Background: Amyloid-β precursor protein is implicated in neural stem cell development.
Results: Neurogenin 2 expression and neuronal differentiation correlated with amyloid-β precursor protein expression in neural stem/progenitor cells.
Conclusion: Amyloid-β precursor protein regulates neuronal differentiation by altering neurogenin 2 expression.
Significance: The study provides a new mechanism to explain the effects of amyloid-β precursor protein on neural stem cell differentiation.
Keywords: Amyloid Precursor Protein (APP), Basic Helix-loop-helix Transcription Factor (bHLH), Neurogenesis, RNA Interference (RNAi), Transgenic Mice
Abstract
Amyloid-β precursor protein (APP) is well studied for its role in Alzheimer disease, although its normal function remains uncertain. It has been reported that APP stimulates the proliferation and neuronal differentiation of neural stem/progenitor cells (NSPCs). In this study we examined the role of APP in NSPC differentiation. To identify proteins that may mediate the effect of APP on NSPC differentiation, we used a gene array approach to find genes whose expression correlated with APP-induced neurogenesis. We found that the expression of neurogenin 2 (Ngn2), a basic helix-loop-helix transcription factor, was significantly down-regulated in NSPCs from APP knock-out mice (APPKO) and increased in APP transgenic (Tg2576) mice. Ngn2 overexpression in APPKO NSPCs promoted neuronal differentiation, whereas siRNA knockdown of Ngn2 expression in wild-type NSPCs decreased neuronal differentiation. The results demonstrate that APP-stimulated neuronal differentiation of NSPCs is mediated by Ngn2.
Introduction
Amyloid-β precursor protein (APP)2 is an integral type I transmembrane protein that is the precursor of amyloid-β protein (Aβ) of Alzheimer disease. Although APP has been well studied for its role in Alzheimer disease, little is known about the normal function of APP. A number of physiological functions have been attributed to APP including synapse formation (1) and stabilization (2, 3), neurite outgrowth promotion (4–8), stimulation of neuronal migration (9), intracellular signaling (10, 11), promotion of cell growth (12–14), neural repair (15–17), and neurogenesis (18–23). However, the mechanism by which APP may regulate these functions is still unknown.
During development, neural stem or progenitor cells (NSPCs) located in germinal zones give rise to both glial and neuronal lineages. There is evidence that APP is able to stimulate the proliferation of NSPCs. For example, J20 mice, which overexpress human APP with the Swedish and Indiana familial AD mutations, have a 2-fold increase in the number of proliferating stem cells in the dentate gyrus and subventricular zone at an age of 3 months (22, 23). Other studies have reported that soluble APPα and soluble APPβ promote the proliferation of NSPCs (24, 25). Furthermore, inhibition of α-secretase was reported to decrease NSPC proliferation, and soluble APPα was able to rescue this effect (24). However, our recent study showed that an APP-induced increase in NSPC proliferation was primarily due to the secretion of cystatin C rather than soluble APPα (19).
APP may also play a role in promoting the differentiation of NSPCs. APP expression increases as NSPCs mature into neurons (26), and soluble APP has been reported to promote neural differentiation (27, 28). We previously found that NSPCs derived from Tg2576 mice, which overexpress APP, possessed a greater potential to differentiate into neurons, whereas cells derived from APP-knock-out (APPKO) mice exhibited decreased neuronal differentiation (19).
In the present study we examined the role of APP in NSPC differentiation further. We demonstrate that the neuronal differentiation of NSPCs, induced by APP, is mediated by neurogenin 2 (Ngn2).
EXPERIMENTAL PROCEDURES
Animals
APPKO mice and their corresponding C57Bl/6 wild-type (WT) control mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Human APP-overexpressing, APPSW Tg2576 mice, and the corresponding background strain controls, C57Bl/6_SJL, were purchased from Taconic Farms (Hudson, NY). Mice were housed in the animal facility at the University of Tasmania. All experiments were approved by the University of Tasmania Animal Ethics Committee.
Materials and Antibodies
Dulbecco's modified Eagle's medium (DMEM), B27 supplement, and poly-l-lysine were from Invitrogen. Penicillin, streptomycin, and human recombinant EGF were all obtained from Sigma. Human recombinant bovine FGF was from PeproTech (Rocky Hill, NJ). Poly-l-lysine was from Sigma. Human recombinant sAPPα was from Sigma, Aβ 1–40 and Aβ 1–42 were from Keck Foundation Biotechnology (New Haven, CT). Anti-rabbit neurogenin2 was from Sapphire Bioscience (Waterloo, Australia), anti-mouse β-actin was from Sigma, anti-mouse βIII-tubulin mAb was from Promega (Alexandria, Australia), anti-mouse 6E10 mAb was from Covance (Sydney, Australia), and anti-rabbit MAP2 mAb was from Sigma. Anti-mouse and anti-rabbit HRP-conjugated secondary antibodies were from Dako Australia Pty. Ltd. (Campbellfield, Australia). Secondary antibodies were goat anti-mouse IgG conjugated to Alexa Fluor-488 and 568 (Invitrogen). 4,6-Diamidino-2-phenylindole (DAPI) was from Sigma. Plasmid pCAG-Ngn2-IRES-GFP for the expression of Ngn2 was a kind gift of François Guillemot (MRC-National Institute for Medical Research, London, UK), and the plasmids pCAG-IRES-GFP control vector and pCAG-IRES-APP695 were from Addgene (Cambridge, MA). The plasmids contained an IRES or IRES-enhanced green fluorescence GFP expression cassette under the control of a CMV enhancer and a chicken β-actin promoter (29). Fluorescently (Cy-3)-labeled Ngn2 siRNA and negative control siRNA oligonucleotides were obtained from Applied Biosystems.
Neurosphere and Isolated NSPC Culture
Primary neurosphere cultures derived from cerebral cortices of postnatal day 0 mice were prepared according to previously described procedures (19). Neurospheres were prepared by growing cells in suspension in 75-cm2 cell culture flasks at a density of 400,000 cells in proliferation medium (DMEM supplemented with 2% (v/v) B27, 100 units/ml penicillin, 100 units/ml streptomycin, 20 ng/ml human bovine FGF, and 20 ng/ml human EGF). After 7 days in culture, neurospheres were dissociated mechanically by trituration, and cells were counted in a hemocytometer and then either reseeded as suspension cultures or replated as adherent cultures. All cultures were incubated in a humidified incubator at 37 °C in an atmosphere containing 5% CO2.
Differentiation of NSPCs
Neurospheres were mechanically dissociated, and then isolated cells were plated at a density of 105 cells/well in 24-well plate. The cells were grown in a differentiation medium (DMEM supplemented with 2% (v/v) B27, 100 units/ml penicillin, 100 units/ml streptomycin, and 1% (v/v) fetal calf serum) for 5 days at 37 °C in an atmosphere containing 5% CO2. The cells were then fixed with 4% (w/v) paraformaldehyde in phosphate-buffered saline (PBS) (8 g/liter NaCl, 0.2 g/liter KCl, 1.44 g/liter Na2HPO4, and 0.24 g/liter KH2PO4, pH 7.2) for 15 min, permeabilized with 0.03% (v/v) Triton X-100 in PBS for 5 min, and incubated in 10% (v/v) sheep serum in PBS for 1 h to block nonspecific binding sites. Fixed cells were stained with a mouse anti-βIII tubulin antibody (1:1,000 diluted in 10% (v/v) sheep serum in PBS) and then incubated with a goat anti-mouse IgG conjugated to Alexa Fluor 488 or -568 (1:1,000 diluted in 10% (v/v) sheep serum in PBS) and DAPI at 1:10,000 dilution. βIII-tubulin+ and DAPI+ cells were counted under the 20× objective using a Zeiss Palm microbeam IV (Carl Zeiss, Sydney, Australia) fluorescence microscope.
