Background: Polβ has been shown to accurately but slowly incorporate nucleotide opposite the cisplatin-1,2-d(GpG) (Pt-GG) cross-link; however, its structural basis is unknown.
Results: Both guanines of the Pt-GG form Watson-Crick base pairing with the primer terminus dC and incoming dCTP.
Conclusion: Polβ accommodates the Pt-GG adduct with a semiopen conformation.
Significance: Our studies explain why polβ is less efficient at bypassing the Pt-GG than polη.
Keywords: DNA Damage, DNA Polymerase, DNA Replication, Enzyme Catalysis, Enzyme Structure
Abstract
Human DNA polymerase β (polβ) has been suggested to play a role in cisplatin resistance, especially in polβ-overexpressing cancer cells. Polβ has been shown to accurately albeit slowly bypass the cisplatin-1,2-d(GpG) (Pt-GG) intramolecular cross-link in vitro. Currently, the structural basis for the inefficient Pt-GG bypass mechanism of polβ is unknown. To gain structural insights into the mechanism, we determined two ternary structures of polβ incorporating dCTP opposite the templating Pt-GG lesion in the presence of the active site Mg2+ or Mn2+. The Mg2+-bound structure shows that the bulky Pt-GG adduct is accommodated in the polβ active site without any steric hindrance. In addition, both guanines of the Pt-GG lesion form Watson-Crick base pairing with the primer terminus dC and the incoming dCTP, providing the structural basis for the accurate bypass of the Pt-GG adduct by polβ. The Mn2+-bound structure shows that polβ adopts a catalytically suboptimal semiclosed conformation during the insertion of dCTP opposite the templating Pt-GG, explaining the inefficient replication across the Pt-GG lesion by polβ. Overall, our studies provide the first structural insights into the mechanism of the potential polβ-mediated cisplatin resistance.
Introduction
Cisplatin (cis-diamminedichloroplatinum(II)) and its related platinum-based chemotherapeutic agents are widely used in the treatment of various human malignancies including testicular, ovarian, cervical, and non-small cell lung cancers (Fig. 1). Cisplatin is activated via intracellular aquation to form cis-[(NH3)2PtCl(OH2)]+ and cis-[(NH3)2Pt(OH2)2]2+ (1). The aquated cisplatin species are potent electrophiles and react with the nucleophilic N7 atoms of purines in DNA to produce a cisplatin-1,2-d(GpG) (Pt-GG2 hereafter) intramolecular cross-link as the major cytotoxic lesion. The bulky cisplatin-DNA adducts can block DNA replication and transcription, thereby triggering cell cycle arrest and apoptosis of rapidly dividing cancer cells (1). The major limitation to cisplatin-based chemotherapy is cisplatin resistance, which arises from various mechanisms including uptake/efflux, nucleotide excision repair, and translesion bypass by DNA polymerases (2). The Y-family human DNA polymerase η (polη) efficiently replicates across the bulky Pt-GG lesion and has been implicated in the accurate bypass of the Pt-GG lesion in vivo (3, 4).
FIGURE 1.
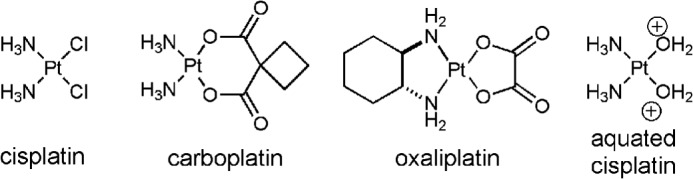
Chemical structures of platinum anticancer drugs.
Several reports suggest that the X-family human DNA polymerase β (polβ) may play a role in cisplatin resistance. Polβ has been shown to accurately bypass the Pt-GG cross-links in vitro (5) and to extend DNA chains of DNA polymerases α, δ, and ϵ that are stalled by the Pt-GG adducts (6). It has been suggested that polβ may contribute to cisplatin resistance in polβ-overexpressing cancer cells (7–9). Overexpression of polβ has been shown to facilitate translesion synthesis of the Pt-GG adduct and increase cisplatin resistance in vivo (10–14). Interestingly, polβ inserts dCTP opposite the Pt-GG adduct with an efficiency of ∼2% relative to unmodified DNA, whereas polη inserts the nucleotide opposite the lesion with an efficiency of ∼40% relative to unmodified DNA (5, 15), indicating that the X-family DNA polymerase is much less efficient at replicating across the Pt-GG lesion than the Y-family DNA polymerase.
Several crystal structures of polη and DNA polymerase IV (Dpo4) complexed with the Pt-GG-containing DNA have revealed the mechanisms for the accurate and efficient bypass of the Pt-GG lesion by the Y-family DNA polymerases (16–20), providing mechanistic insights into DNA polymerase-mediated chemoresistance to the platinum anticancer drugs. The structural basis for the accurate but inefficient catalysis across the Pt-GG lesion by the X-family DNA polymerase polβ is currently unknown. Herein, we report two crystal structures of polβ incorporating a non-hydrolyzable dCTP analog opposite the 5′-dG of the templating Pt-GG in the presence of the active site Mg2+ or Mn2+. These structures provide insights into the mechanism of the accurate but slow bypass of the Pt-GG by polβ. These structures also explain why the X-family DNA polymerase polβ is much less efficient at catalyzing through the bulky Pt-GG adducts than the Y-family DNA polymerase polη.
