Background: PPR (pentatricopeptide repeat) proteins in dimer state can specifically recognize single-stranded RNA.
Results: The different amino-terminal lengths and critical point mutations in PPR protein lead to distinct dimerization state of PPR protein.
Conclusion: PPR proteins are regulated to adopt different dimerization state.
Significance: The biochemical studies advanced our understanding of PPR protein regulations and can facilitate the biotechnological applications of PPR proteins.
Keywords: Cell Biology, Crystal Structure, Protein-Nucleic Acid Interaction, RNA, RNA Binding Protein, RNA-Protein Interaction
Abstract
Pentatricopeptide repeat (PPR) proteins, particularly abundant in plastids and mitochrondria of angiosperms, include a large number of sequence-specific RNA binding proteins that are involved in diverse aspects of organelle RNA metabolisms. PPR proteins contain multiple tandom repeats, and each repeat can specifically recognize a RNA base through residues 2, 5, and 35 in a modular fashion. The crystal structure of PPR10 from maize chloroplast exhibits dimeric existence both in the absence and presence of the 18-nucleotide psaJ RNA element. However, previous biochemical analysis suggested a monomeric shift of PPR10 upon RNA binding. In this report, we show that the amino-terminal segments of PPR10 determine the dimerization state of PPR10. A single amino acid alteration of cysteine to serine within repeat 10 of PPR10 further drives dimerization of PPR10. The biochemical elucidation of the determinants for PPR10 dimerization may provide an important foundation to understand the working mechanisms of PPR proteins underlying their diverse physiological functions.
Introduction
Pentatricopeptide repeat (PPR)3 proteins, prevalent across eukaryotic kingdoms of lives and especially abundant in terrestrial flowering plants, function as sequence-specific single-stranded RNA binding proteins (1–6). PPR proteins mainly exist in chloroplasts and mitochondria, where they are involved in diverse aspects of organelle RNA metabolism processes, including RNA editing, maturation, stability, and translation (3, 4, 7). PPR mutants of important organelle functions frequently failed to survive through to the fetus stage, leading to high lethality (3). Various plant physiological studies on PPR proteins revealed their crucial roles in the ubiquitous plant development processes such as fertility, embryogenesis, circadian response, and organ separation (3, 7, 8). For example, certain Rf (restorer-of-fertility) genes can encode PPR proteins to overcome the cytoplasmic male sterility (9–12). In human mitochondria, PPR protein leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC) is closely correlated with the French-Canadian type of Leigh syndrome and adenocarcinoma (13, 14).
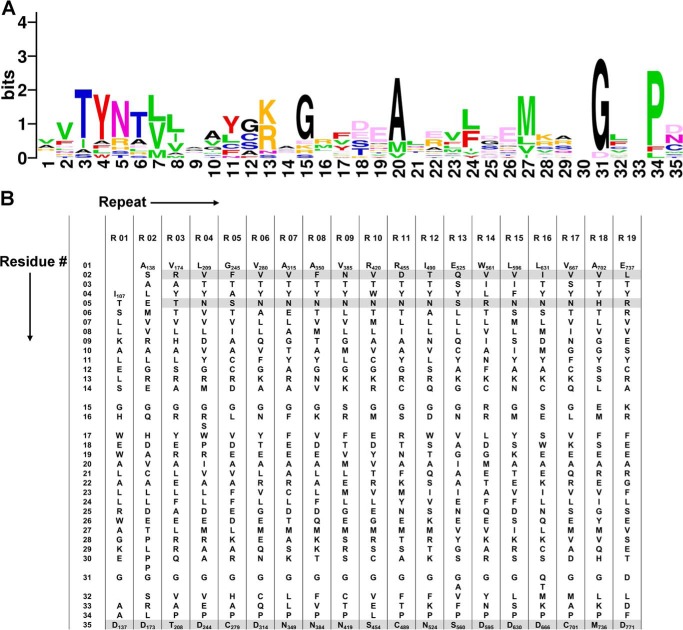
PPR proteins generally contain an array of 2–30 tandem repeats, and each canonical repeat is composed of 35 amino acids arranged into a hairpin of α-helices (15–17). The system of RNA recognition code was tentatively disclosed by bioinformatic analysis in conjunction with biochemical assays and recently corroborated by structural elucidation (see Figs. 1, A and B, and 2A) (6, 15, 17–20). Within an intact PPR repeat, the amino acid residues at the 2nd, 5th, and 35th position are responsible for sequence-specific recognition of single-stranded RNA bases (Fig. 1, A and B) (15, 17, 18).
FIGURE 1.
Sequence alignment of the repeats in PPR10. A, a sequence logo generated using repeat sequences of PPR10 reveals relatively conserved amino acid positions within the repeats. The amino acids in the repeats are colored based on the physiochemical properties of the side chains: small (Gly (G) and Ala (A)) in black, nucleophilic (Ser (S), Thr (T), and Cys (C)) in blue, hydrophobic (Val (V), Leu (L), Ile (I), Met (M), and Pro (P)) in green, aromatic (Phe (F), Tyr (Y), and Trp (W)) in red, acidic (Asp (D) and Glu (G)) in light pink, amide (Asn (N) and Gln (Q)) in dark pink, and basic (His (H), Lys (K), and Arg (R)) in orange (33, 44). B, the principal portion of PPR10 repeats (residues 107–771) is summarized in the table of 19 repeat alignment. The molecular determining residues for RNA recognition at the 2nd, 5th, and 35th positions are shaded light gray.
