Abstract
Short interspersed nuclear elements (SINEs) are non-autonomous non-LTR retroelements that are present in most eukaryotic species. While SINEs have been intensively investigated in humans and other animal systems, they are poorly studied in plants, especially in wheat (Triticum aestivum). We used quantitative PCR of various wheat species to determine the copy number of a wheat SINE family, termed Au SINE, combined with computer-assisted analyses of the publicly available 454 pyrosequencing database of T. aestivum. In addition, we utilized site-specific PCR on 57 Au SINE insertions, transposon methylation display and transposon display on newly formed wheat polyploids to assess retrotranspositional activity, epigenetic status and genetic rearrangements in Au SINE, respectively. We retrieved 3706 different insertions of Au SINE from the 454 pyrosequencing database of T. aestivum, and found that most of the elements are inserted in A/T-rich regions, while approximately 38% of the insertions are associated with transcribed regions, including known wheat genes. We observed typical retrotransposition of Au SINE in the second generation of a newly formed wheat allohexaploid, and massive hypermethylation in CCGG sites surrounding Au SINE in the third generation. Finally, we observed huge differences in the copy numbers in diploid Triticum and Aegilops species, and a significant increase in the copy numbers in natural wheat polyploids, but no significant increase in the copy number of Au SINE in the first four generations for two of three newly formed allopolyploid species used in this study. Our data indicate that SINEs may play a prominent role in the genomic evolution of wheat through stress-induced activation.
Keywords: SINE, transposable elements, allopolyploidy, genome evolution, wheat, methylation
Introduction
Transposable elements (TEs) are sequences of DNA that are capable of replicating themselves independently of the systematic replication of genomic DNA. These sequences are divided into two classes, based on their mode of transposition: class I retroelements (or retrotransposons) transpose by reverse transcribing their own transcripts and reintegrating into the genome, and class II DNA elements transpose by excising themselves from one locus and integrating into another (Wicker et al., 2007). Short interspersed nuclear elements (SINEs) are class I transposons, which range in size from 80 to 500 bp, contain a PolIII promoter at their 5′ end, and may contain a terminator sequence (T+ SINEs) and/or a central ‘body’ sequence (Deragon and Zhang, 2006). SINEs are non-autonomous, as they contain no coding sequences, but rely on long interspersed nuclear elements (LINEs) to supply the enzymes required for their retrotransposition (i.e. reverse transcriptase, RNaseH and endonuclease; Kramerov and Vassetzky, 2011). SINEs resemble non-coding RNAs, such as tRNA, 7SL RNA and 5S RNA (Kapitonov and Jurka, 2003); while primates contain mostly 7SL RNA-like SINEs, other eukaryotes primarily harbor tRNA-like SINEs.
SINEs have frequently been used to explore phylogenetic relationships, based on insertional polymorphism, in humans (Perna et al., 1992; Roy-Engel et al., 2001; Salem et al., 2003; Hedges et al., 2004; Xing et al., 2007), rodents (Churakov et al., 2010), salmon (Murata et al., 1993), reptiles (Piskurek et al., 2006), birds (Watanabe et al., 2006; Kriegs et al., 2007) and cetaceans and artiodactyls (Nikaido et al., 1999). In plants, several SINE families have been discovered, such as in Brassica napus (Deragon et al., 1994), Oryza sativa (Umeda et al., 1991), Nicotiana tabacum (Yoshioka et al., 1993), Myotis daubentoni (Borodulina and Kramerov, 1999) and others (Deragon and Zhang, 2006). The evolutionary history of a SINE element (Au SINE) has been studied in divergent plant species, showing its wide distribution and ancient origin before the divergence between monocots and eudicots (Fawcett et al., 2006). The diversity of the same element in diploid and polyploid Aegilops species has also been investigated (Han-yu and Jian-bo, 2006). The Au SINE family consists of insertions with a structure characteristic of tRNA SINEs: namely a PolIII promoter that includes an A-box and a B-box at the 5′ end and a short poly(T) at the 3′ end (Koval et al., 2011).
