10.1136/gutjnl-2012-303954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guarner F. What is the role of the enteric commensal flora in IBD? Inflamm Bowel Dis. 2008;14 Suppl 2:S83–S84. doi: 10.1002/ibd.20548. [DOI] [PubMed] [Google Scholar]
- 32.Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology. 2014;146:1489–1499. doi: 10.1053/j.gastro.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Fölsch UR, Timmis KN, Schreiber S. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez C, Antolin M, Santos J, Torrejon A, Casellas F, Borruel N, Guarner F, Malagelada JR. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008;103:643–648. doi: 10.1111/j.1572-0241.2007.01592.x. [DOI] [PubMed] [Google Scholar]
- 35.Chassaing B, Darfeuille-Michaud A. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology. 2011;140:1720–1728. doi: 10.1053/j.gastro.2011.01.054. [DOI] [PubMed] [Google Scholar]
- 36.Rowan F, Docherty NG, Murphy M, Murphy B, Calvin Coffey J, O’Connell PR. Desulfovibrio bacterial species are increased in ulcerative colitis. Dis Colon Rectum. 2010;53:1530–1536. doi: 10.1007/DCR.0b013e3181f1e620. [DOI] [PubMed] [Google Scholar]
- 37.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willing B, Halfvarson J, Dicksved J, Rosenquist M, Järnerot G, Engstrand L, Tysk C, Jansson JK. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn’s disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- 40.Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology. 2004;127:412–421. doi: 10.1053/j.gastro.2004.04.061. [DOI] [PubMed] [Google Scholar]
- 41.Erickson AR, Cantarel BL, Lamendella R, Darzi Y, Mongodin EF, Pan C, Shah M, Halfvarson J, Tysk C, Henrissat B, et al. Integrated metagenomics/metaproteomics reveals human host-microbiota signatures of Crohn’s disease. PLoS One. 2012;7:e49138. doi: 10.1371/journal.pone.0049138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McLaughlin SD, Walker AW, Churcher C, Clark SK, Tekkis PP, Johnson MW, Parkhill J, Ciclitira PJ, Dougan G, Nicholls RJ, et al. The bacteriology of pouchitis: a molecular phylogenetic analysis using 16S rRNA gene cloning and sequencing. Ann Surg. 2010;252:90–98. doi: 10.1097/SLA.0b013e3181e3dc8b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castells A, Castellví-Bel S, Balaguer F. Concepts in familial colorectal cancer: where do we stand and what is the future? Gastroenterology. 2009;137:404–409. doi: 10.1053/j.gastro.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 44.Zhu Q, Gao R, Wu W, Qin H. The role of gut microbiota in the pathogenesis of colorectal cancer. Tumour Biol. 2013;34:1285–1300. doi: 10.1007/s13277-013-0684-4. [DOI] [PubMed] [Google Scholar]
- 45.Terzić J, Grivennikov S, Karin E, Karin M. Inflammation and colon cancer. Gastroenterology. 2010;138:2101–2114.e5. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 46.Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, Huso DL, Brancati FL, Wick E, McAllister F, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azcárate-Peril MA, Sikes M, Bruno-Bárcena JM. The intestinal microbiota, gastrointestinal environment and colorectal cancer: a putative role for probiotics in prevention of colorectal cancer? Am J Physiol Gastrointest Liver Physiol. 2011;301:G401–G424. doi: 10.1152/ajpgi.00110.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scanlan PD, Shanahan F, Clune Y, Collins JK, O’Sullivan GC, O’Riordan M, Holmes E, Wang Y, Marchesi JR. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol. 2008;10:789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- 49.Uronis JM, Mühlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shaheen NJ, Hansen RA, Morgan DR, Gangarosa LM, Ringel Y, Thiny MT, Russo MW, Sandler RS. The burden of gastrointestinal and liver diseases, 2006. Am J Gastroenterol. 2006;101:2128–2138. doi: 10.1111/j.1572-0241.2006.00723.x. [DOI] [PubMed] [Google Scholar]
- 51.Simrén M, Barbara G, Flint HJ, Spiegel BM, Spiller RC, Vanner S, Verdu EF, Whorwell PJ, Zoetendal EG. Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut. 2013;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spiller R, Garsed K. Infection, inflammation, and the irritable bowel syndrome. Dig Liver Dis. 2009;41:844–849. doi: 10.1016/j.dld.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Dunlop SP, Jenkins D, Neal KR, Spiller RC. Relative importance of enterochromaffin cell hyperplasia, anxiety, and depression in postinfectious IBS. Gastroenterology. 2003;125:1651–1659. doi: 10.1053/j.gastro.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 54.Swan C, Duroudier NP, Campbell E, Zaitoun A, Hastings M, Dukes GE, Cox J, Kelly FM, Wilde J, Lennon MG, et al. Identifying and testing candidate genetic polymorphisms in the irritable bowel syndrome (IBS): association with TNFSF15 and TNFα. Gut. 2013;62:985–994. doi: 10.1136/gutjnl-2011-301213. [DOI] [PubMed] [Google Scholar]
