Background: Escherichia coli OmpA interacts with FcγRI α-chain (FcγRIa) to invade macrophages.
Results: Lack of three N-glycans in FcγRIa prevents E. coli invasion of macrophages and the onset of meningitis.
Conclusion: OmpA binding to FcγRIa via N-glycans is crucial in the pathogenesis of E. coli meningitis.
Significance: Targeting OmpA and FcγRIa interface may be a therapy for E. coli-induced meningitis.
Keywords: Computer Modeling, Escherichia coli (E. coli), Fc-gamma Receptor, Infectious Disease, Macrophage, N-Glycans, Meningitis, Outer Membrane Protein A
Abstract
Neonatal meningitis, caused by Escherichia coli K1, is a serious central nervous system disease. We have established that macrophages serve as permissive niches for E. coli K1 to multiply in the host and for attaining a threshold level of bacterial load, which is a prerequisite for the onset of the disease. Here, we demonstrate experimentally that three N-glycans in FcγRIa interact with OmpA of E. coli K1 for binding to and entering the macrophages. Adoptive transfer of FcγRIa−/− bone marrow-derived macrophages transfected with FcγRIa into FcγRIa−/− newborn mice renders them susceptible to E. coli K1-induced meningitis. In contrast, mice that received bone marrow-derived macrophages transfected with FcγRIa in which N-glycosylation sites 1, 4, and 5 are mutated to alanines exhibit resistance to E. coli K1 infection. Our molecular dynamics and simulation studies predict that N-glycan 5 exhibits strong binding at the barrel site of OmpA formed by loops 3 and 4, whereas N-glycans 1 and 4 interact with loops 1, 3, and 4 of OmpA at tip regions. Molecular modeling data also suggest no role for the IgG binding site in the invasion process. In agreement, experimental mutations in IgG binding site had no effect on the E. coli K1 entry into macrophages in vitro or on the onset of meningitis in newborn mice. Together, this integration of experimental and computational studies reveals how the N-glycans in FcγRIa interact with the OmpA of E. coli K1 for inducing the disease pathogenesis.
Introduction
In the event of bacterial invasion of host tissues, the intruder encounters an arsenal of host defense mechanisms that result in either resolution of the pathogen by the host or subversion of the defense by the pathogen. Cross-talk between host defense components is critical for successful resolution of the infection. Macrophages are long-lived cells that play a critical role in engulfing pathogenic microorganisms and degrading them. They express a range of receptors that recognize bacteria, including Toll-like receptors, Fcγ receptors, complement receptors, scavenger receptors, and mannose receptors (1, 2). Most microbial structures are recognized by more than one macrophage receptor, and these receptors also interact with each other. Several pathogens subvert the antimicrobial mechanism by exploiting receptor interactions to create their own safe havens inside of which they survive. The bacterial pathogens upon entering the host are coated with complement proteins, enabling them to be recognized by complement receptors on the macrophages (3). Similarly, antibody-coated bacterial recognition involves interaction with Fcγ receptors, which subsequently elicits antimicrobial mechanisms. A large number of bacteria introduce microbial factors that govern macrophage function by type III or IV secretion systems (1). However, very few bacteria control the attack of macrophages at the receptor level. One example is Staphylococcus aureus, which uses protein A to bind to the Fc region of IgG, thus avoiding recognition by Fcγ receptors (4).
Several studies have shown that FcγRI expression increases during septicemia and meningitis caused by a variety of bacteria. The Fc region of IgG recognizes Fcγ receptors (FcγRs),4 enabling it to play an important role in linking the cellular and humoral immune response. FcγR comprises a multigene family divided into three classes (FcγRI, II, and III), which are defined by their affinity for IgG. FcγRI is a transmembrane receptor that binds IgG with high affinity and induces the association of the γ-chain for signal transduction and triggering of effector responses such as macrophage phagocytosis. The ligation of FcγRI with IgG also mediates antibody-dependent cellular cytotoxicity-induced transcription of cytokine genes and release of inflammatory mediators (5). We previously demonstrated that OmpA (outer membrane protein A) of Escherichia coli K1, which causes neonatal meningitis, directly interacts with Fcγ receptor I α-chain (FcγRIa) to bind to and enter macrophages. Indeed, depletion of macrophages or lack of FcγRIa expression in macrophages in newborn mice renders the animals resistant to E. coli K1-induced meningitis. Despite the general requirement of FcγRIa association with the γ-chain for the internalization of the receptor, the interaction of OmpA+ E. coli with FcγRIa and the subsequent entry into macrophages do not require the γ-chain to facilitate E. coli K1 entry into macrophages, which is a novel mechanism. E. coli K1 interaction with macrophages in the absence of FcγRIa induces the expression of complement receptor 3, which elicits antimicrobial mechanisms to kill the intracellular bacteria (6). In addition, macrophages generate biopterin and neopterin upon E. coli K1 infection to suppress the production of nitric oxide and superoxide, respectively (7). We showed previously that mutation of three amino acids in loops 1 and 3 of the extracellular domains of OmpA prevented the bacterial survival in macrophages. Concomitantly, E. coli K1 containing a mutation in loop 1 could not cause meningitis in newborn mouse model (8). However, there has been no molecular level understanding of how OmpA interaction with FcγRIa controls these cellular events.
OmpA has been shown to interact with GlcNAc1–4GlcNAc epitopes of host receptors (9, 10). In addition, our previous molecular modeling predictions of OmpA interaction with GlcNAc1–4GlcNAc epitopes demonstrated that this moiety can bind to OmpA at two sites, one at the tip of loops 1 and 2 and the second at the barrel site formed by loops 3 and 4 (6). We now report investigations on the role of N-glycans in FcγRIa in E. coli K1 entry of macrophages and for the onset of meningitis in the newborn mouse model. The experimental studies show that N-glycosylation sites 1, 4, and 5 of FcγRIa are critical for interacting with OmpA of E. coli K1, both for binding to and entry of macrophages. Adoptive transfer of FcγRIa−/− macrophages transfected with N-glycosylation (NG) mutants of FcγRIa into FcγRIa−/− mice revealed that the presence of FcγRIa with one, four, or five NG sites are important for the onset of meningitis. To determine a structural basis for the experimental results, we conducted computer simulations to predict the atomistic structure for the OmpA protein complexed with the glycosylated FcγRIa, which identified how N-glycans contribute to the E. coli K1 interaction with macrophages.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Antibodies, and Other Reagents
Escherichia coli K1 (OmpA+ E. coli) is a spontaneous rifampicin-resistant mutant of strain RS218 (serotype O18:K1:H7), which was isolated from the cerebrospinal fluid of a newborn with meningitis (12). Bacteria were grown in LB medium (Difco, Detroit, MI) with 100 μg/ml rifampicin. Antibodies to FcγRIa and Myc tag were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Cell Signaling Technology (Danvers, MA), respectively. Secondary antibodies tagged to various fluorophores were purchased from Invitrogen. FuGENE HD reagent from Roche Applied Science was used for plasmid transfection. QuikChange site-directed mutagenesis kit was from Agilent Technologies (Santa Clara, CA). Phagocytosis assay kit (IgG-FITC) was from Cayman chemical company (Ann Arbor, MI).
E. coli Invasion Assays
Bone marrow-derived macrophages (BMDMs) were isolated and cultured as described earlier (6). RAW 264.7 macrophages were grown to confluence (∼105 cells/well) in 24-well plates. RAW 264.7 macrophages were then incubated with 106 colony forming units (CFU) of E. coli K1 in experimental medium (DMEM containing 5% heat-inactivated fetal bovine serum) for 60 min at 37 °C in CO2 incubator. For BMDMs, the cells were incubated for 2 h with 106 CFU of E. coli K1. The monolayers were washed three times with RPMI 1640 and incubated further with experimental medium containing gentamicin (100 μg/ml) for 1 h to kill bound bacteria. The monolayers were washed again and lysed with 0.5% Triton X-100. The intracellular bacteria were determined by plating the dilutions on sheep blood agar. To enumerate the total cell-associated bacteria, the experiments were performed without a gentamicin step.
