Background: NORE1A is a Ras effector with a poorly defined function.
Results: NORE1A forms a direct, Ras-regulated complex with β-TrCP and activates the SCFβ-TrCP ubiquitin ligase complex to promote β-catenin degradation.
Conclusion: Ras controls the activity and specificity of SCFβ-TrCP via NORE1A.
Significance: We identify a novel Ras signaling pathway, defective in many tumors, that allows Ras to control specific protein stability.
Keywords: E3 Ubiquitin Ligase, Oncogene, Ras Protein, Tumor Suppressor Gene, Wnt Signaling
Abstract
Ras is the most frequently activated oncogene found in human cancer, but its mechanisms of action remain only partially understood. Ras activates multiple signaling pathways to promote transformation. However, Ras can also exhibit a potent ability to induce growth arrest and death. NORE1A (RASSF5) is a direct Ras effector that acts as a tumor suppressor by promoting apoptosis and cell cycle arrest. Expression of NORE1A is frequently lost in human tumors, and its mechanism of action remains unclear. Here we show that NORE1A forms a direct, Ras-regulated complex with β-TrCP, the substrate recognition component of the SCFβ-TrCP ubiquitin ligase complex. This interaction allows Ras to stimulate the ubiquitin ligase activity of SCFβ-TrCP toward its target β-catenin, resulting in degradation of β-catenin by the 26 S proteasome. However, the action of Ras/NORE1A/β-TrCP is substrate-specific because IκB, another substrate of SCFβ-TrCP, is not sensitive to NORE1A-promoted degradation. We identify a completely new signaling mechanism for Ras that allows for the specific regulation of SCFβ-TrCP targets. We show that the NORE1A levels in a cell may dictate the effects of Ras on the Wnt/β-catenin pathway. Moreover, because NORE1A expression is frequently impaired in tumors, we provide an explanation for the observation that β-TrCP can act as a tumor suppressor or an oncogene in different cell systems.
Introduction
Genetic mutations leading to activated forms of the Ras oncogene can be detected in almost one-third of human cancers (1). When activated, the Ras oncoprotein drives growth and transformation by binding and activating multiple effectors that stimulate multiple signaling pathways. Two of the best characterized cascades are the Raf and PI3K effector pathways (2). However, Ras can also impact other signaling pathways important to proliferation and development, such as the Wnt/β-catenin signaling pathway (3). The interaction of the Ras and the Wnt/β-catenin signaling pathways is quite complex and appears to occur at multiple levels (4–6). An interesting dichotomy exists because Ras has been reported to exhibit both positive and negative effects on the pathway (4–6). Moreover, although synergistic activation of transformation by Ras and Wnt/β-catenin has been reported, other reports have identified an antagonistic role in transformation (5, 6). How Ras impacts the Wnt/β-catenin pathway is still not fully understood, nor is the balance of cellular factors that dictate whether a net positive or negative effect is observed for Ras on Wnt/β-catenin pathway signaling.
In the canonical Wnt/β-catenin pathway, β-catenin is the terminal executor, serving as both a nuclear transcriptional coregulator and key component of adherens junctions (7). In the absence of Wnt ligand signaling, a multiprotein complex consisting of APC, Axin, and GSK-3β phosphorylates β-catenin. This phosphorylation is necessary for the binding of β-TrCP, the substrate recognition component of the SCFβ-TrCP ubiquitin ligase complex. SCFβ-TrCP-mediated ubiquitination of β-catenin results in its rapid degradation by the 26 S proteasome (8). Upon Wnt signaling, the phosphorylation complex is destabilized by Disheveled family proteins, allowing for unphosphorylated β-catenin levels to quickly increase in the cytoplasm and translocate to the nucleus. Nuclear β-catenin functions as a cofactor for transcription factors of the TCF/LEF family, modulating genes involved in growth and survival (9).
Although cells constitutively synthesize β-catenin to maintain a ready response to incoming Wnt signaling, the high turnover rate of β-catenin via the SCFβ-TrCP ubiquitin ligase complex maintains homeostasis (10). Mechanistic corruption in the regulatory mechanism of β-catenin has been described in many human cancers, and mutated forms of β-catenin that cannot be ubiquitinated by the SCFβ-TrCP ubiquitin ligase are oncogenic (9). By enhancing down-regulation of β-catenin, β-TrCP can serve as a tumor suppressor (11). However, the situation is more complex because β-TrCP has also been reported to have oncogenic potential in some situations (12, 13). This may be due to its role in degrading non-β-catenin targets such as Claspin (14).
Paradoxically, activated forms of Ras are not only powerfully transforming but can also act as powerful stimulators of cell death pathways (15, 16). In part, Ras-induced cell death appears to be mediated by members of the RASSF family of tumor suppressors, which bind Ras and serve as death effectors (16). NORE1A (RASSF5) was the first member of the RASSF family to be identified (17). It connects Ras to the pro-apoptotic Hippo pathway, but this does not seem to be essential for its tumor suppressor function. It also modulates p53 (18, 19), which may explain its ability to promote cell cycle arrest. NORE1A is expressed in most normal tissues, but its expression is lost in many tumors because of epigenetic inactivation or deregulated proteolysis (18, 20). NORE1A is a bona fide tumor suppressor because a hereditary genetic defect in NORE1A predisposes the human carriers to kidney cancer (21). However, the biological functions of the Ras/NORE1A interaction remain mostly uncharacterized.
We identified a direct interaction between NORE1A and β-TrCP in a yeast two-hybrid screen. This suggests that either NORE1A is a substrate for SCFβ-TrCP or, more interestingly, might connect Ras to the control of the complex. Because SCFβ-TrCP modulates the Wnt/β-catenin pathway by targeting β-catenin for degradation by the proteasome (11), NORE1A might serve to link Ras to the control of β-catenin.
We sought to determine the role of NORE1A in the control of the SCFβ-TrCP ubiquitin ligase and Wnt/β-catenin signaling. We show that the binding of NORE1A to β-TrCP is Ras-regulated and that it allows Ras to stimulate the activity of SCFβ-TrCP toward β-catenin.
The activation of the SCFβ-TrCP ubiquitin ligase complex by Ras/NORE1A is substrate-specific because NORE1A had no effect on SCFβ-TrCP substrate IκB. Moreover, NORE1A-deficient lung tumor cells exhibit enhanced steady-state levels of β-catenin and resist the growth inhibitory effects of β-TrCP. Therefore, the cellular levels of NORE1A may dictate how Ras modulates β-catenin and determine the substrate profile of the SCFβ-TrCP ubiquitin ligase. β-TrCP has been reported to be an oncogene and a tumor suppressor in different cell systems. The levels of NORE1A may dictate the ultimate activity of β-TrCP in a cell.