Alamar Blue Assay
The number of viable cells was estimated using an Alamar Blue assay. Dissociated cells cultured adherently on poly-l-lysine-precoated 96-well plates were incubated for up to 5 days, and then 20 μl of Alamar Blue reagent (Invitrogen) was added into each well, and the cells were incubated for a further 4 h. The fluorescence intensity was determined using a FLUOstar Optima microplate fluorescence plate reader at an excitation wavelength of 540 nm and an emission wavelength of 590 nm. Cell number was expressed as the relative fluorescence intensity.
PCR Array
A mouse neurogenesis and neural stem cell PCR array in 384-well plate format (PAMM-404ZG-4; Qiagen, Chadstone, Australia) was used to assay gene expression changes. RNA was extracted from neurospheres cultures derived from n = 3 independent mouse cohorts using a RNeasy mini kit (Qiagen). Each preparation of neurospheres contained ∼106 cells in proliferation medium. cDNA was reverse-synthesized from 400 ng of RNA with a RT2 first strand kit (Qiagen). cDNA samples were added to the reaction plates, and the real-time amplification data (Ct values) were determined using a Roche Diagnostics LightCycler 480. Analysis of gene expression from real-time results was carried out using the RT2 profiler PCR array data analysis v3.5 provided by Qiagen. Expression of the β-actin gene was used as a reference housekeeping gene.
Immunoblotting
The level of Ngn2 in NSPC cultures and in the brain cortex of the WT and APPKO mice was determined by western blotting. Cells were washed with PBS and then lysed as described previously (19). Cortices, 4 per group, derived from WT and APPKO mice were homogenized in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, 0.1% SDS, and protease inhibitor mixture from Roche Diagnostics). Proteins were then separated on 12% sodium dodecyl sulfate-polyacrylamide gels before being transferred electrophoretically onto polyvinylidene difluoride membranes (Merck). The membranes were blocked for 2 h with 2% (w/v) skim milk powder in 50 mm Tris-buffered saline, pH 8, containing 0.05% (v/v) Tween 20 (TBS-Tween) and incubated overnight at 4 °C with either anti-Ngn2 (1:1000 dilution) or anti-β-actin (1:10,000 dilution). Protein expression was detected using HRP-conjugated secondary antibodies (1:10,000 dilution). Chemiluminescence reactions were monitored using a CHEMI-SMART 5000, and images were collected using Chemi-Capt 50001. For quantification of immunoreactivity, images of blots were analyzed using ImageJ Version 1.46r (National Institutes of Health, Bethesda, MD).
Cell Transfection
NSPCs were electroporated with the appropriate plasmid using the Amaxa mouse neural stem cell nucleofector kit (VPG-1004, Lonza Ltd., Germany). Briefly, 2.5 × 106 dissociated cells and 2 μg of plasmid were resuspended in 100 μl of nucleofector solution (Amaxa), then the cell/DNA suspension was transferred into a certified cuvette and electroporated with nucleofector device program A-033. Proliferation medium (500 μl) was added to the cuvette, and the suspension was gently transferred onto poly-l-lysine pre-coated coverslips in a 12-well plate containing 300 μl of proliferation medium prewarmed to 37 °C. After 24 h the medium was changed to differentiation medium, and the cells were incubated for 5 days at 37 °C in an atmosphere containing 5% CO2. For siRNA transfections, 300,000 cells per well were plated onto poly-l-lysine-precoated coverslips maintained in proliferation medium for 48 h. After this time, the medium was changed to differentiation medium and the siRNA:Effectene (Qiagen) complex was added in a ratio of 20 nmol to 4 μl per well, and the cells were incubated for 5 days at 37 °C in an atmosphere containing 5% CO2. Next, the cells were fixed in 4% (w/v) paraformaldehyde in PBS. Fixed cells were stained with a mouse anti-βIII tubulin, anti-mouse 6E10, or anti-rabbit MAP2 antibody (all used at 1:1000 diluted in 10% (v/v) sheep serum in PBS) and then incubated with a goat anti-mouse IgG conjugated to Alexa Fluor 488 or 568 or with a goat anti-rabbit IgG conjugated to Alexa Fluor 568 (1:1000 diluted in 10% (v/v) sheep serum in PBS) and DAPI at 1:10,000 dilution. βIII-tubulin+, MAP2+, and DAPI+ cells were counted under the 20× objective using a Zeiss Palm microbeam IV (Carl Zeiss, Sydney, Australia) fluorescence microscope. Images of fluorescently labeled oligonucleotides were collected using an UltraView confocal microscope with Volocity Software (PerkinElmer Life Sciences).
Statistical Analysis
Statistical analysis was performed with GraphPad Prism software, Version 5.04. Data were tested by Student's t test, χ2, or one-way analysis of variance. Post-hoc comparisons were analyzed using Tukey's test. Differences were considered statistically significant when the probability, p, of the null hypothesis was ≤0.05. Data are presented as the means ± S.E. All results were obtained in at least three independent experiments.
RESULTS
Cystatin C Is Not Involved in Neuronal Differentiation
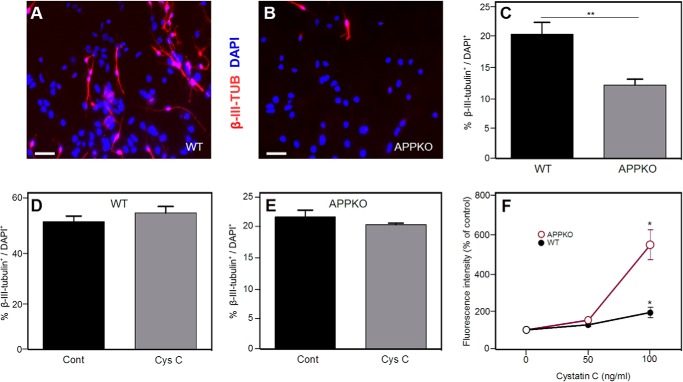
Previous studies from our group (19) showed that NSPCs derived from APPKO mice proliferated less rapidly and differentiated into neurons less readily than NSPCs from WT mice. The effect on proliferation was found to be mediated by the secretion of cystatin C. Therefore, to examine the possibility that cystatin C might also influence NSPC differentiation, we first compared the neural differentiation levels of NSPCs derived from P0 WT and APPKO mice. After 5 days in differentiation medium, the number of β-III-tubulin+ neurons was determined and expressed as a percentage of the total number of cells measured with DAPI. Similar to our previous study (19), we found that a smaller proportion of APPKO NSPCs were positive for β-III-tubulin relative to WT cells (Fig. 1, A–C). This result confirmed our previous finding that there is a decrease in neuronal differentiation in APPKO NSPC cultures compared with WT cultures.
FIGURE 1.