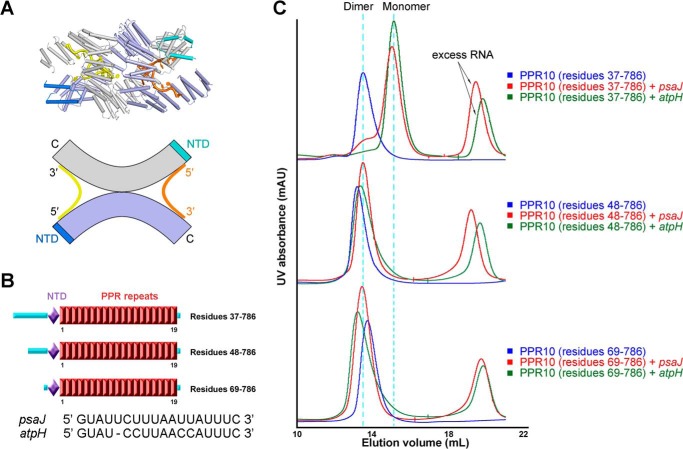
FIGURE 2.
Amino-terminal truncations lead to different dimerization states of PPR10 bound to RNA. A, upper panel: dimeric crystal structure of PPR10 in complex with psaJ RNA, two protomers of PPR10 molecules are colored light purple and gray with their NTDs colored blue and cyan, and the two bound RNA molecules are colored orange and yellow. Lower panel: a schematic illustration for dimerization state of PPR10 with psaJ RNAs. B, schematic representation of successive amino-terminal truncations on PPR10. Red boxes indicate tandem PPR repeats. The NTD is colored light purple, and loop regions are colored cyan. The sequences of two native RNA targets psaJ and atpH for PPR10 are shown below. C, SEC analyses of successive amino-terminal truncated PPR10 proteins in the absence and presence of psaJ or atpH RNA element. Dimer and monomer positions are indicated by cyan dotted lines. All structure figures were prepared with PyMOL (43). mAU, milliabsorbance units.
PPR10 from maize chloroplast has been extensively exploited as a prototype for deciphering RNA recognition code (18, 21, 22). PPR10, containing 19 repeats, targets two native RNA elements from atpI-atpH and psaJ-rpl33 intergenic regions, referred to as “atpH” and “psaJ”, respectively (21, 22). PPR10 is proposed to act as a site-specific barrier to protect target RNA from nuclease degradation and to enhance mRNA translational activation by remodeling the ribosomal binding site (21, 22). We recently determined the crystal structures of PPR10 in the RNA-free and psaJ RNA-bound forms (15). The crystal structure provides the first glance of sequence specific recognition of single-stranded RNA by PPR proteins. Interestingly, whereas analytical ultracentrifugation analysis suggested that atpH-bound PPR10 is a monomer (18), two molecules of PPR10 intertwine into an anti-parallel homodimer both in the absence and presence of psaJ RNA element in the crystal structures (15).
PPR proteins may be monomeric or dimeric. For example, PPR4, PPR5, and THA8 from maize and THA8L from Arabidopsis, exist as monomers (23–26), whereas the PPR protein HCF152 from Arabidopsis was identified to be a homodimer (27). These observations raise the possibility that different dimerization states of PPR proteins may correspond to their diverse functions. Is it possible that PPR10 can be intrinsically regulated to adopt different dimerization states suitably fulfilling their respective physiological roles? If so, what are the determining factors for the dimerization states of PPR10? In this report, we sought to answer the questions through extensive biochemical analysis.
EXPERIMENTAL PROCEDURES
Protein Preparation
The codon-optimized cDNA of full-length PPR10 (gene ID 100302579) from Zea mays was subcloned into pET15b vector (Novagen). Overexpression of PPR10 protein was induced in Escherichia coli BL21(DE3) with 0.2 mm isopropyl-β-d-thiogalactoside at an optical density of 1.2 at 600 nm. After growing for 16 h at 16 °C, the cells were collected and homogenized in a buffer containing 25 mm Tris-HCl, pH 8.0, and 150 mm NaCl. After sonication and centrifugation, the supernatant was applied to Ni2+ affinity resin (nickel-nitrilotriacetic acid, Qiagen) and further fractionated by heparin affinity chromatography (HiPrep Heparin FF 16/10, GE Healthcare) and gel filtration chromatography (Superdex-200 10/30, GE Healthcare). The PPR10 mutants were generated using two-step PCR and subcloned, overexpressed, and purified in the same way as the wild-type protein.
Crystallization
For the crystallization trials of PPR10 (residues 69–786, C256S/C430S/C449S) in complex with psaJ RNA, the protein was purified through Ni2+ affinity resin (nickel-nitrilotriacetic acid, Qiagen), followed by heparin affinity column (HiPrep Heparin FF 16/10, GE Healthcare). The protein was then applied to gel filtration chromatography (Superdex-200 10/30, GE Healthcare) in the buffer containing 25 mm Tris-HCl, pH 8.0, 50 mm NaCl, 5 mm MgCl2, and 10 mm dithiothreitol (DTT). The peak fractions were incubated with target RNA oligonucleotides with a molar ratio of ∼1:1.5 at 4 °C for ∼40 min before crystallization trials. RNA-bound PPR10 protein was crystallized by hanging drop vapor diffusion method at 18 °C. The protein-RNA complex of PPR10 (residues 69–786, C256S/C430S/C449S) and psaJ RNA with the sequence 5′-GUAUUCUUUAAUUAUUUC-3′ gave rise to crystals in the reservoir solution containing 8–10% (w/v) polyethylene glycol 3350, 8% tacsimate, pH 6.0 (Hampton Research), and 0.1 m MES, pH 5.5.
Data Collection, Structure Determination, and Refinement
The diffraction data were collected at Shanghai Synchrotron Radiation Facility, integrated, and scaled with the HKL2000 package (28). Further data processing was carried out using programs from the CCP4 suite (29). To determine the complex structure of triple cysteine mutant PPR10 (residues 69–786, C256S/C430S/C449S), the previous resolved quadruple cysteine mutant PPR10 (residues 69–786, C256S/C279S/C430S/C449S) (Protein Data Bank code 4M59) was selected as the molecular replacement model. The molecular replacements were performed with program PHASER, and manually model rebuilding and refinement were iteratively performed with COOT and Phenix (30, 31). Native data collection and refinement statistics are summarized in Table 1.