EXPERIMENTAL PROCEDURES
Molecular Biology
GFP-β-catenin was generated by PCR amplification of the human β-catenin cDNA (Addgene, catalog no. 16828) (22). The PCR product was TOPO-cloned into pGEM-T-Easy (Promega) and subcloned into pEGFP-C1 (Clonetech) using BamHI and SalI restriction enzymes. β-TrCP1 was obtained from Addgene (catalog no. 4489) and cloned into enhanced GFP in a similar manner. Dominant negative β-TrCP1 was a gift from Tianyan Gao (University of Kentucky). Full-length human NORE1A cDNA was obtained from Origene (Rockville, MD), amplified, and subcloned using BglII/EcoRI into a BamHI/EcoRI restriction digest of pCDNA3 (Invitrogen) containing an in-frame 5′ HA tag (23, 24). We developed a triple point mutant of NORE1A, mutating the 92–94 residues from Arg to Ala using a PCR-based approach. Activated Ras has been described previously (25). NORE1A shRNA constructs (RHS4531-EG83593) were obtained from Open Biosystems (Rockford, IL). pGL3-BAR-Luc was a gift from Randall Moon (University of Washington, WA). The TK-Renilla luciferase control plasmid was obtained from Promega (Madison, WI).
Antibodies
Anti-GFP (Santa Cruz Biotechnology, catalog no. SC-9996), anti-HA (Covance, catalog no. MMS-101P), anti-FLAG (Sigma-Aldrich, St. Louis MO, catalog no. F1804), anti-β-TrCP (Cell Signaling Technology, catalog no. 4394), anti-β-catenin (Cell Signaling Technology, catalog no. 8814), anti-phospho-β-catenin (Cell Signaling Technology, catalog no. 9561), rabbit polyclonal anti-NORE1A (19), and secondary antibodies were from Amersham Biosciences.
Cell Culture and Transfections
Cells were obtained from the ATCC. HEK-293 cells were grown in DMEM supplemented with 10% FBS. HEK-293 NFκB-luciferase cells were provided by Howard Donninger (University of Louisville, KY). NCI-H1299 cells were grown in RPMI medium with 10% FBS. Cells were transfected using jetPRIME® (Polyplus) transfection reagent. In the β-catenin stability studies, the proteasome was inhibited using MG132 (Sigma-Aldrich) at a final concentration of 5 μm and cycloheximide (Sigma-Aldrich) at a final concentration of 10 μm. Cell selections were performed in 500 μg/ml G418 (Sigma). FTI-277 was obtained from Calbiochem (La Jolla, CA).
Immunoprecipitation and Western Blotting
Cellular lysates for immunoprecipitation were prepared using a modified radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.4), 200 mm NaCl, and 1% Nonidet P-40). Precleared lysate was incubated with GFP-Trap® beads (Allele Biotech, San Diego, CA) or with primary antibody or control IgG, followed by protein A/G beads (eBioscience), and washed with lysis buffer. Proteins were run on a 4–15% Tris-glycine gel (Bio-Rad) and transferred to 0.2 μm nitrocellulose (Bio-Rad). Blots were developed using West Pico enhanced ECL (Pierce) or West Femto enhanced ECL (Pierce).
Luciferase Assays
Dual luciferase assays were performed using reagents from the Promega Dual-Luciferase assay kit (catalog no. E1960). Cells were transfected for 24 h before lysing and reading using a Luminat LB 9507 from Berthold Technologies.
RESULTS
NORE1A Forms a Direct, Endogenous, Ras-regulated Complex with β-TrCP
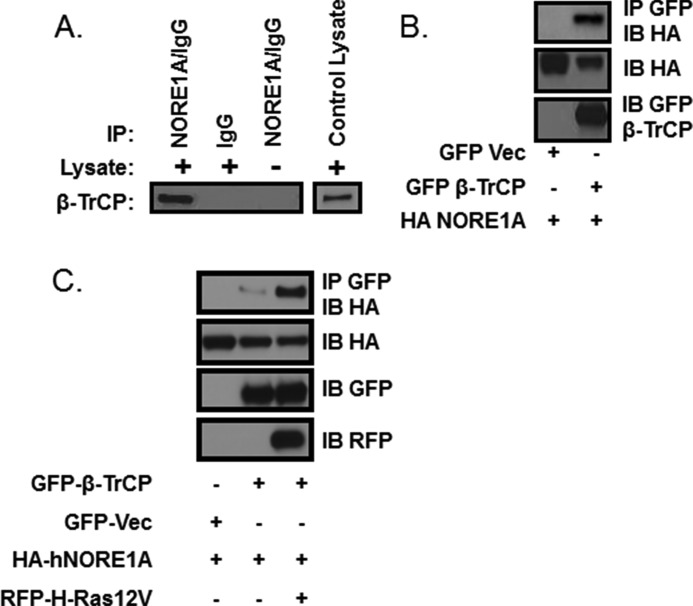
β-TrCP is the substrate recognition component of the SCFβ-TrCP ubiquitin ligase. It is expressed as two closely related isoforms, β-TrCP1 and β-TrCP2. A yeast two-hybrid screen (Myriad Genetics, Salt Lake City, UT) using full-length NORE1A as bait identified β-TrCP1 as a potential direct binding partner. To determine whether this result was physiological, we immunoprecipitated lysates from MCF10A cells for NORE1A and immunoblotted them for the presence of β-TrCP. Fig. 1A) shows that NORE1A and β-TrCP could be detected in an endogenous complex. Further studies were then performed using exogenously expressed proteins. HEK-293 cells were cotransfected with expression constructs for NORE1A and β-TrCP1. The cells were then lysed and immunoprecipitated for β-TrCP1 and immunoblotted for NORE1A. The proteins could be coimmunoprecipitated (Fig. 1B).
FIGURE 1.
A, whole cell lysates were prepared from MCF-10A cells, and the purified protein extracts were immunoprecipitated (IP) with anti-NORE1A for 16 h and then immunoblotted for β-TrCP. IgG incubated with MCF-10A lysate and Ig/NORE1A antibody incubated with lysis buffer served as the negative controls. The blot of NORE1A levels is not shown because of interference from the IgG band. MCF10A lysate served as the positive control. B, HEK-293 cells were transfected with expression constructs for NORE1A and β-TrCP1. Cell lysates were immunoprecipitated with GFP-Trap® beads (Allele Biotech) and immunoblotted (IB) for the presence of β-TrCP1. Vec, vector. C, HEK-293 cells were transfected with expression constructs for NORE1A and β-TrCP1 in the presence or absence of a construct expressing a red fluorescent tagged activated H-Ras (12V). After 24 h, the cells were lysed and immunoprecipitated with GFP-Trap® beads before immunoblotting. A shorter exposure of the immunoprecipitation shows that the presence of activated H-Ras enhances NORE1A binding to β-TrCP.
NORE1A is a Ras effector protein. Consequently we examined the role of Ras in the interaction between NORE1A and β-TrCP1 in similar experiments. The presence of activated H-Ras in the cotransfection enhanced the association between NORE1A and β-TrCP1 proteins (Fig. 1C). These blots were underexposed in comparison with the blots in Fig. 1B.