Western Blotting
RAW 264.7 or BMDMs (WT and FcγRIa−/−) were transfected with FL-FcγRIa or NG-FcγRIa mutants using FuGENE HD according to the manufacturer's instructions and allowed to recover for 24 h, and the total cell lysates were prepared using lysis buffer (50 mm Tris-Cl, 150 mm NaCl, 1 mm EGTA, and 1% Triton X-100). Unbroken cells and cell debris were removed by centrifuging the lysates at 700 × g for 10 min at 4 °C. The protein content was estimated using Pierce BCA protein assay kit. 40 μg of protein fractions were resolved on an 8% gel and transferred to a nitrocellulose membrane. The membrane was blocked with 5% milk in PBS, 0.1% Tween 20 for 1 h at room temperature. The blots were then incubated with anti-Myc or anti-FcγRIa antibodies overnight using appropriate dilutions and counterstained with HRP-conjugated secondary antibody. The membrane was developed with Super Signal chemiluminescence substrate (Pierce) and exposed to x-ray film for protein visualization.
Flow Cytometry
To detect the expression of transfected FcγRIa plasmids using Myc antibody, RAW 264.7 or BMDMs (WT and FcγRIa−/−) were transfected as described earlier and allowed to recover for 24 h. The cells were washed three times with PBS and then detached with TrypLE Express (Invitrogen) from the plates. The cells were fixed using BD Cytofix for 15 min, washed, and preincubated for 30 min with blocking/wash buffer (PBS + 3% normal goat serum) to mask nonspecific binding sites. Cells were then incubated with anti-Myc antibody or an isotype-matched control antibody for 1 h at 4 °C and washed with buffer. Then FITC-conjugated secondary antibody was added, incubated for 30 min at 4 °C, and washed with the buffer. The stained cells were then analyzed by four-color flow cytometry using FACSCalibur Cell Quest Pro software (BD Biosciences, San Diego, CA), and at least 10,000 events were collected for analysis. The results are expressed as bar graphs or histogram overlays with respect to isotype-matched antibody controls. For detection of IgG-FITC uptake, transfected RAW 264.7 or BMDMs were incubated with 25 μl of IgG-FITC beads for 2 h at 37 °C to promote phagocytosis. The extracellular fluorescence from noninternalized beads was quenched with 500 μl of trypan blue (diluted 1:10 in assay buffer) and then subjected to processing for flow cytometry as mentioned above. Alexa 647 secondary antibody was used to detect Myc expression, and histogram overlays were plotted using FL1 (FITC) and FL4 (Alexa 647) channels.
Transmission and Scanning Electron Microscopy
WT BMDMs were transfected with FcγRIa-FL or FcγRIa-NG mutants and incubated with E. coli K1 at a multiplicity of infection of 10 for 1 h, washed, and then fixed with 2% glutaraldehyde in 0.1 m cacodylate buffer, pH 7.1. All samples were washed three times in 0.1 m cacodylate buffer for 15 min each. The cells were then postfixed for 20 min in 1% osmium tetroxide at 4 °C followed by addition of EtOH (60%). Samples were dehydrated through 70, 80, 95, and 100% EtOH (two times, 15 min each) and then into propylene oxide (two time, 15 min each), and into a 1:1 propylene oxide/eponate, left overnight, and capped at room temperature. The propylene oxide/eponate mixture was decanted and replaced with 100% eponate mixture. The samples were polymerized at 70 °C for 48 h. Thin sections (80 nm) were cut using a diamond knife, mounted on uncoated 300 mesh copper grids and stained with 5% uranyl acetate for 20 min. Photographs were taken with transmission electron microscopy (JEOL JEM 2100 LaB6) or scanning electron microscopy (JEOL JSM/6390LV).
Tissue Immunostaining
Sections of paraffinized brain sections from control and infected pups were submitted to the Department of Pathology, Children's Hospital Los Angeles for hematoxylin and eosin (H&E), glial fibrillary acidic protein (GFAP), and myeloperoxidase (MPO) staining.
Newborn Mouse Model of Meningitis
The animal studies were approved by the Institutional Animal Care and Use Committee of Children's Hospital of Los Angeles and followed guidelines for the performance of animal experiments implemented by the National Institutes of Health. FcγRIa−/− mice were described previously (6). FcγRIa−/− BMDMs were cultured in Petri dishes and maintained till the pups were born. On the day of birth, the BMDMs were transferred to 24-well plates and transfected with the respective FcγRIa plasmids after 24 h. Three-day-old mouse pups (n = 5 per group) were infected intranasally with 103 CFU of bacteria in pyrogen-free saline as previously described (6). 5 μl of facial vein blood was collected aseptically from all the pups in the respective groups at 48 h postinfection to determine the bacterial load by plating 10-fold serial dilutions on rifampicin LB agar plates. All the pups were euthanatized at 72 h postinfection, and cerebrospinal fluid was collected from all the pups without traumatic trap and incubated overnight in LB broth with rifampicin to check for bacterial growth. 5 μl of blood was collected by heart puncture and plated on rifampicin LB agar plates after serial dilution. One half of the brain from all the pups was homogenized, serial diluted, and plated on rifampicin LB agar to determine brain bacterial load. The other half of the brain was preserved in formalin, paraffin-embedded, and sectioned for staining.
Protein-Protein Docking Protocol
A hybrid docking strategy was adopted that first considered an ensemble of 15 protein conformations for OmpA. Next, each of the conformations was docked to FcγRIa to obtain an ensemble of 810,000 OmpA-FcγRIa protein-protein poses through exhaustive rotational and translational sampling of the relative position space that satisfies shape complementarity of the two given protein conformations. Then experimental observations and topological constraints were used to dramatically reduce the most likely poses to just a few that could be subjected to more detailed considerations including molecular dynamics. This strategy overcomes the limitations of finding scoring functions that can reliably rank the energies for protein-protein interactions, and it reduces the issues regarding the conformational flexibility of glycosylated amino acids.
First, an ensemble of 15 structures of OmpA that consider the flexibility of its four loops was predicted. To construct this ensemble, (a) we selected as one candidate the PDB code 1BXW x-ray crystal structure of OmpA, which resolves more of the loop regions than the PDB code 1QJP x-ray crystal structure (13, 14). (b) In addition, 2.5-ns MD simulation of the protein in explicit membrane and water (including salt) was carried out, starting from a conformation previously generated from the 1BXW x-ray crystal structure equilibrated with MD at 300 K. Here the protein was embedded in a periodically infinite box containing the POPC lipid system fully solvated with water and salt. We used the CHARMM26 forcefield method for the lipid and OmpA protein and the TIP3P water forcefield (15–17). The visual molecular dynamics membrane plug-in to build the membrane and the visual molecular dynamics solvate plug-in to solvate the protein at both the intracellular and extracellular regions were used (18). All MD simulations used the NAMD program (19) with the isothermal-isobaric ensemble and periodic boundary conditions at 300 K and 1 atm. From this trajectory, 2500 conformations (1 snapshot every 1 ps) have been selected, which we clustered into 13 diverse structures using an root mean square deviation Voronoi criterion of 1.5 Å, from which we selected the 13 family heads. (c) In addition, an ensemble of 10 candidate structures based on distance-geometry fits to NMR (PDB code 1G90) (20) is available, and their energies were minimized in vacuum using the DREIDING force field (21). From this, the one with the lowest total energy (number 4) was selected.
These 15 OmpA conformations have a minimum root mean square deviation diversity of 1.5 Å in the loop regions. Next, we examined the crystal structure of FcγRIa (PDB code 3RJD) (22). The interaction between the D1 and D2 fragments in the crystal is well defined; however, the D3 fragment is interacting with a nearby image of the protein in the crystal packing as visualized in the crystal supercell. This interaction may cause artifacts in the hinge angle between D2 and D3 fragments. Therefore, only the Ig-like domains D1 and D2 of the FcγRIa crystal structure were taken for docking purposes. Then the Man-Manβ1-4GlcNAc β1–4GlcNAcβ1 portion of N-glycans to each of the five N-glycosylation sites on D1 and D2 (residues 59, 78, 152, 159, and 163, predicted by NetNGlyc 1.0 Server) was added. Further, we relaxed the backbone of D1 and D2 with 1-ns MD in a solvent box of explicit water (including 154 mm NaCl or 0.9% w/w NaCl), whereas the side chains including the N-glycans were relaxed with 5 ns of MD in explicit water. The snapshot closest to the average conformation was then chosen for docking.