TABLE 1.
Statistics of data collection and refinement
Values in parentheses are for the highest resolution shell.
| Data collection | PPR10 (C256S/C430S/C449S) |
|---|---|
| Integration package | HKL2000 |
| Space group | P43 |
| Unit cell | α = 83.32, β = 83.32, and γ = 225.50 Å; a = 90, b = 90, and c = 90° |
| Wavelength (Å) | 0.9791 |
| Resolution (Å) | 40 ∼ 2.80 (2.90 ∼ 2.80) |
| Rmerge (%)a | 8.0 (57.5) |
| I/σ | 19.9 (2.8) |
| Completeness (%) | 98.0 (99.2) |
| No. of measured reflections | 125,406 |
| No. of unique reflections | 37,060 |
| Redundancy | 3.4 (3.4) |
| Wilson B factor (Å2) | 59.5 |
| Rwork/Rfree (%) | 26.29/27.76 |
| No. of atoms | |
| Protein | 10,384 |
| Main chain | 5,480 |
| Side chain | 4,904 |
| RNA | 647 |
| Others | 5 |
| Average B value (Å2) | |
| Protein | 79.06 |
| Main chain | 78.84 |
| Side chain | 79.30 |
| RNA | 90.38 |
| Others | 79.17 |
| r.m.s.d.b | |
| Bonds (Å) | 0.015 |
| Angle | 1.050° |
| Ramachandran plot statistics (%) | |
| Most favorable | 88.8 |
| Additionally allowed | 10.3 |
| Generously allowed | 0.9 |
| Disallowed | 0.0 |
a Rmerge = ΣhΣi|Ih,i − Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry-related reflections of h. r = Σ|Fobs − Fcalc|/ΣFobs, where Fcalc is the calculated protein structure factor from the atomic model (Rfree was calculated with 5% of the reflections selected).
b r.m.s.d., root mean square deviation.
Size Exclusion Chromatography (SEC)
The SEC analyses were performed with a SD200 column (Superdex-200 10/30, GE Healthcare) in the buffer containing 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, and 2 mm DTT. To examine the dimerization state of PPR10 and its mutants, 10 μm, 500 μl protein is applied each time onto the Superdex-200 column. To examine the interactions between protein and RNA, the indicated proteins of 10 μm, 500 μl were incubated with target RNA oligonucleotides with a molar ratio of ∼1:1.5 at 4 °C for ∼40 min before injection.
EMSA
The single-stranded RNA oligonucleotides were radiolabeled at the 5′-end with [γ-32P]ATP (PerkinElmer) catalyzed by T4 polynucleotide kinase (Takara). The sequences of single-stranded RNA oligonucleotides used in EMSA were as follows: psaJ, 5′-GUAUUCUUUAAUUAUUUC-3′; atpH, 5′-GUAUCCUUAACCAUUUC-3′.
For EMSA, PPR10 of different amino-terminal boundaries and other variants consisting of the indicated point mutations were incubated with ∼40 pm 32P-labeled RNA probe in the final binding reactions containing 40 mm Tris-HCl, pH 7.5, 100 mm NaCl, 4 mm DTT, 0.1 mg ml−1 BSA, 5 μg ml−1 heparin, and 10% glycerol at room temperature for 20 min (22 °C). Reactions were then resolved on 6% native acrylamide gels (37.5: 1 for acrylamide: bisacrylamide) in 0.5× Tris glycine buffer under an electric field of 15 V/cm for 40 min. Vacuum-dried gels were visualized on a phosphor screen (Amersham Biosciences) with a Typhoon Trio Imager (Amersham Biosciences).
RESULTS
N-terminal Boundaries Determine Dimerization State of PPR10 in Complex with RNA
In our previous structural study, we have exerted a systematic protein engineering effort on PPR10 for crystallization. Considering the oxidation process of surface cysteine residues can sabotage protein homogeneity hampering uniform crystal packing, we generated and combined the cysteines to serine mutants for each of the 18 cysteines in the PPR10 repeat region. After trials on a myriad of truncations, one construct of PPR10, residues 69–786 (C256S/C279S/C430S/C449S) in complex with the psaJ RNA element exhibiting excellent behavior gave rise to crystals, and the dimer formation of two PPR10 molecules in complex with RNA was finally observed in the refined structure (Fig. 2A) (15). The 19 PPR repeats (residues 107–771) constitute the principal portion of PPR10 preceded by three α-helices forming N-terminal domain (NTD, residues 74–106) (Fig. 2B). The amino-terminal fragment of unknown function prior to NTD domain is predicted to be a flexible region containing the chloroplast transit peptide and is removed during structural study to facilitate crystallization. Along with previously reported data (15, 18, 21, 22), these accumulated results describing PPR10 dimerization states aroused our interest. Two provocative, yet unanswered questions in PPR biology are thus introduced. Is it possible that amino-terminal length of PPR10 determines protein dimerization? Or is it possible that an amino acid mutation leads to the dimeric state of PPR10? In seeking the answers, extensive biochemical investigations were performed, respectively.
First, we tested whether the dimerization is associated with protein amino-terminal truncation boundaries. Three successive amino-terminal truncations of distinct biochemical properties were studied: PPR10 residues 37–786, PPR10 residues 48–786, and PPR10 residues 69–786. These protein constructs were referred as “PPR10–37”, “PPR10–48”, and “PPR10–69”, respectively (Fig. 2B). Because PPR10 can specifically target two native RNA element 18-nucleotide psaJ and 17-nucleotide atpH, we investigated the binding properties with both RNAs.