- 55.Ford AC, Spiegel BM, Talley NJ, Moayyedi P. Small intestinal bacterial overgrowth in irritable bowel syndrome: systematic review and meta-analysis. Clin Gastroenterol Hepatol. 2009;7:1279–1286. doi: 10.1016/j.cgh.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 56.Spiegel BM. Questioning the bacterial overgrowth hypothesis of irritable bowel syndrome: an epidemiologic and evolutionary perspective. Clin Gastroenterol Hepatol. 2011;9:461–469; quiz e59. doi: 10.1016/j.cgh.2011.02.030. [DOI] [PubMed] [Google Scholar]
- 57.Tack J. Antibiotic therapy for the irritable bowel syndrome. N Engl J Med. 2011;364:81–82. doi: 10.1056/NEJMe1011211. [DOI] [PubMed] [Google Scholar]
- 58.Halmos EP, Power VA, Shepherd SJ, Gibson PR, Muir JG. A diet low in FODMAPs reduces symptoms of irritable bowel syndrome. Gastroenterology. 2014;146:67–75.e5. doi: 10.1053/j.gastro.2013.09.046. [DOI] [PubMed] [Google Scholar]
- 59.Ringel Y, Maharshak N. Intestinal microbiota and immune function in the pathogenesis of irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2013;305:G529–G541. doi: 10.1152/ajpgi.00207.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carroll IM, Ringel-Kulka T, Siddle JP, Ringel Y. Alterations in composition and diversity of the intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:521–530, e248. doi: 10.1111/j.1365-2982.2012.01891.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maukonen J, Satokari R, Mättö J, Söderlund H, Mattila-Sandholm T, Saarela M. Prevalence and temporal stability of selected clostridial groups in irritable bowel syndrome in relation to predominant faecal bacteria. J Med Microbiol. 2006;55:625–633. doi: 10.1099/jmm.0.46134-0. [DOI] [PubMed] [Google Scholar]
- 62.Parkes GC, Rayment NB, Hudspith BN, Petrovska L, Lomer MC, Brostoff J, Whelan K, Sanderson JD. Distinct microbial populations exist in the mucosa-associated microbiota of sub-groups of irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:31–39. doi: 10.1111/j.1365-2982.2011.01803.x. [DOI] [PubMed] [Google Scholar]
- 63.Tana C, Umesaki Y, Imaoka A, Handa T, Kanazawa M, Fukudo S. Altered profiles of intestinal microbiota and organic acids may be the origin of symptoms in irritable bowel syndrome. Neurogastroenterol Motil. 2010;22:512–519, e114-115. doi: 10.1111/j.1365-2982.2009.01427.x. [DOI] [PubMed] [Google Scholar]
- 64.Martínez C, Lobo B, Pigrau M, Ramos L, González-Castro AM, Alonso C, Guilarte M, Guilá M, de Torres I, Azpiroz F, et al. Diarrhoea-predominant irritable bowel syndrome: an organic disorder with structural abnormalities in the jejunal epithelial barrier. Gut. 2013;62:1160–1168. doi: 10.1136/gutjnl-2012-302093. [DOI] [PubMed] [Google Scholar]
- 65.Duboc H, Rainteau D, Rajca S, Humbert L, Farabos D, Maubert M, Grondin V, Jouet P, Bouhassira D, Seksik P, et al. Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol Motil. 2012;24:513–520, e246-247. doi: 10.1111/j.1365-2982.2012.01893.x. [DOI] [PubMed] [Google Scholar]
- 66.Collins SM, Bercik P. The relationship between intestinal microbiota and the central nervous system in normal gastrointestinal function and disease. Gastroenterology. 2009;136:2003–2014. doi: 10.1053/j.gastro.2009.01.075. [DOI] [PubMed] [Google Scholar]
- 67.Bercik P. The microbiota-gut-brain axis: learning from intestinal bacteria? Gut. 2011;60:288–289. doi: 10.1136/gut.2010.226779. [DOI] [PubMed] [Google Scholar]
- 68.Mayer EA, Savidge T, Shulman RJ. Brain-gut microbiome interactions and functional bowel disorders. Gastroenterology. 2014;146:1500–1512. doi: 10.1053/j.gastro.2014.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ceapa C, Wopereis H, Rezaïki L, Kleerebezem M, Knol J, Oozeer R. Influence of fermented milk products, prebiotics and probiotics on microbiota composition and health. Best Pract Res Clin Gastroenterol. 2013;27:139–155. doi: 10.1016/j.bpg.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 70.Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. The role of the microbiome in rheumatic diseases. Curr Rheumatol Rep. 2013;15:314. doi: 10.1007/s11926-012-0314-y. [DOI] [PubMed] [Google Scholar]