EXPERIMENTAL PROCEDURES
DNA Sequences Used for X-ray Crystallographic Studies
The oligonucleotides used for the crystallographic studies were obtained from Integrated DNA Technologies (Coralville, IA). The template DNA sequence used for co-crystallization was 5′-CCCACGGCCCATCACC-3′ (GG denotes the modification site for the Pt-GG cross-link). The upstream primer sequence was 5′-GGTGATGGGC-3′. The downstream primer sequence was 5′-phosphate-GTGGG-3′. The platinum-modified template was prepared following published procedures (21). In brief, cisplatin (8.3 mm in H2O) was activated by adding 2 molar eq of silver nitrate and incubating the mixture in the dark at room temperature for 14–16 h. The reaction mixture was centrifuged at 13,000 × g for 20 min to collect the supernatant. The unmodified template DNA (1.5 μmol) in 10 mm sodium phosphate buffer, pH 6.8 was mixed with the activated cisplatin (1.7 μmol), and the mixture was incubated at 37 °C for 14–16 h. The Pt-GG-containing template DNA was purified by ion exchange column (Mono Q 5/50 GL, GE Healthcare) using a gradient from 0.1 to 1.0 m NaCl in 10 mm Tris buffer, pH 8.0. The site-specifically platinated template DNA was desalted using Sep-Pak C18 cartridges (Waters) and dried under reduced pressure. The dried Pt-GG template DNA was reconstituted in water and annealed with upstream and downstream primers to give a single nucleotide gapped DNA as described previously (22).
Steady-state Kinetics of Single Nucleotide Incorporation Opposite the Templating Pt-GG by Polβ
Steady-state kinetic parameters were determined using the conditions described previously (23). Oligonucleotides used for kinetic assays (upstream primer, 5′-FAM (fluorescein amidite)-ATGGGGTTGATGTGC-3′; downstream primer, 5′-phosphate-GTAGGGATGTTTGGGTAG-3′; and template, 5′-CTACCCAAACATCCCTACGGCACATCAACTCCAT-3′ where GG denotes the site of cisplatin modification) were purchased from Integrated DNA Technologies. The cisplatinated template was prepared using the method described above. To prepare DNA substrate containing a single nucleotide gap opposite the Pt-GG, the template, upstream primer, and downstream primer were annealed in hybridization buffer containing 10 mm Tris-HCl, pH 7.5 and 1 mm EDTA. Polymerase activities were determined using reaction mixtures containing 50 mm Tris-HCl, pH 7.5, 100 mm KCl, 5 mm MgCl2 or MnCl2, 80 nm single nucleotide gapped DNA, and various concentrations of incoming dCTP. To avoid product inhibition or substrate depletion that could interfere with initial velocity measurements, the polβ concentrations and reaction time intervals were adjusted for every experiment to ensure that the insertion product yield was less than 20%. The polymerase reactions were initiated by adding the enzyme to the single nucleotide gapped DNA substrate at 37 °C and stopped by adding a stop solution containing 95% formamide, 20 mm EDTA, 45 mm Tris borate, 0.1% bromphenol blue, and 0.1% xylene cyanol. The quenched reaction mixtures were separated on 20% denaturing polyacrylamide gels. The gels were analyzed using a PhosphorImager to quantify product formation. The kcat and Km were determined by fitting reaction rates over dCTP concentrations to the Michaelis-Menten equation. Each experiment was repeated three times. The efficiency of nucleotide insertion was calculated as kcat/Km. The relative efficiency of dCTP incorporation opposite the Pt-GG was determined as f = (kcat/Km)[dC:Pt-GG]/(kcat/Km)(dC:dG).
Co-crystallization of the Polβ:GG·dCTP and Polβ:Pt-GG·dCTP Ternary Complexes
Polβ was prepared with minor modifications of the method described previously (22). The binary polβ complex with the templating GG or Pt-GG in a single nucleotide gapped DNA was prepared by methods described previously (22). To prepare a polβ ternary complex with an incoming nucleotide opposite the templating unmodified GG or the Pt-GG, non-hydrolyzable dCMPNPP (dCTP*; 5 mm; Jena Biosciences) was added to the mixture of the polβ single nucleotide gapped binary complex. The ternary polβ-DNA complex co-crystals with dCMPNPP, paired with the templating dG, were grown by sitting drop vapor diffusion at 22 °C over several weeks in a buffer solution containing 50 mm imidazole, pH 7.5, 14–23% PEG3400, 20 mm divalent metal ions (MgCl2 or MnCl2), and 350 mm sodium acetate. The polβ:DNA co-crystals were cryoprotected with 50 mm Tris, pH 7.5, 14–23% PEG3400, 12% ethylene glycol, and 350 mm sodium acetate and flash frozen in liquid nitrogen. Diffraction data were collected at 100 K at the beamline 5.0.3 at the Advanced Light Source, Lawrence Berkeley National Laboratory. All diffraction data were processed using HKL 2000. The co-crystal structures were determined by molecular replacement (24) with a gapped binary complex structure (Protein Data Bank code 1BPX) and a ternary complex structure (Protein Data Bank code 1BPY) as the search models (25). The initial model was built using Coot (26) and refined using CCP4 (27). Ramachandran plots were obtained using MolProbity (28). All the crystallographic figures were generated using the molecular graphics program PyMOL (Schrödinger, LLC).
RESULTS AND DISCUSSION
We co-crystallized a ternary complex of polβ incorporating a non-hydrolyzable dCTP analog dCMPNPP (dCTP* hereafter) opposite the templating 5′-dG of the Pt-GG lesion in the presence of Mg2+ (Fig. 2). The non-hydrolyzable dCTP* was used for the crystallographic studies because it is isosteric to dCTP and coordinates with the active site metal ion in a manner comparable with that of the natural nucleotide (29). Non-hydrolyzable nucleotide analogs such as dUMPNPP (dUTP*) and dAMPCPP have been shown to tightly bind the polβ active site (29, 30), and the use of these analogs does not significantly alter the active site conformation of a polβ ternary complex (29, 31). The dNTP* analogs have been used to obtain ternary complex structures of various DNA polymerases (17, 32, 34, 35) including polη in complex with cisplatin- and phenanthriplatin-modified DNA (32, 35). Unfortunately, our extensive efforts to crystalize a ternary complex of polβ incorporating dCTP* opposite the templating 3′-dG of the Pt-GG lesion failed.