NORE1A Promotes β-Catenin Degradation via β-TrCP
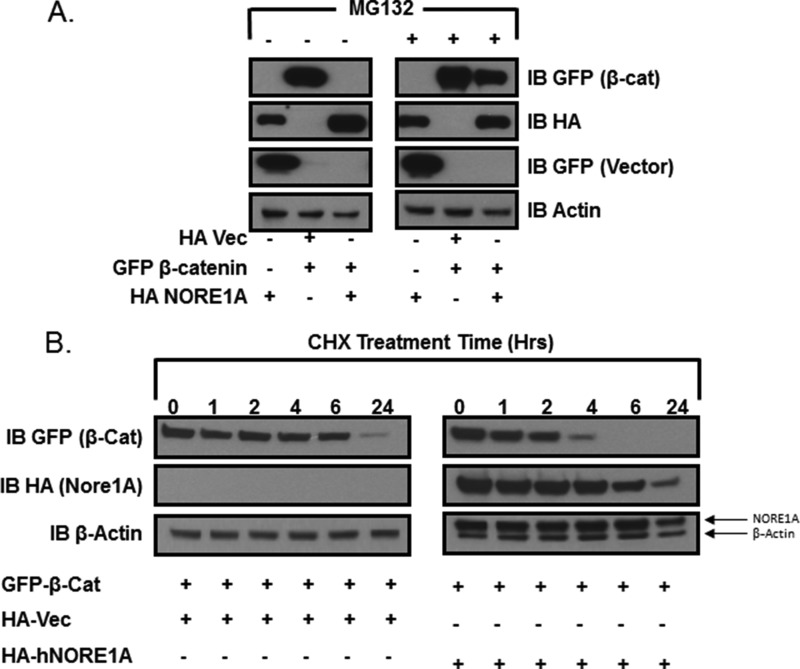
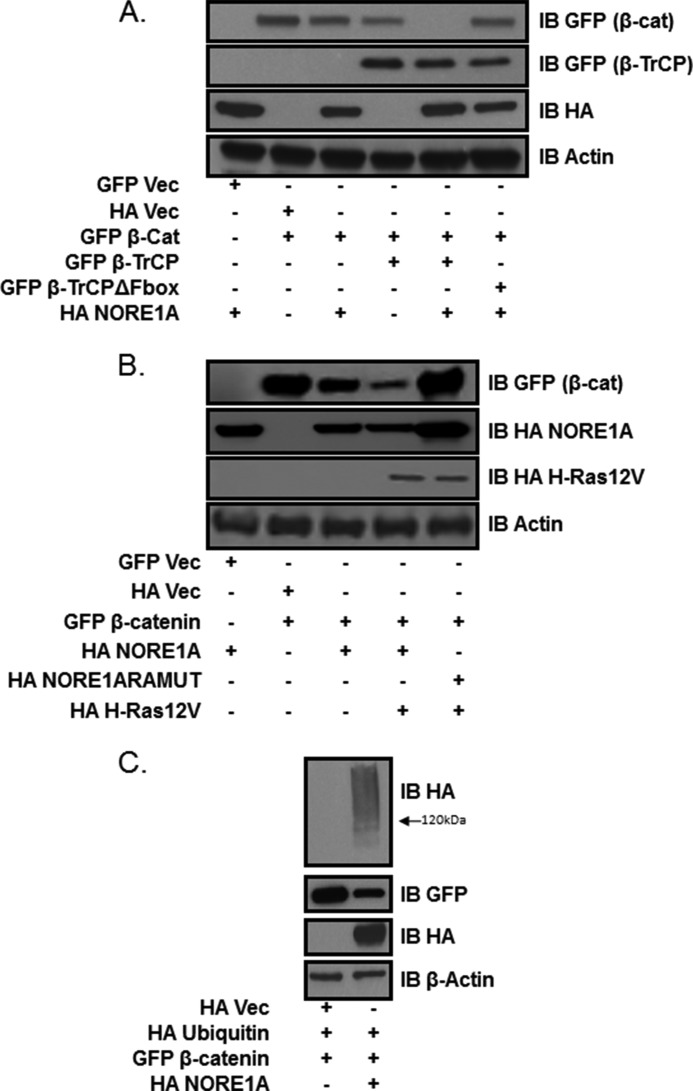
Because β-TrCP is the substrate recognition component of the major ubiquitin ligase complex regulating protein levels of β-catenin (8), we examined the effects of NORE1A expression on β-catenin protein stability using transient transfections performed in HEK-293 cells. Coexpression of NORE1A suppressed the expression of β-catenin to subdetectable levels. This effect was largely blocked by the 26 S proteasome inhibitor MG132 (Fig. 2A). To further support the MG132 experiment, the transient transfections of NORE1A and β-catenin were repeated in HEK-293 cells. 24 h post-transfection, the cells were treated with cycloheximide. Dishes were lysed over a time course of 24 h, and samples were examined by Western blot analysis. The results confirmed that NORE1A was acting at a protein stability level (Fig. 2B). To determine whether NORE1A and β-TrCP1 synergize to promote the degradation of β-catenin, we partially inhibited the proteasome by adding low levels of MG132 for 6 h. Under these conditions, the ability of both NORE1A and β-TrCP1 to suppress β-catenin expression was impaired rather than abolished. This allowed us to detect a synergistic reduction in β-catenin levels when NORE1A and β-TrCP1 were transfected together (Fig. 3A). Furthermore, a dominant negative form of β-TrCP1 (26) blocked NORE1A-mediated degradation of β-catenin (Fig. 3A).
FIGURE 2.
A, HEK-293 cells were transfected with expression constructs for β-catenin and NORE1A. 24 h post-transfection, the cells were trypsinized, split into two groups, and allowed to rest for an additional 24 h. One group was then treated with the proteasome inhibitor MG132 for 5 h, and the other group was treated with dimethyl sulfoxide for 5 h. The cells were then lysed and immunoblotted (IB) for levels of β-catenin (β-cat). Vec, vector. B, HEK-293 cells were transfected with vector or NORE1A and, after 24 h, split into six dishes each. Cycloheximide (CHX) was added after a further 24 h, and dishes were lysed over a time course. Levels of β-catenin protein were measured by Western blot analysis. Shown is a representative blot of two experiments.
FIGURE 3.
A, HEK-293 cells were transfected with expression constructs for NORE1A, β-catenin (β-cat), β-TrCP1, and a dominant negative β-TrCP (ΔFBOX) that is unable to ubiquitinate target substrates. Low levels of MG132 were added to partially inhibit the 26 S proteasome and reveal differences between weak and strong activity. IB, immunoblot; Vec, vector. B, HEK-293 cells were transfected with expression constructs for NORE1A, a NORE1A mutant unable to bind to Ras (RAMUT), β-catenin, and activated H-Ras. Relative levels of β-catenin were assayed by immunoblot analysis. Actin served as a loading control. C, HEK-293 cells were cotransfected with expression constructs for NORE1A, β-catenin, and a HA-tagged ubiquitin. 24 h post-transfection, the cells were lysed and analyzed by Western blotting for HA incorporation into GFP- β-catenin (∼120 kDa).
