Background: BMP signaling regulates expression of the odontogenic gene Msx1 in dental mesenchyme.
Results: Canonical BMP signaling is not operating in early developing tooth, and Smad1/5 but not Smad4 are required for BMP-induced Msx1 expression in dental mesenchymal cells.
Conclusion: BMP/Smads regulation of Msx1 expression is Smad4-independent.
Significance: A novel atypical canonical BMP signaling is identified to regulate tooth development.
Keywords: Bone Morphogenetic Protein (BMP), Cell Signaling, Craniofacial Development, Gene Expression, SMAD Transcription Factor, Dental Mesenchyme, Msx1, Smad4-independent
Abstract
Bone morphogenetic protein (BMP) signaling plays an essential role in early tooth development, evidenced by disruption of BMP signaling leading to an early arrested tooth development. Despite being a central mediator of BMP canonical signaling pathway, inactivation of Smad4 in dental mesenchyme does not result in early developmental defects. In the current study, we investigated the mechanism of receptor-activated Smads (R-Smads) and Smad4 in the regulation of the odontogenic gene Msx1 expression in the dental mesenchyme. We showed that the canonical BMP signaling is not operating in the early developing tooth, as assessed by failed activation of the BRE-Gal transgenic allele and the absence of phospho-(p)Smad1/5/8-Smad4 complexes. The absence of pSmad1/5/8-Smad4 complex appeared to be the consequence of saturation of Smad4 by pSmad2/3 in the dental mesenchyme as knockdown of Smad2/3 or overexpression of Smad4 led to the formation of pSmad1/5/8-Smad4 complexes and activation of canonical BMP signaling in dental mesenchymal cells. We showed that Smad1/5 but not Smad4 are required for BMP-induced expression of Msx1 in dental mesenchymal cells. We further presented evidence that in the absence of Smad4, BMPs are still able to induce pSmad1/5/8 nuclear translocation and their binding to the Msx1 promoter directly in dental mesenchymal cells. Our results demonstrate the functional operation of an atypical canonical BMP signaling (Smad4-independent and Smad1/5/8-dependent) pathway in the dental mesenchyme during early odontogenesis, which may have general implication in the development of other organs.
Introduction
It is well known that bone morphogenetic protein (BMP,3 belonging to the TGF-β superfamily) signaling is a fundamental regulator that controls organogenesis including tooth morphogenesis (1). Several Bmp genes, including Bmp2, Bmp3, Bmp4, and Bmp7, are expressed in either the epithelial or the mesenchymal component of the developing tooth (2). Among these Bmp genes, Bmp4 is the first identified signal mediating the inductive interaction between the epithelium and mesenchyme (3), and it has been suggested to play a central role as a morphogen during early tooth development (4, 5). Bmp4 expression is found initially in the dental epithelium and induces the expression of the odontogenic gene Msx1 in the dental mesenchyme at the initiation stage (E11.5) (3). At the subsequent bud stage, Msx1 further activates Bmp4 expression in the dental mesenchyme where Msx1 and Bmp4 form a positive regulatory loop that is required for the transition of the bud to the cap stage. This is evidenced by the arrested tooth development at the bud stage and loss of Bmp4 expression in the dental mesenchyme of Msx1 mutant and by the fact that ectopic transgenic Bmp4 expression partially rescued the tooth phenotype in the Msx1 mutant background (5–7).
The canonical TGF-β/BMP signaling pathway involves binding of ligands to the type I and type II transmembrane serine/threonine kinase receptor complex. With binding of ligand, the type II receptor activates the type I receptor by phosphorylation. The activated type I receptor phosphorylates receptor-activated Smads (R-Smads, including Smad1, Smad2, Smad3, Smad5, and Smad8) in cytoplasm. The BMP and anti-Müllerian hormone (AMH) type I receptors (ALK1, ALK2, ALK3/BMPRIA, ALK6/BMPRIB) phosphorylate Smad1, Smad5, and Smad8, whereas the type I receptors for TGF-β, activin, nodal, and myostatin (ALK4, ALK5, ALK7) phosphorylate Smad2 and Smad3 (8). These phosphorylated R-Smads bind to common Smad (Co-Smad, Smad4), forming transfer complexes to bring them into the nucleus and regulate target gene expression. In addition to the canonical signaling pathway, BMP can also activate Smad-independent mitogen-activated protein kinase (MAPK) signaling pathway, known as noncanonical signaling, including p38, ERK, and JNK pathways.
Although Smad4 is regarded the central mediator of the canonical BMP signaling pathway, it has been reported that phospho-(p)Smad1/5 are able to accumulate in the nucleus and transduce BMP signaling independently of Smad4 to the downstream target genes (9, 10). In addition, in the development of several organs, including the nervous system, lens, and bone, inactivation of Smad1/5 causes severe defects, but Smad4 inactivation gives rise to no or mild defects (10–13). These observations appear to challenge the current model of the canonical BMP signaling, which considers Smad4 a requisite mediator of this pathway by forming a complex with R-Smad to enter the nucleus and bind to target genes.
The mechanism by which BMP/Smad signaling controls early tooth germ development remains elusive. Although inactivation of BmprIa in the dental mesenchyme resulted in a severe developmental defect associated with dramatically down-regulated Msx1 expression (14), surprisingly, conditional inactivation of Smad4 in the dental mesenchyme by Osr2-Cre resulted in neither altered Msx1 expression nor observable early tooth developmental abnormality (15). These discrepancies prompted us to investigate the role of BMP-related R-Smads and Smad4 during early tooth development.
Here we provide evidence that during early tooth development, the canonical BMP signaling pathway is not operating in the dental mesenchyme. We also provide evidence that BMP-induced pSmad1/5 can enter the nucleus and regulate Msx1 expression directly in the dental mesenchyme in a Smad4-independent manner. We identified an atypical canonical BMP signaling pathway that is functionally operating in the dental mesenchyme during early odontogenesis.
EXPERIMENTAL PROCEDURES
Animals and Embryo Collection
The use of animals in this study was approved by the Institutional Animal Care and Use Committee (IACUC) of Tulane University. The generation of BRE-Gal and Smad4f/f mice has been described previously (16, 17). This Smad4f/f allele, upon Cre recombination, would lack exon 8 and resemble the Smad4-null allele (17, 18). All wild-type mice were on CD-1 background and purchased from Charles River Laboratories. Embryos were collected from timed-mated pregnant mice in ice-cold PBS, and tail sample from each embryo was subjected to PCR-based genotyping.
X-Gal Staining, Immunofluorescence, and in Situ Hybridization
For X-gal staining, samples were fixed in 4% paraformaldehyde, washed in ice-cold PBS, subsequently washed with 30% sucrose/PBS, embedded in O.C.T. (Tissue-Tek), and cryo-sectioned. Standard X-gal staining was conducted as described previously (19). For immunofluorescence and in situ hybridization on tissue section, samples were fixed with 4% paraformaldehyde at 4 °C overnight, dehydrated through graded ethanol series, and then processed for paraffin sectioning. Sections were subjected to standard immunofluorescence staining as described previously (20). For immunofluorescence on cell culture, cells grown on glass coverslips were fixed with 4% paraformaldehyde for 10 min and then permeabilized with 0.5% Triton-X for 15 min. The following primary antibodies were used: anti-Smad4 (Abcam), anti-pSmad1/5/8 (Cells Signaling), anti-pSmad2/3 (Santa Cruz Biotechnology), anti-pp38 (R&D Systems), anti-pERK (R&D Systems), anti-pJNK (R&D Systems), and anti-β-galactosidase (Abcam). Alexa Fluor 568 (Invitrogen) was used as secondary antibody. DAPI (Invitrogen) was used as nuclear counterstain. Section or whole-mount in situ hybridization using nonradioactive probes was performed as described previously (21).