It was reported that RNA-free PPR10 exists in dimeric state, whereas atpH-bound PPR10 is a monomer (18, 21, 22, 32). In these studies, the initial 36 amino acids were absent in the recombinant protein and designated as PPR10–37 construct. Indeed, when RNA-free PPR10–37 was applied upon the SEC (Superdex 200 10/30), the dimeric protein peak position was 13.5 ml. However PPR10–37 incubated with atpH or psaJ RNA both eluted at 15.0 ml, indicative of monomeric state of RNA-bound PPR10–37 (Fig. 2C). Intriguingly, the SEC profile analyses on PPR10–48 and PPR10–69, yielded distinct results from those of PPR10–37. For PPR10 proteins alone, in agreement with PPR10–37, PPR10–48, and PPR10–69 also both appear in dimeric states in SEC chromatogram. Nevertheless, in the presence of atpH or psaJ RNA, disparate with those of PPR10–37, elution peaks of PPR10–48 and PPR10–69 appear at dimer positions (Fig. 2C). Additionally, the dimerization formation of PPR10–69 with psaJ RNA in SEC characterization is in consistency with the PPR10–69 homodimer construction in crystallographic observation (Fig. 2, A and C) (15).
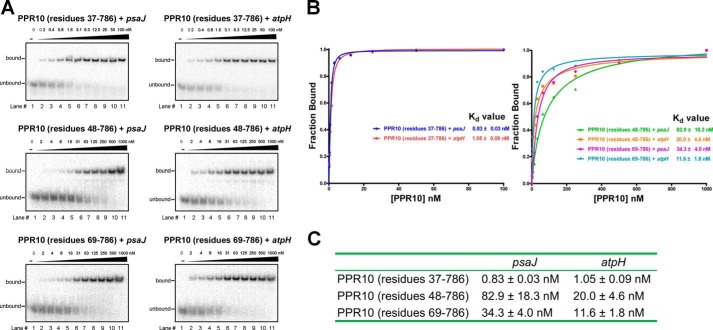
Simultaneously, we attempted to test the binding capacity of amino-terminal length variant constructs of PPR10 with its native target RNA elements by the EMSA. PPR10–37 binds to the 18-nucleotide psaJ and 17-nucleotide atpH elements with dissociation constant (Kd) values of ∼0.8 nm and 1.0 nm, respectively, indicative of strong interaction with the RNA elements (Fig. 3, A–C). For PPR10–48 or PPR10–69, drastically different from those of PPR10–37, the Kd values increased by over 10-fold, implying a much weaker interactions in comparison with those of PPR10–37 (PPR10–48, ∼83 nm with psaJ, ∼20 nm with atpH; PPR10–69, ∼34 nm with psaJ, ∼12 nm with atpH) (Fig. 3, A–C). Taken together, these results argue the crucial role of amino-terminal fragments of PPR10, both in dimerization state regulation and target RNA binding affinity.
FIGURE 3.
Binding affinity assays of different amino-terminal truncations of PPR10 with RNA. A, EMSAs are performed for different amino-terminal PPR10 with RNA elements. The RNA sequences of psaJ and atpH are indicated in Fig. 1B. Binding reactions contain ∼40 pm radiolabeled RNAs and PPR10 proteins at the indicated concentrations. B, the PPR10 fractions of RNA bound and concentrations in each lane of Fig. 2B are plotted and fitted to the calculate equilibrium Kd values. C, the summary table of equilibrium Kd values for EMSA reactions of different amino-terminal PPR10 with psaJ and atpH RNA elements.
A Single Amino Acid Alteration Determines Dimerization State of PPR10-RNA Complex
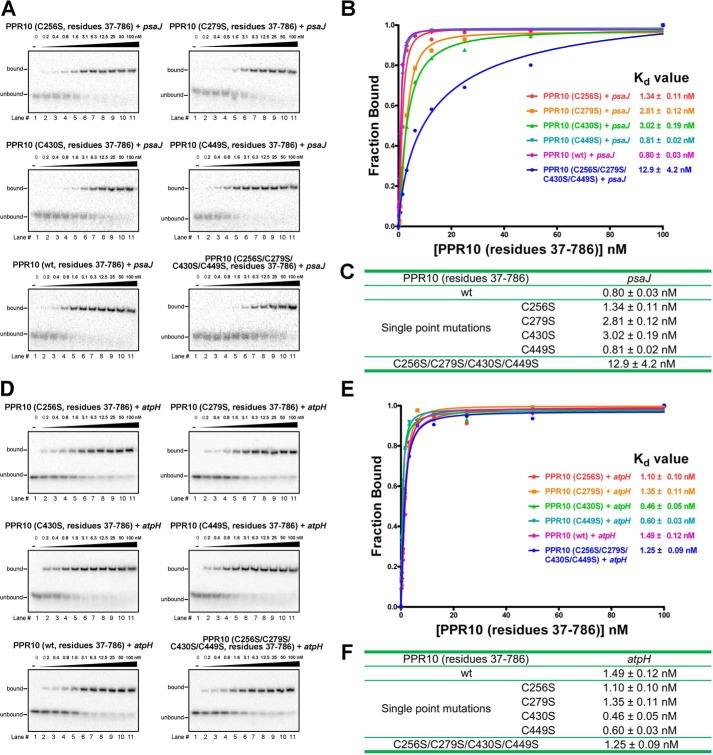
Previous structural study aroused our interest to discover whether these four cysteine to serine mutations played a role in the dimerization state formation of PPR10 for target RNAs (Fig. 4A). Accordingly, we have conducted SEC analyses for wild type PPR10 (residues 37–786) and a series of PPR10 mutants (residues 37–786): (i) C256S single mutant; (ii) C279S single mutant; (iii) C430S single mutant; (iv) C449S single mutant; and (v) C256S/C279S/C430S/C449S quadruple mutant as a control.