To describe the glycosylated Asn consistently with natural amino acids in the MD, the AMBER ff99SBildn (improved side chain torsion potentials for the Amber ff99SB) forcefield for the natural residues in the protein and the general AMBER force field (GAFF) for the glycosylated Asn were used (23, 24). The Antechamber auxiliary program was used to prepare the GAFF library of the glycosylated Asn (25). The TIP3P water model and the AmberTools (26) including a terminal interface called tLEAP were used to add the ions and the water solvation box.
Next, all 15 OmpA conformations were docked to the FcγRIa D1 and D2 structure using only the protein-protein complex pose generation part of ZDOCK (27, 28). The ZDOCK shape complementarity algorithm generated 54,000 complex poses with exhaustive rotational and translational sampling for each configuration of OmpA. Combining this with the 15 separate docking procedures yielded 810,000 complex poses. We then eliminated all poses in which FcγRIa would have a likely clash with the virtual membrane where OmpA is buried. This virtual membrane screening left us 496,100 poses. These poses were analyzed to determine which glycans and loops are involved in binding and the number of interprotein salt bridges within each pose. After eliminating poses that did not agree with the experimental evidence, we obtained 326 poses.
Poses Matching Experimental Input
These 326 hits were then ranked by their number of interprotein salt bridges, leading to one pose with a maximum of eight salt bridges. This pose of OmpA-FcγRIa (D1-D2 only) was selected as the structure likely to have the strongest protein-protein structure that is consistent with all experimental data. Then the D3 domain of FcγRIa as it is in the crystal structure was added, and the consequent clashes between several loops were resolved by resampling the loop conformations using the DREIDING forcefield, suggesting that the D2-D3 hinge angle may not be affected much by the crystal packing as we initially speculated. This new structure is referred to as OmpA-FcγRIa for the remainder of the manuscript. We then subjected this OmpA-FcγRIa structure to 50 ns of isothermal-isobaric ensemble at 300 K and 1 atm MD simulation in a periodic box with explicit lipid membrane (POPC) and explicit water with 154 mm NaCl, where the position of OmpA in the membrane is calculated according to the implicit solvent model of the lipid bilayer in the Orientations of Proteins in Membranes (OPM) database (29). To allow the predicted pose to relax any strains that might arise in the newly formed protein-protein interface from our rigid body docking procedure, we carried out MD for 50 ns on the full system including explicit water and ions. We found that all eight salt bridges and the other polar interactions between the subunits are maintained during the 50 ns of MD, indicating that the predicted protein-protein complex structure is stable.
Statistical Analysis
All experimental data were derived from at least three independent experiments. Statistical analyses were conducted using GraphPad prism online calculator, and the values are presented as means ± S.D. Significant differences (p < 0.01) between the groups were determined using the unpaired Student's t test.
RESULTS
N-Glycosylation Sites 1, 4, and 5 in the Extracellular Domain of FcγRIa Are Important for E. coli K1 Binding to and Invasion of RAW 264.7 Macrophages
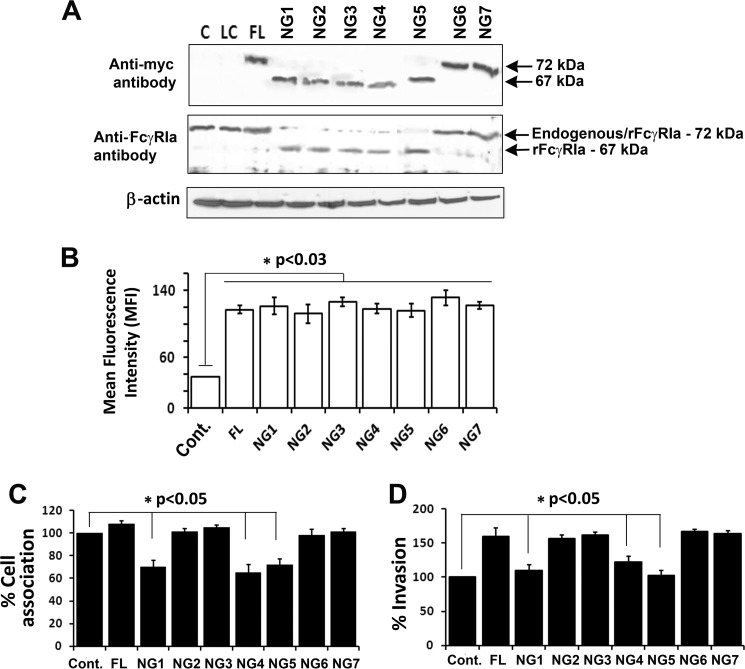
We previously demonstrated experimentally that OmpA of E. coli K1 interacts with GlcNAc1–4GlcNAc epitopes of glycoproteins and that FcγRIa expression is critical for the entry into macrophages (6, 9, 10). Therefore, we sought to understand the role of NG sites in FcγRIa for E. coli K1 invasion into macrophages. FcγRIa contains seven glycosylation sites attached to asparagine residues as analyzed by the NetNGlyc 1.0 program. We mutated each of these to alanines in the FcγRIa sequence that contains a Myc tag. Primers sequences used in the mutation of these asparagine residues are given in Table 1. The change in the amino acid from Asn to Ala was verified by nucleotide sequencing. In addition, loss of one glycosylation site changes the molecular mass of a glycoprotein by ∼3 kDa. Therefore, the recombinant proteins were subjected to SDS-PAGE and Western blotting with anti-Myc and anti-FcγRIa antibodies. As shown in Fig. 1A, NG mutants 1–5 of FcγRIa migrated lower than wild type FcγRIa, whereas NG mutants 6 and 7 showed similar molecular mass to that of wild type protein. This suggests that the consensus NG sequence sites 6 and 7 identified by NetNGlyc 1.0 program were not really glycosylated in FcγRIa. Next, the five FcγRIa NG mutants were overexpressed in RAW 264.7 macrophages, and the expression was verified using anti-Myc antibody by flow cytometry (Fig. 1B). Transfection efficiency in these cells was ∼80%, which were then subjected to total cell association and invasion assays. Total cell associated and intracellular bacteria were significantly higher with full-length FcγRIa (FL-FcγRIa), FcγRIa/NG2,/NG3,/NG6, and/NG7. However, overexpression of FcγRIa/NG1, FcγRIa/NG4, and FcγRIa/NG5 showed ∼40% decrease in the total cell associated and 60% in the invasion of E. coli K1 compared with FL-FcγRIa transfected cells (Fig. 1, C and D). All these constructs are referred hereafter as FL, NG1, NG2, NG3, NG4, NG5, NG6, and NG7. The difference in the invasion in these cells is not due to differences in expression levels of recombinant FcγRIa because flow cytometry of the transfected cells revealed similar levels of expression.
TABLE 1.
Primer sequences used to generate mutations in FcγRIa
| Forward | Reverse | |
|---|---|---|
| NG1 | CAGTGGTTTCTCGCAGGCACAGCCAC | GTGGCTGTGCCTGCGAGAAACCACTG |
| NG2 | CTCTGCCAGTGTCGCAGACAGTGGTGAATAC | GTATTCACCACTGTCTGCGACACTGGCAGAG |
| NG3 | CCACTGGAATTCTGCCCTCACCATTCTG | CAGAATGGTGAGGGCAGAATTCCAGTGG |
| NG4 | CCATTCTGAAAACCGGCATAAGTCACAATGGC | GCCATTGTGACTTATGGCGGTTTTCAGAATGG |
| NG5 | CCAACATAAGTCACGCCGGCACCTACCATTG | CAATGGTAGGTGCCGGCGTGACTTATGTTGG |
| NG6 | GCTCCAGTGCTGGCAGCATCTGTGAC | GTCACAGATGCTGCCAGCACTGGAGC |
| NG7 | CTGCGAGGCAGGGCAACATCCTCTGAATAC | GTATTCAGAGGATGTTGCCCTGCCTCGCAG |
| IgG1 | CAGGCATGGGACAGCATCGCTACACATC | GATGTGTAGCGATGCTGTCCCATGCCTG |
| IgG2 | GGCATGGGAAAGCATCAGTACACATCAGCAG | CTGCTGATGTGTACTGATGCTTTCCCATGCC |
FIGURE 1.