Co-immunoprecipitation
Cultured cells were lysed in radioimmunoprecipitation assay buffer (Cell Signaling Technology) with protease inhibitor (Roche Diagnostics) for 15 min at 4 °C. For molar germs, tissue was digested with 0.25% trypsin (Life Technologies) at 37 °C for 10 min prior to lysis. Lysates were centrifuged at 12,000 rpm at 4 °C for 20 min to remove cellular debris. The supernatant was precleared with 40 μl of Dynabeads protein A/G (Life Technologies) at 4 °C for 30 min. Anti-Smad4 antibody or negative control IgG (Santa Cruz Biotechnology, 3 μg of each) was added to the precleared lysates following incubation at 4 °C overnight and then with 50 μl of Dynabeads protein A/G for 2 h to precipitate immunocomplexes. Beads were washed three times with wash buffer (10 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10% glycerol, 1% Nonidet P-40, and 2 mm EDTA). 50 μl of sample buffer were added to the beads, and samples were heated to 95 °C for 10 min. pSmad1/5/8 and pSmad2/3 were detected in the protein complex by Western blot.
Western Blot
Proteins were separated with SDS-PAGE gel and transferred to nitrocellulose membrane. The membrane was blocked in 5% nonfat milk for 30 min at room temperature with constant rocking and then incubated with anti-pSmad1/5/8, anti-pSmad2/3, anti-Smad4, anti-Smad1 (GeneTex), anti-Smad5 (GeneTex), anti-Smad8 (Santa Cruz Biotechnology), anti-Msx1 (R&D Systems), or anti-GAPDH (Santa Cruz Biotechnology) antibodies overnight at 4 °C with constant rocking. The membrane was then washed three times and incubated with IRDye® 800 secondary antibody (LI-COR) for 1 h. Immunoreactive bands were visualized with Odyssey® imaging system (LI-COR).
In Situ Proximity Ligation Assay (PLA)
The presence of endogenous pSmad1/5/8-Smad4 and pSmad2/3-Smad4 complexes were detected in situ using the in situ proximity ligation assay kit (Duolink kit, Sigma-Aldrich). Tissue sections or fixed and permeabilized cells cultured on glass chamber slides were blocked with Duolink blocking reagent. Samples were incubated with anti-Smad4 (mouse monoclonal) and anti-pSmad1/5/8 (rabbit monoclonal) or anti-pSmad2/3 (goat polyclonal) antibodies overnight at 4 °C. Samples were washed and incubated with secondary antibodies conjugated with oligonucleotides (anti-mouse PLA probe Minus and anti-rabbit PLA probe Plus or anti-goat PLA Plus) in a humidity chamber for 1 h at 37 °C. Samples were then incubated with a ligation solution at 37 °C for 30 min. Ligation of the oligonucleotides probes was followed by amplification reaction at 37 °C for 100 min. Slides were mounted with mounting medium containing DAPI nuclear stain (Sigma-Aldrich). PLA signals were detected using a fluorescence microscope.
Organ Culture
Mandibular molar germs were carefully isolated from staged embryos and placed in semisolid culture medium as described previously (22). For small molecule inhibition experiments, dorsomorphin (Sigma) or SB203580 (Cell Signaling) and U0126 (Cell Signaling) were added into the medium, respectively, at a final concentration of 20 μm. DMSO was used in control group.
Primary Dental Mesenchymal Cell Culture and Lentivirus Infection
Embryonic mandibular molar germs were dissected out and incubated with 2 units/ml Dispase (Life Technologies) in PBS for 30 min at room temperature. Dental epithelium was removed from mesenchyme with the aid of fine forceps. The isolated mesenchyme was then digested with 0.25% trypsin at 37 °C for 10 min and dissociated into a single-cell suspension by gentle pipetting. Cells were plated onto a 6-well plate and cultured in DMEM (Life Technologies), supplemented with 20% FBS (Life Technologies), at 37 °C and 5% CO2. The dental mesenchymal cells isolated from Smad4f/f (referred as Smad4f/f cells) and wild-type mice were infected with Cre-lentivirus particles (GenTarget Inc.) and mSmad4-lentivirus (Abm Inc.), respectively, according to the manufacturer's instructions. Smad4f/f cells infected with Cre-lentivirus (referred as Cre-Smad4f/f cells) were blasticidin-resistant. Seventy-two hours after infection, Cre-Smad4f/f cells were subjected to selection with blasticidin (Life Technologies). For stimulating experiments, cell were starved overnight in FBS-free medium and then were treated with recombinant mouse BMP-4 protein (R&D Systems) at a final concentration of 100 ng/ml and/or recombinant mouse TGFβ1 (R&D Systems) at a final concentration of 1 ng/ml.
RNA Interference
siRNA against mouse Smad1, Smad2, Smad3, Smad4, Smad5, Smad8, or negative control siRNA (all from Invitrogen) were transfected into cells at a final concentration of 30 nm using Lipofectamine® RNAiMAX (Invitrogen). Forty-eight hours after transfection, Western blot or immunofluorescence staining was performed to confirm interference efficiency, and infected cells were subjected to further experiments.
RT-PCR and Quantitative RT-PCR
Molar germs isolated from E12.5 or E13.5 embryos were subjected to RNA extraction with the RNase mini kit (Qiagen) and reversely transcribed using the SuperScript III first strand synthesis system (Invitrogen). RT-PCR was performed to examine the expression of Smad1 (forward: 5′-TCAGCAGAGGAGATGTTCAGGCAGTT-3′, and reverse: 5′-CCAAGGCAGAAGCGGTTCTTATTGTT-3′), Smad5 (forward: 5′-GTCCAGTCTTACCTCCAGTATTAGTGCC-3′, and reverse: 5′-CTCCTCATAGGCGACAGGCTGAACAT-3′), and Smad8 (forward: 5′-CGGCTTCACCGACCCTTCCAATAACA-3′, and reverse: 5′-TCGCTCACGCACTCCGCATACACCTC-3′). Quantitative RT-PCR was used to quantify the expression of Tgfβ1 (forward: 5′-CCTGAGTGGCTGTCTTTTGA-3′, and reverse: 5′-CGTGGAGTTTGTTATCTTTGCTG-3′), Tgfβ2 (forward: 5′-TGCTAACTTCTGTGCTGGG-3′, and reverse: 5′-GCTTCGGGATTTATGGTGTTG-3′), and Bmp4 (forward: 5′-ATTGCAGCTTTCTAGAGGTCC-3′, and reverse: 5′-GGGAGCCAATCTTGAACAAAC-3′) using SYBR Green (Applied Biosystems). GAPDH (forward: 5′-CGCCTGGAGAAACCTGCCAAGTATGA-3′, and reverse: 5′-TGGAAGAGTGGGAGTTGCTG-3′) was included as internal control.
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation assay was carried out according to the manufacturer's instructions using the ChIP assay kit (Millipore, Billerica, MA). Cells were cross-linked with 1% formaldehyde for 10 min, washed with cold PBS, and lysed in lysis buffer. Lysates were sonicated to shear DNA using an ultrasonic processor (Fisher Scientific). Supernatant was diluted 10-fold in ChIP dilution buffer. 10% of the diluted supernatant was kept as input DNA. After preclearing for 30 min with Dynabeads protein A/G/Salmon Sperm DNA, samples were incubated overnight at 4 °C with 3 μg of anti-pSmad1/5/8 antibody or negative control IgG. Dynabeads protein A/G/salmon sperm DNA was then added for 1 h at 4 °C to collect the immune complexes, which were then sequentially washed with low salt, high salt, and LiCl washing buffers and Tris-EDTA buffer for 5 min each with rotation at 4 °C. Immune complexes were eluted by the addition of elution buffer for 30 min with rotation at room temperature, and cross-links were reversed by the addition of 5 m NaCl and heating at 65 °C for 4 h. Samples were then incubated with 0.5 m EDTA, 1 m Tris-HCl, pH 6.5, and proteinase K for 1 h at 45 °C. DNA was recovered by phenol/chloroform extraction, precipitated with ethanol, and resuspended in 25 μl of water. The recovered DNA was analyzed by PCR using the following primers: forward, 5′-TTGGTAGAATCCACATCCAGGAGTGT-3′; reverse, 5′-GAGACAGCCCTGGATCATGGGTTTCG-3′, which covered the Msx1 promoter region from −537 to −8.