Neuronal differentiation of NSPCs from WT and APPKO mice. Panels A and B show representative immunofluorescence images demonstrating that there are more β-III-tubulin+ (TUB) cells in WT cultures (A) than in APPKO cultures (B) when NSPCs are incubated for 5 days in differentiation conditions. β-III-Tubulin staining (red) and DAPI staining (blue) are shown. C, quantification of results from immunofluorescence analysis. The panel shows % β-III-tubulin+/DAPI+ of ∼300 cells per group in three independent experiments. Bars show the means ± S.E. D and E, percentage of β-III-tubulin+ cells (β-III-tubulin+/DAPI+ × 100%) after cystatin C treatment (100 ng/ml) in WT cultures (D) and APPKO cultures (E). F, effect of cystatin C on NSPC proliferation. Dissociated neurosphere-derived cells were incubated for 5 days in proliferation medium. The number of viable cells was calculated from the fluorescence intensity in an Alamar Blue assay. Values are the means ± S.E. (n = 4) and shown as the percentage of the control value (no added cystatin C) (* = p < 0.05 as assessed by analysis of variance with post hoc Tukey's test; ** = p < 0.01 as assessed by χ2 test). Scale bars = 30 μm. Cont, control; Cys C, cystatin C.
To examine the possibility that cystatin C is a mediator of APP-induced neuronal differentiation, cultures of WT and APPKO NSPCs were treated with cystatin C (100 ng/ml), and the cells were incubated for 5 days in differentiation medium. After this time, the percentage of β-III-tubulin+ cells present in the cultures was determined. We did not find a significant difference between the number of β-III-tubulin+ cells in untreated WT and APPKO cultures when compared with those treated with cystatin C (Fig. 1, D and E). To ensure that the cystatin C was active, we conducted a parallel study in which we exposed proliferating NSPCs to the same concentration of cystatin C that we used in our differentiation assay. Using a fluorescence (Alamar Blue) assay to measure the number of viable cells (Fig. 1F), we observed an increase in the number of cells when the cells were incubated with 100 ng/ml cystatin C. This confirmed that although cystatin C could stimulate NSPC proliferation, the effect of APP on NSPC differentiation was not due to cystatin C.
Expression of Ngn2 Is Decreased in APPKO NSPCs
To identify a factor(s) that mediates APP-induced neuronal differentiation, the expression of neurogenesis-related genes in NSPCs derived from APPKO mice and from Tg2576 mice (which overexpress human APP) was analyzed using a mouse neurogenesis and neural stem cell-specific PCR array (RT2 Profiler PCR). RNA was extracted from NSPCs of APPKO and Tg2576 mice and from the corresponding WT control mice.
The expression levels of 84 genes that regulate neurogenesis including genes related to apoptosis, cell cycle, growth, and transcription were compared. We found that 18 of the 84 genes had expression levels that were either up- or down-regulated more than 2-fold in APPKO cells when compared with the corresponding WT cells (Table 1, Fig. 2). As expected, the measured App gene expression was at least 1800-fold lower in NSPCs of APPKO mice. Expression of the genes Bmp8, Mdk, Nf1, Nrp1, Odz1, Shh, and Th was significantly higher in the APPKO cells, whereas expression of Fgf2, Gdnf, Hey1, Hey2, Neurog2, Pax3, Pax5, and Pou3f3 was significantly lower. Ngn2 expression was 42-fold down-regulated in the APPKO relative to WT cells, which was a considerably lower level than for the other genes with the exception of App. Furthermore, Ngn2 expression was 2.5-fold up-regulated in the APP overexpressing Tg2576 cells relative to WT cells (Table 2, Fig. 2).
TABLE 1.
Analysis of gene expression in NSPCs from APPKO using a mouse neurogenesis and neural stem cell PCR array
Mouse PCR neurogenesis array results show the expression of genes in APPKO NSPCs that are significantly up-regulated or down-regulated relative to NSPCs from the corresponding background strain WT mice. RNA was extracted from NSPC cultures derived from three independent mouse cohorts. For the data analysis, the ΔΔCt method was used (64). For a -fold change of >1, the result is reported as a -fold up-regulation. For a -fold change <1, the negative inverse of the result is shown as a -fold down-regulation. Values of -fold up- or down-regulation >2 or <−2 were assumed to be significant.
| Gene name | Expression level (relative to β-actin) |
-Fold up- or down- regulation in KO | |
|---|---|---|---|
| APPKO | WT | ||
| Amyloid-β (A4) precursor protein | 1.2 × 10−6 | 2.2 × 10−3 | −1803.60 |
| Bone morphogenetic protein 8b | 1.8 × 10−6 | 6.8 × 10−7 | 2.60 |
| Fibroblast growth factor 2 | 2.2 × 10−5 | 5.1 × 10−5 | −2.35 |
| Glial cell line-derived neurotrophic factor | 3.5 × 10−6 | 8.9 × 10−6 | −2.57 |
| Hairy/enhancer-of-split related with YRPW motif1 | 3.0 × 10−4 | 6.7 × 10−4 | −2.26 |
| Hairy/enhancer-of-split related with YRPW motif2 | 5.2 × 10−6 | 1.2 × 10−5 | −2.38 |
| Midkine | 1.2 × 10−4 | 3.4 × 10−5 | 3.37 |
| Neurogenin 2 | 3.5 × 10−7 | 1.4 × 10−5 | −41.69 |
| Neurofibromatosis 1 | 9.3 × 10−4 | 2.3 × 10−4 | 4.02 |
| Noggin | 1.2 × 10−5 | 3.3 × 10−5 | −2.69 |
| Neuropilin 1 | 2.3 × 10−4 | 8.9 × 10−5 | 2.57 |
| Odd Oz/ten-m homolog 1 | 2.3 × 10−6 | 6.5 × 10−7 | 3.50 |
| Paired box gene 3 | 3.0 × 10−6 | 1.6 × 10−5 | −5.33 |
| Paired box gene 5 | 2.6 × 10−3 | 7.4 × 10−3 | −2.83 |
| POU domain, class 3, transcription factor 3 | 2.2 × 10−3 | 1.9 × 10−2 | −8.79 |
| Sonic hedgehog | 3.9 × 10−5 | 1.8 × 10−5 | 2.14 |
| Tyrosine hydroxylase | 2.0 × 10−6 | 5.9 × 10−7 | 3.44 |
FIGURE 2.
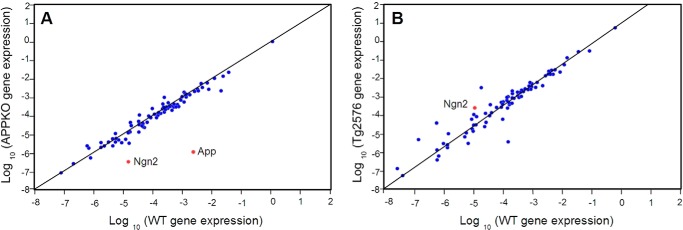
Two-dimensional plot of gene expression levels in APPKO NSPCs (A) or Tg2576 NSPCs (B) versus WT NSPCs. Gene expression was calculated relative to the expression in the corresponding background strain (Tables 1 and 2) and is shown on a log10 scale. Eighty-four genes were analyzed. Gene expression relative to β-actin (reference gene) is shown. Analysis of gene expression from n = 3 independent experiments was carried out using the RT2 profiler PCR array data analysis v3.5 provided by Qiagen.
TABLE 2.