N-Glycosylation sites 1, 4, and 5 in the extracellular domain of FcγRIa are important for E. coli K1 binding to and invasion of RAW 264.7 macrophages. A, RAW 264.7 macrophages were transfected with FL or with different NG mutants of FcγRIa were subjected to Western blotting analysis using anti-Myc or anti-FcγRIa antibody. Reduction in the size of the bands indicates loss of one NG site. The NG mutation sites in FcγRIa are sequentially labeled as NG1 to NG7. B, the efficiency of plasmid transfection was analyzed concurrently using flow cytometry with the anti-Myc antibody, and the fluorescence intensity is graphed. The increase in fluorescence intensity in transfected cells was statistically significant compared with untransfected control cells. C and D, cell association (C) and invasion (D) of E. coli K1 in transfected RAW 264.7 macrophages were performed as described under “Experimental Procedures.” Decrease in cell association/invasion in FcγRIa/NG1,/NG4 and/NG5 transfected RAW 264.7 macrophages was statistically significant. All experiments were performed at least three times. Cont., control.
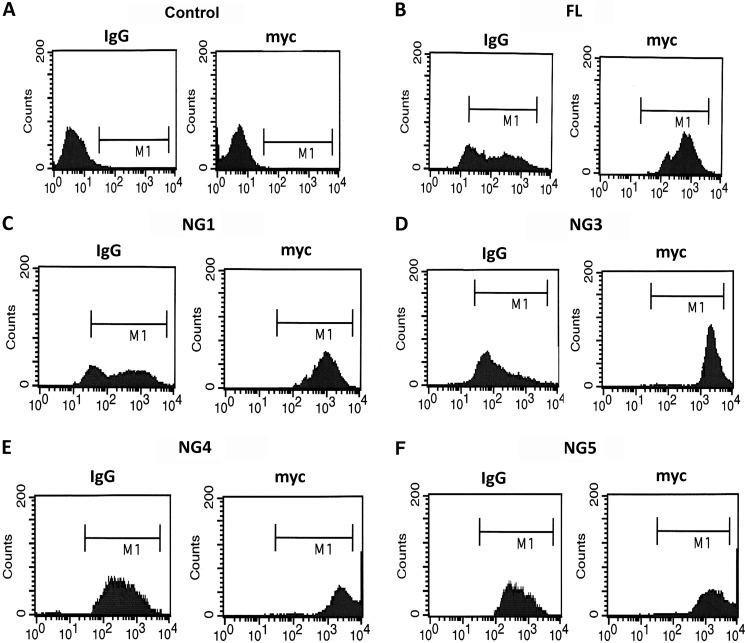
In addition, we also verified whether the overexpression of NG mutations alter the Fc-mediated uptake of IgG, FITC-dextran beads coated with IgG were added to RAW 264.7 macrophages. After 30 min of incubation, the extracellular fluorescence was quenched with Trypan blue, and the cells were then subjected to flow cytometry to identify the intracellular fluorescence for FITC-IgG, as well as Myc tag expression. The data revealed that NG mutations in FcγRIa did alter the Fc-mediated uptake of FITC-beads. In addition, the expression of FcγRIa and its NG mutants was similar in all the transfectants, indicating that NG mutations in FcγRIa did not alter the phagocytic capacity of the macrophages (Fig. 2). These results suggest that glycosylation sites 1, 4, and 5 may play an important role in FcγRIa for E. coli K1 invasion of RAW 264.7 macrophages, and the invasion process might be independent of Fc-mediated phagocytosis.
FIGURE 2.
Overexpression of FL or NG mutants of FcγRIa does not affect IgG uptake in RAW 264.7 macrophages. Untransfected, FL, or NG mutants of FcγRIa transfected RAW 264.7 macrophages were incubated with FITC-IgG for 2 h at 37 °C or incubated with trypan blue to quench extracellular fluorescence. The cells were then incubated with anti-Myc antibody followed by Alexa 647 secondary antibody and then subjected to flow cytometry to detect FITC, as well as Myc expression. Cells were gates (M1) based on fluorescent output from isotype-matched control. A, control. B, full length. C, NG1. D, NG3. E, NG4. F, NG5.
Lack of Specific N-Glycosylation Sites in FcγRIa Also Prevents E. coli K1 Entry into Bone Marrow-derived Macrophages
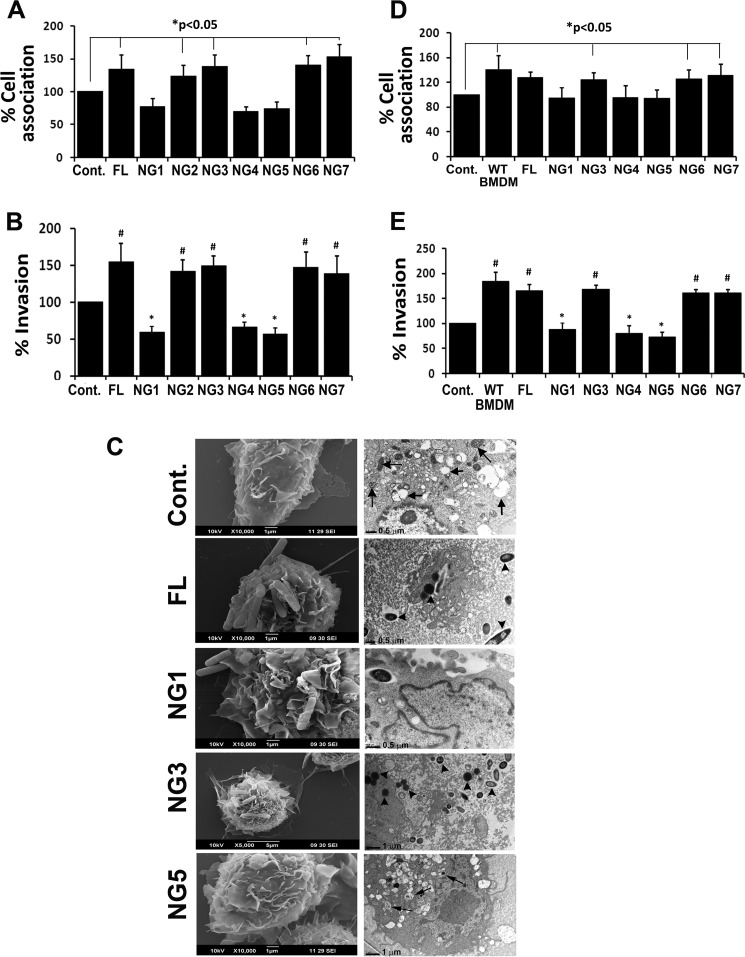
The role of NG sites 1, 4, and 5 in FcγRIa in the entry of E. coli K1 in primary macrophages was also examined. BMDMs from mice were cultured and transfected with various constructs of FcγRIa containing NG mutations and then subjected to total cell association and invasion assays. Flow cytometry analysis of transfected BMDMs revealed that Myc expression was similar in all the transfections (data not shown). Similar to RAW264.7 macrophages, overexpression of NG1, NG4, and NG5 in BMDMs significantly inhibited both total cell-associated and intracellular bacteria compared with FL, NG2, NG3, NG6, or NG7 (Fig. 3, A and B). To examine whether the added bacteria entered BMDMs transfected with the mutant FcγRIa plasmids, scanning and transmission electron microscopy were performed. As shown in Fig. 3C, uninfected BMDMs showed spread out morphology in S.E., whereas transmission electron microscopy showed several vacuoles (black arrows) and mitochondria (yellow arrows) in the cells. Overexpression of FL revealed efficient binding of bacteria on the surface of BMDMs, and a number of bacteria entered into the cells as shown in transmission electron microscopy, which was also observed in NG3 transfected BMDMs. In contrast, NG1 and NG5 transfected cells had very few bacteria attached to the cell surface, and those that entered the cells appeared to be subsequently killed inside vacuoles (black arrows). NG4 transfected BMDMs also showed results similar to those for NG1 (data not shown). In addition, FcγRIa−/− macrophages were also transfected with FL and mutant FcγRIa plasmids and used in E. coli cell association and invasion assays. FcγRIa−/− macrophages inhibited bacterial invasion, evident from a 1.5/2-fold decrease in cell association/invasion, respectively, when compared with WT BMDMs, which is consistent with previous data (Fig. 3, D and E). However, transfection with FL overexpression in these cells renders them susceptible to E. coli entry. In contrast, overexpression of NG1, NG4, and NG5 showed lesser intracellular E. coli compared with NG2-, NG3-, NG6-, and NG7-expressing cells. These results suggest that NG1, NG4, and NG5 play a significant role in the binding of E. coli to FcγRIa and subsequent entry into BMDMs but that NG2 and NG3 are not important.