Plants may undergo speciation by polyploidization through one of two mechanisms: (i) autopolyploidy, which involves doubling of the plant genome to form a new species with twice the number of chromosome sets, and (ii) allopolyploidy, which is hybridization of two related plant species and doubling of the genome to produce a new species that includes both the parental genomes (Feldman and Levy, 2005). Allopolyploid species exhibit rapid and reproducible genetic and epigenetic changes soon after hybridization (Yaakov and Kashkush, 2011b). These changes include activation of miniature transposons (Yaakov and Kashkush, 2012), transcriptional activation of retrotransposons, changes in the cytosine methylation status of transposons, and DNA rearrangements in transposon sequences (Yaakov and Kashkush, 2011b). The ‘genomic stress’ caused by inter-specific hybridization is only one of many biotic and abiotic stresses that may activate transposable elements (Mansour, 2007). In addition, DNA hypomethylation in rice has been shown to change the expression of endogenous genes via activation of promoters from LTR retrotransposable elements antisense to the gene (Kashkush and Khasdan, 2007). Thus, allopolyploidy-induced changes in genomic function may have an epigenetic basis.
Hexaploid wheat (Triticum aestivum) is a relatively recent allopolyploid, and is the result of the combination of three related genomes: first, Triticum urartu (genome AA; 2n = 2x = 14) hybridized with an as yet undiscovered species from section Sitopsis, probably similar to Aegilops speltoides (genome BB; 2n = 2x = 14) to produce Triticum dicoccoides (genome AABB; 2n = 4x = 28); second, Aegilops tauschii (genome DD; 2n = 2x = 14) hybridized with T. dicoccoides to produce T. aestivum (genome AABBDD; 2n = 6x = 42) (Feldman and Levy, 2005). This allopolyploid wheat system is useful in the study of the impact of ‘genomic stress’ on TE activity. Furthermore, the availability of newly formed wheat polyploids, mimicking the evolution of wheat, allow us to examine very early events that influence TEs immediately after polyploidization events.
In this study, we retrieved 3706 Au SINE insertions using the 454 pyrosequencing database of the wheat line Chinese Spring, and analyzed them in detail. In addition, we provide compelling evidence for clear mobilization of Au SINE between the first and second generations of a newly formed allohexaploid. Furthermore, we quantified this element, based on an in silico analysis of a 454 database and quantitative PCR in 40 species and accessions of Triticum and Aegilops, and found huge differences in Au SINE copy number among species and accessions. Finally, we showed a correlation between Au SINE retrotranspositional activity and hypermethylation in newly formed allopolyploids.
Results and discussion
In silico analysis of Au SINE
It is known that SINEs are one of the most abundant retrotransposons in humans, e.g. Alu elements reach up to a million copies (Xing et al., 2009). Thus, SINEs appear to play a prominent role in human evolution (Xing et al., 2009). SINEs in plants and specifically in wheat have been studied to a limited extent. In this study, in order to evaluate the level of proliferation of SINEs in the wheat genome using mak software (Yang and Hall, 2003), we searched the publicly available 454 pyrosequencing database of Chinese Spring for the presence of a SINE family, termed Au SINE, which was previously characterized in Aegilops umbellulata (Yasui et al., 2001), and found 3706 different intact insertions. The mean length of all retrieved Au SINEs was 172.8 ± 8.4 bp, compared to 181 bp for the consensus sequence that was used as input in MAK, and the sequence similarity among the Au SINE insertions was approximately 80% (Figure S1a). Multiple sequence alignment of the retrieved Au SINEs showed higher conservation of the elements at their termini compared to the internal sequence, and particular conservation of the A and B box sequences, which are required for transcription by RNA polymerase III, and a 3′ end poly(T), which is required for template-primed reverse transcription (Figure S1a) (Kajikawa and Okada, 2002). In addition, in silico retrieval and analysis of Au SINE elements was also performed for a 454 pyrosequencing database of Ae. tauschii (see ‘Experimental procedures’), which resulted in 191 retrieved elements.