To examine the effects of Ras on the system, we included activated H-Ras in the transfections, again with attenuation of the proteasome with low levels of MG132. Activated Ras enhances NORE1A activity against β-catenin (Fig. 3B). As further support for the role of Ras in directly modifying the action of NORE1A, we included a NORE1A point mutant that is defective for Ras binding in the assay (NORE1ARAMUT) (27). This experiment showed that the effects of Ras were due to its interaction with NORE1A (Fig. 3B).
NORE1A appears to regulate β-catenin protein levels through the proteasome by interacting with the ubiquitin ligase, β-TrCP. If this is indeed the mechanism of action, it would require β-catenin to be polyubiquitinated by β-TrCP, which would mark it for proteasomal degradation. To confirm this, HEK-293 cells were transfected with expression constructs for NORE1A and GFP-β-catenin along with a HA-tagged ubiquitin and treated with MG132. Incorporation of ubiquitin into GFP-β-catenin could be detected (Fig. 3C).
NORE1A Suppresses β-Catenin Signaling
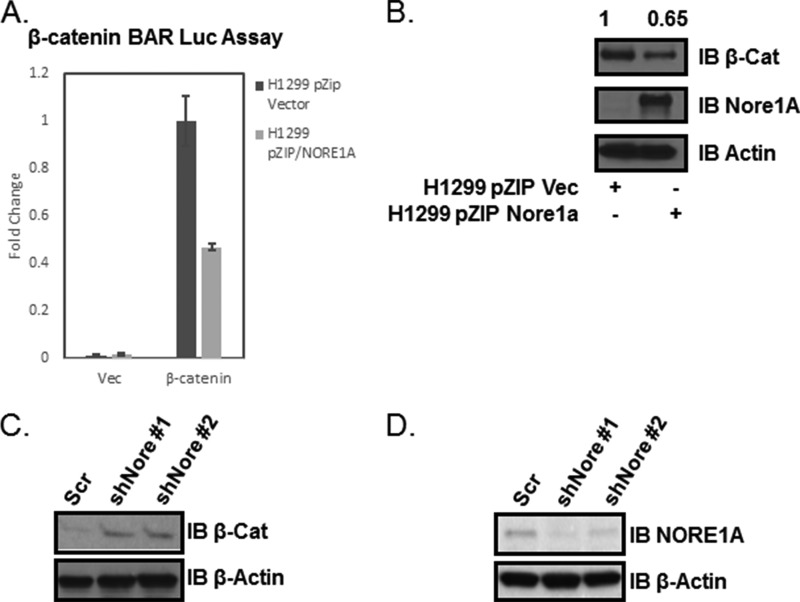
Active β-catenin translocates to the nucleus, where it binds the transcriptional repressors TCF/LEF, thereby activating specific gene transcription. An artificial promoter sequence linked to the luciferase gene has been developed as a transcriptional reporter for β-catenin activity (28). This reporter, containing a β-catenin activation region (BAR-Luc), was cotransfected with an expression construct for β-catenin and a Renilla luciferase internal control into a matched pair of NCI-H1299 cells stably transfected with vector or NORE1A expression vector. The NORE1A-expressing cells showed reduced β-catenin activity (Fig. 4A).
FIGURE 4.
A, NCI-H1299 cells stably transfected with vector (Vec) or NORE1A were transfected with an expression construct for β-catenin along with a β-catenin-responsive luciferase reporter (BAR-Luc) and a TK-Renilla internal control. Dual-Luciferase assays were performed, and results were standardized with the Renilla readout (M1/M2). n = 2 independent experiments. Error bars show mean ± S.E.; p = 0.02. B, immunoblot (IB) analysis confirming expression of HA-NORE1A. NCI-H1299 ± for NORE1A expression show differences in steady-state expression of β-catenin (β-cat). Whole cell lysates were prepared, and endogenous levels of β-catenin were analyzed by immunoblotting. Quantification was performed by densitometry using QuantityOne software by Bio-Rad and is shown in arbitrary units. C, HEK-293 cells were transfected with shRNA constructs against NORE1A. Endogenous levels of β-catenin were analyzed by Western blotting. Scr, scrambled. D, stable NORE1A shRNA constructs were validated against endogenous NORE1A in HBEC-3KT cells.
Loss of NORE1A expression enhances steady-state levels of β-Catenin
The above results suggest that the loss of NORE1A that is so frequently found in human tumors should uncouple Ras from the negative regulation of β-catenin. Therefore, the levels of β-catenin in a Ras-driven tumor cell should inversely correlate with NORE1A expression. To test this hypothesis, we examined the levels of endogenous β-catenin in the NCI-H1299 NORE1A ± matched pair. These cells carry an activated Ras mutation and undetectable levels of endogenous NORE1A (29). Fig. 4B shows that the NORE1A-negative cells exhibited higher levels of endogenous β-catenin than the cells stably transfected with NORE1A. As further confirmation, we transfected HEK-293 cells with shRNA constructs against NORE1A and assayed for the levels of endogenous β-catenin protein. β-catenin was elevated in the shRNA-transfected cells but not in the scrambled shRNA-transfected cells (Fig. 4C).
NORE1A Differentially Regulates β-TrCP Targets
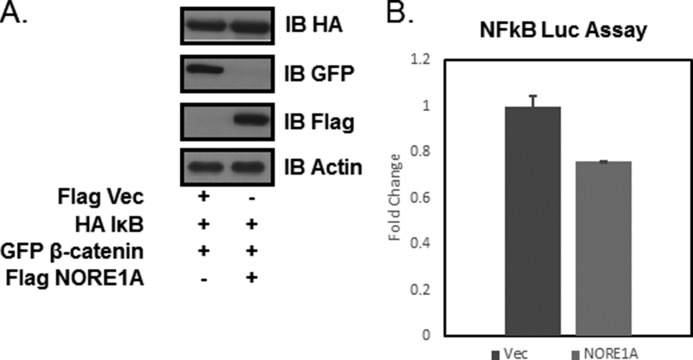
To determine whether the effect of NORE1A on SCFβ-TrCP is a general activation or a specific activation toward a particular subset of targets, a second degradation target of β-TrCP1, IκB (30), was chosen for analysis. HEK-293 cells were transfected with expression constructs for IκB, β-catenin, and NORE1A. Western blot analysis of the protein levels showed that, although IκB levels remained the same in the presence of NORE1A, NORE1A suppressed β-catenin, (Fig. 5A). IκB is a negative regulator of NFκB (30). If NORE1A promotes its degradation, we should expect NFκB signaling to increase. Luciferase assays using HEK-293 cells stably transfected with an NFκB luciferase reporter showed that NORE1A does not promote the activation of NFκB (Fig. 5B).
FIGURE 5.
A, HEK-293 cells were transfected with expression constructs for NORE1A, β-catenin, and IκB. After 24 h, the cells were lysed and assayed for differential protein expression by Western blot analysis. Shown is a representative blot of at least two experiments. IB, immunoblot; Vec, vector. B, HEK-293 cells stably expressing an NFκB luciferase (Luc) reporter were transfected with HA-empty vector and HA-NORE1A. A slight suppression of activity was observed because of NORE1A, but quantification showed that this was not significant (p = 0.12). n = 2 independent experiments. Error bars show mean ± S.E.