FIGURE 3.
Lack of specific N-glycosylation sites in FcγRIa also prevents entry into bone marrow-derived macrophages. A and B, bone marrow-derived macrophages from C57BL/6 mice (WT BMDMs) were transfected with FL or NG mutants of FcγRIa and subjected to cell association (A) and invasion (B) assays. C, scanning and transmission electron microscopy analysis of WT BMDMs transfected with FL or NG1, NG3, and NG5 mutants of FcγRIa. D and E, FcγRIa−/− BMDMs were transfected with FL or NG mutants and subjected to cell association (D) and invasion (E) assays. Untransfected FcγRIa−/− BMDMs and BMDMs from C57BL/6 mice served as controls. The cell association/invasion assays were performed at least three times, and the increase (*) or decrease (#) in cell association/invasion patterns was statistically significant (p < 0.05). Arrowheads, bacteria; black arrows, vacuoles or mitochondria. Cont., control.
Adoptive Transfer of FL, but Not NG1-, NG4-, or NG5-transfected BMDMs into FcγRIa Knock-out Newborn Mice Restores the Susceptibility to E. coli K1 Meningitis
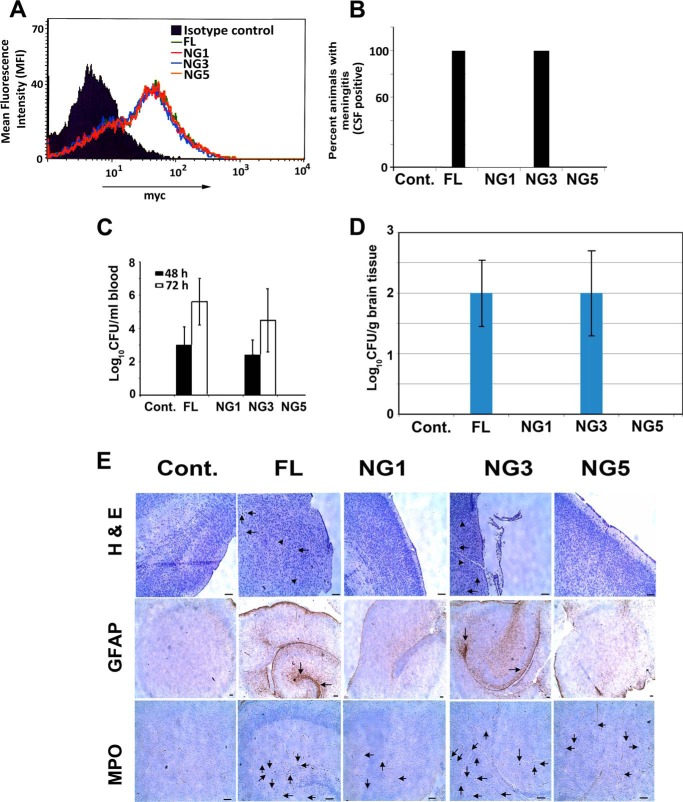
We successfully used adoptive transfer of BMDMs into macrophage-depleted or FcγRIa−/− mice and demonstrated that the presence of macrophages is critical for the onset of meningitis (6). Therefore, to determine whether NG sites in FcγRIa are important for E. coli K1-induced meningitis in newborn mice, we adoptively transferred the BMDMs transfected with FL or NG FcγRIa into 3-day-old FcγRIa−/− mice and then challenged with E. coli K1 by intranasal instillation on day 3. We conducted the animal experiments with FL, NG1, NG3, NG4, or NG5 glycosylation mutant FcγRIa transfected BMDMs, but only the data from NG1, NG3, and NG5 experiments are presented. The transfected FcγRIa−/− macrophages expressed similar levels of recombinant proteins on the surface as analyzed by flow cytometry (Fig. 4A). As shown previously, FcγRIa−/− newborn mice were resistant to E. coli K1 induced meningitis, whereas adoptive transfer of BMDMs transfected with FL or NG3 rendered the animals susceptible for E. coli K1 infection as measured by positive cerebrospinal fluid cultures (Fig. 4B). In contrast, adoptive transfer of NG1 and NG5 transfected BMDMs still showed resistance to E. coli K1 infection. In agreement with the onset of meningitis, bacteremia levels and brain bacterial load of FL or NG3 BMDMs transferred mice were significantly higher than the levels of E. coli K1 in NG1 and NG5 BMDMs transferred mice (Fig. 4, C and D). Moreover, histological examination of brain sections of the pups from respective groups was performed using standard H&E, MPO, and GFAP staining. Brains of newborn mice adoptively transferred with FL or NG3 BMDMs showed loss of tissue integrity, neutrophil infiltration, and glial cell migration, which are hallmarks of E. coli K1 meningitis, whereas pups transferred with NG1 and NG5 BMDMs had intact brain architecture with the absence of neutrophils and glial cells (Fig. 4E).
FIGURE 4.
Adoptive transfer of FL and NG3, but not NG1- or NG5-transfected FcγRIa−/− BMDMs into FcγRIa−/− newborn mice restores the susceptibility to E. coli K1 meningitis. A, transfection efficiency of plasmids in FcγRIa−/− BMDMs was confirmed by flow cytometry using anti-Myc tag antibody. FcγRIa−/− newborn mice (n = 5) were adoptively transferred with FcγRIa−/− BMDMs containing different plasmid constructs at 66 h after birth and then infected intranasally with 103 E. coli K1 at 72 h. B, all five pups carrying FL or NG3 transfected BMDMs had E. coli K1-positive cerebrospinal fluid (CSF) cultures 72 h postinfection. C, bacterial load in blood at 48 and 72 h postinfection from each pup was collected and plated on LB + rifampicin to enumerate bacteremia levels. D, homogenates from half of the brain from all pups were plated on LB + rifampicin after euthanatizing them 72 h postinfection. E, the other half of the brain was preserved in formalin, embedded in paraffin, sectioned, and stained for brain pathology (H&E), GFAP, and neutrophil infiltration (MPO). Arrows represent neuronal loss in H&E stained sections or glial cells in GFAP-stained sections. Neutrophil infiltration is represented by arrowheads in H&E-stained sections and arrows in MPO-stained sections. Cont., control.