RESULTS
Canonical BMP Signaling Is Not Operating in the Early Developing Tooth
A previous study utilizing a canonical BMP pathway reporter transgenic allele, which harbors the BMP-response element (BRE) from the Id1 promoter and Gfp reporter, reported a lack of canonical BMP signaling activity in the early developing tooth germ (23). We revisited this observation by using another reporter mouse line harboring seven copies of BRE and the Id3 minimal promoter with LacZ reporter (BRE-Gal) that has been shown to serve as a faithful indicator of BMP canonical signaling activity in developing mouse embryo (16). However, this BRE-Gal reporter also failed to show canonical BMP activity in the early developing tooth (Fig. 1, A and B), but exhibited a high level of canonical BMP activity in the eye (Fig. 1B′), which was shown as a positive control, despite equally abundant levels of Smad4 and pSmad1/5/8 in both developing tooth germ and developing eye (Fig. 1, C–F′). These results further confirmed the lack of functional canonical BMP signaling in the early developing tooth.
FIGURE 1.
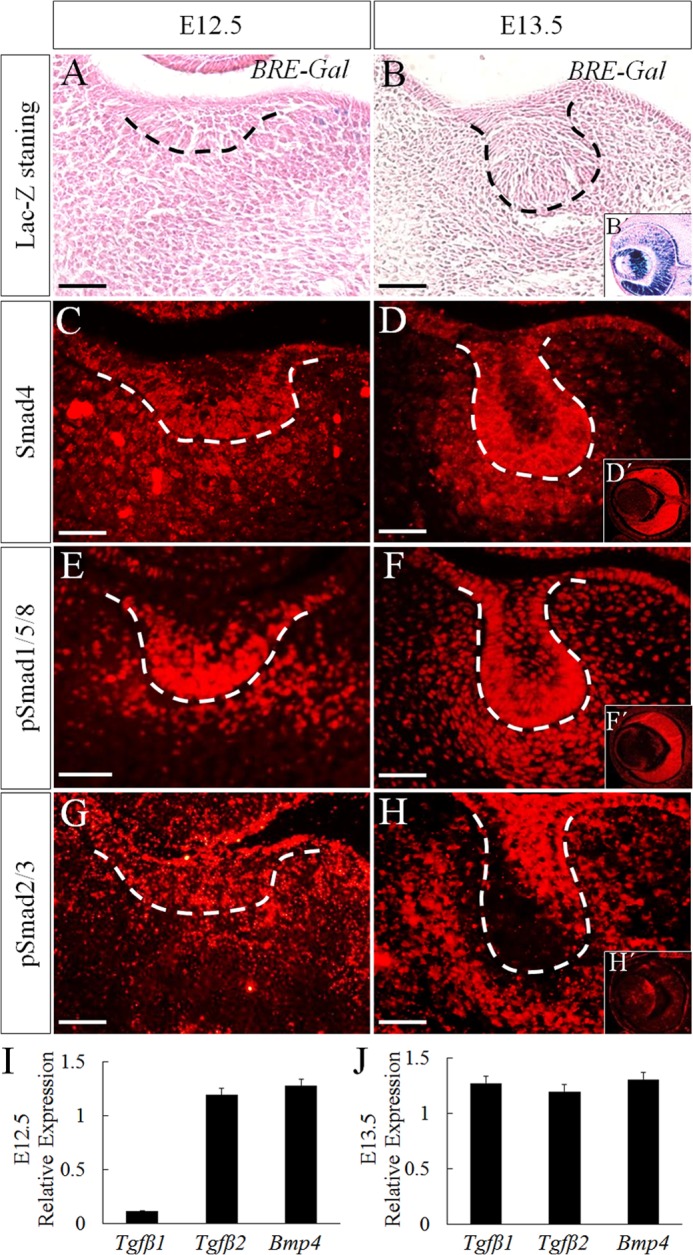
Canonical BMP signaling is not activated in early tooth development. A and B, x-gal staining of molar germs from BRE-Gal reporter embryos at E12.5 (A) and E13.5 (B). C–H, immunofluorescence shows expressions of Smad4 (C and D), pSmad1/5/8 (E and F), and pSmad2/3 (G and H) in wild-type tooth germs at E12.5 (C, E, and G) and E13.5 (D, F, and H). B′, D′, F′, and H′, eye tissue at E13.5. Bar = 50 μm. I and J, quantitative RT-PCR shows the relative expression levels of Tgfβ1, Tgfβ2, and Bmp4 at E12.5 (I) and E13.5 (J) tooth germ. Error bars indicate mean ± S.D.
Because activation of BRE needs the formation of pSmad1/5/8-Smad4 complex (16), we set out to determine whether the failed BRE-Gal activation in the tooth germ can be attributed to a lack of pSmad1/5/8-Smad4 complex by conducting a co-immunoprecipitation assay using E12.5 tooth germ and eye. Because TGFβ signaling is active in the developing tooth germ (24, 25) at comparable expression level with Bmp4 (Fig. 1, I and J), and pSmad2/3 are present abundantly in the tooth germ as well as in the developing eye, although at a relatively lower level (Fig. 1, G, H, and H′), we included pSmad2/3-Smad4 complexes as positive controls in the co-immunoprecipitation assay. Indeed, using Smad4 antibody, we were able to pull down abundant pSmad1/5/8 from the eye tissue, but the amounts of pSmad1/5/8 pulled down from the tooth germ were barely detectable on Western blot (Fig. 2A). However, abundant pSmad2/3-Smad4 complexes were detected in the tooth germ as compared with those in the eye (Fig. 2A). To further confirm these results, formation of pSmad1/5/8-Smad4 and pSmad2/3-Smad4 complexes was determined on tissue section using in situ PLA, a technology capable of detecting protein-protein interaction with high specificity and sensitivity in vivo (26, 27). PLA signals are detectable when two interacting proteins are in close proximity (<40 nm separation) (28). Although a small amount of pSmad1/5/8-Smad4 complexes was detected in the dental epithelium, no signals at all were seen in the mesenchyme (Fig. 2B, panel a). However, abundant pSmad2/3-Smad4 complexes were found in both dental epithelium and dental mesenchyme (Fig. 2B, panel b). Consistent with the expression of BRE-Gal and the presence of pSmad1/5/8-Smad4 and pSmad2/3-Smad4 complexes in the developing eye, PLA signals were constantly detected there (Fig. 2B, panels a′ and b′). These results suggest that Smad4 has a higher binding affinity with pSmad2/3 than pSmad1/5/8.
FIGURE 2.
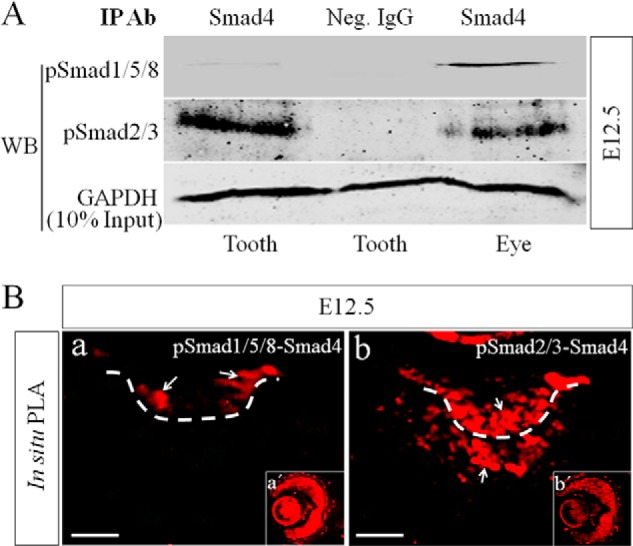
Lack of pSmad1/5/8-Smad4 complex but abundant pSmad2/3-Smad4 complexes are present in early dental mesenchyme. A, Western blot assay shows barely detectable pSmad1/5/8-Smad4 complexes but abundant pSmad2/3-Smad4 complexes in the precipitated immunocomplexes from E12.5 tooth germs. E12.5 eye samples were included as positive controls. IP Ab, immunoprecipitation antibody; Neg. IgG, negative control IgG; WB, Western blot. B, in situ PLA shows the presence of a small amount of pSmad1/5/8-Smad4 complexes in the dental epithelium (panel a) and abundant pSmad2/3-Smad4 (panel b) complexes in E12.5 tooth germ. Eyes were used as positive controls (panels a′ and b′). White arrows point to PLA signals (red fluorescence). Bar = 50 μm.