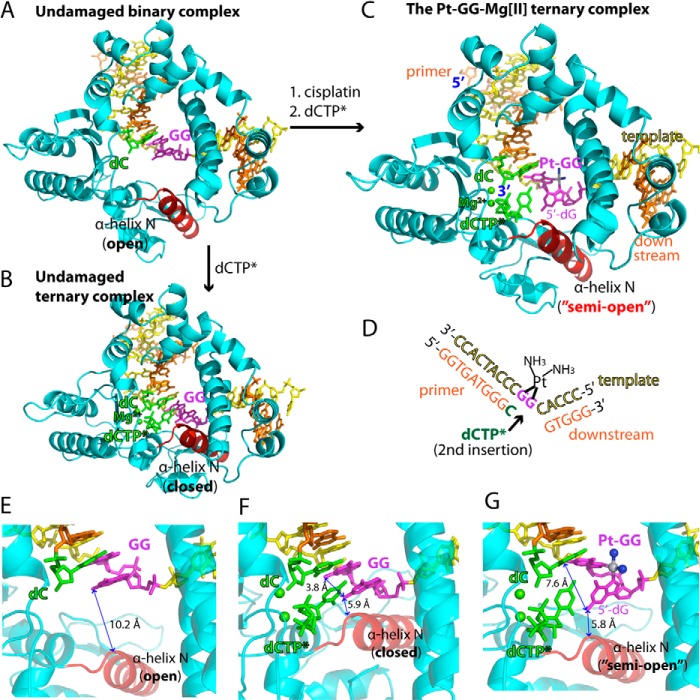
FIGURE 2.
Structures of the non-modified GG and the Pt-GG-Mg2+ complexes. A, overall structure of the unmodified GG gapped binary complex (Protein Data Bank code 4TUP). The template and primer strands are shown in yellow and orange, respectively. The α-helix N that contains the minor groove recognition amino acids is shown in red. The two guanines modified by cisplatin are shown in magenta. The primer terminus dC is shown in green. B, overall structure of the unmodified GG·dCTP* ternary complex (Protein Data Bank code 4TUQ). The incoming non-hydrolyzable dCTP* is shown in green. The active site Mg2+ ions are shown in green spheres. C, overall structure of the Pt-GG·dCTP*-Mg2+ ternary complex (Protein Data Bank code 4TUR). D, DNA sequences used for crystallographic studies. The site opposite the 5′-dG of the Pt-GG is indicated by an arrow. E, active site of the GG gapped binary complex structure. The distance between N2 of the templating dG and the α-helix N is indicated. F, active site of the GG·dCTP* ternary complex structure. The distance between N2(5′-dG) and N2(3′-dG) is indicated. G, active site of the Pt-GG·dCTP*-Mg2+ ternary complex structure. Platinum and NH3 of the Pt-GG lesion are shown in gray and blue spheres, respectively. The cisplatination of the GG increases the distance between N2(5′-dG) and N2(3′-dG).
For comparison, we determined crystal structures of a single nucleotide gapped binary complex and a ternary complex with the templating GG-containing DNA (Fig. 2D). As expected, the overall structures of the gapped binary GG complex and the ternary GG·dCTP* complex are essentially identical to those of the published gapped binary and ternary polβ complexes, respectively (Fig. 2, A and B; see Table 1 for refinement statistics) (31). The root mean square deviation (r.m.s.d.) between our GG·dCTP* ternary complex (Protein Data Bank code 4TUQ) and the published GG·dCTP* ternary complex (Protein Data Bank code 4KLE (31)) is 0.325 Å, indicating that polβ ternary structures are not sequence-dependent. The unmodified GG gapped binary complex adopts an open protein conformation where the α-helix N that contains the minor groove edge recognition amino acids is ∼10 Å away from the active site (Fig. 2E). The incorporation of dCTP opposite the templating unmodified dG induces a large conformational change of the protein to form the GG·dCTP* ternary complex with a closed protein conformation where the replicating base pair is sandwiched between the primer terminus base pair and the α-helix N (Fig. 2F). In contrast, the Pt-GG·dCTP*-Mg2+ ternary complex structure, refined to 2.2-Å resolution, shows that the incorporation of dCTP opposite the templating 5′-dG of the Pt-GG induces only a minor conformational change in the protein. The Pt-GG·dCTP*-Mg2+ complex adopts a “semiopen” protein conformation (Fig. 2, C and G) where the α-helix N shifts toward the active site ∼2 Å relative to the position observed in the GG gapped binary structure. The Pt-GG·dCTP*-Mg2+ ternary complex is structurally more similar to the unmodified GG binary complex (r.m.s.d. = 0.556 Å) than the GG·dCTP* ternary complex (r.m.s.d. = 1.396 Å; see Fig. 3E).
TABLE 1.