Analysis of gene expression in NSPCs from Tg2576 using a mouse neurogenesis and neural stem cell PCR array
Mouse PCR neurogenesis array results showing the expression of genes in Tg2576 NSPCs that are significantly up-regulated or down-regulated relative to NSPCs from the corresponding background strain WT mice. RNA was extracted from NSPC cultures derived from three independent mouse cohorts. Analysis and presentation of the data is the same as Table 1.
| Gene name | Expression level (relative to β-actin) |
-Fold up- or down- regulation in Tg | |
|---|---|---|---|
| Tg2576 | WT | ||
| Bone morphogenetic protein 4 | 3.1 × 10−3 | 6.4 × 10−3 | −2.05 |
| Bone morphogenetic protein 8 | 1.9 × 10−6 | 4.0 × 10−6 | −2.07 |
| CDK5 regulatory subunit associated protein 2 | 4.6 × 10−4 | 1.7 × 10−4 | 2.76 |
| Chemokine (C-X-C motif) ligand1 | 1.0 × 10−5 | 3.4 × 10−5 | −3.40 |
| Dopamine receptor D2 | 1.5 × 10−5 | 1.1 × 10−6 | 13.24 |
| Filamin, α | 1.9 × 10−3 | 7.7 × 10−4 | 2.41 |
| Glial cell line-derived neurotrophic factor | 1.8 × 10−4 | 5.1 × 10−5 | 3.46 |
| Hairy and enhancer of split1 | 1.1 × 10−4 | 3.9 × 10−4 | −3.42 |
| Hairy/enhancer-of-split related with YRPW motif1 | 2.6 × 10−5 | 8.7 × 10−6 | 3.00 |
| Myocyte enhancer factor 2C | 2.6 × 10−3 | 8.8 × 10−5 | 29.24 |
| Neurogenic differentiation 1 | 5.5 × 10−5 | 1.1 × 10−4 | −2.05 |
| Neurogenin 1 | 8.4 × 10−7 | 2.5 × 10−7 | 3.33 |
| Neurogenin 2 | 1.1 × 10−4 | 4.5 × 10−5 | 2.43 |
| Noggin | 3.4 × 10−4 | 5.8 × 10−5 | 5.92 |
| Nuclear receptor subfamily 2, group E, member 3 | 7.6 × 10−5 | 3.9 × 10−6 | 19.45 |
| POU domain, class 4, transcription factor 1 | 1.1 × 10−5 | 5.7 × 10−4 | −51.63 |
| Slit homolog 2 | 1.5 × 10−4 | 6.3 × 10−5 | 2.35 |
Taking into account the fact that Ngn2 expression correlated positively with APP expression in both the APPKO and Tg2576 mice, we decided to examine the role of Ngn2 in mediating APP-induced NSPC neuronal commitment and differentiation. We focused our study on APPKO mice in order to exclude the possibility of nonspecific transgene effects as might be expected from studying the effects of human mutant APP in Tg2576.
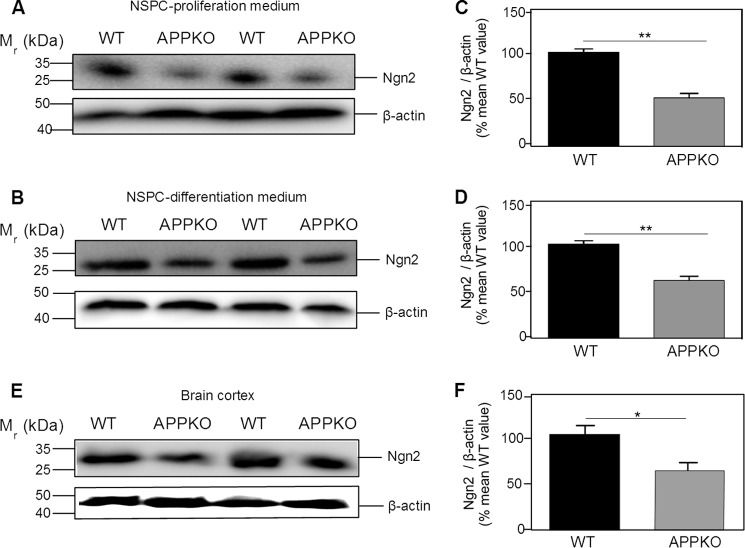
To confirm that the change in Ngn2 mRNA expression correlated with the level of Ngn2 (protein), we quantified the amount of Ngn2 by western blotting of NSPC lysates cultured for 5 days in proliferation medium. There was an ∼50% lower level of Ngn2 in NSPCs derived from APPKO compared with the level in WT NSPCs (Fig. 3, A and C). When NSPCs were cultured in differentiation medium (Fig. 3, B and D), again we observed a lower level (40%) of Ngn2 in the APPKO cells.
FIGURE 3.
Western blotting analysis of Ngn2 in NSPC culture and brain cortex derived from WT and APPKO mice. The figure shows western blots (A, B, and E) and the corresponding quantification of immunoreactivity (C, D, and F). NSPCs from WT and APPKO mice were incubated in proliferation medium (A and C) or differentiation medium (B and D). Cells were lysed after 5 days, and the lysates were analyzed for Ngn2 by western blotting and for β-actin (loading control). E and F, analysis of in vivo expression. Brain cortices from WT and APPKO mice were homogenized, and the lysates were analyzed for Ngn2 by western blotting and for β-actin (loading control). Bars show the means ± S.E. (* = p < 0.05; ** p < 0.01 as determined by Student's t test).
To examine whether the expression of Ngn2 was also affected in vivo, we quantified the amount of Ngn2 by western blotting of brain cortex derived from APPKO and WT mice. We observed a 60% lower level of Ngn2 immunoreactivity in the brain cortex of APPKO mice compared with that of WT mice (Fig. 3, E and F). These results were consistent with the results obtained with NSPCs. We concluded that in the absence of APP there was a lower level of Ngn2 (protein) due to a decrease in the level of Ngn2 mRNA expression.
Effect of Ngn2 on Neuronal Differentiation
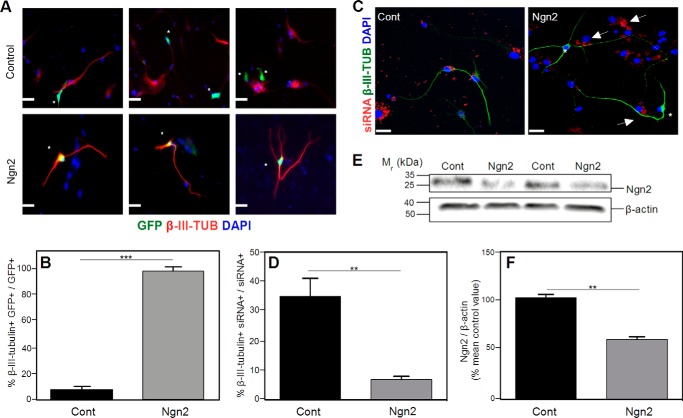
As NSPCs derived from APPKO mice had lower levels of Ngn2 and a decreased propensity to differentiate into neurons than NSPCs derived from WT mice, this suggested that APP might enhance neuronal differentiation through Ngn2. To confirm this hypothesis, we examined whether the expression of APP in NSPCs derived from APPKO mice could induce neuronal differentiation. The cells were transiently transfected with a pCAG-IRES-APP or a pCAG-IRES-GFP plasmid in order to express APP or GFP into the NSPCs. Then cells were cultured for 5 days in differentiation medium. After this, the percentage of transfected cells (APP+ or GFP+) that were also positive for MAP2 were analyzed, and the significance of the difference was determined with a χ2 test (n = 34 cells counted). The results showed that 79 ± 5% of cells were APP+MAP2+ compared with 15 ± 6% that were GFP+MAP2+. The difference was statistically significant (p = 0.027), confirming that APP expression resulted in a recovery of neuronal differentiation of APPKO NSPCs.