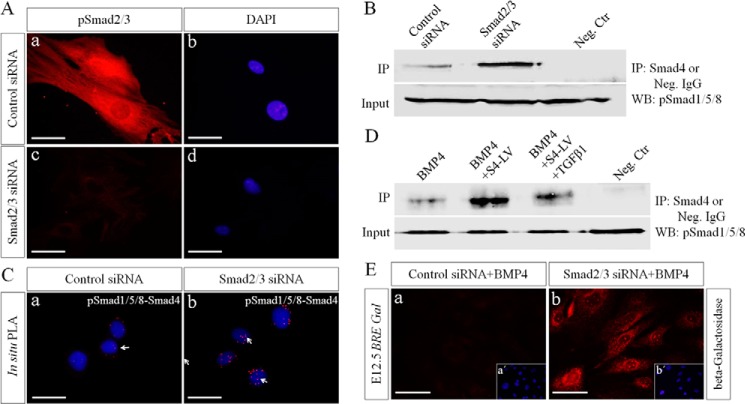
It was reported previously that pSmad2/3 were able to compete with pSmad1/5/8 for Smad4 (29–31). The absence of pSmad1/5/8-Smad4 complex in the early developing tooth germ could likely be the consequence of saturated Smad4 by pSmad2/3. To test this possibility, we knocked down Smad2 and Smad3 simultaneously using Smad2/3 siRNAs in primary dental mesenchymal cells from E12.5 molar germs. The knockdown efficiency of these siRNAs was confirmed by immunofluorescence (Fig. 3A). As shown in Fig. 3B, abundant pSmad1/5/8 proteins were immunoprecipitated by antibodies against Smad4 in cells transfected with Smad2/3 siRNAs, as compared with that in cells transfected with control siRNA. This result was further confirmed by in situ PLA (Fig. 3C). Because pSmad2/3 mainly mediates TGFβ signaling, Smad4 was overexpressed using lentivirus to confirm whether TGFβ was able to inhibit canonical BMP signaling by consuming Smad4 in dental mesenchymal cells. As shown in Fig. 3D, treatment with BMP4 alone was not able to lead to formation of pSmad1/5/8-Smad4 complexes, but overexpression of Smad4 did lead to formation of pSmad1/5/8-Smad4 complexes, which was reduced significantly by the addition of TGFβ1.
FIGURE 3.
Inhibition of Smad2/3 or overexpression of Smad4 leads to formation of pSmad1/5/8-Smad4 complexes and activation of BRE-Gal reporter in dental mesenchymal cells. Primary dental mesenchymal cells from E12.5 mouse embryos were transfected with control siRNA or Smad2/3 siRNA. A, immunofluorescence shows the expression of pSmad2/3 in cells transfected with control siRNA (panels a and b) and Smad2/3 siRNA (panels c and d). B, Western blot shows dramatically increased level of pSmad1/5/8-Smad4 complexes after silencing of Smad2/3 in dental mesenchymal cells. IP, immunoprecipitation; Neg. Ctr, negative control; Neg. IgG, negative control IgG; WB, Western blot. C, in situ PLA fails to detect pSmad1/5/8-smad4 complex in cells transfected with control siRNA (panel a) but shows the presence of pSmad1/5/8-Smad4 complexes in cells transfected with Smad2/3 siRNA (panel b). White arrows point to PLA signals (red fluorescence). D, Western blot shows dramatically increased level of pSmad1/5/8-Smad4 complexes after overexpression of Smad4 and significantly reduced level of these complexes after the addition of TGFβ1. S4-LV, Smad4 lentivirus. E, immunofluorescence using antibody against β-galactosidase shows absent BRE-Gal reporter activity in primary dental mesenchymal cells from E12.5 BRE-Gal in the presence of exogenous BMP4 (panel a), but indeed detects BRE-Gal expression in cells with Smad2/3knockdown (panel b). Insets (panels a′ and b′) are nuclear staining of these cells with DAPI. Bar = 50 μm.
To further elucidate whether these pSmad1/5/8-Smad4 complexes are able to activate canonical BMP signaling in dental mesenchymal cells, dental mesenchymal cells from E12.5 BRE-Gal mice were transfected with Smad2/3 siRNAs or control siRNAs followed by BMP4 induction. As determined by immunofluorescence shown in Fig. 3E, the expression of β-galactosidase driven by BRE, an indication of activated canonical BMP signaling, was constantly detected in cells with Smad2/3 knockdown, but not in cells only transfected with control siRNAs. All these results indicate that the canonical BMP signaling is inhibited by Smad2/3 in the dental mesenchyme during early tooth development.
Expression of Dental Mesenchymal Marker Msx1 Is Regulated by BMP/Smad1/5/8 Signaling Pathway
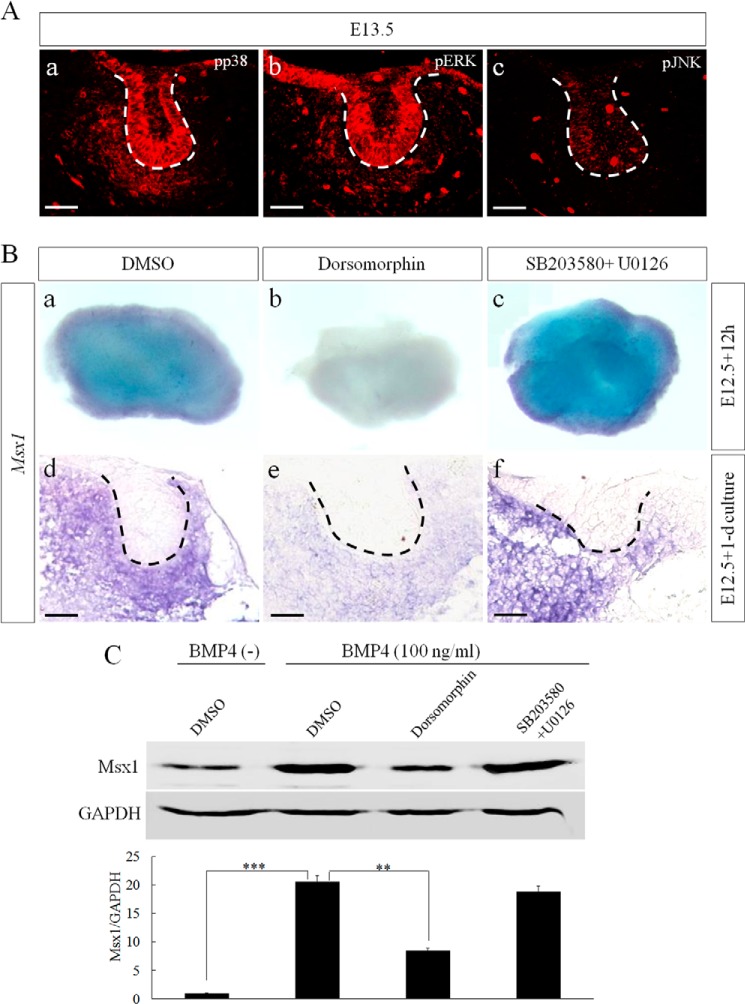
BMP signal transduces through Smads and MAPK signaling pathways. During early tooth development, both pp38 and pERK were highly expressed in the dental epithelium and mesenchyme, whereas pJNK was only expressed in the dental epithelium (Fig. 4A). Msx1 is a well characterized BMP signaling target gene and is required in the dental mesenchyme for early tooth development (5, 6, 32). However, the specific pathway that mediates the regulation of Msx1 by BMP signaling remains unknown. Dorsomorphin inhibits BMP-induced phosphorylation of Smad1/5/8 (33) and has been used to block BMP-Smad1/5/8 signaling during tooth development spatially and temporally (34). As shown in Fig. 4B, dorsomorphin-treated tooth germs exhibited dramatically down-regulated expression of Msx1 (Fig. 4B, panels b and e), whereas SB203580 (a specific p38 inhibitor)- and U0126 (a specific ERK inhibitor)-treated tooth germs (Fig. 4B, panels c and f) showed an Msx1 expression level comparable with that treated with DMSO (Fig. 4B, panels a and d). This inhibition of Msx1 expression by dorsomorphin but not SB203580 and U0126 was further confirmed in cultured primary dental mesenchymal cells induced by exogenous BMP4 (Fig. 4C). These results indicate that BMP-induced Msx1 expression is mediated by Smad1/5/8 pathway but not by MAPK.