Data collection and refinement statistics
| Protein Data Bank code | GG gapped binary | GG·dCTP Mg2+ ternary | Pt-GG·dCTP Mg2+ ternary | Pt-GG·dCTP Mn2+ ternary |
|---|---|---|---|---|
| 4TUP | 4TUQ | 4TUR | 4TUS | |
| Data collection | ||||
| Space group | P21 | P21 | P21 | P21 |
| Cell constants | ||||
| a (Å) | 54.354 | 50.820 | 54.785 | 54.878 |
| b (Å) | 79.257 | 80.542 | 79.335 | 78.417 |
| c (Å) | 54.927 | 55.535 | 54.786 | 54.803 |
| α (°) | 90.00 | 90.00 | 90.00 | 90.00 |
| β (°) | 105.42 | 107.69 | 107.81 | 112.80 |
| γ (°) | 90.00 | 90.00 | 90.00 | 90.00 |
| Resolution (Å)a | 20-1.80 (1.83-1.80) | 20-2.37 (2.41-2.37) | 20-2.17 (2.21-2.17) | 20-2.42 (2.42-2.46) |
| Rmergeb (%) | 0.068 (0.347) | 0.105 (0.485) | 0.095 (0.476) | 0.101 (0.544) |
| I/σ | 26.4 (2.89) | 15.7 (2.30) | 22.8 (2.18) | 17.7 (1.96) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (99.8) | 99.9 (99.1) |
| Redundancy | 4.7 (4.1) | 4.4 (4.2) | 5.4 (4.3) | 4.6 (4.0) |
| Refinement | ||||
| Rworkc/Rfreed (%) | 20.5/23.6 | 20.1/25.9 | 20.2/25.6 | 20.0/24.3 |
| Unique reflections | 41,437 | 17,394 | 23,644 | 16,369 |
| Mean B factor (Å2) | ||||
| Protein | 24.29 | 29.41 | 39.48 | 42.65 |
| Ligand | 25.35 | 36.79 | 36.33 | 44.28 |
| Solvent | 27.48 | 31.66 | 37.64 | 40.01 |
| Ramachandran plot | ||||
| Most favored (%) | 97.2 | 98.1 | 95.7 | 96.9 |
| Add.e allowed (%) | 2.8 | 1.9 | 4.3 | 3.1 |
| r.m.s.d. | ||||
| Bond lengths (Å) | 0.004 | 0.004 | 0.004 | 0.004 |
| Bond angles (°) | 0.822 | 1.044 | 1.130 | 1.120 |
a Values in parentheses are for the highest resolution shell.
b Rmerge = Σ|I − I|/ΣI where I is the integrated intensity of a given reflection.
c Rwork = Σ|F(obs) − F(calc)|/ΣF(obs).
d Rfree = Σ|F(obs) − F(calc)|/ΣF(obs), calculated using 5% of the data.
e Additionally.
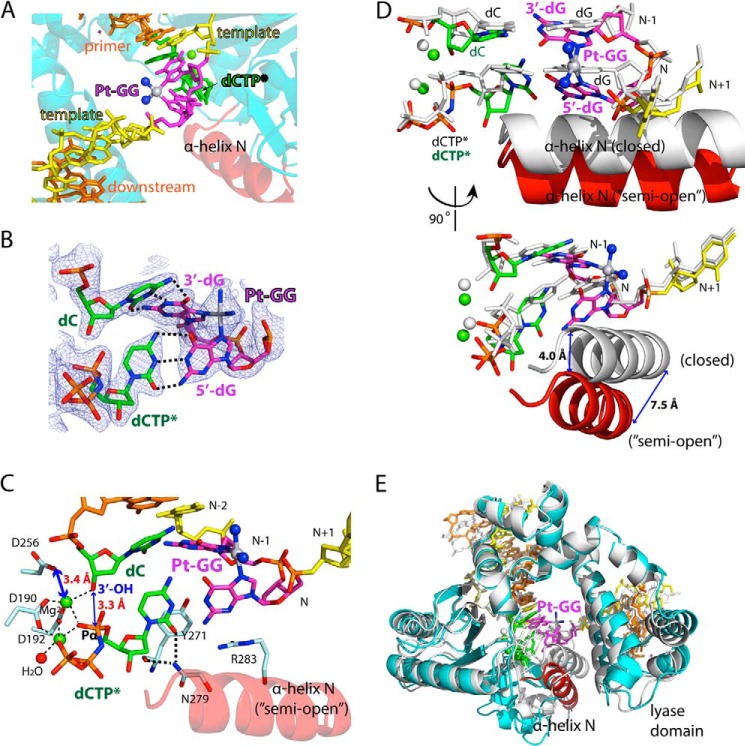
FIGURE 3.
Structure of the Pt-GG·dCTP*-Mg2+ ternary complex (Protein Data Bank code 4TUR). A, close-up view of the Pt-GG lesion in the polβ:Pt-GG-Mg2+ complex. Platinum and NH3 are shown in gray and blue spheres, respectively. B, a 2Fo − Fc electron density map contoured at 1σ around the primer terminus and the replicating base pairs of the Pt-GG lesion. C, close-up view of the active site of the Pt-GG-Mg2+ complex. The three catalytic aspartates (Asp-190, Asp-192, and Asp-256) and the minor groove-interacting amino acids (Tyr-271, Asn-279, and Arg-283) are indicated. D, overlay of the active sites of the Pt-GG·dCTP*-Mg2+ (multicolored) and the unmodified GG·dCTP*-Mg2+ (white) ternary structures. E, overlay of the overall structures of the Pt-GG·dCTP*-Mg2+ complex (multicolored; Protein Data Bank code 4TUR) and the unmodified GG·dCTP*-Mg2+ complex (white; Protein Data Bank code 4TUQ).