The next step was to study whether Ngn2 was involved in the APPKO NSPC neuronal differentiation produced by APP. NSPCs derived from APPKO mice were transiently transfected with a pCAG-Ngn2-IRES-GFP or a p-CAG-IRES-GFP control vector and cultured for 5 days in differentiation medium. Analysis of the pCAG-Ngn2-IRES-GFP-transfected cells showed that 95.6 ± 4.3% of the GFP-positive cells also stained positively for β-III-tubulin (300 cells counted across ×3 independent cultures). However, only 7.0% ± 0.6% of the cells that were transfected with pCAG-IRES-GFP (control plasmid) were β-III-tubulin-positive (300 cells counted across ×3 independent cultures) (Fig. 4, A and B). The percentage of β-III-tubulin+ cells after the transfection with the GFP-control vector was similar to the percentage of β-III-tubulin+ cells obtained in NSPC culture derived from APPKO (Fig. 1B), confirming that the expression of GFP had no significant effect.
FIGURE 4.
Transfection of NSPCs with a Ngn2 cDNA induces neuronal differentiation. Cells were cultured for 5 days in differentiation medium. A, NSPCs derived from APPKO mice were transfected either with empty vector pCAG-IRES-GFP (Control) or with pCAG-Ngn2-IRES-GFP (Ngn2). The figure shows 3 independent representative immunofluorescence images for each group. Transfected cells (green) are marked with asterisks. Cells were stained for β-III-tubulin (red). No double-stained GFP+ β-III-tubulin+ cells were observed in empty vector incubation, indicating that cells transfected with the empty vector are not neurons. In cells transfected with the Ngn2 plasmid, there was co-staining of GFP and β-III-tubulin, indicating that each cell transfected with Ngn2 was neuronally differentiated. B, quantification of the results in panel A shows the % of β-III-tubulin+ GFP+ cells to total GFP+ cells. C, representative immunofluorescence images of NSPCs derived from WT mice transfected with siRNA-control-Cy3 vector or siRNA-Ngn2-Cy3 vector (Ngn2). The figure shows siRNA (red), β-III-tubulin (green), and DAPI (blue) staining. Cells transfected with the control plasmid (61) showed co-staining of siRNA and β-III-tubulin. Cells transfected with the Ngn2 siRNA (Ngn2), marked with arrows, were not β-III-tubulin+ (asterisk). D, quantification of the results in C showing the % β-III-tubulin+ siRNA+ cells to total siRNA+ cells. F, western blotting analysis of Ngn2 levels. The figure shows a representative western blot (E) and quantification (F). Decreased expression of Ngn2 was observed in NSPCs after siRNA interference. Cells were cultured for 5 days in differentiation medium. β-Actin was used as a control for protein loading. Scale bars = 50 μm. Data are expressed as the means ± S.E. (** = p < 0.001; *** = p < 0.0001 as determined by Student's t test). Cont, control; Ngn2, neurogenin 2.
Role of Neurogenin 2 in Neuronal Differentiation
As our results suggested that Ngn2 might be a mediator of APP-induced neuronal differentiation, we next examined the effect of Ngn2 knockdown on differentiation. NSPCs derived from WT mice were transfected either with a RNA interference plasmid (siRNA-Ngn2-Cy3) or with a control plasmid (siRNA-control-Cy3). After transfection, the cells were cultured for 5 days in differentiation medium. Under these conditions ∼70% of cells were transfected with the siRNA. Approximately 40 ± 6% of the cells transfected with a control Cy3-labeled siRNA expressed β-III tubulin. However, when Ngn2 expression was knocked down by transfection with a Cy3-labeled Ngn2 siRNA, only 9 ± 1% of the transfected cells were β-III tubulin-positive (Fig. 4, C and D).
The levels of Ngn2 (protein) in NSPCs transfected with each siRNA plasmid were assessed after culturing for 5 days in differentiation medium. There was a lower level of Ngn2 in NSPCs transfected with siRNA-Ngn2-Cy3 compared with NSPCs transfected with a control siRNA (Fig. 4, E and F). The results showed that even in the presence of normal expression levels of APP, knockdown of Ngn2 was sufficient to decrease neuronal differentiation. This result confirmed that Ngn2 mediates APP-induced differentiation of NSPCs into neurons.
Effect of sAPPα and Aβ Peptides on Neuronal Differentiation
Porayette et al. (30) previously reported that APP cleavage was necessary for an effect of APP on the differentiation of embryonic stem cells. To examine this possibility in our cells, we treated the NSPCs from APPKO mice with 10 nm sAPPα (Fig. 5, A–C), 1 μm Aβ 1–40 (Fig. 5, D–F), and 1 μm Aβ 1–42 (Fig. 5, G–I) and cultured in differentiation medium for 5 days. The concentrations of sAPPα and Aβ were chosen because they were similar to concentrations in our previous study (19) and equal to or greater than concentrations reported to affect stem cell differentiation by Porayette et al. (30). After this, the percentage of β-III-tubulin+ cells present in the cultures was determined and expressed as a percentage of the total number of DAPI+ cells. We did not find a significant difference between the number of β-III-tubulin+ cells in the untreated cultures and the number of β-III-tubulin+ cells in the cultures treated with APP fragments (Fig. 5). Thus the increase in neuronal differentiation of NSPCs observed in association with APP expression was unlikely to have been due to an increase in production of sAPPα or Aβ peptides.
FIGURE 5.
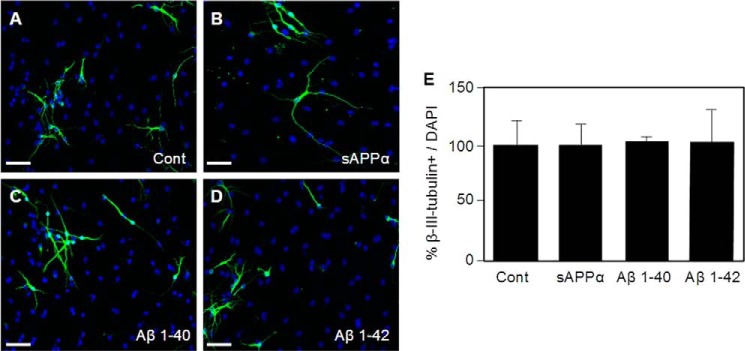
Neuronal differentiation of NSPCs is not increased by APP cleavage products. Panels A–D show representative immunofluorescence images of NSPCs stained for β-III-tubulin (green) and DAPI (blue). The cultures were either untreated (Control, panel A) or treated with 10 nm sAPPα (B), 1 μm Aβ 1–40 (C), and 1 μm Aβ 1–42 (D). E, quantification of results from immunofluorescence analysis showing % β-III-tubulin+/DAPI+. Approximately 300 cells per group in 3 independent experiments were counted. Bars show means ± S.E. Scale bars = 30 μm.
DISCUSSION
The present study shows that neuronal differentiation is lower in NSPCs derived from APPKO mice and that this effect is due to decreased expression of Ngn2. Levels of Ngn2 correlated with APP expression. Furthermore, knockdown of Ngn2 in wild-type cells lowered neuronal differentiation, whereas transfection of APPKO NSPCs with either APP or Ngn2 resulted in recovery of neuronal differentiation. Taken together, the results demonstrate that APP contributes actively to neurogenesis during development.
Despite a large number of published studies on APP (31), the normal function of APP is largely unknown. Emerging evidence suggests that APP plays a role in neuronal growth and neural repair, although the precise mechanisms have been unclear (31). The expression of APP is regulated developmentally (28, 32) and is increased in association with neurite outgrowth and synaptogenesis (8, 26). Similarly, overexpression or knock-out of APP has been reported to disrupt a number of developmental functions including neuronal migration (9) and cell growth (33, 34). Secreted forms of APP are reported to have neurotrophic and neuroprotective effects (7, 8, 35, 36) and to stimulate proliferation of adult neural progenitors (37, 38). Additionally, there are reports that infusion of soluble APPα after traumatic brain injury can improve neuronal survival and recovery (24, 39, 40). Collectively, these studies provide evidence that APP has a trophic function.