FIGURE 4.
BMP/Smad1/5/8 signaling pathway regulates Msx1 expression. A, immunofluorescence shows expressions of pp38 (panel a), pERK (panel b), and pJNK (panel c) in E13.5 molar tooth germ. B, in situ hybridization shows Msx1 expression in E12.5 molar germs after 12 h or 1 day in organ culture in the presence of DMSO (panels a and d), dorsomorphin (panels b and e), or SB203580 and U0126 (panels c and f). Panels a–c, whole mount in situ hybridization; panels d–f, tissue section in situ hybridization. C, a Western blot assay shows attenuation of Msx1 expression by dorsomorphin but not SB203580 and U0126 in BMP-induced primary dental mesenchymal cells from E13.5 embryos. GAPDH was used as internal control. Quantitative analysis is shown in the lower part. **, p < 0.01; ***, p < 0.001. Bar = 50 μm. Error bars indicate mean ± S.D.
BMP-induced pSmad1/5/8 Nuclear Translocation Is Smad4-independent in Dental Mesenchymal Cells
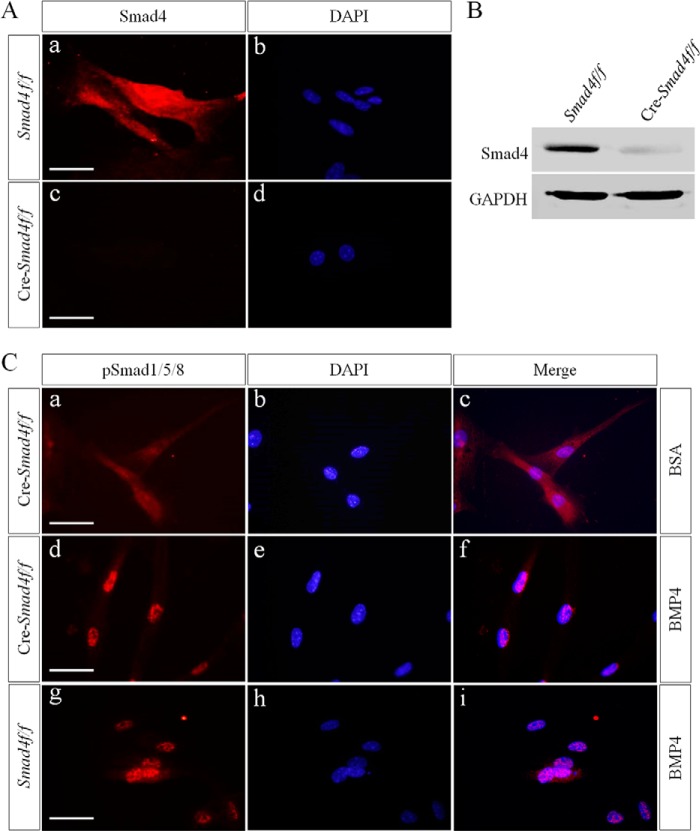
We showed that Smad1/5/8-mediated BMP signaling induces Msx1 expression in the dental mesenchyme and functional canonical BMP signaling and that pSmad1/5/8-Smad4 complexes are absent in the early developing tooth. These results prompted us to test the possibility that pSmad1/5/8 function to transduce BMP signaling in a Smad4-independent manner in the dental mesenchyme. As the BMP signaling effectors, activated Smad1/5/8 must be shuttled into the nucleus where they bind to the promoter of target genes and regulate gene expression. We first examined whether pSmad1/5/8 are able to enter the nucleus without Smad4 under BMP induction. To delete Smad4, suspended molar mesenchymal cells from E13.5 Smad4f/f embryos were cultured in tissue culture plates and infected with Cre-lentivirus particles. Knock-out efficiency was confirmed by immunofluorescence and Western blot, respectively (Fig. 5, A and B). In cells lacking Smad4 (Cre-Smad4f/f), pSmad1/5/8 proteins appeared primarily in the cytoplasm and faintly in the nucleus without exogenous BMP induction, whereas BMP treatment increased not only the expression level of pSmad1/5/8, but also their nuclear localization, similar to that in Smad4f/f cells (Fig. 5C). These results demonstrate that BMP-induced pSmad1/5/8 nuclear translocation is Smad4-independent in the dental mesenchymal cells, indicating operation of a functional atypical canonical BMP signaling pathway in the dental mesenchymal cells.
FIGURE 5.
BMP4-induced pSmad1/5/8 nuclear translocation is Smad4-independent in dental mesenchymal cells. Primary dental mesenchymal cells from R13.5 Smad4f/f mouse embryo were infected with Cre-lentivirus. A and B, knock-out efficiency of Smad4 was confirmed by immunofluorescence (A) and Western blot (B). C, immunofluorescence shows nuclear translocation of pSmad1/5/8 in cells lacking Smad4 after BMP4 stimulation for 30 min.
Smad4 Is Not Required but pSmad1/5 Are Required for BMP-induced Msx1 Expression in Dental Mesenchymal Cells
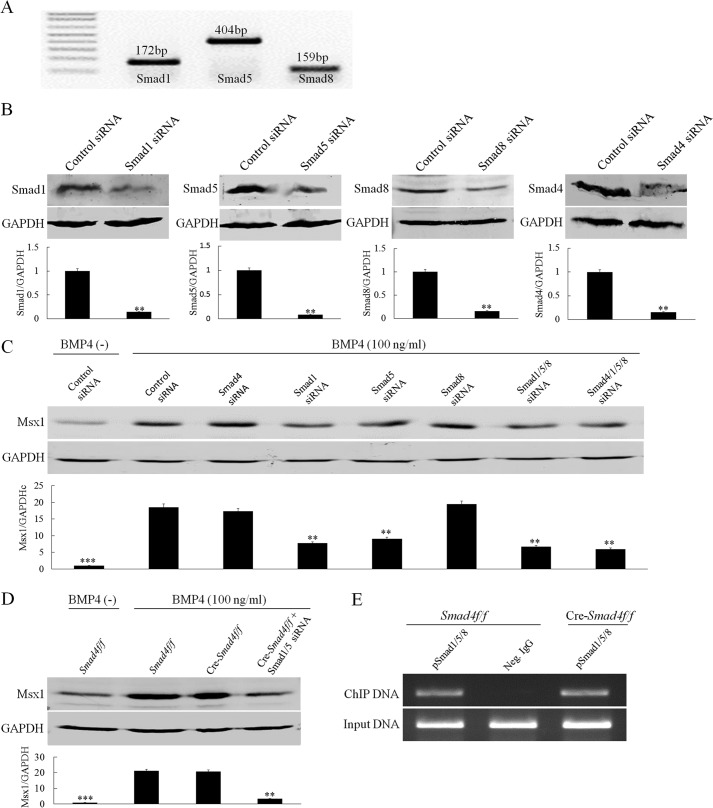
The application of pSmad1/5/8 inhibitor dorsomorphin dramatically down-regulated Msx1 expression in the dental mesenchyme in organ culture, indicating that Msx1 expression is regulated by Smad-dependent BMP signaling. We further tested whether pSmad1/5/8-mediated BMP signaling indeed regulates Msx1 expression independent of Smad4 in the dental mesenchyme. Although abundant pSmad1/5/8 were detected in the early developing tooth germ (Fig. 1, E and F), the antibody we used did not distinguish pSmad1, pSmad5, and pSmad8. To determine which R-Smad is involved in mediating BMP signaling in the developing dental mesenchyme, we conducted an RT-PCR assay and confirmed the expression of Smad1, Smad5, and Smad8 in the early tooth germ (Fig. 6A). We then conducted siRNA knockdown experiments in E13.5 molar mesenchymal cells using siRNAs for Smad1, Smad5, Smad8, and Smad4, respectively. The knockdown efficiencies of these siRNAs to their respective target genes are presented in Fig. 6B. As shown in Fig. 6C, Smad4 knockdown did not affect BMP4-induced Msx1 expression, but Smad1 or Smad5 silencing, respectively, did lead to down-regulation of Msx1 expression. However, Smad8 knockdown had little effect on Msx1 expression, suggesting that Smad1/5 are the major players in regulating Msx1 expression. Furthermore, simultaneously silencing of both Smad1 and Smad5 exhibited synergistic repressive effect on Msx1 expression. To further confirm that Smad4 was not required for BMP-induced Msx1 expression, Cre-Smad4f/f cells were used. As shown in Fig. 6D, BMP4-induced Msx1 expression was not affected by Smad4 deficiency in cells, but was dramatically reduced by Smad1/5 knockdown with siRNAs.