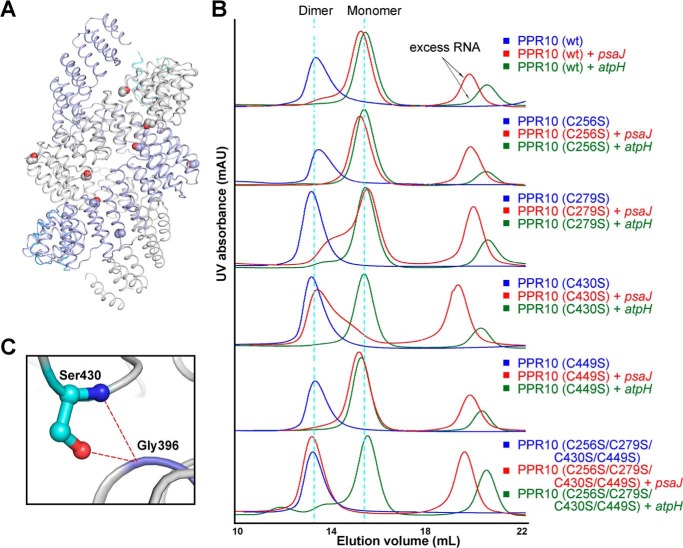
FIGURE 4.
Cysteine-to-serine mutations can affect the dimerization states of PPR10 in the presence of RNA. A, in the dimeric crystal structure of PPR10 (C256S/C279S/C430S/C449S), two protomers of PPR10 are colored light purple and gray with their NTDs colored blue and cyan. The four cysteine-to-serine mutation sites in each PPR protomer are highlighted as spheres. B, SEC analyses of wild type PPR10, PPR10 variants containing each single cysteine to serine mutation, and PPR10 with quadruple cysteine-to-serine mutations. The SEC analyses were performed in the absence and presence of psaJ or atpH RNA element. C, a close-up view of Ser-430. Ser-403 can form hydrogen bonds with Gly-396 from the adjacent repeat and is crucial for the dimerization of PPR10. The hydrogen bonds are shown as red dotted lines. mAU, milliabsorbance units.
In the absence of RNA, the wild type protein and all mutants consistently appeared as dimers on the chromatograms (Fig. 4B). In the presence of atpH RNA, the wild type protein and mutants uniformly eluted at monomer positions. Nevertheless, notable discrepancies were observed in the presence of psaJ RNA. The distinct elution volume value of 15.0 ml and 13.5 ml for wild type protein and quadruple mutants with psaJ RNA indicated the distinctive monomer and dimer formation, respectively (Fig. 4B), consistent with the previous sedimentation equilibrium analytical ultracentrifugation (SE-AUC) results (15). For single cysteine mutants with psaJ RNA, C256S, C279S, and C449S all eluted at similar positions as the wild type protein, illustrating cysteine to serine mutations at these three cysteine sites exert little impact on protein dimerization. Although it is noteworthy that the peak position of single mutant C430S with psaJ RNA shifted to the similar position as the quadruple mutants (Fig. 4B). This underscores the notion that Cys-430 is an important determinant residue in dimeric state formation, and C430S mutation can cause monomer-dimer transition of PPR10 in the presence of psaJ RNA element.
In the previously reported crystal structure, hydrogen bonds were observed between Ser-430 residue (side chain hydroxyl group) in repeat 10 and Gly-396 in repeat 9 (Fig. 4C) (15), whereas Cys-430 (side chain thiol group) may not efficiently generate polar contacts due to the lower electronegativity of sulfur atom in thiol group. This intramolecular interaction with the adjacent repeat suggests the intricately complicated role of Ser-430 in dimerization determination.
Concomitantly, we set out to test the RNA binding activity of wild type protein and a series of mutants. In the presence of psaJ RNA, the four single mutants of C256S, C279S, C430S, and C449S exhibit similar profiles as the wild type protein in EMSA experiments. The Kd values ∼0.8–3.0 nm of C256S, C279S, C430S, and C449S single mutants resembled the value ∼0.8 nm of the wild type protein (Fig. 5, A–C). Notably, for quadruple mutants, the Kd value ∼13 nm estimated from gel shift results is >10-fold higher than that of the wild type protein (Fig. 5, A–C). When binding to atpH RNA, the wild type protein, four single mutants, and quadruple mutants all exhibited similar EMSA profile and numerically comparable Kd values ranging from ∼0.5–1.5 nm (Fig. 5, D–F). The results illustrate that cysteine to serine mutations in the indicated PPR10 single mutants or quadruple mutants bring little influence on the binding affinity with atpH RNA element. Taken together, these results indicate that any of the single cysteine mutation fails to exert obvious impact on protein binding affinity with RNAs, whereas the multiple cysteine mutation combination can collectively reduce protein binding affinity with psaJ RNA element but impose no impact on the binding capacity with atpH RNA element.
FIGURE 5.
Binding affinity assays of PPR10 containing cysteine to serine mutations with RNA element. A and D, EMSAs are performed on wild type PPR10, PPR10 variants containing each single cysteine to serine mutation, and PPR10 with quadruple cysteine-to-serine mutations in the presence of psaJ or atpH RNA. The RNA sequences of psaJ and atpH are indicated in Fig. 1B. Binding reactions contain ∼40 pm radiolabeled RNAs and PPR10 at the indicated concentrations. B and E, the PPR10 fractions of RNA bound and concentrations in each lane of Fig. 4, A and D, are plotted and fitted to the calculate equilibrium Kd values. C and F, the summary table of equilibrium Kd values for reactions of different PPR10 mutants with psaJ or atpH RNA elements. In the presence of psaJ RNA, the quadruple mutant displays >10-fold higher Kd value than others, indicative of much weaker binding affinity of quadruple mutant for psaJ RNA. In the presence of atpH RNA, the wild type, four single mutants, and the quadruple mutants exhibit uniform EMSA profiles and similar Kd values.