Some studies suggest that APP may play a role in the regulation of stem cell proliferation or differentiation. For example, APP is expressed in developing neuroblasts at the time of cell proliferation and differentiation (26, 31, 41). In the present study we found that in the absence of APP, there was a decrease in NSPC differentiation into neurons. These findings strongly support the idea that APP is involved in the regulation of NSPC neurogenesis. Indeed, APP is processed in a manner that is very similar to the protein Notch, which regulates NSPC proliferation (42). Therefore, APP may have a similar or related developmental function to that of Notch (43).
Previously we found that NSPC proliferation was regulated by APP through the secretion of cystatin C (19). However, in the present study we found that the APP-induced neuronal differentiation of NSPC was not due to cystatin C but rather to Ngn2. Ngn2 is a bHLH transcription factor that was first identified for its ability to promote neuronal differentiation in brain and spinal cord (44–46). Ngn2 also specifies phenotypic features of neurons (47–50), regulates axonal guidance (51) as well as dendritic morphologies of cortical neurons (52) and cortical neuron migration (53), and plays a crucial role along with other bHLH transcription factors during development (48, 54–56). Additionally, Ngn2 has an important function in stem-cell differentiation during neurogenesis driving neuronal differentiation (57–60). In this context, our present findings support the idea that the APP-induced differentiation of NSPCs is mediated by Ngn2.
In primary cultures of NSPCs derived from APPKO and WT mice, we found that Ngn2 mRNA expression and protein levels were decreased in APPKO cells consistent with the view that Ngn2 is involved in NSPC neuronal differentiation. We also analyzed the Ngn2 protein levels in brain cortices derived from APPKO mice and found a decrease in Ngn2 levels compared with cortices derived from WT mice, consistent with the results obtained in NSPC cultures. In support of the view that APP regulates neuronal differentiation of NSPCs via Ngn2, we found that transfection of a Ngn2 cDNA into NSPCs recovered the neuronal differentiation phenotype of the APPKO NSPCs. Furthermore, we found that knocking down the expression of Ngn2 gene in NSPCs derived from WT mice using a siRNA approach led to a decrease in neuronal differentiation. Taken together, the studies support the conclusion that APP increases neuronal differentiation by regulating Ngn2 expression.
Interestingly, Ngn2 expression was higher in APP-expressing cells than in APPKO cells when they were cultured in proliferation medium (i.e. before differentiation) (Fig. 3A). As the cells did not differentiate in proliferation medium, this implies that APP-induced Ngn2 expression is not in itself a sufficient condition for neuronal differentiation.
Previously, Porayette et al. (30) reported that in human embryonic stem cells, proliferation and differentiation were mediated via APP cleavage (i.e. sAPPα). Our previous study (19) found no effect of sAPPα or Aβ peptides on NSPC proliferation. In the present study we similarly found no effect of sAPPα or Aβ peptides on neuronal differentiation. The reason for the differences between the previous study (30) and our own are unclear, but it seems possible that mechanisms of regulation of embryonic stem cells differ from those of the NSPCs used in our study. However, despite the finding that APP fragments did not have an effect on neuronal differentiation, this does not exclude the possibility that they may have effects on other events such as migration (9) or neurite outgrowth (4–8). Further studies are needed to examine this possibility.
It is not yet clear how APP regulates Ngn2 to increase NSPC neurogenesis. Some studies suggest that Ngn2 can be regulated through multisite phosphorylation of Ngn2 by kinases. Li et al. (46) reported that the capacity of Ngn2 to induce differentiation is regulated by glycogen synthase kinase 3β. For this reason, glycogen synthase kinase 3β may be a good candidate for further study because it may be regulated by APP as reported by Zhou et al. (61). On the other hand, Ngn2 expression can be regulated by other transcription factors such as Pax6, Nurr, and Mash1 (46, 55, 58, 62). As a consequence of this mechanism, the increased activity and expression of Ngn2 can induce stem cell differentiation. Studies have shown that Ngn2 expression can be regulated by retinoic acid, sonic hedgehog, and fibroblast growth factor (63). Whether these factors are involved in mediating the effect of APP on Ngn2 expression remains to be determined, as further studies will be required to define a specific mechanism through which APP regulates Ngn2.
Acknowledgments
We thank François Guillemot (MRC-National Institute for Medical Research, London, UK) for the kind donation of the Ngn2 plasmid, Julian Heng (Monash University, Melbourne, Australia) for discussions regarding Ngn2, and Meredith Roberts-Thompson for work on the APPKO colony.
This work was supported by project grants from the National Health and Medical Research Council of Australia, BUPA/Alzheimer's Society UK and Multiple Sclerosis Research Australia.
- APP
- amyloid-β precursor protein
- sAPPα
- secreted amyloid-β protein precursor-α
- Aβ
- amyloid-β protein
- NSPC
- neural stem or progenitor cells
- APPKO
- APP-knock-out
- IRES
- internal ribosome entry site
- bHLH
- basic helix-loop-helix.
REFERENCES
- 1. Wang Z., Wang B., Yang L., Guo Q., Aithmitti N., Songyang Z., Zheng H. (2009) Presynaptic and postsynaptic interaction of the amyloid precursor protein promotes peripheral and central synaptogenesis. J. Neurosci. 29, 10788–10801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tyan S. H., Shih A. Y., Walsh J. J., Maruyama H., Sarsoza F., Ku L., Eggert S., Hof P. R., Koo E. H., Dickstein D. L. (2012) Amyloid precursor protein (APP) regulates synaptic structure and function. Mol. Cell Neurosci. 51, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Laßek M., Weingarten J., Einsfelder U., Brendel P., Müller U., Volknandt W. (2013) Amyloid precursor proteins are constituents of the presynaptic active zone. J. Neurochem. 127, 48–56 [DOI] [PubMed] [Google Scholar]
- 4. Young-Pearse T. L., Chen A. C., Chang R., Marquez C., Selkoe D. J. (2008) Secreted APP regulates the function of full-length APP in neurite outgrowth through interaction with integrin β1. Neural Dev. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Small D. H., Clarris H. L., Williamson T. G., Reed G., Key B., Mok S. S., Beyreuther K., Masters C. L., Nurcombe V. (1999) Neurite-outgrowth regulating functions of the amyloid protein precursor of Alzheimer's disease. J. Alzheimers Dis. 1, 275–285 [DOI] [PubMed] [Google Scholar]
- 6. Leyssen M., Ayaz D., Hébert S. S., Reeve S., De Strooper B., Hassan B. A. (2005) Amyloid precursor protein promotes post-developmental neurite arborization in the Drosophila brain. EMBO J. 24, 2944–2955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Milward E. A., Papadopoulos R., Fuller S. J., Moir R. D., Small D., Beyreuther K., Masters C. L. (1992) The amyloid protein precursor of Alzheimer's disease is a mediator of the effects of nerve growth factor on neurite outgrowth. Neuron 9, 129–137 [DOI] [PubMed] [Google Scholar]