NORE1A Levels Dictate the Biological Activity of β-TrCP
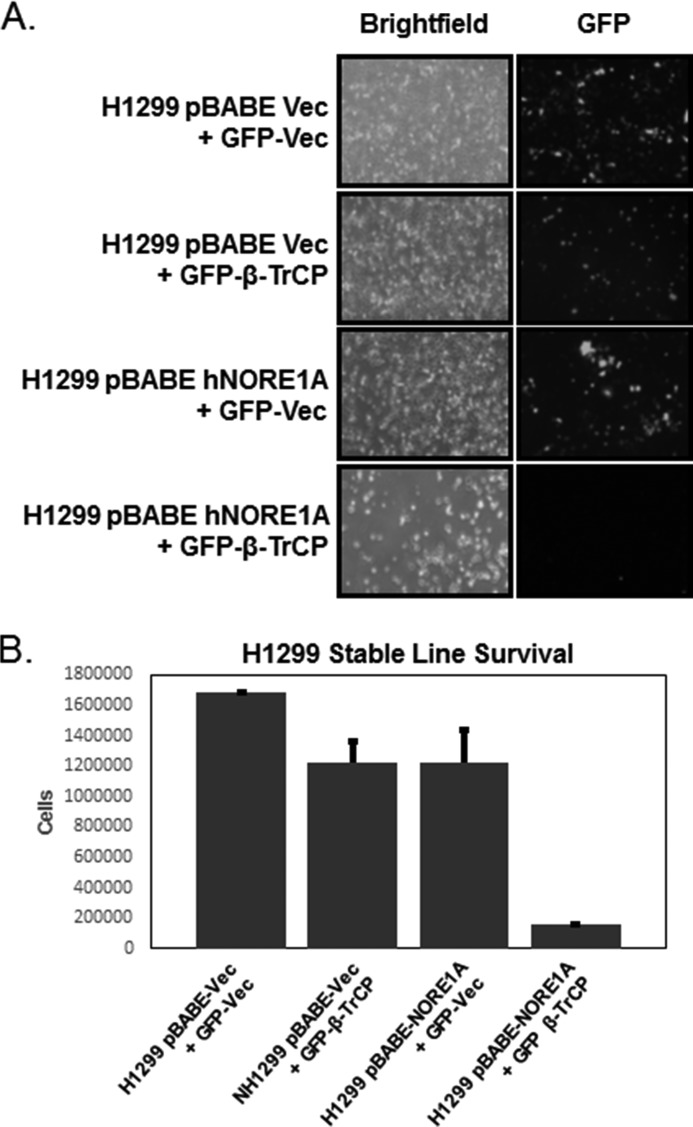
β-TrCP has a range of targets and can act as an oncogene or a tumor suppressor, depending upon the particular cellular milieu (11–14). The cofactors determining whether the net effect on growth of β-TrCP is positive or negative are not known. To determine whether NORE1A may be one of these factors, we transfected the NORE1A ± NCI-H1299 cell system with β-TrCP1 and selected for the marker carried by the β-TrCP1 construct. Fig. 6A shows that β-TrCP1 was highly growth-inhibitory in the NORE1A-positive cells but only weakly inhibitory in the NORE1A-negative cells. The quantification is shown in Fig. 6B.
FIGURE 6.
A, NCI-H1299 cells stably expressing NORE1A or an empty vector were transfected with an expression construct for β-TrCP1 and selected with G418. Shown is a representative experiment. Vec, vector. B, quantification of live cells after 2 weeks. Data are the average of two separate assays performed in duplicate. Error bars represent mean ± S.E. (p = 0.03).
NORE1A Is a Key Node in the Regulation of β-Catenin via Ras
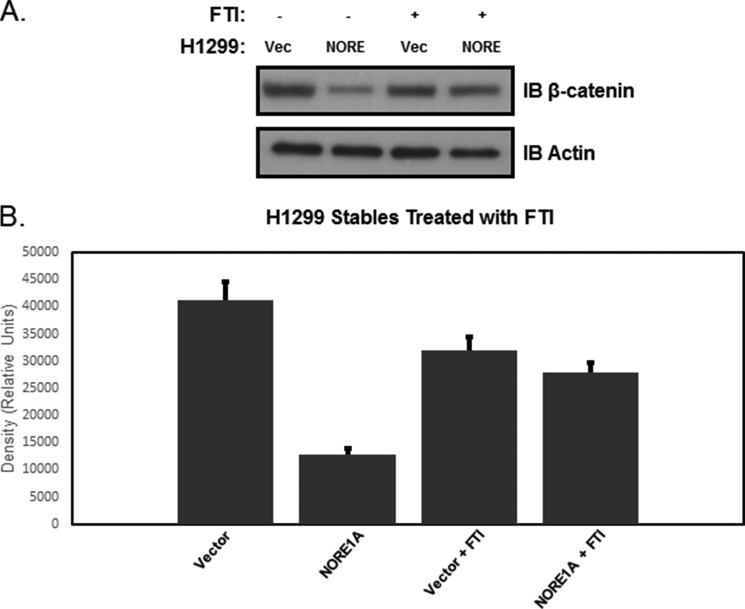
Ras has been reported to exhibit both negative and positive effects on β-catenin protein levels (31, 32). The factors that determine the net effect of Ras on β-catenin are not clear. To determine whether NORE1A dictates the effects of Ras on β-catenin, we treated the NCI-NCI-H1299 NORE1A ± cell system with a farnesyl transferase inhibitor (FTI)2 (Fig. 7A) that inactivates Ras (33). NCI-H1299 cells contain an activated form of Ras, which allowed us to examine the effects of endogenous Ras activation on endogenous β-catenin expression in a NORE1A-positive or -negative background. A “normal” cell is positive for NORE1A expression and does not have an activated Ras signaling pathway. The levels of β-catenin under these circumstances are shown in the fourth column of Fig. 7B. In the presence of a competent activated Ras (no FTI) in these NORE1A-positive cells, the levels of β-catenin go down (Fig. 7B, second column). However, when we remove the FTI from NORE1A-negative cells, we observed that the levels of β-catenin increased (Fig. 7B, first column) relative to the levels in the normal cells (Fig. 7B, fourth column). Therefore, the presence or absence of NORE1A may dictate the effects of Ras, positive or negative, on β-catenin protein levels.
FIGURE 7.
A, NCI-H1299 cells stably transfected with NORE1A or empty vector (Vec) were treated with a farnesyl transferase inhibitor for 24 h to modulate Ras. Whole cell lysates were harvested and probed for total levels of endogenous β-catenin. A representative blot is shown. B, quantification of two experiments. Vector, Ras+/NORE1A−; NORE1A, Ras+/NORE1A+; Vector + FTI, Ras−/NORE1A−; NORE1A + FTI, Ras−/NORE1A+. A normal cell would be expected to be negative for activated Ras and positive for NORE1A. Therefore, the fourth column is the reference column. Quantification using densitometry was calculated on QuantityOne software from Bio-Rad. Values from two separate assays were combined for quantification. The graph shows these data as mean ± S.E. (p = 0.02).