The Pt-GG·dCTP*-Mg2+ ternary complex structure explains why polβ accurately but slowly incorporates nucleotide opposite the bulky Pt-GG intrastrand cross-link adduct (Fig. 3). The DNA-distorting Pt-GG lesion is located in an open region and is accommodated in the polβ active site without any steric clashes with the protein (Fig. 3, A and B). The conformational perturbation of DNA caused by the Pt-GG lesion is mainly confined to the primer terminus and the replicating base pairs (Fig. 3, C and E), which have a large roll angle (∼60°) and an increased distance between N2 of 3′-dG and N2 of 5′-dG (7.6 Å) compared with that in the unmodified complex (3.8 Å). The large roll angle induced by the Pt-GG adduct destacks the two guanine bases. The most striking feature of the Pt-GG·dCTP*-Mg2+ structure is that, despite the large roll angle, both the 3′-dG and 5′-dG of the Pt-GG form Watson-Crick base pairing with the primer terminus dC and the incoming dCTP*, respectively. The primer terminus and the replicating base pairs have large propeller twist angles (22° and 17°, respectively), which promote the accommodation of the bulky Pt-GG lesion with a relaxed conformation in the polβ active site while retaining Watson-Crick base pairing. To base pair with the templating 5′-dG, the incoming nucleotide does not stack with the primer terminus. The platinum is positioned near N7 of the 5′-dG found in the GG ternary complex (Fig. 3D), pushing the templating dG of the Pt-GG lesion toward the minor groove. Tyr-271 and Arg-283, which typically interact with the minor groove edges of the primer terminus and the templating base, respectively, in the polβ structure with correct insertion (22), do not engage in the minor groove interaction (Fig. 3C). The primer terminus 3′-OH is 3.3 Å away from the Pα of the incoming dCTP* and is aligned with the Pα of dCTP* at an angle of ∼160° for the nucleotidyl transfer reaction. The Pt-GG·dCTP*-Mg2+ complex shows the presence of the two active site metal ions, but the catalytic carboxylate Asp-256 is not coordinated to the catalytic metal ion (3.4 Å), which will decrease the insertion efficiency of dCTP opposite the Pt-GG lesion. Hence, the Pt-GG·dCTP*-Mg2+ complex structure with a semiopen protein conformation explains why insertion opposite the templating 5′-dG of the Pt-GG lesion is much less efficient than the dCTP insertion opposite unmodified dG. Taken together, the Pt-GG·dCTP*-Mg2+ structure provides mechanistic insights into the accurate yet inefficient nucleotide incorporation opposite the Pt-GG lesion by polβ.
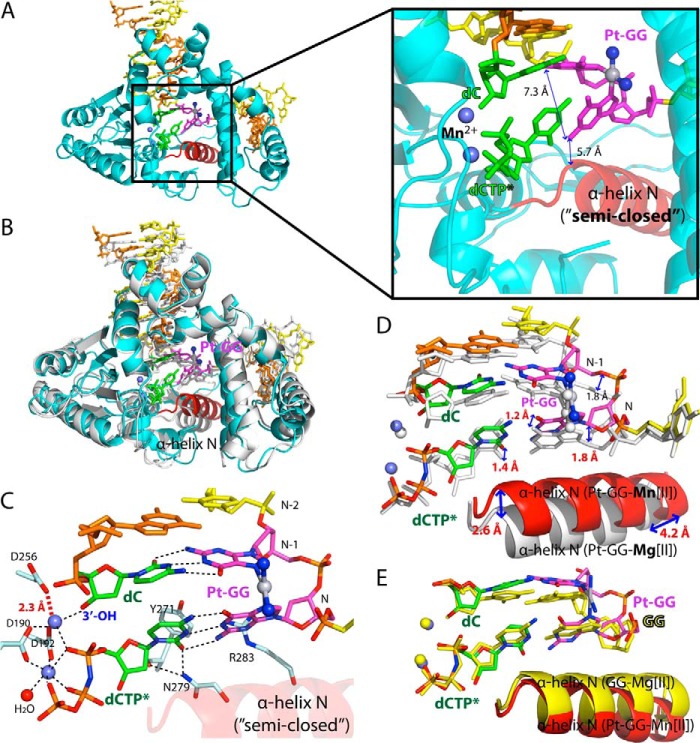
The Pt-GG·dCTP*-Mg2+ structure with a semiopen protein conformation most likely represents a ground state that requires a further conformational change to adopt a catalytically favorable conformation. To gain further insight into how polβ inserts dCTP opposite the 5′-dG of the Pt-GG lesion, we determined a ternary structure of the Pt-GG·dCTP* complex in the presence of the active site Mn2+, which has been shown to increase the insertion efficiency for various DNA polymerases (23, 31, 36). The Pt-GG·dCTP*-Mn2+ structure, refined to 2.4-Å resolution, is substantially different from the Pt-GG·dCTP*-Mg2+ structure (r.m.s.d. = 1.418 Å; Fig. 4A) and is quite similar to the GG·dCTP* ternary structure (r.m.s.d. = 0.672 Å). In the Mn2+-bound structure, Asp-256 is now coordinated to the catalytic metal ion (2.3 Å), completing the coordination of the catalytic metal ion. In contrast to the Pt-GG·dCTP*-Mg2+ structure, the protein adopts a nearly closed or slightly open conformation (Fig. 4B). In particular, the α-helix N shifts toward the nascent base pair ∼2.6–4.2 Å from the position observed in the Pt-GG·dCTP*-Mg2+ structure (Fig. 4D), sandwiching the replicating base pair between the primer terminus base pair and the α-helix N. The conformational reorganization of α-helix N triggers an upstream shift (∼1.5 Å) of the template:primer duplex DNA including the replicating and the primer terminus base pairs but does not induce any significant conformational change of the Pt-GG adduct (Fig. 4D). As observed in the Pt-GG·dCTP*-Mg2+ structure, the roll angle for the Pt-GG base pairs is ∼60°, and both guanines of the Pt-GG engage in Watson-Crick base pairing with the primer terminus dC and the incoming nucleotide dCTP*. The propeller twist angles for the primer terminus and the nascent base pairs (17° and 3°, respectively) are smaller than those observed in the Pt-GG·dCTP*-Mg2+ structure (22° and 17°, respectively). The non-stacked conformation of the Pt-GG lesion appears to prevent the formation of the fully closed protein conformation (Fig. 4E) and the engagement of Tyr-271 and Arg-283 in the minor groove edge interaction. The primer terminus 3′-OH is 3.6 Å away from the Pα of the incoming nucleotide, which is poised for the nucleotidyl transfer reaction. Taken together, the Pt-GG·dCTP*-Mn2+ structure suggests that the polβ catalysis across the Pt-GG lesion may occur through a “semiclosed” protein conformation, which is suboptimal for nucleotidyl transfer.