- 71.Ebel B, Lemetais G, Beney L, Cachon R, Sokol H, Langella P, Gervais P. Impact of probiotics on risk factors for cardiovascular diseases. A review. Crit Rev Food Sci Nutr. 2014;54:175–189. doi: 10.1080/10408398.2011.579361. [DOI] [PubMed] [Google Scholar]
- 72.Shanahan F. The gut microbiota in 2011: Translating the microbiota to medicine. Nat Rev Gastroenterol Hepatol. 2012;9:72–74. doi: 10.1038/nrgastro.2011.250. [DOI] [PubMed] [Google Scholar]
- 73.Gupta A, Dhiman RK, Kumari S, Rana S, Agarwal R, Duseja A, Chawla Y. Role of small intestinal bacterial overgrowth and delayed gastrointestinal transit time in cirrhotic patients with minimal hepatic encephalopathy. J Hepatol. 2010;53:849–855. doi: 10.1016/j.jhep.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 74.Szabo G, Bala S, Petrasek J, Gattu A. Gut-liver axis and sensing microbes. Dig Dis. 2010;28:737–744. doi: 10.1159/000324281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Victor DW, Quigley EM. Hepatic encephalopathy involves interactions among the microbiota, gut, brain. Clin Gastroenterol Hepatol. 2014;12:1009–1011. doi: 10.1016/j.cgh.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 76.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 77.Vajro P, Paolella G, Fasano A. Microbiota and gut-liver axis: their influences on obesity and obesity-related liver disease. J Pediatr Gastroenterol Nutr. 2013;56:461–468. doi: 10.1097/MPG.0b013e318284abb5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quigley EM, Stanton C, Murphy EF. The gut microbiota and the liver. Pathophysiological and clinical implications. J Hepatol. 2013;58:1020–1027. doi: 10.1016/j.jhep.2012.11.023. [DOI] [PubMed] [Google Scholar]
- 79.Noverr MC, Huffnagle GB. The ‘microflora hypothesis’ of allergic diseases. Clin Exp Allergy. 2005;35:1511–1520. doi: 10.1111/j.1365-2222.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- 80.Riedler J, Braun-Fahrländer C, Eder W, Schreuer M, Waser M, Maisch S, Carr D, Schierl R, Nowak D, von Mutius E. Exposure to farming in early life and development of asthma and allergy: a cross-sectional survey. Lancet. 2001;358:1129–1133. doi: 10.1016/S0140-6736(01)06252-3. [DOI] [PubMed] [Google Scholar]
- 81.Ege MJ, Mayer M, Normand AC, Genuneit J, Cookson WO, Braun-Fahrländer C, Heederik D, Piarroux R, von Mutius E. Exposure to environmental microorganisms and childhood asthma. N Engl J Med. 2011;364:701–709. doi: 10.1056/NEJMoa1007302. [DOI] [PubMed] [Google Scholar]
- 82.Omland Ø, Hjort C, Pedersen OF, Miller MR, Sigsgaard T. New-onset asthma and the effect of environment and occupation among farming and nonfarming rural subjects. J Allergy Clin Immunol. 2011;128:761–765. doi: 10.1016/j.jaci.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 83.Ouwehand AC, Isolauri E, He F, Hashimoto H, Benno Y, Salminen S. Differences in Bifidobacterium flora composition in allergic and healthy infants. J Allergy Clin Immunol. 2001;108:144–145. doi: 10.1067/mai.2001.115754. [DOI] [PubMed] [Google Scholar]
- 84.Gollwitzer ES, Marsland BJ. Microbiota abnormalities in inflammatory airway diseases - Potential for therapy. Pharmacol Ther. 2014;141:32–39. doi: 10.1016/j.pharmthera.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 85.Lynch SV, Goldfarb KC, Wild YK, Kong W, De Lisle RC, Brodie EL. Cystic fibrosis transmembrane conductance regulator knockout mice exhibit aberrant gastrointestinal microbiota. Gut Microbes. 2013;4:41–47. doi: 10.4161/gmic.22430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schippa S, Iebba V, Santangelo F, Gagliardi A, De Biase RV, Stamato A, Bertasi S, Lucarelli M, Conte MP, Quattrucci S. Cystic fibrosis transmembrane conductance regulator (CFTR) allelic variants relate to shifts in faecal microbiota of cystic fibrosis patients. PLoS One. 2013;8:e61176. doi: 10.1371/journal.pone.0061176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Duytschaever G, Huys G, Bekaert M, Boulanger L, De Boeck K, Vandamme P. Cross-sectional and longitudinal comparisons of the predominant fecal microbiota compositions of a group of pediatric patients with cystic fibrosis and their healthy siblings. Appl Environ Microbiol. 2011;77:8015–8024. doi: 10.1128/AEM.05933-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, Moore JH, Guill MF, Morrison HG, Sogin ML, et al. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. MBio. 2012;3:pii: e00251–12. doi: 10.1128/mBio.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Forsythe P. Probiotics and lung diseases. Chest. 2011;139:901–908. doi: 10.1378/chest.10-1861. [DOI] [PubMed] [Google Scholar]