DISCUSSION
NORE1A binds activated Ras and serves as a proapoptotic Ras effector (18, 19). Furthermore, NORE1A suppresses tumor cell growth and is frequently inactivated in human tumors, and its inactivation is implicated in a hereditary cancer syndrome (16, 21, 23). Therefore, it is likely to serve as an important human tumor suppressor. NORE1A binds to the MST kinases and may connect Ras to the proapoptotic Hippo signaling pathway (18). However, deletion mutants of NORE1A that cannot interact with MST kinases retain the ability to inhibit cell growth and suppress the tumorigenic phenotype (34). Therefore, unknown tumor suppressor pathways independent of the classic Hippo pathway must also be modulated by NORE1A. β-TrCP is the substrate recognition component for the SCFβ-TrCP ubiquitin ligase complex and exists as two closely related isoforms, β-TrCP1 and β-TrCP2. Under the influence of Wnt signaling, it binds and regulates the degradation of the proto-oncogene β-catenin (11). β-catenin has a dual role in the cell, serving both as a component of adherens junctions in the cytoplasm and as a transcriptional coregulator in the nucleus, modulating the function of the TCF/LEF transcription factors (35). The Wnt/β-catenin pathway plays key roles in development and tumorigenesis, and mutations in the system leading to the excessive accumulation of β-catenin have been identified at high frequency in human tumors (9, 35, 36).
Aberrant activation of the Ras pathway is one of the most common defects observed in human cancers (37). Over a decade of study has revealed multiple, subtle links between the Ras and Wnt/β-catenin pathways. The links are complex and remain poorly understood because they invoke both synergistic and antagonistic relationships (6). Indeed, Ras has been reported to suppress β-catenin levels but also to promote β-catenin transcription (31, 32). The net effect appears to be context-dependent. However, in vivo studies have confirmed that defects in the two pathways can synergize to promote cancer (3).
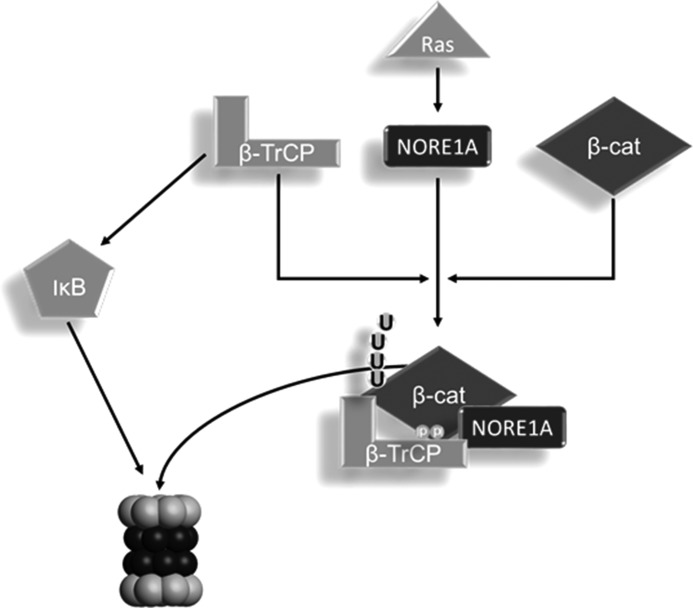
Here we show that Ras promotes the binding of NORE1A to β-TrCP1, the substrate recognition component of the SCFβ-TrCP ubiquitin ligase, and that this interaction promotes the degradation of β-catenin (Fig. 8). Because we identified NORE1A and β-TrCP1 in a yeast two hybrid interaction screen, this interaction is likely to be direct. The interaction of NORE1A with β-TrCP1 explains how Ras can negatively regulate β-catenin and how such regulation may be defective in tumor cells that have suffered inactivation of NORE1A. By using FTIs to inhibit the endogenous activated Ras in our NCI-H1299 cell system, we were able to show that Ras down-regulates endogenous β-catenin protein levels in the presence of NORE1A but enhances them in its absence. Therefore, the levels of NORE1A in a cell may play a major role in determining the net effect of Ras on β-catenin. However, it is not impossible that these experiments also impacted non-Ras farnesylated targets. Therefore, further experiments to examine the biological effects of the Ras/NORE1A/β-TrCP1 interaction may be warranted.
FIGURE 8.
Diagram of the new Ras/NORE1A/Wnt signaling pathway. We interpret our data to suggest a novel Ras/NORE1A signaling pathway that activates β-TrCP toward a subset of its substrates, such as β-catenin, but not IκB. This results in differential degradation of SCFβ-TrCP substrates by the proteasome in the presence of Ras/NORE1A signaling.
The mechanism by which NORE1A specifically stimulates SCFβ-TrCP via β-TrCP1 remains unclear. We hypothesize that NORE1A may be acting as a scaffold for other regulatory components such as kinases.
β-TrCP1 can act as a tumor suppressor (11), as expected, because it can serve as a negative regulator of β-catenin, but it has also been reported to exhibit oncogenic functions in some cellular environments (12, 13). This may be due to its effects on substrates in addition to β-catenin. These include a range of proteins involved in cell cycle regulation and transcriptional control, such as IκB (38, 39), NFκB (40), GLI2 (41, 42), REST (43, 44), ATF4 (45), CDC25A (46, 47), CDC25B (47), and Claspin (48, 49). It seems likely that mechanisms exist to target β-TrCP to particular substrates under particular conditions. NORE1A may be part of such a mechanism because it activates β-TrCP1 toward β-catenin but not toward IκB. Because we found that the ability of β-TrCP1 to suppress growth of tumor cells is heavily dependent upon the presence of NORE1A, it may be the NORE1A status of a cell that dictates whether or not β-TrCP1 is oncogenic or tumor-suppressive. It will be interesting to determine against which other substrates NORE1A activates β-TrCP1.
NORE1A is not the first member of the RASSF family to be identified as binding to β-TrCP1. A splice variant of the RASSF1A protein, RASSF1C, can also bind (50). However, unlike NORE1A, RASSF1C acts to somehow stabilize β-catenin protein levels and enhance β-catenin signaling. Perhaps it acts by competing with NORE1A for binding.
Another pathway with which RASSF family members have been associated is the Hippo pathway. This pathway is a major signaling mechanism involved in controlling organ size and apoptosis (51). The canonical Hippo signaling pathway involves a kinase cascade where MST kinases phosphorylate LATS kinases, which then act on the transcriptional coregulators YAP and TAZ (52). RASSF proteins such as NORE1A can bind and activate the MST/Hippo kinases (18) and, therefore, may regulate the activity the Hippo pathway executors YAP and TAZ by phosphorylation. However, in addition to β-catenin, β-TrCP1 also binds and regulates the stability of YAP and TAZ (53). Moreover, β-catenin has been shown to bind to YAP and TAZ in a transcription complex (54). Therefore, the interaction of NORE1A with β-TrCP1 may allow NORE1A to invoke powerful non-canonical Hippo pathway regulation, which may also be controlled by Ras. This may prove to be a potent additional mechanism where the RASSF family proteins contribute to growth and survival control.