FIGURE 4.
Structure of the polβ:Pt-GG·dCTP*-Mn2+ ternary complex (Protein Data Bank code 4TUS). A, overall structure of the Pt-GG·dCTP*-Mn2+ ternary complex. B, overlay of the Pt-GG·dCTP*-Mg2+ (white) and the Pt-GG·dCTP*-Mn2+ (cyan) structures. C, close-up view of the active site region of the Pt-GG·dCTP*-Mn2+ structure. D, overlay of the active site region of the Pt-GG·dCTP*-Mg2+ (white) and the Pt-GG·dCTP*-Mn2+ (multicolored) structures. E, comparison of the unmodified GG·dCTP*-Mg2+ (yellow) and the Pt-GG·dCTP*-Mn2+ (multicolored) structures.
To evaluate the effect of the active site metal ion on the efficiency of dCTP insertion opposite the Pt-GG lesion, we determined kinetic parameters for dCTP incorporation opposite the templating dG and Pt-GG (Table 2). In the presence of the active site Mg2+, the relative efficiency for the dCTP insertion opposite the Pt-GG was ∼90-fold lower than that opposite dG, which is consistent with the Pt-GG·dCTP*-Mg2+ structure with the semiopen conformation and the incomplete active site metal ion coordination. The substitution of Mn2+ for Mg2+ increased the relative efficiency 8-fold, which is consistent with the Pt-GG·dCTP*-Mn2+ structure with the nearly closed conformation and the complete active site metal ion coordination. Mn2+ is known to be mutagenic (25, 37) and has been shown to increase the misincorporation rate and replication infidelity of various DNA polymerases (32, 33).
TABLE 2.
Steady-state kinetic parameters for dCTP insertion opposite the templating dG by polβ
| Template·dCTP | Metal ion | Km | kcat | kcat/Km | f a |
|---|---|---|---|---|---|
| μm | 10−3 s−1 | 10−3 s−1 μm−1 | |||
| dG·dCTP | Mg2+ | 0.67 ± 0.03 | 164.30 ± 6.65 | 245.2 | 1 |
| Pt-GG·dCTP | Mg2+ | 5.22 ± 1.01 | 15.76 ± 1.24 | 3.0 | 1.2 × 10−2 |
| Pt-GG·dCTP | Mn2+ | 1.14 ± 0.05 | 27.60 ± 1.62 | 24.2 | 9.9 × 10−2 |
a f = (kcat/Km)[Pt-GG·dCTP]/(kcat/Km)(dG·dCTP).
Comparison of the polβ:Pt-GG structures with other DNA polymerase:Pt-GG structures reveals that the replication across the 5′-dG of the Pt-GG adduct by the X-family DNA polymerase and the Y-family DNA polymerase occurs through distinct mechanisms. Whereas the conformation of a polη:Pt-GG·dCTP-Mg2+ ternary complex and the normal polη ternary complexes are similar (r.m.s.d. = 0.3–0.4 Å) (17), the conformations of the polβ:Pt-GG·dCTP-Mg2+ ternary complex and the polβ:GG·dCTP-Mg2+ ternary complex are dissimilar (r.m.s.d. = 1.396 Å), which partially explains why polη is more efficient at bypassing the Pt-GG than polβ. The structural comparison also reveals that the conformations of the Pt-GG adduct in the complexes of the Y-family DNA polymerases polη (17) and Dpo4 (18) are significantly different from that of the X-family DNA polymerase polβ (Fig. 5). The roll angles for the Pt-GG base pairs in the Sulfolobus solfataricus Dpo4 (∼20°) and human polη (∼30°) complexes are smaller than that in the polβ complex (∼60°), and the distance between N2(5′-dG) and N2(3′-dG) of the Pt-GG lesion for the Dpo4 (4.6 Å) and polη (5.2 Å) structures is shorter than that for the polβ structure (7.3 Å). In the Y-family DNA polymerase:Pt-GG complex structures, the incoming nucleotide stacks with the primer terminus, which is in contrast to the polβ:Pt-GG complex structures. The conformational differences in the Pt-GG lesions of the X-family and Y-family DNA polymerases appear to stem from the difference in the local environment of the Pt-GG lesion. In the case of the Y-family DNA polymerase, the Pt-GG lesion resides in an open cleft created by the finger domain and the polymerase-associated domain (also known as little finger) (19). The finger domain interacts with the replicating base pair, and the polymerase-associated domain interacts with the phosphate backbone of the templating Pt-GG lesion, thereby promoting the stacked Pt-GG conformation with a relatively small roll angle in the active site of the enzyme. In the case of the X-family DNA polymerase polβ, which lacks the polymerase-associated domain, the templating Pt-GG lesion does not experience any significant geometric constraints imposed by the protein. In addition, whereas the finger domains of polη and Dpo4 are relatively rigid, the α-helix N in the finger domain of polβ is flexible, which enables the polβ active site to accommodate the Pt-GG cross-link with the relaxed conformation and a large roll angle.