FIGURE 6.
Smad1/5 but not Smad4 are required for BMP-induced Msx1 expression in dental mesenchymal cells. A, RT-PCR shows the mRNA expressions of Smad1, Smad5, and Smad8 in E13.5 molar germs. B, Western blot shows knockdown efficiency of each siRNAs on their respective target genes in primary dental mesenchymal cells from E13.5 embryos. Quantitative analyses are shown in the lower part of each panel. C, a representative Western blot assay shows Msx1 expression in primary dental mesenchymal cells from E13.5 embryos. These cells were transfected with Smad1, Smad5, Smad8, or Smad4 siRNAs or control siRNA for 48 h followed by BMP4 treatment for additional 6 h. GAPDH was used as internal control. Quantitative analysis is shown in the lower part, in which the average value of Msx1/GAPDH from each individual group was compared with that from BMP4-induced control siRNA group. D, Western blot shows Msx1 protein levels in E13.5 Smad4f/f or Cre-Smad4f/f dental mesenchymal cells with or without transfection of Smad1/5 siRNAs after BMP4 induction for 6 h. Quantitative analysis is shown in the lower part, in which the average value of Msx1/GAPDH from Smad4f/f cells after BMP stimulation is set as the reference. E, a ChIP assay shows binding of pSmad1/5/8 to the Msx1 promoter in E13.5 dental mesenchymal cells in either the presence or the absence of Smad4. Neg. IgG, negative control IgG. **, p < 0.01; ***, p < 0.001. Error bars indicate mean ± S.D.
The mouse Msx1 promoter contains three potential Smad binding motifs (35). One of the binding motifs (from −234 to −240) locates very near to the transcription start site, a region designated as the basal promoter, and was confirmed to allow Smad1/5 binding directly (36). To determine whether pSmad1/5 binding to the Msx1 promoter is Smad4-independent in dental mesenchymal cells, we performed a ChIP assay. As expected, using primers covering the Msx1 promoter region from −537 to −8 that covers the proximal Smad binding motif, we were able to amplify the 520-bp PCR products from the DNA fragments immunoprecipitated by anti-pSmad1/5 antibody both in wild-type cells and in Cre-Smad4f/f cells (Fig. 6E), indicating that pSmad1/5 bind to the Msx1 promoter directly in a Smad4-independent manner.
DISCUSSION
In the present investigation, we provide compelling evidence that the canonical BMP signaling pathway is not operating during early tooth development due to the absence of Smad1/5/8-Smad4 complex. Most importantly, we demonstrate that Smad1/5/8 are able to transduce BMP signal independently of Smad4 to regulate the expression of downstream target genes, such as Msx1, in the dental mesenchyme during early tooth development.
The Msx1 homeobox gene is expressed in several developing organs in vertebrates, particularly at the site where epithelial-mesenchymal interactions occur during organogenesis (37). The expression of Msx1 is restricted in the dental mesenchyme from the tooth initiation stage and is believed to be induced by the dental epithelial signal BMP4 and subsequently to be maintained by the mesenchymally expressed BMP4 (6, 38). However, it remained elusive as to which signaling pathway BMP4 regulates Msx1 in the developing tooth germ. In the present investigation, we showed that inhibition of BMP/Smad1/5/8 signaling pathway by dorsomorphin dramatically reduces Msx1 expression in the dental mesenchyme, and knockdown of either Smad1 or Smad5 or both attenuates the induction of Msx1 by BMP4, indicating the regulation of Msx1 by Smad-mediated BMP signaling. However, knockdown by siRNA or knock-out by gene inactivation of the common Smad Smad4 had no effect on BMP-induced Msx1 expression in dental mesenchymal cells. Taken together, our results reveal a novel Smad-mediated BMP signaling pathway in the regulation of Msx1 expression in the dental mesenchyme, which is Smad1/5-dependent but Smad4-independent. This conclusion is further strengthened by the fact that BMP-induced nuclear translocation of pSmad1/5/8 in dental mesenchymal cells is Smad4-independent and that pSmad1/5/8 can bind to the Msx1 promoter directly in the absence of Smad4.
The presence of BMP ligand or BMP signaling components is not an unequivocal indicator of BMP activity (16). Previous studies have identified BRE as a regulatory sequence activated by BMP signal in various BMP target genes, including the Xenopus id3 and ventx2 genes and the Drosophila brk gene (39, 40). BRE is recognized by the complex of Smad1/5/8 and Smad4 in response to the activation of the BMP type I receptor (41), which makes BRE a faithful indicator for the transcriptional response of cells to the canonical BMP signaling. In this study, we confirmed that no BRE is activated in early tooth germ and attributed this failure of BRE activation to the absence of pSmad1/5-Smad4 complexes. However, Smad1/5 are indeed activated by BMP signaling and play critical functions (such as activation of Msx1) in the absence of Smad4 in the dental mesenchyme. We therefore name this novel BMP signaling the atypical canonical BMP signaling pathway.
The presence of this atypical canonical BMP signaling pathway has been implied in several other developing organs. During nervous system development, conditional inactivation of Smad1 and Smad5 resulted in cerebellar hypoplasia, reduced granule cell numbers, and disorganized Purkinje neuron migration during embryonic development, whereas conditional inactivation of Smad4 produced only very mild cerebellar defects (10). Furthermore, during lens development, cell death occurred associated with inactivation of Smad1/5, but not with inactivation of Smad4 (12). In addition, Smad1 and Smad5 have been proven essential for bone development, but inactivation of Smad4 led to only minor bone defects (42). The expression of Nkx2.5 requires R-Smad during initial heart development, but inactivation of Smad4 does not abolish Nkx2.5 expression (43). These findings, together with our present results, challenge the current perception that Smad4 is essential for BMP/Smad signaling pathway. In some cellular processes that depend on low levels of BMP signaling, Smad4 is essential for R-Smads to maximize their full functions, probably by forming an obligate heterotrimer with R-Smads. However, in other cellular processes that have high levels of BMP signaling, abundant phosphorylated R-Smads can be formed. Under these conditions, R-Smads can still activate their downstream targets, and Smad4 appears to be less important (10). This might explain why pSmad1/5/8 is able to function without Smad4 during early tooth development, in which high expression level of BMP4 is evident in the dental mesenchyme (44).