- 8. Small D. H., Nurcombe V., Reed G., Clarris H., Moir R., Beyreuther K., Masters C. L. (1994) A heparin-binding domain in the amyloid protein precursor of Alzheimer's disease is involved in the regulation of neurite outgrowth. J. Neurosci. 14, 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Young-Pearse T. L., Bai J., Chang R., Zheng J. B., LoTurco J. J., Selkoe D. J. (2007) A critical function for β-amyloid precursor protein in neuronal migration revealed by in utero RNA interference. J. Neurosci. 27, 14459–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Octave J. N., Pierrot N., Ferao Santos S., Nalivaeva N. N., Turner A. J. (2013) From synaptic spines to nuclear signaling: nuclear and synaptic actions of the amyloid precursor protein. J. Neurochem. 126, 183–190 [DOI] [PubMed] [Google Scholar]
- 11. Vogt D. L., Thomas D., Galvan V., Bredesen D. E., Lamb B. T., Pimplikar S. W. (2011) Abnormal neuronal networks and seizure susceptibility in mice overexpressing the APP intracellular domain. Neurobiol. Aging 32, 1725–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Itoh T., Satou T., Nishida S., Tsubaki M., Hashimoto S., Ito H. (2009) Expression of amyloid precursor protein after rat traumatic brain injury. Neurol Res. 31, 103–109 [DOI] [PubMed] [Google Scholar]
- 13. Blumbergs P. C., Scott G., Manavis J., Wainwright H., Simpson D. A., McLean A. J. (1995) Topography of axonal injury as defined by amyloid precursor protein and the sector scoring method in mild and severe closed head injury. J. Neurotrauma 12, 565–572 [DOI] [PubMed] [Google Scholar]
- 14. Gentleman S. M., Graham D. I., Roberts G. W. (1993) Molecular pathology of head trauma: altered β APP metabolism and the aetiology of Alzheimer's disease. Prog. Brain Res. 96, 237–246 [DOI] [PubMed] [Google Scholar]
- 15. Tabaton M., Cammarata S., Mandybur T., Richey P., Kawai M., Perry G., Gambetti P. (1992) Senile plaques in cerebral amyloid angiopathy show accumulation of amyloid precursor protein without cytoskeletal abnormalities. Brain Res. 593, 299–303 [DOI] [PubMed] [Google Scholar]
- 16. Cras P., Kawai M., Lowery D., Gonzalez-DeWhitt P., Greenberg B., Perry G. (1991) Senile plaque neurites in Alzheimer disease accumulate amyloid precursor protein. Proc. Natl. Acad. Sci. U.S.A. 88, 7552–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cochran E., Bacci B., Chen Y., Patton A., Gambetti P., Autilio-Gambetti L. (1991) Amyloid precursor protein and ubiquitin immunoreactivity in dystrophic axons is not unique to Alzheimer's disease. Am. J. Pathol 139, 485–489 [PMC free article] [PubMed] [Google Scholar]
- 18. Small D. H., Hu Y., Bolós M., Dawkins E., Foa L., Young K. M. (2014) β-Amyloid precursor protein: function in stem cell development and Alzheimer's disease brain. Neurodegener. Dis. 13, 96–98 [DOI] [PubMed] [Google Scholar]
- 19. Hu Y., Hung A. C., Cui H., Dawkins E., Bolós M., Foa L., Young K. M., Small D. H. (2013) Role of cystatin C in amyloid precursor protein-induced proliferation of neural stem/progenitor cells. J. Biol. Chem. 288, 18853–18862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou Z. D., Chan C. H., Ma Q. H., Xu X. H., Xiao Z. C., Tan E. K. (2011) The roles of amyloid precursor protein (APP) in neurogenesis: implications to pathogenesis and therapy of Alzheimer disease. Cell Adh. Migr. 5, 280–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. López-Toledano M. A., Shelanski M. L. (2004) Neurogenic effect of β-amyloid peptide in the development of neural stem cells. J. Neurosci. 24, 5439–5444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jin K., Galvan V., Xie L., Mao X. O., Gorostiza O. F., Bredesen D. E., Greenberg D. A. (2004) Enhanced neurogenesis in Alzheimer's disease transgenic (PDGF-APPSw,Ind) mice. Proc. Natl. Acad. Sci. U.S.A. 101, 13363–13367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. López-Toledano M. A., Shelanski M. L. (2007) Increased neurogenesis in young transgenic mice overexpressing human APP(Sw, Ind). J. Alzheimers Dis. 12, 229–240 [DOI] [PubMed] [Google Scholar]
- 24. Demars M. P., Bartholomew A., Strakova Z., Lazarov O. (2011) Soluble amyloid precursor protein: a novel proliferation factor of adult progenitor cells of ectodermal and mesodermal origin. Stem Cell Res. Ther. 2, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hayashi Y., Kashiwagi K., Ohta J., Nakajima M., Kawashima T., Yoshikawa K. (1994) Alzheimer amyloid protein precursor enhances proliferation of neural stem cells from fetal rat brain. Biochem. Biophys. Res. Commun. 205, 936–943 [DOI] [PubMed] [Google Scholar]
- 26. Clarris H. J., Key B., Beyreuther K., Masters C. L., Small D. H. (1995) Expression of the amyloid protein precursor of Alzheimer's disease in the developing rat olfactory system. Brain Res. Dev. Brain Res. 88, 87–95 [DOI] [PubMed] [Google Scholar]
- 27. Freude K. K., Penjwini M., Davis J. L., LaFerla F. M., Blurton-Jones M. (2011) Soluble amyloid precursor protein induces rapid neural differentiation of human embryonic stem cells. J. Biol. Chem. 286, 24264–24274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee J. A., Cole G. J. (2007) Generation of transgenic zebrafish expressing green fluorescent protein under control of zebrafish amyloid precursor protein gene regulatory elements. Zebrafish 4, 277–286 [DOI] [PubMed] [Google Scholar]
- 29. Megason S. G., McMahon A. P. (2002) A mitogen gradient of dorsal midline Wnts organizes growth in the CNS. Development 129, 2087–2098 [DOI] [PubMed] [Google Scholar]
- 30. Porayette P., Gallego M. J., Kaltcheva M. M., Bowen R. L., Vadakkadath Meethal S., Atwood C. S. (2009) Differential processing of amyloid-β precursor protein directs human embryonic stem cell proliferation and differentiation into neuronal precursor cells. J. Biol. Chem. 284, 23806–23817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dawkins E., Small D. H. (2014) Insights into the physiological function of the β-amyloid precursor protein: beyond Alzheimer's disease. J. Neurochem. 129, 756–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Small D. H., Nurcombe V., Moir R., Michaelson S., Monard D., Beyreuther K., Masters C. L. (1992) Association and release of the amyloid protein precursor of Alzheimer's disease from chick brain extracellular matrix. J. Neurosci. 12, 4143–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hornsten A., Lieberthal J., Fadia S., Malins R., Ha L., Xu X., Daigle I., Markowitz M., O'Connor G., Plasterk R., Li C. (2007) APL-1, a Caenorhabditis elegans protein related to the human β-amyloid precursor protein, is essential for viability. Proc. Natl. Acad. Sci. U.S.A. 104, 1971–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Joshi P., Liang J. O., DiMonte K., Sullivan J., Pimplikar S. W. (2009) Amyloid precursor protein is required for convergent-extension movements during Zebrafish development. Dev. Biol. 335, 1–11 [DOI] [PubMed] [Google Scholar]