- 90.Margolles A, Ruas-Madiedo P, de los Reyes-Gavilán CG, Sánchez B, Gueimonde M. Bifidobacterium. In: Liu D, editor. Molecular Detection of human bacterial pathogens. Florida: CRC Press, Taylor & Francis Group; 2011. pp. 45–57. [Google Scholar]
- 91.Turroni F, Marchesi JR, Foroni E, Gueimonde M, Shanahan F, Margolles A, van Sinderen D, Ventura M. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 2009;3:745–751. doi: 10.1038/ismej.2009.19. [DOI] [PubMed] [Google Scholar]
- 92.Masco L, Huys G, De Brandt E, Temmerman R, Swings J. Culture-dependent and culture-independent qualitative analysis of probiotic products claimed to contain bifidobacteria. Int J Food Microbiol. 2005;102:221–230. doi: 10.1016/j.ijfoodmicro.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 93.Gueimonde M, Ouwehand A, Pitkälä K, Strandberg T, Finne-Soveri H, Salminen S. Fecal Bifidobacterium levels in elderly nursing home patients-Are levels as expected? Biosci Microflora. 2010;29:111–113. [Google Scholar]
- 94.He F, Ouwehand AC, Isolauri E, Hashimoto H, Benno Y, Salminen S. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. FEMS Immunol Med Microbiol. 2001;30:43–47. doi: 10.1111/j.1574-695X.2001.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 95.Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 96.Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373–382. doi: 10.1111/j.1572-0241.2005.40312.x. [DOI] [PubMed] [Google Scholar]
- 97.Kerckhoffs AP, Samsom M, van der Rest ME, de Vogel J, Knol J, Ben-Amor K, Akkermans LM. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol. 2009;15:2887–2892. doi: 10.3748/wjg.15.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Favier C, Neut C, Mizon C, Cortot A, Colombel JF, Mizon J. Fecal beta-D-galactosidase production and Bifidobacteria are decreased in Crohn’s disease. Dig Dis Sci. 1997;42:817–822. doi: 10.1023/a:1018876400528. [DOI] [PubMed] [Google Scholar]
- 99.Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Doré J. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut. 2003;52:237–242. doi: 10.1136/gut.52.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Macfarlane S, Furrie E, Cummings JH, Macfarlane GT. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin Infect Dis. 2004;38:1690–1699. doi: 10.1086/420823. [DOI] [PubMed] [Google Scholar]
- 101.Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:481–487. doi: 10.1097/01.mib.0000159663.62651.4f. [DOI] [PubMed] [Google Scholar]
- 102.Gueimonde M, Ouwehand A, Huhtinen H, Salminen E, Salminen S. Qualitative and quantitative analyses of the bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J Gastroenterol. 2007;13:3985–3989. doi: 10.3748/wjg.v13.i29.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sanz Y, Sánchez E, Marzotto M, Calabuig M, Torriani S, Dellaglio F. Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunol Med Microbiol. 2007;51:562–568. doi: 10.1111/j.1574-695X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- 104.Collado MC, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalances in faecal and duodenal Bifidobacterium species composition in active and non-active coeliac disease. BMC Microbiol. 2008;8:232. doi: 10.1186/1471-2180-8-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Collado MC, Isolauri E, Laitinen K, Salminen S. Effect of mother’s weight on infant’s microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr. 2010;92:1023–1030. doi: 10.3945/ajcn.2010.29877. [DOI] [PubMed] [Google Scholar]
- 106.World Health Organization-Food and Agriculture Organization. Probiotics in foods. Health and nutritional properties and guidelines for evaluation, FAO Food and Nutritional Paper. Rome: FAO/WHO; 2006. pp. No. 8592–5-105513-0. [Google Scholar]
- 107.Tissier H. Traitement des infections intestinales par la méthod de la flore bactérienne de línstein. Crit Rev Soc Biol. 1906;60:359–361. [Google Scholar]
- 108.Vasiljevic T, Shah NP. Probiotics - from Metchnikoff to bioactives. Int Dairy J. 2008;18:714–728. [Google Scholar]