Molecular Simulation of OmpA and Glycosylated FcγRIa Interaction Supports the Experimental Results that NG1, NG4, and NG5 Sites Are Important for Binding
Based on the experimental data that NG sites 1, 4, and 5 in FcγRIa are critical to interacting with OmpA, simulation studies were performed to unravel molecular bases for this interaction. As described under “Experimental Procedures,” the FcγRIa structure was assembled initially using D1-D2 domains, and the D3 domain was added finally by resolving loop clashes (Fig. 5A). The orientation of NG sites 1–5 obtained from the centroid conformation of 5-ns MD trajectory is shown in Fig. 5B. The modeling predicted a large number (810,000) of docking poses for the protein-protein complex, which was done with no inclusion of experimental data. These structures were then analyzed on the basis of the experiments to obtain 326 poses. One pose had eight salt bridges, whereas four poses that had seven salt bridges were identified, and all others had fewer. Four of these five structures were similar, and we chose the one with eight salt bridges to examine in detail. The resulting three-dimensional structure supports interpretations of the experiments that N-glycan sites 1, 4, and 5 interact with OmpA. The transmembrane region of OmpA was then embedded in the POPC membrane with the rest of the complex and solvated in water. Then protein-protein complex was then subjected to 50 ns of MD simulation. The 50-ns MD trajectory on the OmpA-FcγRIa complex led to stable and reasonable interactions for the five important salt bridges, each of which remained stable during 50 ns of MD (Fig. 5B). In addition to many new water-mediated hydrogen bonds showing up in the MD, we found that NG1 and NG5 formed direct noncovalent interactions with OmpA (Fig. 5C). It should be noted here that the waters in the x-ray structure were removed before doing the protein-protein docking to allow the protein side chains full freedom to interact with the other protein and prevent water-mediated effects on side chains that might interfere with the docking. Some of the internal waters from our MD might not find the same positions as in the x-ray. Although NG4 is close to OmpA, the 50-ns MD simulation of OmpA-FcγRIa did not lead to any direct interaction between the glycan of NG4 and OmpA (Fig. 5D). Instead, the predicted NG4 conformation was locked into a hydrogen bond within FcγRIa between the side chain carbonyl group of Asn-159 and the backbone amino group of Ser-161. This hydrogen bond remained stable during 50 ns of MD trajectory. This lack of interaction of NG4 with OmpA was inconsistent with the experimental data because a lack of NG4 also prevented the bacterial invasion into macrophages. However, we speculated that the strong interaction with Asn-159 and Ser-161 might be accidental, caused by our initial configuration. To test this, we rotated the side chain torsional angles of Asn-159 (the χ1 angle from 66 to 145 ° and the χ2 angle from 75 to 30 °) to form direct interactions between NG4 and OmpA. This caused the hydrogen bond between Asn-159/Ser-161 to break, forming instead three new hydrogen bonds (one between carbonyl of Asn-159 side chain and backbone amino group of Ile-160 within FcγRIa and another two between the second GlcNAc moiety in NG4 and Asn-109 on loop 3 of OmpA). This new conformation of NG4 does not clash with any of the protein-protein interfacial peptides. We carried out 50 ns of simulation with this new structure and found it to be stable (Fig. 5E).
FIGURE 5.
Modeling of N-glycosylated FcγRIa and OmpA interaction shows that NG1, NG4, and NG5 bind to loop and barrel sites of OmpA. A, N-glycan precursors (Man-GlcNAc-GlcNAc) are added to the five N-glycosylation sites in D1 and D2 domains of the FcγRIa crystal structure. Glycan conformations are taken from the centroid conformation of 5ns MD trajectory. B, glycans 1, 4 and 5 (shown as silver sticks) of D1 and D2 of FcγRIa (cyan) are involved in binding OmpA (green). Glycan 5 sticks into the β-barrel mouth of OmpA and forms salt bridges in all corners with FcγRIa, stabilizing the protein-protein interface. Positively charged residues are represented as blue spheres, whereas negatively charged residues are shown as red spheres. The IgG-binding loop of FcγRIa (orange) is not involved in binding OmpA. C, glycans 1 forms a hydrogen bond with Asp-149 of OmpA and is in proximity to Asn-146 and Arg-156 of OmpA to form water mediated hydrogen bonds. Glycan 5 is in the barrel binding pocket between loops 3 and 4 of OmpA and forms direct noncovalent interaction with Trp-102, Ser-120, Ala-150, His-151, Thr-155, and Pro-157 of OmpA. Glycan 5 could also form water-mediated interaction with Asp-116, Thr-106, and Lys-107 of OmpA. Glycan 2 is too far away from OmpA and does not have any possibility to form direct or indirect interaction with OmpA. D, NG4 does not form any direct noncovalent interactions with OmpA but is in proximity to OmpA to form indirect interaction like water-mediated hydrogen bonds. E, rotating the torsional angles χ1 and χ2 of Asn-159 in FcγRIa by 79 ° and −45 °, respectively, replaces the hydrogen bond between glycan 4 (silver conformation) and Ser-161 of FcγRIa (colored cyan) by a new hydrogen bond between glycan 4 (salmon conformation) and Ile-160 of FcγRIa, while also forming two more new hydrogen bonds that contribute to the direct interaction between glycan 4 and Asn-109 in loop 3 of OmpA (colored green).
The NG5 binding site in the barrel between loops 3 and 4 of OmpA predicted here coincides with the barrel binding site of GlcNAc1–4GlcNAc (chitobiose) epitopes predicted in our previous work, in which we docked small ligand GlcNAc1-4GlcNAc (chitobiose) to OmpA (9). To test how sensitive this barrel binding site is to the sugar unit when attached to the protein, we added the common N-glycan core sugar sequence Manβ1–4GlcNAcβ1–4GlcNAcβ1 to D1 and D2 domains of FcγRIa. In this predicted binding mode of the three-sugar unit Man1-GlcNAc2 at the NG5 site, the two GlcNAc moieties maintain their interaction with OmpA as in GlcNAcβ1-4GlcNAc binding mode (Fig. 5C). Subsequently, the D3 domain and Manα1–6(Manα1–3)-Manβ1–4GlcNAcβ1–4GlcNAcβ1 were also included in the minimization, which did not change the interaction of GlcNAcβ1–4GlcNAc with the OmpA (data not shown). This suggests that mannoses play a less important role than GlcNAc epitopes in N-glycan binding to OmpA. These data are consistent with the previous experimental observations in which removal of mannose residues from N-glycosylation sites using different mannosidases did not alter the binding of E. coli to endothelial cells (10). Taken together, the simulation studies support the experimental evidence that NG1, NG4, and NG5 interact with loops 1, 3, and 4 of OmpA for binding, and provide a detailed three-dimensional structure that can be used to better understand the nature of the protein-protein complex, which will help in developing molecules that might block formation of this complex and hence prevent neonatal meningitis.
Mutation of Fc-binding Region in FcγRIa Did Not Affect the E. coli Binding to or Entry in Macrophages
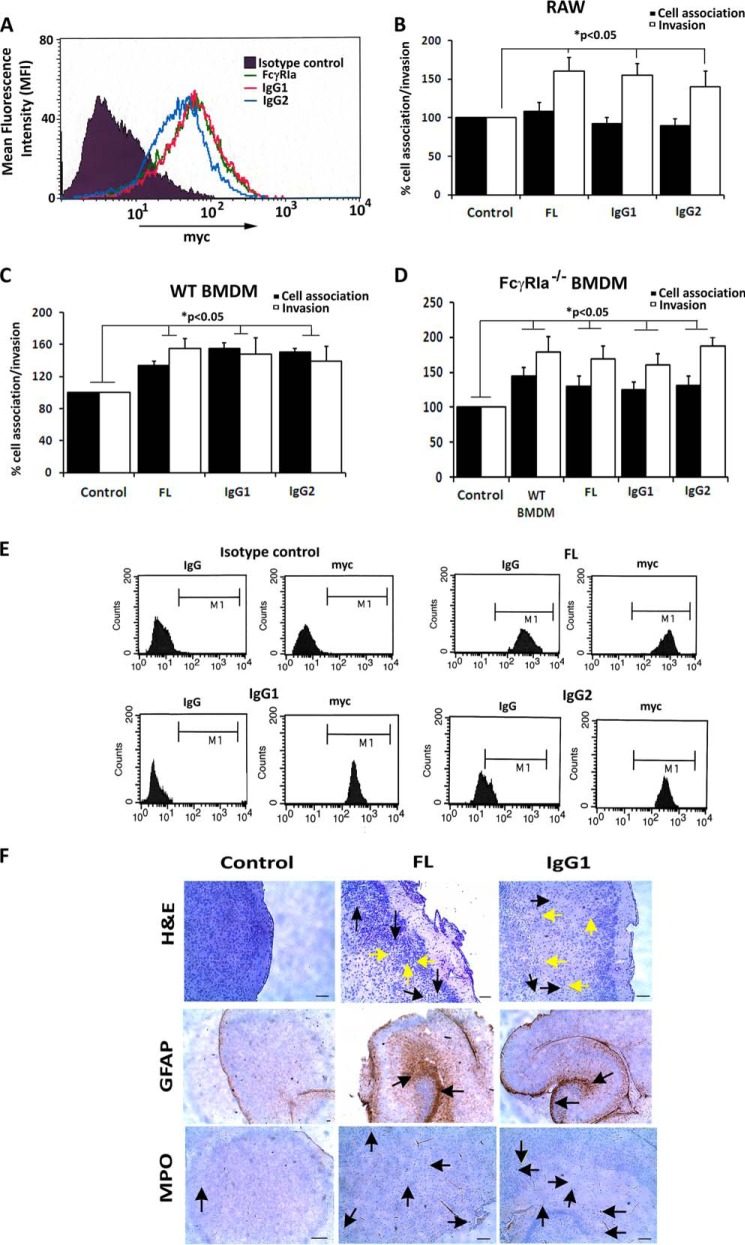
The crystal structure of the N-terminal region of FcγRIa was reported recently (22). We note that the NG sites 1, 2, and 3 are upstream of the Fc-binding region, whereas the other NG sites were downstream of it (Fig. 5A). Our previous experimental studies showed that OmpA+ E. coli, but not OmpA− E. coli, was able to displace FcγRIa-bound IgG in RAW 264.7 macrophages (6). However, it was unclear whether OmpA displaced IgG by competitive binding to the Fc-binding site or by an allosteric effect in which OmpA binds to a different region on FcγRIa. Because the Fc-binding region was strategically placed between active binding sites of E. coli K1 OmpA, we next experimentally ascertained the role of IgG-binding site (sequence MGKHRY) of FcγRIa in OmpA binding. To do this, we performed site-directed mutagenesis at two residues, a lysine to glutamine and an arginine to glutamine. Briefly, MGKHRY was mutated to MGQHRY (IgG1) and MGKHQY (IgG2) in FL-FcγRIa-Myc plasmid. RAW 264.7 macrophages were transfected with mutated IgG1 or IgG2 plasmids, expression efficiency was verified by Myc expression using flow cytometry (Fig. 6A), and the macrophages were subsequently subjected to total cell-associated and invasion assays. We found that E. coli K1 bound and invaded IgG1 and IgG2 transfected RAW 264. 7 macrophages at the same frequency as that of FL, implying that mutation of critical residues in Fc-binding region did not alter bacterial binding and invasion (Fig. 6, B and C). We also observed similar results when FcγRIa−/− BMDMs were transfected with IgG1 and IgG2 plasmids (Fig. 6D). This led us to conclude that OmpA does not bind directly to the Fc region and that OmpA may displace IgG by binding to flanking NG1, 4, and 5 sites.