We identify a function for NORE1A acting as a Ras-regulated scaffold for β-TrCP1 that activates the ubiquitin ligase complex toward specific targets. These results may explain, mechanistically, many of the apparent contradictions in the relationship between the Ras and Wnt/β-catenin pathways.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA133171-01A2 (to G. J. C.).
- FTI
- farnesyl transferase inhibitor.
REFERENCES
- 1. Malumbres M., Barbacid M. (2003) RAS oncogenes: the first 30 years. Nat. Rev. Cancer 3, 459–465 [DOI] [PubMed] [Google Scholar]
- 2. Campbell S. L., Khosravi-Far R., Rossman K. L., Clark G. J., Der C. J. (1998) Increasing complexity of Ras signaling. Oncogene 17, 1395–1413 [DOI] [PubMed] [Google Scholar]
- 3. Pacheco-Pinedo E. C., Morrisey E. E. (2011) Wnt and Kras signaling: dark siblings in lung cancer. Oncotarget 2, 569–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jang J. W., Boxer R. B., Chodosh L. A. (2006) Isoform-specific ras activation and oncogene dependence during MYC- and Wnt-induced mammary tumorigenesis. Mol. Cell. Biol. 26, 8109–8121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jeong W. J., Yoon J., Park J. C., Lee S. H., Lee S. H., Kaduwal S., Kim H., Yoon J. B., Choi K. Y. (2012) Ras stabilization through aberrant activation of Wnt/β-catenin signaling promotes intestinal tumorigenesis. Sci. Signal. 5, ra30. [DOI] [PubMed] [Google Scholar]
- 6. Kim S. E., Yoon J. Y., Jeong W. J., Jeon S. H., Park Y., Yoon J. B., Park Y. N., Kim H., Choi K. Y. (2009) H-Ras is degraded by Wnt/β-catenin signaling via β-TrCP-mediated polyubiquitylation. J. Cell Sci. 122, 842–848 [DOI] [PubMed] [Google Scholar]
- 7. Saifo M. S., Rempinski D. R., Jr., Rustum Y. M., Azrak R. G. (2010) Targeting the oncogenic protein β-catenin to enhance chemotherapy outcome against solid human cancers. Mol. Cancer 9, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Orford K., Crockett C., Jensen J. P., Weissman A. M., Byers S. W. (1997) Serine phosphorylation-regulated ubiquitination and degradation of β-catenin. J. Biol. Chem. 272, 24735–24738 [DOI] [PubMed] [Google Scholar]
- 9. Giles R. H., van Es J. H., Clevers H. (2003) Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta 1653, 1–24 [DOI] [PubMed] [Google Scholar]
- 10. Xu M. X., Zhao L., Deng C., Yang L., Wang Y., Guo T., Li L., Lin J., Zhang L. (2013) Curcumin suppresses proliferation and induces apoptosis of human hepatocellular carcinoma cells via the Wnt signaling pathway. Int. J. Oncol. [DOI] [PubMed] [Google Scholar]
- 11. Liu C., Kato Y., Zhang Z., Do V. M., Yankner B. A., He X. (1999) β-Trcp couples β-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc. Natl. Acad. Sci. U.S.A. 96, 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Karin M., Cao Y., Greten F. R., Li Z. W. (2002) NF-κB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2, 301–310 [DOI] [PubMed] [Google Scholar]
- 13. Richmond A. (2002) NF-κB, chemokine gene transcription and tumour growth. Nat. Rev. Immunol. 2, 664–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mamely I., van Vugt M. A., Smits V. A., Semple J. I., Lemmens B., Perrakis A., Medema R. H., Freire R. (2006) Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 16, 1950–1955 [DOI] [PubMed] [Google Scholar]
- 15. Cox A. D., Der C. J. (2003) The dark side of Ras: regulation of apoptosis. Oncogene 22, 8999–9006 [DOI] [PubMed] [Google Scholar]
- 16. Donninger H., Vos M. D., Clark G. J. (2007) The RASSF1A tumor suppressor. J. Cell Sci. 120, 3163–3172 [DOI] [PubMed] [Google Scholar]
- 17. Schmidt L., C. G. (2011) RASSF5 (Ras association (RalGDS/AF-6) domain family member 5). Atlas Genet. Cytogenet. Oncol. Haematol. [Google Scholar]
- 18. Khokhlatchev A., Rabizadeh S., Xavier R., Nedwidek M., Chen T., Zhang X. F., Seed B., Avruch J. (2002) Identification of a novel Ras-regulated proapoptotic pathway. Curr. Biol. 12, 253–265 [DOI] [PubMed] [Google Scholar]
- 19. Vos M. D., Martinez A., Ellis C. A., Vallecorsa T., Clark G. J. (2003) The pro-apoptotic Ras effector Nore1 may serve as a Ras-regulated tumor suppressor in the lung. J. Biol. Chem. 278, 21938–21943 [DOI] [PubMed] [Google Scholar]
- 20. van der Weyden L., Adams D. J. (2007) The Ras-association domain family (RASSF) members and their role in human tumourigenesis. Biochim. Biophys. Acta 1776, 58–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen J., Lui W. O., Vos M. D., Clark G. J., Takahashi M., Schoumans J., Khoo S. K., Petillo D., Lavery T., Sugimura J., Astuti D., Zhang C., Kagawa S., Maher E. R., Larsson C., Alberts A. S., Kanayama H. O., Teh B. T. (2003) The t(1;3) breakpoint-spanning genes LSAMP and NORE1 are involved in clear cell renal cell carcinomas. Cancer Cell 4, 405–413 [DOI] [PubMed] [Google Scholar]
- 22. Kolligs F. T., Hu G., Dang C. V., Fearon E. R. (1999) Neoplastic transformation of RK3E by mutant β-catenin requires deregulation of Tcf/Lef transcription but not activation of c-myc expression. Mol. Cell. Biol. 19, 5696–5706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clark G. J., Cox A. D., Graham S. M., Der C. J. (1995) Biological assays for Ras transformation. Methods Enzymol. 255, 395–412 [DOI] [PubMed] [Google Scholar]
- 24. Fiordalisi J. J., Johnson R. L., 2nd, Ulkü A. S., Der C. J., Cox A. D. (2001) Mammalian expression vectors for Ras family proteins: generation and use of expression constructs to analyze Ras family function. Methods Enzymol. 332, 3–36 [DOI] [PubMed] [Google Scholar]
- 25. Vos M. D., Ellis C. A., Elam C., Ulku A. S., Taylor B. J., Clark G. J. (2003) RASSF2 is a novel K-Ras-specific effector and potential tumor suppressor. J. Biol. Chem. 278, 28045–28051 [DOI] [PubMed] [Google Scholar]
- 26. Schulman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386 [DOI] [PubMed] [Google Scholar]
- 27. Ortiz-Vega S., Khokhlatchev A., Nedwidek M., Zhang X. F., Dammann R., Pfeifer G. P., Avruch J. (2002) The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene 21, 1381–1390 [DOI] [PubMed] [Google Scholar]
- 28. Biechele T. L., Adams A. M., Moon R. T. (2009) Transcription-based reporters of Wnt/β-catenin signaling. Cold Spring Harbor protocols 2009, pdb prot5223 [DOI] [PubMed] [Google Scholar]