- 109.Allen SJ, Martinez EG, Gregorio GV, Dans LF. Probiotics for treating acute infectious diarrhoea. Cochrane Database Syst Rev. 2010;(11):CD003048. doi: 10.1002/14651858.CD003048.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnston BC, Goldenberg JZ, Vandvik PO, Sun X, Guyatt GH. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev. 2011;(11):CD004827. doi: 10.1002/14651858.CD004827.pub3. [DOI] [PubMed] [Google Scholar]
- 111.Bernaola Aponte G, Bada Mancilla CA, Carreazo NY, Rojas Galarza RA. Probiotics for treating persistent diarrhoea in children. Cochrane Database Syst Rev. 2013;8:CD007401. doi: 10.1002/14651858.CD007401.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pillai A, Nelson R. Probiotics for treatment of Clostridium difficile-associated colitis in adults. Cochrane Database Syst Rev. 2008;(1):CD004611. doi: 10.1002/14651858.CD004611.pub2. [DOI] [PubMed] [Google Scholar]
- 113.Goldenberg JZ, Ma SS, Saxton JD, Martzen MR, Vandvik PO, Thorlund K, Guyatt GH, Johnston BC. Probiotics for the prevention of Clostridium difficile-associated diarrhea in adults and children. Cochrane Database Syst Rev. 2013;5:CD006095. doi: 10.1002/14651858.CD006095.pub3. [DOI] [PubMed] [Google Scholar]
- 114.Alfaleh K, Anabrees J, Bassler D, Al-Kharfi T. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev. 2011;(3):CD005496. doi: 10.1002/14651858.CD005496.pub3. [DOI] [PubMed] [Google Scholar]
- 115.Mallon P, McKay D, Kirk S, Gardiner K. Probiotics for induction of remission in ulcerative colitis. Cochrane Database Syst Rev. 2007;(4):CD005573. doi: 10.1002/14651858.CD005573.pub2. [DOI] [PubMed] [Google Scholar]
- 116.Holubar SD, Cima RR, Sandborn WJ, Pardi DS. Treatment and prevention of pouchitis after ileal pouch-anal anastomosis for chronic ulcerative colitis. Cochrane Database Syst Rev. 2010;(6):CD001176. doi: 10.1002/14651858.CD001176.pub2. [DOI] [PubMed] [Google Scholar]
- 117.Naidoo K, Gordon M, Fagbemi AO, Thomas AG, Akobeng AK. Probiotics for maintenance of remission in ulcerative colitis. Cochrane Database Syst Rev. 2011;(12):CD007443. doi: 10.1002/14651858.CD007443.pub2. [DOI] [PubMed] [Google Scholar]
- 118.Rolfe VE, Fortun PJ, Hawkey CJ, Bath-Hextall F. Probiotics for maintenance of remission in Crohn’s disease. Cochrane Database Syst Rev. 2006;(4):CD004826. doi: 10.1002/14651858.CD004826.pub2. [DOI] [PubMed] [Google Scholar]
- 119.Butterworth AD, Thomas AG, Akobeng AK. Probiotics for induction of remission in Crohn’s disease. Cochrane Database Syst Rev. 2008;(3):CD006634. doi: 10.1002/14651858.CD006634.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Doherty G, Bennett G, Patil S, Cheifetz A, Moss AC. Interventions for prevention of post-operative recurrence of Crohn’s disease. Cochrane Database Syst Rev. 2009;(4):CD006873. doi: 10.1002/14651858.CD006873.pub2. [DOI] [PubMed] [Google Scholar]
- 121.Lirussi F, Mastropasqua E, Orando S, Orlando R. Probiotics for non-alcoholic fatty liver disease and/or steatohepatitis. Cochrane Database Syst Rev. 2007;(1):CD005165. doi: 10.1002/14651858.CD005165.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McGee RG, Bakens A, Wiley K, Riordan SM, Webster AC. Probiotics for patients with hepatic encephalopathy. Cochrane Database Syst Rev. 2011;(11):CD008716. doi: 10.1002/14651858.CD008716.pub2. [DOI] [PubMed] [Google Scholar]
- 123.Senok AC, Verstraelen H, Temmerman M, Botta GA. Probiotics for the treatment of bacterial vaginosis. Cochrane Database Syst Rev. 2009;(4):CD006289. doi: 10.1002/14651858.CD006289.pub2. [DOI] [PubMed] [Google Scholar]
- 124.Othman M, Neilson JP, Alfirevic Z. Probiotics for preventing preterm labour. Cochrane Database Syst Rev. 2007;(1):CD005941. doi: 10.1002/14651858.CD005941.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Osborn DA, Sinn JK. Probiotics in infants for prevention of allergic disease and food hypersensitivity. Cochrane Database Syst Rev. 2007;(4):CD006475. doi: 10.1002/14651858.CD006475.pub2. [DOI] [PubMed] [Google Scholar]
- 126.Boyle RJ, Bath-Hextall FJ, Leonardi-Bee J, Murrell DF, Tang ML. Probiotics for treating eczema. Cochrane Database Syst Rev. 2008;(4):CD006135. doi: 10.1002/14651858.CD006135.pub2. [DOI] [PubMed] [Google Scholar]
- 127.Hao Q, Lu Z, Dong BR, Huang CQ, Wu T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst Rev. 2011;(9):CD006895. doi: 10.1002/14651858.CD006895.pub2. [DOI] [PubMed] [Google Scholar]
- 128.World Gastroenterology Organisation. World Gastroenterology Organisation Global Guidelines: Probiotics and Prebiotics, 2011. Available from: http://www.worldgastroenterology.org/probiotics-prebiotics.html.