FIGURE 6.
Adoptive transfer of BMDMs transfected with IgG1 and IgG2 mutants of FcγRIa into FcγRIa−/− newborn mice restores the susceptibility to E. coli meningitis. A, transfection efficiency of FL, IgG1, and IgG2 plasmids in FcγRIa−/− BMDMs was confirmed by flow cytometry using anti-Myc tag antibody. B–D, cell association and invasion assays of E. coli K1 were performed in RAW 264.7 macrophages (B), WT BMDMs (C), or FcγRIa−/− BMDMs (D) transfected with FL, IgG1, or IgG2. The increase in cell association and/or invasion was statistically significant. E, FL, IgG1, or IgG2 transfected RAW 264.7 macrophages were incubated with IgG-FITC for 2 h at 37 °C, incubated with trypan blue to quench extracellular fluorescence and then subjected to flow cytometry to detect FITC, as well as Myc expression using anti-Myc antibody and Alexa 647 secondary antibody. Cells were gated (M1) based on fluorescent output from isotype-matched control. F, brain sections of FcγRIa−/− pups that were adoptively transferred with FcγRIa−/− BMDMs carrying FL, IgG1, or IgG2 plasmids and then infected with E. coli K1 were subjected to H&E, GFAP, and MPO staining as described previously. Yellow arrows represent neutrophil infiltration, and black arrows represent neuronal apoptosis in H&E-stained sections. Arrows in GFAP or MPO stained sections show the accumulation of glial cells or neutrophils.
Adoptive Transfer of IgG1 and IgG2 Transfected BMDMs into FcγRIa−/− Newborn Mice Also Restores the Susceptibility to E. coli Meningitis
To confirm the insignificant role of IgG mutations in the onset of meningitis in the newborn mouse model, we performed adoptive transfer experiments using BMDMs transfected with FL, IgG1, and IgG2 in 3-day-old FcγRIa−/− pups. Prior to adoptively transferring the BMDMs into newborn mice, the transfected cells were subjected to FITC-IgG uptake assay by flow cytometry. As shown in Fig. 6E, both mutations in the IgG region significantly prevented the entry of FITC-IgG compared with FL transfected cells despite the fact that the expression levels of the mutant proteins were similar as determined with anti-Myc antibody staining. After adoptive transfer into 3-day-old mice, these BMDMs (IgG1 and IgG2 transfected cells) restored the susceptibility of newborn pups similar to FL transfected cells to E. coli K1. Bacteremia levels, brain bacterial load, and cerebrospinal fluid positive cultures in IgG1 and IgG2 were of the same magnitude as FL-FcγRIa, reiterating our in vitro and ex vivo observations that Fc-binding region of FcγRIa does not play a role in E. coli K1 binding and invasion of macrophages. Similarly, histological examination of brain sections of the pups transferred with IgG1 and IgG2 showed loss of tissue integrity, neutrophil infiltration, and glial cell migration, similar to FL transferred pups (Fig. 6F; only IgG1 data shown). These results clearly show that the Fc-binding region in FcγRIa is not critical for E. coli K1-induced onset of meningitis in newborn mice.
DISCUSSION
This study demonstrates three important pieces of evidence to confirm that E. coli K1 OmpA interaction with FcγRIa in macrophages is critical for the onset of meningitis in newborn mice: First, we showed clearly that three N-glycans (NG1, NG4, and NG5) present in the extracellular domains of FcγRIa interact with OmpA for binding to and invasion of macrophages and that there is no role for IgG binding region in this interaction. Second, lack of these NG sites in FcγRIa of macrophages that were adoptively transferred into newborn FcγRIa−/− mice renders the animals resistant to E. coli K1-induced meningitis. Third, computer simulation studies of OmpA interacting with N-glycans added to FcγRIa show that OmpA interacts with the NG sites of FcγRIa for binding.
This mechanism of direct binding of OmpA to FcγRIa NG sites is quite contrary to the mechanisms used by several other bacteria, which utilize a “Trojan horse” mechanism to enter the central nervous system (30). S. aureus avoids macrophage recognition by binding to IgG via protein A, whereas other microbes manipulate macrophage activity by secreting effector proteins (1, 4). Thus, OmpA interaction with FcγRIa via the three extracellular N-glycans represents a novel phenomenon in E. coli K1-induced neonatal meningitis.
Although OmpA is conserved throughout evolution, recent studies have identified that the ompA gene exists in two allelic forms, ompA1 and ompA2. Specific subsets of OmpA may be more or less invasive depending on the sequence of loop 2 of OmpA (31). In analyzing the roles of these loops in the pathogenesis of neonatal meningitis, we observed that loops 1 and 2 are critical for the onset of the disease in newborn mouse model. However, loops 1 and 3 are shown to be necessary for the invasion of E. coli K1 into cultured macrophages, whereas loops 1 and 2 are critical to cross the blood-brain barrier (8). Our current results suggest that mutation of NG sites 1, 4, and 5 show a significantly greater reduction in the invasion of E. coli K1 into macrophages compared with other mutations. Taken together, these results suggest that OmpA uses its four flexible loops to interact with N-glycans of FcγRIa in macrophages or gp96 in brain endothelial cells.
Our experimental observations were utilized in the computational protein-protein docking-based protocol to predict an all-atom protein-protein complex structure that explains all the experimental results in terms of the interactions at the atomic level, revealing where the three N-glycan binding sites are in OmpA. The overlap between two of these three N-glycan binding sites with the previously predicted two chitobiose binding sites by a completely different computational method suggests that OmpA may use these specific loop and barrel sites to recognize the carbohydrate moieties in the glycosylated proteins. Our model is that, after OmpA brings the protein target closer through binding the glycans that extend out from the target protein surface, it then forms salt bridges and hydrogen bonds with the target protein to stabilize the protein-protein interface. In addition, our previous results demonstrated that E. coli K1 binding to FcγRIa displaces bound IgG to macrophages, indicating that OmpA may also interact or form salt bridges with IgG binding regions. However, the current study on the mutations in IgG binding sites in FcγRIa show that this region plays no role in the invasion of E. coli K1 in macrophages. This was predicted by the computed structure of the complex prior to the mutation experiments. Additional studies will be needed to examine whether the peptide regions of FcγRIa interacting with OmpA, which are not involved in binding IgG, play any role in altering the signaling mechanisms induced by FcγRIa.