The relatively short sequence of Au SINE allowed us to identify and characterize intact elements from the unassembled 454 pyrosequencing databases. This means that the number of insertions identified may be under-estimated because some elements may be truncated. In addition, we investigated target-site duplications for all elements from both databases, and did not find a significant target-site preference (based on comparison of the target-site duplications of the various insertions of Au SINE, see ‘Experimental procedures’), indicating that the endonuclease used for nicking the target sites of Au SINE may not have sequence specificity (Figure S1b). However, when we carefully examined the flanking sequence of all elements, we found that, in most cases, Au SINEs were inserted in A/T-rich regions (Figures S2 and S3), similar to what has been reported for SINEs in both plant and animal systems (Jurka, 1997; Tatout et al., 1998; Lenoir et al., 2001). Given that these elements also have a poly(T) tail at their 3′ end, this finding may indicate that Au SINE has an insertional preference for poly(T) sites.
In order to test whether Au SINE is associated with wheat genes, we annotated the flanking sequence of each of the 3706 insertions from the T. aestivum database (up to several hundred base pairs on both sides of the element) using the EST and mRNA databases from the National Center for Biotechnology Information. We found unique ESTs flanking 1075 insertions and unique mRNA molecules flanking 344 insertions, while the remaining 2287 flanking sequences showed no similarity to transcribed sequences. These data indicate that approximately 38% of the Au SINEs were inserted in transcribed regions. In addition, we searched for Au SINE insertions in the limited National Center for Biotechnology Information database of annotated genomic sequences for wheat and found four insertions: three were inserted in introns, including a zinc finger protein (GQ422824.1), a VRN-B1 gene (AY747602.1) and a 5-methylcytosine DNA glycosylase (JF683316.1), while one was inserted 328 bp downstream of a putative protein kinase gene (AY368673.1) (Figure S4). The results indicate that hundreds, if not thousands, of Au SINEs may be inserted into or adjacent to wheat genes. Surprisingly, we found 62 unique ESTs and 13 unique mRNA molecules that were similar to the Au SINE sequence, indicating that approximately 2% of the SINE insertions may be transcribed under normal conditions. It is known that retrotransposon promoters retain activity under normal conditions and initiate transcription (Vicient et al., 2001; Kashkush et al., 2002, 2003; Nigumann et al., 2002; Kashkush and Khasdan, 2007).
Activity of Au SINE in newly formed allopolyploids and throughout wheat evolution
In order to assess the dynamics of Au SINE in vivo, we used site-specific PCR and transposon display assays. For site-specific PCR analysis, we used a pair of primers that flank each Au SINE insertion; thus, a site containing an Au SINE insertion (termed ‘full site’) will yield a longer PCR product than a site lacking this element (termed an ‘empty site’). Unlike DNA elements, RNA elements move via a ‘copy and paste’ mechanism, thus it is impossible to observe retrotranspositions in the offspring when analyzing an insertion (full site) that is present in one or both parental lines. For example, if we analyze a ‘full site’ in one or both parental lines using site-specific PCR, we cannot determine whether this element transposed in the offspring because it will not leave an ‘empty site’ behind. However, because DNA elements