- 129.Corrêa NB, Péret Filho LA, Penna FJ, Lima FM, Nicoli JR. A randomized formula controlled trial of Bifidobacterium lactis and Streptococcus thermophilus for prevention of antibiotic-associated diarrhea in infants. J Clin Gastroenterol. 2005;39:385–389. doi: 10.1097/01.mcg.0000159217.47419.5b. [DOI] [PubMed] [Google Scholar]
- 130.Weizman Z, Asli G, Alsheikh A. Effect of a probiotic infant formula on infections in child care centers: comparison of two probiotic agents. Pediatrics. 2005;115:5–9. doi: 10.1542/peds.2004-1815. [DOI] [PubMed] [Google Scholar]
- 131.Kajander K, Myllyluoma E, Rajilić-Stojanović M, Kyrönpalo S, Rasmussen M, Järvenpää S, Zoetendal EG, de Vos WM, Vapaatalo H, Korpela R. Clinical trial: multispecies probiotic supplementation alleviates the symptoms of irritable bowel syndrome and stabilizes intestinal microbiota. Aliment Pharmacol Ther. 2008;27:48–57. doi: 10.1111/j.1365-2036.2007.03542.x. [DOI] [PubMed] [Google Scholar]
- 132.Guyonnet D, Chassany O, Ducrotte P, Picard C, Mouret M, Mercier CH, Matuchansky C. Effect of a fermented milk containing Bifidobacterium animalis DN-173 010 on the health-related quality of life and symptoms in irritable bowel syndrome in adults in primary care: a multicentre, randomized, double-blind, controlled trial. Aliment Pharmacol Ther. 2007;26:475–486. doi: 10.1111/j.1365-2036.2007.03362.x. [DOI] [PubMed] [Google Scholar]
- 133.Agrawal A, Houghton LA, Morris J, Reilly B, Guyonnet D, Goupil Feuillerat N, Schlumberger A, Jakob S, Whorwell PJ. Clinical trial: the effects of a fermented milk product containing Bifidobacterium lactis DN-173 010 on abdominal distension and gastrointestinal transit in irritable bowel syndrome with constipation. Aliment Pharmacol Ther. 2009;29:104–114. doi: 10.1111/j.1365-2036.2008.03853.x. [DOI] [PubMed] [Google Scholar]
- 134.Guyonnet D, Schlumberger A, Mhamdi L, Jakob S, Chassany O. Fermented milk containing Bifidobacterium lactis DN-173 010 improves gastrointestinal well-being and digestive symptoms in women reporting minor digestive symptoms: a randomised, double-blind, parallel, controlled study. Br J Nutr. 2009;102:1654–1662. doi: 10.1017/S0007114509990882. [DOI] [PubMed] [Google Scholar]
- 135.Marteau P, Guyonnet D, Lafaye de Micheaux P, Gelu S. A randomized, double-blind, controlled study and pooled analysis of two identical trials of fermented milk containing probiotic Bifidobacterium lactis CNCM I-2494 in healthy women reporting minor digestive symptoms. Neurogastroenterol Motil. 2013;25:331–e252. doi: 10.1111/nmo.12078. [DOI] [PubMed] [Google Scholar]
- 136.Tillisch K, Labus J, Kilpatrick L, Jiang Z, Stains J, Ebrat B, Guyonnet D, Legrain-Raspaud S, Trotin B, Naliboff B, et al. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144:1394–1401, 1401.e1-4. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Waller PA, Gopal PK, Leyer GJ, Ouwehand AC, Reifer C, Stewart ME, Miller LE. Dose-response effect of Bifidobacterium lactis HN019 on whole gut transit time and functional gastrointestinal symptoms in adults. Scand J Gastroenterol. 2011;46:1057–1064. doi: 10.3109/00365521.2011.584895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lin HC, Hsu CH, Chen HL, Chung MY, Hsu JF, Lien RI, Tsao LY, Chen CH, Su BH. Oral probiotics prevent necrotizing enterocolitis in very low birth weight preterm infants: a multicenter, randomized, controlled trial. Pediatrics. 2008;122:693–700. doi: 10.1542/peds.2007-3007. [DOI] [PubMed] [Google Scholar]