Amino Acid Combination of “Phe-Ser-Ser (FSS)” and “Phe-Ser-Cys (FSC)” in PPR10 Can Both Target Adenine in RNA
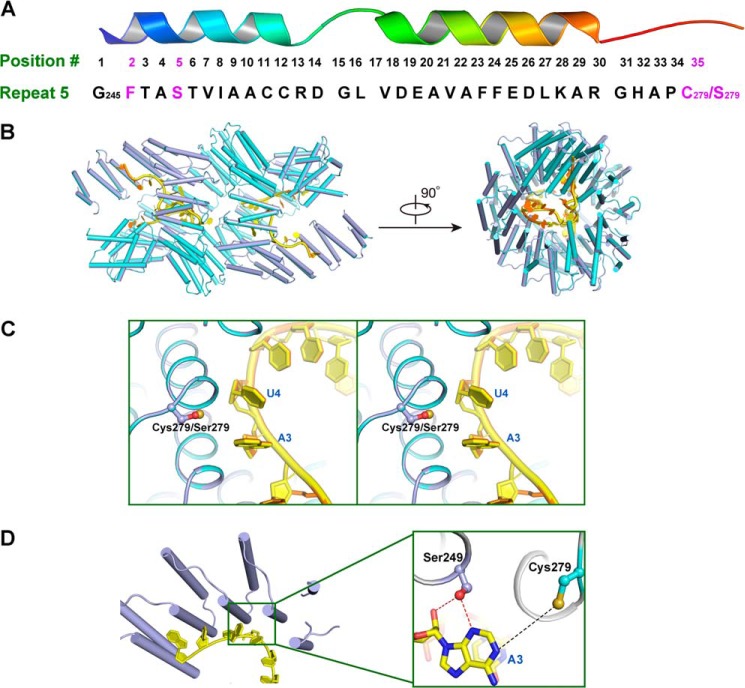
In each PPR repeat, residues 2, 5, and 35 can specifically target a certain RNA nucleotide as a recognition code (18, 19). Repeat 5 in PPR10 can recognize adenine in RNA strand as elucidated through biochemical and computational studies (6, 15). In the prior study, quadruple cysteine to serine mutations C256S/C279S/C430S/C449S (residues 69–786) are present in the recently determined crystal structure of PPR10 in complex with the psaJ RNA element (15). This structure revealed, in repeat 5 of PPR10, Phe-246 in the 2nd position, Ser-249 in the 5th position, and Ser-279 in the 35th position, constituting FSS combination for discrimination of adenine RNA nucleotide (Fig. 6A). The results validated the previously proposed combination of FSS targeting adenine (6, 19).
FIGURE 6.
Dimeric crystal structure of PPR10 (C256S/C430S/C449S) with RNA. A, amino acid residues in repeat 5 of PPR10 spatially organized into a hairpin of α-helices. Residues Phe-246, Ser-249, and Cys-279/Ser-279 involved in recognition of adenine base in psaJ RNA in the crystal structure are highlighted in magenta. B, superimposition of PPR10 (C256S/C430S/C449S) colored light purple with RNA in yellow and PPR10 (C256S/C279S/C430S/C449S) colored cyan with RNA in orange (Protein Data Bank code 4M59) revealed almost identical recognition pattern toward RNA. The left and right panels are related by a 90° rotation around a horizontal axis. C, regional structure comparison between PPR10 (C256S/C279S/C430S/C449S) and PPR10 (C256S/C430S/C449S) in the vicinity of Cys-279/Ser-279 residue. The side chain of Cys-279/Ser-279 is colored by atoms with sulfur in Cys-279 colored beige and oxygen in Ser-279 colored red. Stereo view of superimposition is shown. D, recognition interface between PPR10 (C256S/C430S/C449S) and RNA. A close-up view of residues in repeat 5 recognizing adenine in psaJ RNA is depicted on the right. Polar contacts and van der Waals interactions are shown as red and black dotted lines, respectively.
Due to the limited impact of C279S single mutation on protein dimerization and RNA binding capacity (Figs. 4B and 5), we proceeded to explore whether the FSC combination in the context of PPR10 repeat 5 can recognize adenine via structural biology study of PPR10 triple mutations C256S/C430S/C449S (residue 69–786) with the RNA element. In this case, the 35th position of repeat 5 became a cysteine instead of a serine (Fig. 6A). Although the triple mutant was biochemically less stable and prone to precipitate in solution, we finally determined the crystal structure of the triple mutant with psaJ RNA at 2.8 Å resolution (Table 1). Superimposition of two structures of quadruple mutant C256S/C279S/C430S/C449S and triple mutant C256S/C430S/C449S revealed an RMSD of only 0.041Å over 1368 aligned Cα atoms, indicative of almost identical spatial arrangement (Fig. 6B). A structure comparison between Ser-279 in quadruple mutant and Cys-279 in triple mutant yielded minor differences for recognizing RNA bases (Fig. 6C). Scrutinizing the detailed microenvironment of Cys-279, through van der Waals interactions, the side chain thiol group can contribute to stabilization of the target adenine RNA base conformation in the RNA strand (Fig. 6D). Based on the dimeric crystal structure observation and comparison, repeat 5 of PPR10 can specifically target adenine in RNA element. With a phenylalanine at the 2nd position and a serine at the 5th position as a major recognition determinant, the 35th position in the repeat is suggested to be a cysteine, serine, or an amino acid with a small or nucleophilic side chain.