leave empty sites, we are able to track transpositions in offspring using the site-specific PCR assay (Yaakov and Kashkush, 2012). In this case, site-specific PCR analysis is informative only when analyzing a ‘full site’ in the offspring and determining whether this specific insertion is present in one or both parental lines. To this end, we examined 57 randomly selected Au SINE insertions, present in the 454 database, in four generations of a newly formed wheat allohexaploid (S1–S4) and their parental species, Triticum turgidum ssp. durum (accession TTR19) and Ae. tauschii (accession TQ27). We designed primers from the regions flanking each Au SINE insertion, and performed PCR using genomic DNA of newly formed allohexaploid plants and their parental lines as template. The PCR analysis revealed that 40 of the 57 Au SINE insertions examined were not present in the parental lines or in the S1–S4 generations of the newly formed allohexaploid, while 14 insertions were present in one of the two parental lines, and, as expected, an additive pattern (both full and empty site) was detected in the S1–S4 generations. Surprisingly, we observed three cases where the Au SINE insertion was not present in either parental line or the S1 generation, but appeared in the S2–S4 generations. Note that the experiment was repeated many times and the data were reproducible. The results indicate retrotransposition events, all occurring between the first and second generations of the allohexaploid, resulting in the appearance of a full site band on the agarose gel (Figure 1). Examination of the flanking sequences following retrotransposition showed one target-site duplication (Figure 1a) and two target-site deletions (Figure 1b and Figure S5). Sequence analysis of the bands showed the insertion of nearly complete elements, including the PolIII promoter region, into areas rich in poly(T) (Figure S2). These surprising results may be explained by three possible scenarios: (i) these are true retrotransposition events that occurred in natural hexaploid wheat (as the primers were designed based on the 454 database of natural hexaploid wheat) and in the newly formed allohexaploid at the same position, (ii) the Au SINE insertions are present in different accessions of the parental lines, and different accessions were mistakenly used for the PCR analysis, and (iii) the newly formed polyploids (S2–S4) were contaminated by a natural hexaploid wheat species.
Figure 1.
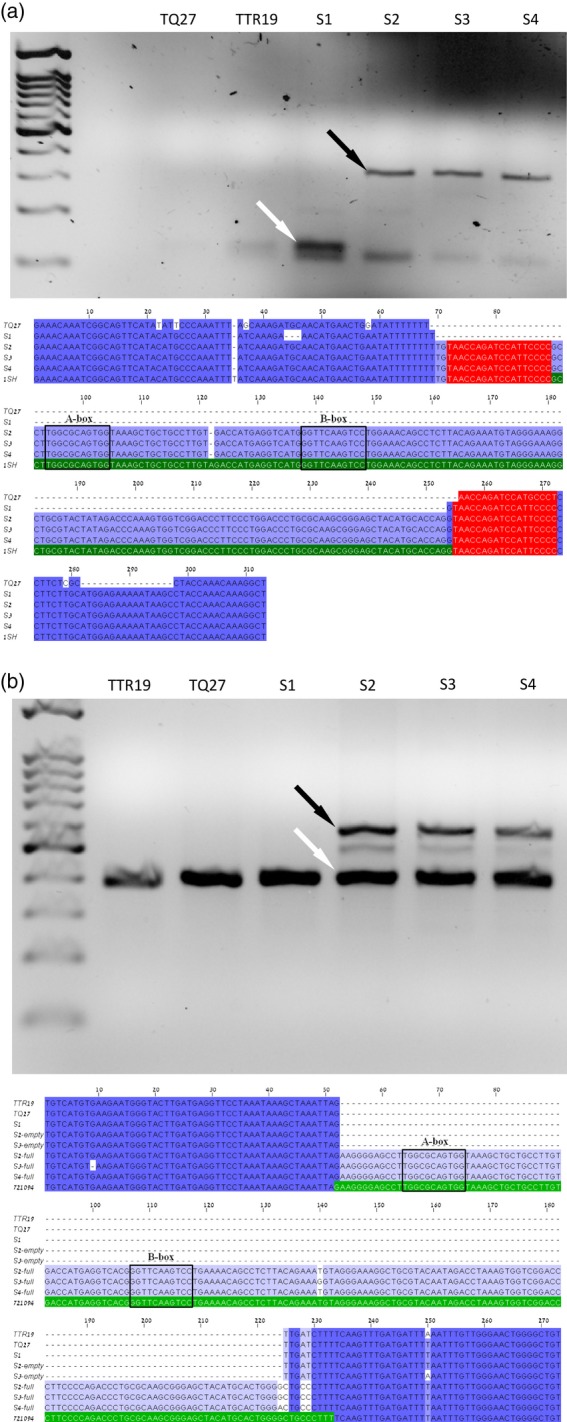
Retrotransposition of Au SINE in newly formed allohexaploid wheat. Site-specific PCR amplification and multiple sequence alignment of Au SINE insertion sites from a 454 pyrosequencing database of Chinese Spring for (a) PCR analysis of read GIABLPU08JR1SH showing a 133 bp empty site (white arrow) and a 314 bp full site (black arrow), and (b) PCR analysis of contig 721094 showing a 376 bp empty site (white arrow) and a 557 bp full site (black arrow). Note that the read and contig numbers indicate the sequence code from the 454 database. Each gel has a lane of DNA markers on the left. The multiple sequence alignment includes the original database sequence, with Au SINE colored green. The A- and B-box promoter sequences are indicated. T. turgidum ssp. durum (accession TTR19), Ae. tauschii (accession TQ27) and four generations of their hexaploid progeny (S1–S4) were used as templates. Read GIABLPU08JR1SH (a) includes target-site duplications (colored orange), and contig 721094 (b) includes a target-site deletion, shown as a mis-aligned 5 bp sequence between nucleotides 225 and 229.