FIGURE 5.
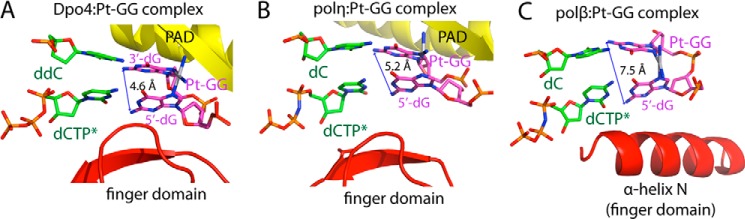
Structural comparison of the polymerase:Pt-GG·dCTP complexes. A, structure of Dpo4 in complex with the Pt-GG-containing DNA. The finger domain and the polymerase-associated domain (PAD) are shown. The distance between N2(5′-dG) and N2(3′-dG) of the Pt-GG lesion is indicated. B, structure of human polη in complex with the Pt-GG DNA. C, structure of polβ in complex with the Pt-GG DNA in the presence of the active site Mn2+. The α-helix N that interacts with the replicating base pair is shown.
In summary, our studies provide the structural basis for the mechanism by which polβ performs the accurate but slow replication across the major cisplatin-DNA adduct. Polβ accommodates the bulky templating Pt-GG lesion within its active site without any steric hindrance and allows the formation of Watson-Crick base pairing between the Pt-GG and the incoming dCTP, thereby accurately bypassing the Pt-GG lesion. When complexed with the templating Pt-GG, polβ adopts a catalytically suboptimal conformation, resulting in the slow replication across the Pt-GG adduct. Overall, the polβ:Pt-GG·dCTP* complex structures presented here may contribute to understanding of the mechanism of the potential polβ-mediated cisplatin resistance in polβ-overexpressing cancer cells.
Acknowledgments
Instrumentation and technical assistance for this work were provided by the Macromolecular Crystallography Facility with financial support from the College of Natural Sciences, the Office of the Executive Vice President and Provost, and the Institute for Cellular and Molecular Biology at the University of Texas at Austin. The Berkeley Center for Structural Biology is supported in part by the National Institute of General Medical Sciences. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences of the United States Department of Energy under Contract DE-AC02-05CH11231.
This work was supported by the College of Pharmacy at the University of Texas at Austin and Cancer Prevention and Research Institute of Texas Grant RP130219.
The atomic coordinates and structure factors (codes 4TUP, 4TUQ, 4TUR, and 4TUS) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Pt-GG
- cisplatin-1,2-d(GpG)
- pol
- polymerase
- Dpo4
- DNA polymerase IV
- dCTP* or dCMPNPP
- 2′-deoxycytidine 5′-[(α,β)-imido]triphosphate
- dUMPNPP
- 2′-deoxyuridine 5′-[(α,β)-imido]triphosphate
- dAMPCPP
- α,β-methylene-dATP
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1. Jung Y., Lippard S. J. (2007) Direct cellular responses to platinum-induced DNA damage. Chem. Rev. 107, 1387–1407 [DOI] [PubMed] [Google Scholar]
- 2. Rabik C. A., Dolan M. E. (2007) Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat. Rev. 33, 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chaney S. G., Campbell S. L., Bassett E., Wu Y. (2005) Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit. Rev. Oncol. Hematol. 53, 3–11 [DOI] [PubMed] [Google Scholar]
- 4. Xie K., Doles J., Hemann M. T., Walker G. C. (2010) Error-prone translesion synthesis mediates acquired chemoresistance. Proc. Natl. Acad. Sci. U.S.A. 107, 20792–20797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vaisman A., Chaney S. G. (2000) The efficiency and fidelity of translesion synthesis past cisplatin and oxaliplatin GpG adducts by human DNA polymerase β. J. Biol. Chem. 275, 13017–13025 [DOI] [PubMed] [Google Scholar]
- 6. Hoffmann J. S., Pillaire M. J., Maga G., Podust V., Hübscher U., Villani G. (1995) DNA polymerase β bypasses in vitro a single d(GpG)-cisplatin adduct placed on codon 13 of the HRAS gene. Proc. Natl. Acad. Sci. U.S.A. 92, 5356–5360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Srivastava D. K., Husain I., Arteaga C. L., Wilson S. H. (1999) DNA polymerase β expression differences in selected human tumors and cell lines. Carcinogenesis 20, 1049–1054 [DOI] [PubMed] [Google Scholar]
- 8. Canitrot Y., Frechet M., Servant L., Cazaux C., Hoffmann J. S. (1999) Overexpression of DNA polymerase β: a genomic instability enhancer process. FASEB J. 13, 1107–1111 [DOI] [PubMed] [Google Scholar]
- 9. Canitrot Y. (2000) Nucleotide excision repair DNA synthesis by excess DNA polymerase β: a potential source of genetic instability in cancer cells. FASEB J. 14, 1765–1774 [DOI] [PubMed] [Google Scholar]
- 10. Bergoglio V., Canitrot Y., Hogarth L., Minto L., Howell S. B., Cazaux C., Hoffmann J. S. (2001) Enhanced expression and activity of DNA polymerase β in human ovarian tumor cells: impact on sensitivity towards antitumor agents. Oncogene 20, 6181–6187 [DOI] [PubMed] [Google Scholar]
- 11. Canitrot Y., Cazaux C., Fréchet M., Bouayadi K., Lesca C., Salles B., Hoffmann J. S. (1998) Overexpression of DNA polymerase β in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs. Proc. Natl. Acad. Sci. U.S.A. 95, 12586–12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kraker A. J., Moore C. W. (1988) Elevated DNA polymerase β activity in a cis-diamminedichloroplatinum(II) resistant P388 murine leukemia cell line. Cancer Lett. 38, 307–314 [DOI] [PubMed] [Google Scholar]