- 139.Kajander K, Hatakka K, Poussa T, Färkkilä M, Korpela R. A probiotic mixture alleviates symptoms in irritable bowel syndrome patients: a controlled 6-month intervention. Aliment Pharmacol Ther. 2005;22:387–394. doi: 10.1111/j.1365-2036.2005.02579.x. [DOI] [PubMed] [Google Scholar]
- 140.Ishikawa H, Matsumoto S, Ohashi Y, Imaoka A, Setoyama H, Umesaki Y, Tanaka R, Otani T. Beneficial effects of probiotic bifidobacterium and galacto-oligosaccharide in patients with ulcerative colitis: a randomized controlled study. Digestion. 2011;84:128–133. doi: 10.1159/000322977. [DOI] [PubMed] [Google Scholar]
- 141.Almeida CC, Lorena SL, Pavan CR, Akasaka HM, Mesquita MA. Beneficial effects of long-term consumption of a probiotic combination of Lactobacillus casei Shirota and Bifidobacterium breve Yakult may persist after suspension of therapy in lactose-intolerant patients. Nutr Clin Pract. 2012;27:247–251. doi: 10.1177/0884533612440289. [DOI] [PubMed] [Google Scholar]
- 142.O’Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, O’Sullivan GC, Kiely B, Collins JK, Shanahan F, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. 2005;128:541–551. doi: 10.1053/j.gastro.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 143.Whorwell PJ, Altringer L, Morel J, Bond Y, Charbonneau D, O’Mahony L, Kiely B, Shanahan F, Quigley EM. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am J Gastroenterol. 2006;101:1581–1590. doi: 10.1111/j.1572-0241.2006.00734.x. [DOI] [PubMed] [Google Scholar]
- 144.Groeger D, O’Mahony L, Murphy EF, Bourke JF, Dinan TG, Kiely B, Shanahan F, Quigley EM. Bifidobacterium infantis 35624 modulates host inflammatory processes beyond the gut. Gut Microbes. 2013;4:325–339. doi: 10.4161/gmic.25487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lin HC, Su BH, Chen AC, Lin TW, Tsai CH, Yeh TF, Oh W. Oral probiotics reduce the incidence and severity of necrotizing enterocolitis in very low birth weight infants. Pediatrics. 2005;115:1–4. doi: 10.1542/peds.2004-1463. [DOI] [PubMed] [Google Scholar]
- 146.Bin-Nun A, Bromiker R, Wilschanski M, Kaplan M, Rudensky B, Caplan M, Hammerman C. Oral probiotics prevent necrotizing enterocolitis in very low birth weight neonates. J Pediatr. 2005;147:192–196. doi: 10.1016/j.jpeds.2005.03.054. [DOI] [PubMed] [Google Scholar]
- 147.Miele E, Pascarella F, Giannetti E, Quaglietta L, Baldassano RN, Staiano A. Effect of a probiotic preparation (VSL#3) on induction and maintenance of remission in children with ulcerative colitis. Am J Gastroenterol. 2009;104:437–443. doi: 10.1038/ajg.2008.118. [DOI] [PubMed] [Google Scholar]
- 148.Guandalini S, Magazzù G, Chiaro A, La Balestra V, Di Nardo G, Gopalan S, Sibal A, Romano C, Canani RB, Lionetti P, et al. VSL#3 improves symptoms in children with irritable bowel syndrome: a multicenter, randomized, placebo-controlled, double-blind, crossover study. J Pediatr Gastroenterol Nutr. 2010;51:24–30. doi: 10.1097/MPG.0b013e3181ca4d95. [DOI] [PubMed] [Google Scholar]
- 149.Sood A, Midha V, Makharia GK, Ahuja V, Singal D, Goswami P, Tandon RK. The probiotic preparation, VSL#3 induces remission in patients with mild-to-moderately active ulcerative colitis. Clin Gastroenterol Hepatol. 2009;7:1202–1209, 1209.e1. doi: 10.1016/j.cgh.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 150.Tursi A, Brandimarte G, Papa A, Giglio A, Elisei W, Giorgetti GM, Forti G, Morini S, Hassan C, Pistoia MA, et al. Treatment of relapsing mild-to-moderate ulcerative colitis with the probiotic VSL#3 as adjunctive to a standard pharmaceutical treatment: a double-blind, randomized, placebo-controlled study. Am J Gastroenterol. 2010;105:2218–2227. doi: 10.1038/ajg.2010.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Gionchetti P, Rizzello F, Helwig U, Venturi A, Lammers KM, Brigidi P, Vitali B, Poggioli G, Miglioli M, Campieri M. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202–1209. doi: 10.1016/s0016-5085(03)00171-9. [DOI] [PubMed] [Google Scholar]