We have performed several experiments to rule out scenarios 2 and 3, which indicate technical errors in the experimental system. In our system, we used only TQ27 (genome DD) and TTR19 (genome BBAA) accessions as the true parental lines (Figure S6). However, to rule out scenario 2, we performed the PCR analysis in ten accession of Ae. tauschii (genome DD) and 10–33 accessions of T. turgidum [genome BBAA, including durum and dicoccoides (wild emmer)] and found that none of the accessions contain any of the three Au SINE insertions (see examples in Figure S7). This experiment prompted us to test whether any of the AA or the BB species contain any of the three Au SINEs, thus we performed the PCR analysis on three accessions of T. urartu (the donor of AA genome to wheat), 17 accessions of BB species (eight accessions of Aegilops searsii and nine accessions of Ae. speltoides, the best candidates to donate the BB genome to wheat), and found that none of the AA and BB accessions contain any of the three Au SINE insertions. This analysis indicates that the three Au SINE insertions most probably occurred in hexaploid wheat after allohexaploidization, approximately 10 000 years ago. The question that arises is whether all natural hexaploid wheat species contain these three Au SINE insertions. To answer this, we performed the PCR analysis on 14 accessions of T. aestivum collected from all over the world (Figure S8). The first Au SINE insertion (Figure 1a) was present in nine of the 14 T. aestivum accessions (Figures S8 and S9), the second Au SINE insertion (Figure 1b) was present in three of the 14 T. aestivum accessions (Figures S8 and S9), and the third Au SINE insertion (Figure S5) was present in nine of the 14 T. aestivum accessions (Figures S8 and S9). These data, together with the finding that none of the diploid and tetraploid donors contain the three insertions, indicate that independent retrotransposition events occurred in some accessions of natural T. aestivum at the same position. Interestingly, only two of the 14 T. aestivum accessions (TAA01 and PI436506) contained the three Au SINE insertions. Note that the hexaploid accession (CS42) that was used to generate the 454 database (Brenchley et al., 2012) also contains the three Au SINE insertions.
Furthermore, in order to rule out scenario 3 (contamination of the newly formed polyploids by natural wheat species) and to verify the reproducibility of the retrotransposition events, PCR was performed for each of the transposed Au SINE sites using the first and second generations of two independent crosses of the newly formed polyploid. The results showed complete reproducibility of all retrotranspositions (Figure S10). Note that the only natural hexaploid that may contaminate our samples or hybridize with the S1 generation of the newly formed allohexaploid in the greenhouse is TAA01, as this is the only accession grown in our greenhouses. The rest of the hexaploid seed material was supplied by the US Department of Agriculture and was grown in a special growth room. TAA01 contains the three Au SINE insertions (Figure S9). We used PCR-based DNA markers and showed that TAA01 cannot be a parental line of the S2–S4 plants. Detailed analysis was performed using transposon display (TD) markers. Because of the dominant nature of the TD markers, any band that is present in the homozygous TAA01 plant and absent in the S1 generation must be present in the S2–S4 generations if TAA01 hybridizes with S1 plants. Analysis of approximately 280 bands (four primer combinations of TD reactions, Figure S11) revealed that TAA01 cannot be a parent of S2–S4 plants.