DISCUSSION
PPR proteins, initially discovered in Arabidopsis thaliana chloroplasts and mitochondria, constitute a large protein family. Ascribing to their sequence-specific RNA-binding activity, PPR proteins retain a range of essential functions in organelle gene expression in plants involving RNA editing, splicing, cleavage, and translational activation (3, 5, 6, 33). It is worth mentioning that PPR proteins of distinct RNA binding properties with their target RNA undertake their respective diversified roles (6). In the process of RNA stabilization and translational activation, PPR tracts invariably exert tight binding affinity and complete sequestration on the non-coding RNA element. On another occasion for PPR protein-mediated RNA editing or other related functions, the low-affinity binding activity of PPR proteins allows the timely dissociation from RNA for subsequent translation execution (6). In the recent study, utilizing maize chloroplast protein PPR10 as the model protein target, we determined the crystal structure of PPR protein in complex with its native RNA target psaJ and validated the RNA recognition code involving residues by structure-guided mutagenesis studies (15). Intrigued by the intertwined homodimer configuration of PPR10 in the crystal with RNA and other reported PPR10 dimerization results, we sought to understand the important determining factors for dimeric state formation. In this study, we attempted to unveil the molecular mechanisms underpinning the dimerization state determination.
Biochemical results presented here define a critical role of the variable lengths of amino-terminal fragment not only in determining PPR10 dimerization state but also in RNA binding capacity. Albeit it is proposed the mature portion of PPR10 commence roughly from residues ∼30–40 (21), disparate prediction results for chloroplast transit peptide cleavage site of residues ∼50–90 prior to PPR repeat region (residues 107–771) are retrieved from softwares such as ChloroP Server, PrediSi Software, SignalP Server, or TargetP Server (34–37). The identification of PPR10 mature form in plant still awaits further in vivo appraisals. In plant chloroplasts, a large variety of proteases, including stromal processing peptidases, may alternatively excise a PPR protein, leaving variable lengths of amino termini as divergent mature functional forms. It is previously reported that the amino-terminal portions in a series of tetratricopeptide repeat (TPR) proteins are closely related to protein stability, dimerization formation, and interacting capacity with other protein partners (38–40). Similarly, a strong emphasis can be hence rationally placed on the amino termini of PPR proteins.
Interestingly, the subtle alteration of a single amino acid (C430S) can significantly impact the dimerization state of PPR10. A single amino missense mutation as the least phylogenetic change can result in the distinct conformation, dimerization, and even functional diversity. In fact, a series of possible post-translational modifications in plants, including phosphorylation, sumoylation (41), and nitrosylation (42), leads to distinct mature protein forms. Thus, the strategy of variable length the amino-terminal fragments or alteration of key residues in PPR proteins, corresponding to distinct dimerization states and RNA binding affinities, explicitly implies not only an intrinsic regulatory mechanism for PPR proteins involved pathways in plants but also a tantalizing clue for potential PPR protein engineering in biotechnological application.
Within the PPR protein family, different members may adopt certain dimerization states to undertake their respective functional roles, exemplified by the fact that THA8, THA8L, PPR4, and PPR5 are monomers, whereas HCF152 exists as a homodimer (23–27). It is even more rational to speculate that a plant cell is capable to utilize different PPR proteins of distinctive dimerization states to trigger the corresponding downstream signal responses.
In sum, we have identified the amino-terminal boundaries to be the determinants of the dimerization state of PPR10 in this study. In addition, we found a single cysteine to serine amino acid alteration in repeat 10 of PPR10 favors the shift toward dimerization of the protein. Furthermore, structural comparison shows that in a PPR repeat, in cooperation with other key residues, either cysteine or serine at 35th position can recognize adenine RNA nucleotide in the overall dimer conformation.
Acknowledgments
We acknowledge the China National Center for Protein Sciences Beijing for providing the facility support. We thank F. Yu and J. He at the Shanghai Synchrotron Radiation Facility for on-site assistance.
This work was supported by Ministry of Science and Technology Grant 2011CB910501 (to N. Y.) and Projects 91017011 (to N. Y.), 31070644 (to N. Y.), and 31200567 (to P. Y.) of the National Natural Science Foundation of China and Project 2662014PY026 (to P. Y.) of the Fundamental Research Funds for the Central Universities.
The atomic coordinates and structure factors (code 4OE1) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- PPR
- pentatricopeptide repeat
- SEC
- size exclusion chromatography
- NTD
- N-terminal domain.