- 152.Mimura T, Rizzello F, Helwig U, Poggioli G, Schreiber S, Talbot IC, Nicholls RJ, Gionchetti P, Campieri M, Kamm MA. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut. 2004;53:108–114. doi: 10.1136/gut.53.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pronio A, Montesani C, Butteroni C, Vecchione S, Mumolo G, Vestri A, Vitolo D, Boirivant M. Probiotic administration in patients with ileal pouch-anal anastomosis for ulcerative colitis is associated with expansion of mucosal regulatory cells. Inflamm Bowel Dis. 2008;14:662–668. doi: 10.1002/ibd.20369. [DOI] [PubMed] [Google Scholar]
- 154.Selinger CP, Bell A, Cairns A, Lockett M, Sebastian S, Haslam N. Probiotic VSL#3 prevents antibiotic-associated diarrhoea in a double-blind, randomized, placebo-controlled clinical trial. J Hosp Infect. 2013;84:159–165. doi: 10.1016/j.jhin.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 155.van der Aa LB, van Aalderen WM, Heymans HS, Henk Sillevis Smitt J, Nauta AJ, Knippels LM, Ben Amor K, Sprikkelman AB. Synbiotics prevent asthma-like symptoms in infants with atopic dermatitis. Allergy. 2011;66:170–177. doi: 10.1111/j.1398-9995.2010.02416.x. [DOI] [PubMed] [Google Scholar]
- 156.Perrin Y, Nutten S, Audran R, Berger B, Bibiloni R, Wassenberg J, Barbier N, Aubert V, Moulin J, Singh A, et al. Comparison of two oral probiotic preparations in a randomized crossover trial highlights a potentially beneficial effect of Lactobacillus paracasei NCC2461 in patients with allergic rhinitis. Clin Transl Allergy. 2014;4:1. doi: 10.1186/2045-7022-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Singh A, Hacini-Rachinel F, Gosoniu ML, Bourdeau T, Holvoet S, Doucet-Ladeveze R, Beaumont M, Mercenier A, Nutten S. Immune-modulatory effect of probiotic Bifidobacterium lactis NCC2818 in individuals suffering from seasonal allergic rhinitis to grass pollen: an exploratory, randomized, placebo-controlled clinical trial. Eur J Clin Nutr. 2013;67:161–167. doi: 10.1038/ejcn.2012.197. [DOI] [PubMed] [Google Scholar]
- 158.Xiao JZ, Kondo S, Yanagisawa N, Takahashi N, Odamaki T, Iwabuchi N, Iwatsuki K, Kokubo S, Togashi H, Enomoto K, et al. Effect of probiotic Bifidobacterium longum BB536 [corrected] in relieving clinical symptoms and modulating plasma cytokine levels of Japanese cedar pollinosis during the pollen season. A randomized double-blind, placebo-controlled trial. J Investig Allergol Clin Immunol. 2006;16:86–93. [PubMed] [Google Scholar]
- 159.Odamaki T, Xiao JZ, Iwabuchi N, Sakamoto M, Takahashi N, Kondo S, Miyaji K, Iwatsuki K, Togashi H, Enomoto T, et al. Influence of Bifidobacterium longum BB536 intake on faecal microbiota in individuals with Japanese cedar pollinosis during the pollen season. J Med Microbiol. 2007;56:1301–1308. doi: 10.1099/jmm.0.47306-0. [DOI] [PubMed] [Google Scholar]
- 160.Ouwehand AC, Nermes M, Collado MC, Rautonen N, Salminen S, Isolauri E. Specific probiotics alleviate allergic rhinitis during the birch pollen season. World J Gastroenterol. 2009;15:3261–3268. doi: 10.3748/wjg.15.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]