- 29. Shinmura K., Tao H., Nagura K., Goto M., Matsuura S., Mochizuki T., Suzuki K., Tanahashi M., Niwa H., Ogawa H., Sugimura H. (2011) Suppression of hydroxyurea-induced centrosome amplification by NORE1A and down-regulation of NORE1A mRNA expression in non-small cell lung carcinoma. Lung Cancer 71, 19–27 [DOI] [PubMed] [Google Scholar]
- 30. Baeuerle P. A. (1998) IκB-NF-κB structures: at the interface of inflammation control. Cell 95, 729–731 [DOI] [PubMed] [Google Scholar]
- 31. Espada J., Pérez-Moreno M., Braga V. M., Rodriguez-Viciana P., Cano A. (1999) H-Ras activation promotes cytoplasmic accumulation and phosphoinositide 3-OH kinase association of β-catenin in epidermal keratinocytes. J. Cell Biol. 146, 967–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim D., Rath O., Kolch W., Cho K. H. (2007) A hidden oncogenic positive feedback loop caused by crosstalk between Wnt and ERK pathways. Oncogene 26, 4571–4579 [DOI] [PubMed] [Google Scholar]
- 33. Reuter C. W., Morgan M. A., Bergmann L. (2000) Targeting the Ras signaling pathway: a rational, mechanism-based treatment for hematologic malignancies? Blood 96, 1655–1669 [PubMed] [Google Scholar]
- 34. Aoyama Y., Avruch J., Zhang X. F. (2004) Nore1 inhibits tumor cell growth independent of Ras or the MST1/2 kinases. Oncogene 23, 3426–3433 [DOI] [PubMed] [Google Scholar]
- 35. Morin P. J. (1999) β-Catenin signaling and cancer. BioEssays 21, 1021–1030 [DOI] [PubMed] [Google Scholar]
- 36. Horst D., Chen J., Morikawa T., Ogino S., Kirchner T., Shivdasani R. A. (2012) Differential WNT activity in colorectal cancer confers limited tumorigenic potential and is regulated by MAPK signaling. Cancer Res. 72, 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bos J. L. (1989) Ras oncogenes in human cancer: a review. Cancer Res. 49, 4682–4689 [PubMed] [Google Scholar]
- 38. Yaron A., Hatzubai A., Davis M., Lavon I., Amit S., Manning A. M., Andersen J. S., Mann M., Mercurio F., Ben-Neriah Y. (1998) Identification of the receptor component of the IκBα-ubiquitin ligase. Nature 396, 590–594 [DOI] [PubMed] [Google Scholar]
- 39. Hatakeyama S., Kitagawa M., Nakayama K., Shirane M., Matsumoto M., Hattori K., Higashi H., Nakano H., Okumura K., Onoé K., Good R. A., Nakayama K. (1999) Ubiquitin-dependent degradation of IκBα is mediated by a ubiquitin ligase Skp1/Cul 1/F-box protein FWD1. Proc. Natl. Acad. Sci. U.S.A. 96, 3859–3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Orian A., Gonen H., Bercovich B., Fajerman I., Eytan E., Israël A., Mercurio F., Iwai K., Schwartz A. L., Ciechanover A. (2000) SCF(β)(-TrCP) ubiquitin ligase-mediated processing of NF-κB p105 requires phosphorylation of its C-terminus by IκB kinase. EMBO J. 19, 2580–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huntzicker E. G., Estay I. S., Zhen H., Lokteva L. A., Jackson P. K., Oro A. E. (2006) Dual degradation signals control Gli protein stability and tumor formation. Genes Dev. 20, 276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pan Y., Wang B. (2007) A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J. Biol. Chem. 282, 10846–10852 [DOI] [PubMed] [Google Scholar]
- 43. Guardavaccaro D., Frescas D., Dorrello N. V., Peschiaroli A., Multani A. S., Cardozo T., Lasorella A., Iavarone A., Chang S., Hernando E., Pagano M. (2008) Control of chromosome stability by the β-TrCP-REST-Mad2 axis. Nature 452, 365–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Westbrook T. F., Hu G., Ang X. L., Mulligan P., Pavlova N. N., Liang A., Leng Y., Maehr R., Shi Y., Harper J. W., Elledge S. J. (2008) SCFβ-TRCP controls oncogenic transformation and neural differentiation through REST degradation. Nature 452, 370–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lassot I., Ségéral E., Berlioz-Torrent C., Durand H., Groussin L., Hai T., Benarous R., Margottin-Goguet F. (2001) ATF4 degradation relies on a phosphorylation-dependent interaction with the SCF(βTrCP) ubiquitin ligase. Mol. Cell. Biol. 21, 2192–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Busino L., Donzelli M., Chiesa M., Guardavaccaro D., Ganoth D., Dorrello N. V., Hershko A., Pagano M., Draetta G. F. (2003) Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature 426, 87–91 [DOI] [PubMed] [Google Scholar]
- 47. Kanemori Y., Uto K., Sagata N. (2005) β-TrCP recognizes a previously undescribed nonphosphorylated destruction motif in Cdc25A and Cdc25B phosphatases. Proc. Natl. Acad. Sci. U.S.A. 102, 6279–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peschiaroli A., Dorrello N. V., Guardavaccaro D., Venere M., Halazonetis T., Sherman N. E., Pagano M. (2006) SCFβTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 23, 319–329 [DOI] [PubMed] [Google Scholar]
- 49. Mailand N., Bekker-Jensen S., Bartek J., Lukas J. (2006) Destruction of Claspin by SCFβTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 23, 307–318 [DOI] [PubMed] [Google Scholar]
- 50. Estrabaud E., Lassot I., Blot G., Le Rouzic E., Tanchou V., Quemeneur E., Daviet L., Margottin-Goguet F., Benarous R. (2007) RASSF1C, an isoform of the tumor suppressor RASSF1A, promotes the accumulation of β-catenin by interacting with βTrCP. Cancer Res. 67, 1054–1061 [DOI] [PubMed] [Google Scholar]
- 51. Zhao B., Lei Q. Y., Guan K. L. (2008) The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 20, 638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhao B., Li L., Lei Q., Guan K. L. (2010) The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 24, 862–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu C. Y., Zha Z. Y., Zhou X., Zhang H., Huang W., Zhao D., Li T., Chan S. W., Lim C. J., Hong W., Zhao S., Xiong Y., Lei Q. Y., Guan K. L. (2010) The hippo tumor pathway promotes TAZ degradation by phosphorylating a phosphodegron and recruiting the SCFβ-TrCP E3 ligase. J. Biol. Chem. 285, 37159–37169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Konsavage W. M., Jr., Yochum G. S. (2013) Intersection of Hippo/YAP and Wnt/β-catenin signaling pathways. Acta Biochim. Biophys. Sinica 45, 71–79 [DOI] [PubMed] [Google Scholar]