- 35. Williamson T. G., Mok S. S., Henry A., Cappai R., Lander A. D., Nurcombe V., Beyreuther K., Masters C. L., Small D. H. (1996) Secreted glypican binds to the amyloid precursor protein of Alzheimer's disease (APP) and inhibits APP-induced neurite outgrowth. J. Biol. Chem. 271, 31215–31221 [DOI] [PubMed] [Google Scholar]
- 36. Mattson M. P., Cheng B., Culwell A. R., Esch F. S., Lieberburg I., Rydel R. E. (1993) Evidence for excitoprotective and intraneuronal calcium-regulating roles for secreted forms of the β-amyloid precursor protein. Neuron 10, 243–254 [DOI] [PubMed] [Google Scholar]
- 37. Chasseigneaux S., Allinquant B. (2012) Functions of Aβ, sAPPα and sAPPβ: similarities and differences. J. Neurochem. 120, 99–108 [DOI] [PubMed] [Google Scholar]
- 38. Caillé I., Allinquant B., Dupont E., Bouillot C., Langer A., Müller U., Prochiantz A. (2004) Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult subventricular zone. Development 131, 2173–2181 [DOI] [PubMed] [Google Scholar]
- 39. Thornton E., Vink R., Blumbergs P. C., Van Den Heuvel C. (2006) Soluble amyloid precursor protein α reduces neuronal injury and improves functional outcome following diffuse traumatic brain injury in rats. Brain Res. 1094, 38–46 [DOI] [PubMed] [Google Scholar]
- 40. Corrigan F., Vink R., Blumbergs P. C., Masters C. L., Cappai R., van den Heuvel C. (2012) sAPPα rescues deficits in amyloid precursor protein knockout mice following focal traumatic brain injury. J. Neurochem. 122, 208–220 [DOI] [PubMed] [Google Scholar]
- 41. Salbaum J. M., Ruddle F. H. (1994) Embryonic expression pattern of amyloid protein precursor suggests a role in differentiation of specific subsets of neurons. J. Exp. Zool. 269, 116–127 [DOI] [PubMed] [Google Scholar]
- 42. Ables J. L., Breunig J. J., Eisch A. J., Rakic P. (2011) Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 12, 269–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kimberly W. T., Zheng J. B., Guénette S. Y., Selkoe D. J. (2001) The intracellular domain of the β-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J. Biol. Chem. 276, 40288–40292 [DOI] [PubMed] [Google Scholar]
- 44. Ma Q., Kintner C., Anderson D. J. (1996) Identification of neurogenin, a vertebrate neuronal determination gene. Cell 87, 43–52 [DOI] [PubMed] [Google Scholar]
- 45. Sommer L., Ma Q., Anderson D. J. (1996) neurogenins, a novel family of atonal-related bHLH transcription factors, are putative mammalian neuronal determination genes that reveal progenitor cell heterogeneity in the developing CNS and PNS. Mol. Cell. Neurosci. 8, 221–241 [DOI] [PubMed] [Google Scholar]
- 46. Li S., Mattar P., Zinyk D., Singh K., Chaturvedi C. P., Kovach C., Dixit R., Kurrasch D. M., Ma Y. C., Chan J. A., Wallace V., Dilworth F. J., Brand M., Schuurmans C. (2012) GSK3 temporally regulates neurogenin 2 proneural activity in the neocortex. J. Neurosci. 32, 7791–7805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Florio M., Leto K., Muzio L., Tinterri A., Badaloni A., Croci L., Zordan P., Barili V., Albieri I., Guillemot F., Rossi F., Consalez G. G. (2012) Neurogenin 2 regulates progenitor cell-cycle progression and Purkinje cell dendritogenesis in cerebellar development. Development 139, 2308–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kele J., Simplicio N., Ferri A. L., Mira H., Guillemot F., Arenas E., Ang S. L. (2006) Neurogenin 2 is required for the development of ventral midbrain dopaminergic neurons. Development 133, 495–505 [DOI] [PubMed] [Google Scholar]
- 49. Ma Y. C., Song M. R., Park J. P., Henry Ho H. Y., Hu L., Kurtev M. V., Zieg J., Ma Q., Pfaff S. L., Greenberg M. E. (2008) Regulation of motor neuron specification by phosphorylation of neurogenin 2. Neuron 58, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Scardigli R., Schuurmans C., Gradwohl G., Guillemot F. (2001) Crossregulation between Neurogenin2 and pathways specifying neuronal identity in the spinal cord. Neuron 31, 203–217 [DOI] [PubMed] [Google Scholar]
- 51. Hand R., Polleux F. (2011) Neurogenin2 regulates the initial axon guidance of cortical pyramidal neurons projecting medially to the corpus callosum. Neural Dev. 6, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hand R., Bortone D., Mattar P., Nguyen L., Heng J. I., Guerrier S., Boutt E., Peters E., Barnes A. P., Parras C., Schuurmans C., Guillemot F., Polleux F. (2005) Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron 48, 45–62 [DOI] [PubMed] [Google Scholar]
- 53. Heng J. I., Nguyen L., Castro D. S., Zimmer C., Wildner H., Armant O., Skowronska-Krawczyk D., Bedogni F., Matter J. M., Hevner R., Guillemot F. (2008) Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature 455, 114–118 [DOI] [PubMed] [Google Scholar]
- 54. Ross S. E., Greenberg M. E., Stiles C. D. (2003) Basic helix-loop-helix factors in cortical development. Neuron 39, 13–25 [DOI] [PubMed] [Google Scholar]
- 55. Wilkinson G., Dennis D., Schuurmans C. (2013) Proneural genes in neocortical development. Neuroscience 253, 256–273 [DOI] [PubMed] [Google Scholar]
- 56. Galichet C., Guillemot F., Parras C. M. (2008) Neurogenin 2 has an essential role in development of the dentate gyrus. Development 135, 2031–2041 [DOI] [PubMed] [Google Scholar]
- 57. Nieto M., Schuurmans C., Britz O., Guillemot F. (2001) Neural bHLH genes control the neuronal versus glial fate decision in cortical progenitors. Neuron 29, 401–413 [DOI] [PubMed] [Google Scholar]
- 58. Ali F., Hindley C., McDowell G., Deibler R., Jones A., Kirschner M., Guillemot F., Philpott A. (2011) Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development 138, 4267–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kageyama R., Ohtsuka T., Hatakeyama J., Ohsawa R. (2005) Roles of bHLH genes in neural stem cell differentiation. Exp. Cell Res. 306, 343–348 [DOI] [PubMed] [Google Scholar]
- 60. Sun Y., Nadal-Vicens M., Misono S., Lin M. Z., Zubiaga A., Hua X., Fan G., Greenberg M. E. (2001) Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell 104, 365–376 [DOI] [PubMed] [Google Scholar]
- 61. Zhou F., Gong K., Song B., Ma T., van Laar T., Gong Y., Zhang L. (2012) The APP intracellular domain (AICD) inhibits Wnt signalling and promotes neurite outgrowth. Biochim. Biophys. Acta 1823, 1233–1241 [DOI] [PubMed] [Google Scholar]
- 62. Yi S. H., Jo A. Y., Park C. H., Koh H. C., Park R. H., Suh-Kim H., Shin I., Lee Y. S., Kim J., Lee S. H. (2008) Mash1 and neurogenin 2 enhance survival and differentiation of neural precursor cells after transplantation to rat brains via distinct modes of action. Mol. Ther. 16, 1873–1882 [DOI] [PubMed] [Google Scholar]
- 63. Ribes V., Stutzmann F., Bianchetti L., Guillemot F., Dollé P., Le Roux I. (2008) Combinatorial signalling controls Neurogenin2 expression at the onset of spinal neurogenesis. Dev. Biol. 321, 470–481 [DOI] [PubMed] [Google Scholar]
- 64. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real- time quantitative PCR and the 2ΔΔCt method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]