In fact, Smad4-independent signaling pathway was found not only in BMP/Smad1/5/8 but also in TGFβ/Smad2/3 signaling pathways. Using quantitative ChIP analysis, Smad2 and Smad3 were found to directly target the Snail promoter, but no significant enrichment of Smad4 was detected (45). It has also been reported that, during keratinocyte proliferation and differentiation, the nuclear accumulation of activated Smad2/3 is Smad4-independent, and IκB kinase α (IKKα) but not Smad4 serves as a nuclear cofactor for Smad2/3 recruitment to Mad1 chromatin in epidermis (46). C-terminal phosphorylation of R-Smad proteins by TGFβ receptor kinases is a critical event in signal transduction because it creates a docking site for Smad4, allowing the assembly of signaling complexes (47). Previously, Smad4 was known as the only factor to occupy this key position in the pathway. However, new partners of activated Smads have been identified. For example, TIF1γ was found as an alternative effector of TGFβ-activated Smad2/3 (48). TIF1γ competitively shares the pool of TGFβ-activated Smad2/3 with Smad4 and mediates erythroid differentiation of CD34+ hematopoietic stem/progenitor cells in response to TGFβ (48).
In this investigation, we demonstrate that the absence of pSmad1/5/8-Smad4 complexes in the dental mesenchyme is the consequence of Smad4 saturation by forming pSmad2/3-Smad4 complexes, evidenced by the formation of pSmad1/5/8-Smad4 complexes and activation of BRE-Gal reporter in dental mesenchymal cells with Smad2/3 knockdown or Smad4 overexpression. In line with these observations are several previous studies on the competitive binding of Smad4 between pSmad1/5/8 and pSmad2/3. It was shown that pSmad1 inhibited TGFβ-ALK5/pSmad2 pathway, and overexpression of Smad4 reversed the inhibition completely, indicating a competition between pSmad2 and pSmad1 for limited amounts of their common transcriptional cofactor Smad4 (29). TGFβ stimulation could result in a decrease in pSmad1/5-Smad4 complexes formed in response to BMP and an increase in pSmad2/3-Smad4 complexes (30). This notion is consistent with our present finding that TGFβ1 was able to reverse the formation of pSmad1/5-Smad4 complexes in Smad4-overexpressed cells, indicating that TGFβ inhibits canonical BMP signaling pathway by competing limited Smad4 in the dental mesenchymal cells. Activin/nodal ligands induce activation of Smad2 and Smad3. It was also reported that BMP and activin/nodal signaling pathways antagonize each other through competition between Smad1 and Smad2 for their binding to the common component Smad4 (29, 49, 50).
Taken together, the present studies provide compelling evidence for the existence of a functional atypical canonical BMP signaling pathway that regulates the expression of the odontogenic gene Msx1 and determines the fate of dental mesenchyme during early tooth development.
This work was supported, in whole or in part, by National Institutes of Health Grants R01DE014044 and R01DE024152 (to Y. C.). This work was also supported by National Natural Science Foundation of China Grants 81371105 (to G. Yang) and 81470708 (to G. Yuan).
- BMP
- bone morphogenetic protein
- R-Smad
- receptor-activated Smad
- BRE
- BMP-response element
- PLA
- proximity ligation assay
- DMSO
- dimethyl sulfoxide
- E
- embryonic day
- p
- phospho.
REFERENCES
- 1. Jernvall J., Thesleff I. (2000) Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 92, 19–29 [DOI] [PubMed] [Google Scholar]
- 2. Nie X., Luukko K., Kettunen P. (2006) BMP signalling in craniofacial development. Int. J. Dev. Biol. 50, 511–521 [DOI] [PubMed] [Google Scholar]
- 3. Vainio S., Karavanova I., Jowett A., Thesleff I. (1993) Identification of BMP-4 as a signal mediating secondary induction between epithelial and mesenchymal tissues during early tooth development. Cell 75, 45–58 [PubMed] [Google Scholar]
- 4. Jia S., Zhou J., Gao Y., Baek J. A., Martin J. F., Lan Y., Jiang R. (2013) Roles of Bmp4 during tooth morphogenesis and sequential tooth formation. Development 140, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhang Y., Zhang Z., Zhao X., Yu X., Hu Y., Geronimo B., Fromm S. H., Chen Y. P. (2000) A new function of BMP4: dual role for BMP4 in regulation of Sonic hedgehog expression in the mouse tooth germ. Development 127, 1431–1443 [DOI] [PubMed] [Google Scholar]
- 6. Chen Y., Bei M., Woo I., Satokata I., Maas R. (1996) Msx1 controls inductive signaling in mammalian tooth morphogenesis. Development 122, 3035–3044 [DOI] [PubMed] [Google Scholar]
- 7. Zhao X., Zhang Z., Song Y., Zhang X., Zhang Y., Hu Y., Fromm S. H., Chen Y. (2000) Transgenically ectopic expression of Bmp4 to the Msx1 mutant dental mesenchyme restores downstream gene expression but represses Shh and Bmp2 in the enamel knot of wild type tooth germ. Mech. Dev. 99, 29–38 [DOI] [PubMed] [Google Scholar]
- 8. Massagué J., Seoane J., Wotton D. (2005) Smad transcription factors. Genes Dev. 19, 2783–2810 [DOI] [PubMed] [Google Scholar]
- 9. Park J. E., Shao D., Upton P. D., Desouza P., Adcock I. M., Davies R. J., Morrell N. W., Griffiths M. J., Wort S. J. (2012) BMP-9 induced endothelial cell tubule formation and inhibition of migration involves Smad1 driven endothelin-1 production. PLoS One 7, e30075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tong K. K., Kwan K. M. (2013) Common partner Smad-independent canonical bone morphogenetic protein signaling in the specification process of the anterior rhombic lip during cerebellum development. Mol. Cell. Biol. 33, 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang J., Tan X., Li W., Wang Y., Wang J., Cheng X., Yang X. (2005) Smad4 is required for the normal organization of the cartilage growth plate. Dev. Biol. 284, 311–322 [DOI] [PubMed] [Google Scholar]
- 12. Rajagopal R., Huang J., Dattilo L. K., Kaartinen V., Mishina Y., Deng C. X., Umans L., Zwijsen A., Roberts A. B., Beebe D. C. (2009) The type I BMP receptors, Bmpr1a and Acvr1, activate multiple signaling pathways to regulate lens formation. Dev. Biol. 335, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Retting K. N., Song B., Yoon B. S., Lyons K. M. (2009) BMP canonical Smad signaling through Smad1 and Smad5 is required for endochondral bone formation. Development 136, 1093–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li L., Lin M., Wang Y., Cserjesi P., Chen Z., Chen Y. (2011) BmprIa is required in mesenchymal tissue and has limited redundant function with BmprIb in tooth and palate development. Dev. Biol. 349, 451–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li J., Huang X., Xu X., Mayo J., Bringas P., Jr., Jiang R., Wang S., Chai Y. (2011) SMAD4-mediated WNT signaling controls the fate of cranial neural crest cells during tooth morphogenesis. Development 138, 1977–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Javier A. L., Doan L. T., Luong M., Reyes de Mochel N. S., Sun A., Monuki E. S., Cho K. W. (2012) Bmp indicator mice reveal dynamic regulation of transcriptional response. PLoS One 7, e42566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang X., Li C., Herrera P. L., Deng C. X. (2002) Generation of Smad4/Dpc4 conditional knockout mice. Genesis 32, 80–81 [DOI] [PubMed] [Google Scholar]
- 18. Yang X., Li C., Xu X., Deng C. (1998) The tumor suppressor SMAD4/DPC4 is essential for epiblast proliferation and mesoderm induction in mice. Proc. Natl. Acad. Sci. U.S.A. 95, 3667–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ito Y., Yeo J. Y., Chytil A., Han J., Bringas P., Jr., Nakajima A., Shuler C. F., Moses H. L., Chai Y. (2003) Conditional inactivation of Tgfbr2 in cranial neural crest causes cleft palate and calvaria defects. Development 130, 5269–5280 [DOI] [PubMed] [Google Scholar]