- 13. Horton J. K., Srivastava D. K., Zmudzka B. Z., Wilson S. H. (1995) Strategic down-regulation of DNA polymerase β by antisense RNA sensitizes mammalian cells to specific DNA damaging agents. Nucleic Acids Res. 23, 3810–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boudsocq F., Benaim P., Canitrot Y., Knibiehler M., Ausseil F., Capp J. P., Bieth A., Long C., David B., Shevelev I. (2005) Modulation of cellular response to cisplatin by a novel inhibitor of DNA polymerase β. Mol. Pharmacol. 67, 1485–1492 [DOI] [PubMed] [Google Scholar]
- 15. Vaisman A., Masutani C., Hanaoka F., Chaney S. G. (2000) Efficient translesion replication past oxaliplatin and cisplatin CpG adducts by human DNA polymerase η. Biochemistry 39, 4575–4580 [DOI] [PubMed] [Google Scholar]
- 16. Alt A., Lammens K., Chiocchini C., Lammens A., Pieck J. C., Kuch D., Hopfner K.-P., Carell T. (2007) Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase η. Science 318, 967–970 [DOI] [PubMed] [Google Scholar]
- 17. Zhao Y., Biertümpfel C., Gregory M. T., Hua Y.-J., Hanaoka F., Yang W. (2012) Structural basis of human DNA polymerase η-mediated chemoresistance to cisplatin. Proc. Natl. Acad. Sci. U.S.A. 109, 7269–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wong J. H., Brown J. A., Suo Z., Blum P., Nohmi T., Ling H. (2010) Structural insight into dynamic bypass of the major cisplatin-DNA adduct by Y-family polymerase Dpo4. EMBO J. 29, 2059–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ummat A., Rechkoblit O., Jain R., Roy Choudhury J., Johnson R. E., Silverstein T. D., Buku A., Lone S., Prakash L., Prakash S. (2012) Structural basis for cisplatin DNA damage tolerance by human polymerase η during cancer chemotherapy. Nat. Struct. Mol. Biol. 19, 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reissner T., Schneider S., Schorr S., Carell T. (2010) Crystal structure of a cisplatin-(1,3-GTG) cross-link within DNA polymerase η. Angew. Chem. Int. Ed. Engl. 49, 3077–3080 [DOI] [PubMed] [Google Scholar]
- 21. Wang D., Hara R., Singh G., Sancar A., Lippard S. J. (2003) Nucleotide excision repair from site-specifically platinum-modified nucleosomes. Biochemistry 42, 6747–6753 [DOI] [PubMed] [Google Scholar]
- 22. Sawaya M. R., Prasad R., Wilson S. H., Kraut J., Pelletier H. (1997) Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry 36, 11205–11215 [DOI] [PubMed] [Google Scholar]
- 23. Koag M. C., Lee S. (2014) Metal-dependent conformational activation explains highly promutagenic replication across O6-methylguanine by human DNA polymerase β. J. Am. Chem. Soc. 136, 5709–5721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vagin A., Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 25. Pelletier H., Sawaya M. R., Wolfle W., Wilson S. H., Kraut J. (1996) Crystal structures of human DNA polymerase β complexed with DNA: implications for catalytic mechanism, processivity, and fidelity. Biochemistry 35, 12742–12761 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Batra V. K., Beard W. A., Shock D. D., Krahn J. M., Pedersen L. C., Wilson S. H. (2006) Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure 14, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chamberlain B. T., Batra V. K., Beard W. A., Kadina A. P., Shock D. D., Kashemirov B. A., McKenna C. E., Goodman M. F., Wilson S. H. (2012) Stereospecific formation of a ternary complex of (S)-α,β-fluoromethylene-dATP with DNA pol β. Chem. Bio. Chem. 13, 528–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Freudenthal B. D., Beard W. A., Shock D. D., Wilson S. H. (2013) Observing a DNA polymerase choose right from wrong. Cell 154, 157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Batra V. K., Beard W. A., Shock D. D., Pedersen L. C., Wilson S. H. (2008) Structures of DNA polymerase β with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell 30, 315–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tabor S., Richardson C. C. (1989) Effect of manganese ions on the incorporation of dideoxynucleotides by bacteriophage T7 DNA polymerase and Escherichia coli DNA polymerase I. Proc. Natl. Acad. Sci. U.S.A. 86, 4076–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Clausen A. R., Murray M. S., Passer A. R., Pedersen L. C., Kunkel T. A. (2013) Structure-function analysis of ribonucleotide bypass by B family DNA replicases. Proc. Natl. Acad. Sci. U.S.A. 110, 16802–16807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gregory M. T., Park G. Y., Johnstone T. C., Lee Y. S., Yang W., Lippard S. J. (2014) Structural and mechanistic studies of polymerase η bypass of phenanthriplatin DNA damage. Proc. Natl. Acad. Sci. U.S.A. 111, 9133–9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Vaisman A., Ling H., Woodgate R., Yang W. (2005) Fidelity of Dpo4: effect of metal ions, nucleotide selection and pyrophosphorolysis. EMBO J. 24, 2957–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. El-Deiry W. S., Downey K. M., So A. G. (1984) Molecular mechanisms of manganese mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 81, 7378–7382 [DOI] [PMC free article] [PubMed] [Google Scholar]