REFERENCES
- 1. Small I. D., Peeters N. (2000) The PPR motif – a TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47 [DOI] [PubMed] [Google Scholar]
- 2. Kotera E., Tasaka M., Shikanai T. (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330 [DOI] [PubMed] [Google Scholar]
- 3. Schmitz-Linneweber C., Small I. (2008) Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends Plant Sci. 13, 663–670 [DOI] [PubMed] [Google Scholar]
- 4. Lurin C., Andrés C., Aubourg S., Bellaoui M., Bitton F., Bruyère C., Caboche M., Debast C., Gualberto J., Hoffmann B., Lecharny A., Le Ret M., Martin-Magniette M. L., Mireau H., Peeters N., Renou J. P., Szurek B., Taconnat L., Small I. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16, 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nakamura T., Yagi Y., Kobayashi K. (2012) Mechanistic insight into pentatricopeptide repeat proteins as sequence-specific RNA-binding proteins for organellar RNAs in plants. Plant Cell Physiol. 53, 1171–1179 [DOI] [PubMed] [Google Scholar]
- 6. Barkan A., Small I. (2014) Pentatricopeptide Repeat Proteins in Plants. Annu. Rev. Plant. Biol. 65, 415–442 [DOI] [PubMed] [Google Scholar]
- 7. Saha D., Prasad A. M., Srinivasan R. (2007) Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 45, 521–534 [DOI] [PubMed] [Google Scholar]
- 8. Cushing D. A., Forsthoefel N. R., Gestaut D. R., Vernon D. M. (2005) Arabidopsisemb175 and other ppr knockout mutants reveal essential roles for pentatricopeptide repeat (PPR) proteins in plant embryogenesis. Planta 221, 424–436 [DOI] [PubMed] [Google Scholar]
- 9. Hu J. (2012) The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 24, 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Desloire S. (2003) Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep. 4, 588–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bentolila S., Alfonso A. A., Hanson M. R. (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. U.S.A. 99, 10887–10892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Z. (2006) Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18, 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mootha V. K., Lepage P., Miller K., Bunkenborg J., Reich M., Hjerrild M., Delmonte T., Villeneuve A., Sladek R., Xu F., Mitchell G. A., Morin C., Mann M., Hudson T. J., Robinson B., Rioux J. D., Lander E. S. (2003) Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc. Natl. Acad. Sci. U.S.A. 100, 605–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tian T., Ikeda J., Wang Y., Mamat S., Luo W., Aozasa K., Morii E. (2012) Role of leucine-rich pentatricopeptide repeat motif-containing protein (LRPPRC) for anti-apoptosis and tumourigenesis in cancers. Eur. J. Cancer 48, 2462–2473 [DOI] [PubMed] [Google Scholar]
- 15. Yin P., Li Q., Yan C., Liu Y., Liu J., Yu F., Wang Z., Long J., He J., Wang H. W., Wang J., Zhu J. K., Shi Y., Yan N. (2013) Structural basis for the modular recognition of single-stranded RNA by PPR proteins. Nature 504, 168–171 [DOI] [PubMed] [Google Scholar]
- 16. Ban T., Ke J., Chen R., Gu X., Tan M. H., Zhou X. E., Kang Y., Melcher K., Zhu J. K., Xu H. E. (2013) Structure of a PLS-class pentatricopeptide repeat protein provides insights into mechanism of RNA recognition. J. Biol. Chem. 288, 31540–31548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ke J., Chen R. Z., Ban T., Zhou X. E., Gu X., Tan M. H., Chen C., Kang Y., Brunzelle J. S., Zhu J. K., Melcher K., Xu H. E. (2013) Structural basis for RNA recognition by a dimeric PPR-protein complex. Nat. Struct. Mol. Biol. 20, 1377–1382 [DOI] [PubMed] [Google Scholar]
- 18. Barkan A., Rojas M., Fujii S., Yap A., Chong Y. S., Bond C. S., Small I. (2012) A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 8, e1002910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yagi Y., Hayashi S., Kobayashi K., Hirayama T., Nakamura T. (2013) Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS One 8, e57286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yagi Y., Nakamura T., Small I. (2014) The potential for manipulating RNA with pentatricopeptide repeat proteins. Plant J. 78, 772–782 [DOI] [PubMed] [Google Scholar]
- 21. Pfalz J., Bayraktar O. A., Prikryl J., Barkan A. (2009) Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28, 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prikryl J., Rojas M., Schuster G., Barkan A. (2011) Mechanism of RNA stabilization and translational activation by a pentatricopeptide repeat protein. Proc. Natl. Acad. Sci. U.S.A. 108, 415–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khrouchtchova A., Monde R. A., Barkan A. (2012) A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA 18, 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmitz-Linneweber C., Williams-Carrier R. E., Williams-Voelker P. M., Kroeger T. S., Vichas A., Barkan A. (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18, 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B., Barkan A. (2008) The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell Biol. 28, 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Williams-Carrier R., Kroeger T., Barkan A. (2008) Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14, 1930–1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Meierhoff K., Felder S., Nakamura T., Bechtold N., Schuster G. (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15, 1480–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Otwinowski Z., Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 29. Collaborative Computational, P. (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Adams P. D. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 32. Nakamura T., Meierhoff K., Westhoff P., Schuster G. (2003) RNA-binding properties of HCF152, an Arabidopsis PPR protein involved in the processing of chloroplast RNA. Eur. J. Biochem. 270, 4070–4081 [DOI] [PubMed] [Google Scholar]
- 33. Filipovska A., Rackham O. (2012) Modular recognition of nucleic acids by PUF, TALE and PPR proteins. Mol. Biosyst. 8, 699–708 [DOI] [PubMed] [Google Scholar]
- 34. Emanuelsson O., Nielsen H., von Heijne G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hiller K., Grote A., Scheer M., Münch R., Jahn D. (2004) PrediSi: prediction of signal peptides and their cleavage positions. Nucleic Acids Res. 32, W375–W379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nielsen H., Engelbrecht J., Brunak S., von Heijne G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10, 1–6 [DOI] [PubMed] [Google Scholar]
- 37. Emanuelsson O., Nielsen H., Brunak S., von Heijne G. (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 38. Main E. R., Xiong Y., Cocco M. J., D'Andrea L., Regan L. (2003) Design of stable alpha-helical arrays from an idealized TPR motif. Structure 11, 497–508 [DOI] [PubMed] [Google Scholar]
- 39. Thebault P., Chirgadze D. Y., Dou Z., Blundell T. L., Elowe S., Bolanos-Garcia V. M. (2012) Structural and functional insights into the role of the N-terminal Mps1 TPR domain in the SAC (spindle assembly checkpoint). Biochem. J. 448, 321–328 [DOI] [PubMed] [Google Scholar]
- 40. Kang C., Ye H., Chia J., Choi B. H., Dhe-Paganon S., Simon B., Schütz U., Sattler M., Yoon H. S. (2013) Functional role of the flexible N-terminal extension of FKBP38 in catalysis. Sci. Rep. 3, 2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park H. J., Kim W. Y., Park H. C., Lee S. Y., Bohnert H. J., Yun D. J. (2011) SUMO and SUMOylation in plants. Mol. Cells 32, 305–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Feng J., Wang C., Chen Q., Chen H., Ren B., Li X., Zuo J. (2013) S-nitrosylation of phosphotransfer proteins represses cytokinin signaling. Nat. Commun. 4, 1529. [DOI] [PubMed] [Google Scholar]
- 43. DeLano W. L. (2002) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
- 44. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]