Recent developments in the application of dynamics and simulation have helped unravel temporal progress of bacterial-host interactions. Simulation studies have identified novel mechanisms of “molecular mimicry” by pathogens that modify their virulence factors to resemble host ligands on receptors (32). Moreover, simulation of receptor N-glycosylation has strengthened our understanding of host cell manipulation by parasites such as Trypanosoma brucei (11). However, little information is available on the interaction of bacterial virulence factors with N-glycosylated host receptors. Indeed, molecular simulation studies of an N-glycosylated protein with a protein ligand had not been attempted previously. Thus, our study introduces a novel method of integrating molecular modeling and experimental evidence to predict the protein-protein interface of OmpA with glycosylated FcγRIa. This predicted structure presents an atomistic detail of the interactions that provide a mechanistic understanding of one aspect of the bacterial invasion process in neonatal meningitis. It can also be used to formulate further experimental and computational tests to obtain deeper understanding of the interactions of OmpA with the peptide regions of FcγRIa beyond the glycosylation portions. This would provide the basis for structure-based design of small molecule inhibitors to prevent E. coli K1 interaction with FcγRIa, thereby preventing the bacterial multiplication in macrophages.
Acknowledgment
We thank Rahul Mittal for helping in scanning and transmission electron microscopy using University of Southern California School of Medicine core facilities.
This work was supported, in whole or in part, by National Institutes of Health Grants NS73115 and AI40567 (to N. V. P. and W. A. G.). This work was also supported by Defense University Research Instruments Program and National Science Foundation Caltech Science and Engineering of Materials.
- FcγR
- Fcγ receptor
- NG
- N-glycosylation
- BMDM
- bone marrow-derived macrophage
- H&E
- hematoxylin and eosin
- GFAP
- glial fibrillary acidic protein
- MPO
- myeloperoxidase
- POPC
- 1-palmitoyl-2-oleoylphosphatidylcholine.
REFERENCES
- 1. Rosenberger C. M., Finlay B. B. (2003) Phagocyte sabotage: disruption of macrophage signalling by bacterial pathogens. Nat. Rev. Mol. Cell Biol. 4, 385–396 [DOI] [PubMed] [Google Scholar]
- 2. Taylor P. R., Martinez-Pomares L., Stacey M., Lin H. H., Brown G. D., Gordon S. (2005) Macrophage receptors and immune recognition. Annu. Rev. Immunol. 23, 901–944 [DOI] [PubMed] [Google Scholar]
- 3. Hornef M. W., Wick M. J., Rhen M., Normark S. (2002) Bacterial strategies for overcoming host innate and adaptive immune responses. Nat. Immunol. 3, 1033–1040 [DOI] [PubMed] [Google Scholar]
- 4. Foster T. J. (2005) Immune evasion by staphylococci. Nat. Rev. Microbiol. 3, 948–958 [DOI] [PubMed] [Google Scholar]
- 5. García-García E., Rosales C. (2002) Signal transduction during Fc receptor-mediated phagocytosis. J. Leukoc. Biol. 72, 1092–1108 [PubMed] [Google Scholar]
- 6. Pascal T. A., Abrol R., Mittal R., Wang Y., Prasadarao N. V., Goddard W. A., 3rd (2010) Experimental validation of the predicted binding site of Escherichia coli K1 outer membrane protein A to human brain microvascular endothelial cells: identification of critical mutations that prevent E. coli meningitis. J. Biol. Chem. 285, 37753–37761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shanmuganathan M. V., Krishnan S., Fu X., Prasadarao N. V. (2014) Escherichia coli K1 induces pterin production for enhanced expression of Fcγ receptor I to invade RAW 264.7 macrophages. Microbes Infect. 16, 134–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mittal R., Krishnan S., Gonzalez-Gomez I., Prasadarao N. V. (2011) Deciphering the roles of outer membrane protein A extracellular loops in the pathogenesis of Escherichia coli K1 meningitis. J. Biol. Chem. 286, 2183–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Datta D., Vaidehi N., Floriano W. B., Kim K. S., Prasadarao N. V., Goddard W. A., 3rd (2003) Interaction of E. coli outer-membrane protein A with sugars on the receptors of the brain microvascular endothelial cells. Proteins 50, 213–221 [DOI] [PubMed] [Google Scholar]
- 10. Krishnan S., Prasadarao N. V. (2014) Identification of minimum carbohydrate moiety in N-glycosylation sites of brain endothelial cell glycoprotein 96 for interaction with Escherichia coli K1 outer membrane protein A. Microbes Infect. 16, 540–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mehlert A., Wormald M. R., Ferguson M. A. (2012) Modeling of the N-glycosylated transferrin receptor suggests how transferrin binding can occur within the surface coat of Trypanosoma brucei. PLoS Pathog. 8, e1002618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weiser J. N., Gotschlich E. C. (1991) Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect. Immun. 59, 2252–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pautsch A., Schulz G. E. (1998) Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 5, 1013–1017 [DOI] [PubMed] [Google Scholar]
- 14. Pautsch A., Schulz G. E. (2000) High-resolution structure of the OmpA membrane domain. J. Mol. Biol. 298, 273–282 [DOI] [PubMed] [Google Scholar]
- 15. Klauda J. B., Venable R. M., Freites J. A., O'Connor J. W., Tobias D. J., Mondragon-Ramirez C., Vorobyov I., MacKerell A. D., Jr., Pastor R. W. (2010) Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 114, 7830–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 17. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 18. Humphrey W., Dalke A., Schulten K. (1996) VMD: visual molecular dynamics. J. Mol. Graph. 14, 27–38 [DOI] [PubMed] [Google Scholar]
- 19. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arora A., Abildgaard F., Bushweller J. H., Tamm L. K. (2001) Structure of outer membrane protein A transmembrane domain by NMR spectroscopy. Nat. Struct. Biol. 8, 334–338 [DOI] [PubMed] [Google Scholar]
- 21. Mayo S. L., Olafson B. D., Goddard W. A. (1990) Dreiding: a generic force-field for molecular simulations. J. Phys. Chem-Us 94, 8897–8909 [Google Scholar]
- 22. Lu J., Ellsworth J. L., Hamacher N., Oak S. W., Sun P. D. (2011) Crystal structure of Fcγ receptor I and its implication in high affinity γ-immunoglobulin binding. J. Biol. Chem. 286, 40608–40613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., Shaw D. E. (2010) Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang J., Wolf R. M., Caldwell J. W., Kollman P. A., Case D. A. (2004) Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 [DOI] [PubMed] [Google Scholar]
- 25. Wang J., Wang W., Kollman P. A., Case D. A. (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 25, 247–260 [DOI] [PubMed] [Google Scholar]
- 26. Salomon-Ferrer R., Case D. A., Walker R. C. (2013) An overview of the Amber biomolecular simulation package. Wires Comput. Mol. Sci. 3, 198–210 [Google Scholar]
- 27. Chen R., Li L., Weng Z. P. (2003) ZDOCK: An initial-stage protein-docking algorithm. Proteins 52, 80–87 [DOI] [PubMed] [Google Scholar]
- 28. Pierce B. G., Hourai Y., Weng Z. P. (2011) Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One 6, e24657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lomize M. A., Lomize A. L., Pogozheva I. D., Mosberg H. I. (2006) OPM: orientations of proteins in membranes database. Bioinformatics 22, 623–625 [DOI] [PubMed] [Google Scholar]
- 30. Drevets D. A., Leenen P. J., Greenfield R. A. (2004) Invasion of the central nervous system by intracellular bacteria. Clin. Microbiol. Rev. 17, 323–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith S. G., Mahon V., Lambert M. A., Fagan R. P. (2007) A molecular Swiss army knife: OmpA structure, function and expression. FEMS Microbiol. Lett. 273, 1–11 [DOI] [PubMed] [Google Scholar]
- 32. Drayman N., Glick Y., Ben-nun-shaul O., Zer H., Zlotnick A., Gerber D., Schueler-Furman O., Oppenheim A. (2013) Pathogens use structural mimicry of native host ligands as a mechanism for host receptor engagement. Cell Host Microbe 14, 63–73 [DOI] [PubMed] [Google Scholar]