- 20. He F., Xiong W., Wang Y., Matsui M., Yu X., Chai Y., Klingensmith J., Chen Y. (2010) Modulation of BMP signaling by Noggin is required for the maintenance of palatal epithelial integrity during palatogenesis. Dev. Biol. 347, 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y., Li L., Zheng Y., Yuan G., Yang G., He F., Chen Y. (2012) BMP activity is required for tooth development from the lamina to bud stage. J. Dent Res. 91, 690–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hu B., Nadiri A., Kuchler-Bopp S., Perrin-Schmitt F., Peters H., Lesot H. (2006) Tissue engineering of tooth crown, root, and periodontium. Tissue Eng. 12, 2069–2075 [DOI] [PubMed] [Google Scholar]
- 23. Monteiro R. M., de Sousa Lopes S. M., Bialecka M., de Boer S., Zwijsen A., Mummery C. L. (2008) Real time monitoring of BMP Smads transcriptional activity during mouse development. Genesis 46, 335–346 [DOI] [PubMed] [Google Scholar]
- 24. Chai Y., Mah A., Crohin C., Groff S., Bringas P., Jr., Le T., Santos V., Slavkin H. C. (1994) Specific transforming growth factor-β subtypes regulate embryonic mouse Meckel's cartilage and tooth development. Dev. Biol. 162, 85–103 [DOI] [PubMed] [Google Scholar]
- 25. Pacheco M. S., Reis A. H., Aguiar D. P., Lyons K. M., Abreu J. G. (2008) Dynamic analysis of the expression of the TGFβ/SMAD2 pathway and CCN2/CTGF during early steps of tooth development. Cells Tissues Organs 187, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gustafsdottir S. M., Schallmeiner E., Fredriksson S., Gullberg M., Söderberg O., Jarvius M., Jarvius J., Howell M., Landegren U. (2005) Proximity ligation assays for sensitive and specific protein analyses. Anal. Biochem. 345, 2–9 [DOI] [PubMed] [Google Scholar]
- 27. Söderberg O., Leuchowius K. J., Gullberg M., Jarvius M., Weibrecht I., Larsson L. G., Landegren U. (2008) Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods 45, 227–232 [DOI] [PubMed] [Google Scholar]
- 28. Söderberg O., Gullberg M., Jarvius M., Ridderstråle K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 29. Furtado M. B., Solloway M. J., Jones V. J., Costa M. W., Biben C., Wolstein O., Preis J. I., Sparrow D. B., Saga Y., Dunwoodie S. L., Robertson E. J., Tam P. P., Harvey R. P. (2008) BMP/SMAD1 signaling sets a threshold for the left/right pathway in lateral plate mesoderm and limits availability of SMAD4. Genes Dev. 22, 3037–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grönroos E., Kingston I. J., Ramachandran A., Randall R. A., Vizán P., Hill C. S. (2012) Transforming growth factor β inhibits bone morphogenetic protein-induced transcription through novel phosphorylated Smad1/5-Smad3 complexes. Mol. Cell. Biol. 32, 2904–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Candia A. F., Watabe T., Hawley S. H., Onichtchouk D., Zhang Y., Derynck R., Niehrs C., Cho K. W. (1997) Cellular interpretation of multiple TGF-β signals: intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development 124, 4467–4480 [DOI] [PubMed] [Google Scholar]
- 32. Satokata I., Maas R. (1994) Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat. Genet. 6, 348–356 [DOI] [PubMed] [Google Scholar]
- 33. Yu P. B., Hong C. C., Sachidanandan C., Babitt J. L., Deng D. Y., Hoyng S. A., Lin H. Y., Bloch K. D., Peterson R. T. (2008) Dorsomorphin inhibits BMP signals required for embryogenesis and iron metabolism. Nat. Chem. Biol. 4, 33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shin J. O., Kim E. J., Cho K. W., Nakagawa E., Kwon H. J., Cho S. W., Jung H. S. (2012) BMP4 signaling mediates Zeb family in developing mouse tooth. Histochem. Cell Biol. 137, 791–800 [DOI] [PubMed] [Google Scholar]
- 35. Alvarez Martinez C. E., Binato R., Gonzalez S., Pereira M., Robert B., Abdelhay E. (2002) Characterization of a Smad motif similar to Drosophila Mad in the mouse Msx 1 promoter. Biochem. Biophys. Res. Commun. 291, 655–662 [DOI] [PubMed] [Google Scholar]
- 36. Binato R., Alvarez Martinez C. E., Pizzatti L., Robert B., Abdelhay E. (2006) SMAD 8 binding to mice Msx1 basal promoter is required for transcriptional activation. Biochem. J. 393, 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davidson D. (1995) The function and evolution of Msx genes: pointers and paradoxes. Trends Genet. 11, 405–411 [DOI] [PubMed] [Google Scholar]
- 38. Bei M., Maas R. (1998) FGFs and BMP4 induce both Msx1-independent and Msx1-dependent signaling pathways in early tooth development. Development 125, 4325–4333 [DOI] [PubMed] [Google Scholar]
- 39. von Bubnoff A., Peiffer D. A., Blitz I. L., Hayata T., Ogata S., Zeng Q., Trunnell M., Cho K. W. (2005) Phylogenetic footprinting and genome scanning identify vertebrate BMP response elements and new target genes. Dev. Biol. 281, 210–226 [DOI] [PubMed] [Google Scholar]
- 40. Yao L. C., Blitz I. L., Peiffer D. A., Phin S., Wang Y., Ogata S., Cho K. W., Arora K., Warrior R. (2006) Schnurri transcription factors from Drosophila and vertebrates can mediate Bmp signaling through a phylogenetically conserved mechanism. Development 133, 4025–4034 [DOI] [PubMed] [Google Scholar]
- 41. Katagiri T., Imada M., Yanai T., Suda T., Takahashi N., Kamijo R. (2002) Identification of a BMP-responsive element in Id1, the gene for inhibition of myogenesis. Genes Cells 7, 949–960 [DOI] [PubMed] [Google Scholar]
- 42. Tan X., Weng T., Zhang J., Wang J., Li W., Wan H., Lan Y., Cheng X., Hou N., Liu H., Ding J., Lin F., Yang R., Gao X., Chen D., Yang X. (2007) Smad4 is required for maintaining normal murine postnatal bone homeostasis. J. Cell Sci. 120, 2162–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chu G. C., Dunn N. R., Anderson D. C., Oxburgh L., Robertson E. J. (2004) Differential requirements for Smad4 in TGFβ-dependent patterning of the early mouse embryo. Development 131, 3501–3512 [DOI] [PubMed] [Google Scholar]
- 44. Tucker A. S., Al Khamis A., Sharpe P. T. (1998) Interactions between Bmp-4 and Msx-1 act to restrict gene expression to odontogenic mesenchyme. Dev. Dyn. 212, 533–539 [DOI] [PubMed] [Google Scholar]
- 45. Smith A. P., Verrecchia A., Fagà G., Doni M., Perna D., Martinato F., Guccione E., Amati B. (2009) A positive role for Myc in TGFβ-induced Snail transcription and epithelial-to-mesenchymal transition. Oncogene 28, 422–430 [DOI] [PubMed] [Google Scholar]
- 46. Descargues P., Sil A. K., Sano Y., Korchynskyi O., Han G., Owens P., Wang X. J., Karin M. (2008) IKKα is a critical coregulator of a Smad4-independent TGFβ-Smad2/3 signaling pathway that controls keratinocyte differentiation. Proc. Natl. Acad. Sci. U.S.A. 105, 2487–2492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shi Y., Massagué J. (2003) Mechanisms of TGF-β signaling from cell membrane to the nucleus. Cell 113, 685–700 [DOI] [PubMed] [Google Scholar]
- 48. He W., Dorn D. C., Erdjument-Bromage H., Tempst P., Moore M. A., Massagué J. (2006) Hematopoiesis controlled by distinct TIF1γ and Smad4 branches of the TGFβ pathway. Cell 125, 929–941 [DOI] [PubMed] [Google Scholar]
- 49. Yamamoto M., Beppu H., Takaoka K., Meno C., Li E., Miyazono K., Hamada H. (2009) Antagonism between Smad1 and Smad2 signaling determines the site of distal visceral endoderm formation in the mouse embryo. J. Cell Biol. 184, 323–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Katsu K., Tatsumi N., Niki D., Yamamura K., Yokouchi Y. (2013) Multi-modal effects of BMP signaling on Nodal expression in the lateral plate mesoderm during left-right axis formation in the chick embryo. Dev. Biol. 374, 71–84 [DOI] [PubMed] [Google Scholar]