Additionally, we used TD to assess rearrangements of Au SINEs in the newly formed allohexaploid, similarly to previously reported analyses (Kraitshtein et al., 2010; Yaakov and Kashkush, 2011a). TD allows us to search for rearrangements (absence of Au SINE-containing bands or novel Au SINE-containing bands in newly formed allohexaploid versus parental lines). Of the 280 Au SINE-containing bands analyzed by TD, 22 were novel bands (present in the newly formed allohexaploid and absent in both parental lines), indicating that these are new insertions of Au SINE, and 36 bands were present in one or both parental lines and absent in the newly formed allohexaploid (see examples in Figure S11), indicating that these Au SINE-containing sequences underwent elimination or other rearrangements, as shown previously (Kraitshtein et al., 2010). Note that the presence or absence of bands was seen in the first generations of the newly formed allohexaploid. Interestingly, in some cases, the novel TD bands in the newly formed allohexaploid were also present in the natural hexaploid, indicating that same insertion occurred when the natural hexaploid was created approximately 10 000 years ago. These data support the three specific retrotrasposition events observed in this study (Figure 1 and Figure S5), in which we analyzed Au SINE insertion loci in the natural hexaploid and found clear retrotransposition in the newly formed allohexaploid. Note that the combination of parental lines that was used here produces a newly formed allohexaploid that resembles natural hexaploid wheat. The occurrence of transposition events that occur in a natural hexaploid in newly formed allohexaploid species was previously demonstrated for MITEs (Yaakov and Kashkush, 2012). In addition, previous reports have shown that similar reproducible genetic rearrangements that occurred in newly formed wheat allopolyploids also occurred in a natural hexaploid (Kashkush et al., 2002; Kraitshtein et al., 2010). This may indicate the value of using newly formed allopolyploids to study the rapid, short term changes of evolution through allopolyploidization.
Au SINE dynamics in natural wheat species
In order to study the dynamics of SINEs throughout evolution of wheat, we analyzed the relative quantity of Au SINEs in 40 accessions of ten wheat species by quantitative PCR. The relative quantities were then compared to the copy number derived from the 454 database of T. aestivum (see details in ‘Experimental procedures’), by assuming that this copy number was identical to the relative quantity of T. aestivum (accession TAA01; Figure 2). The results showed a huge amplification burst of Au SINE between the diploid species (Ae. searsii, Ae. speltoides, Ae. tauschii, T. urartu, Aegilops monococcum, Aegilops sharonensis and Aegilops longissima), with a mean copy number of 540 ± 323, and the polyploid species (T. turgidum ssp. dicoccoides, T. turgidum ssp. durum and T. aestivum), with a mean copy number of 3289 ± 362, which cannot be explained by the additive values of any accessions from any parental species. Furthermore, the coefficient of variation for Au SINE copy number was highest in Ae. speltoides (0.38, compared to 0.2 for Ae. searsii, 0.17 for Ae. tauschii and 0.19 for T. urartu), suggesting that SINEs are active in the B-genome. In order to verify the accuracy of relative quantification by quantitative PCR, we compared the ratio of T. aestivum Au SINE amplification by quantitative PCR and the mean Au SINE amplification by quantitative PCR of all Ae. tauschii accessions to the ratio of Au SINE copy number (based on the retrieved elements from the 454 pyrosequencing databases) from T. aestivum and Ae. tauschii. Based on quantitative PCR analysis, the mean copy number of Au SINE in Ae. tauschii was estimated to be 318, while the number of retrieved elements from the Ae. tauschii 454 database was 191. This confirms that the copy numbers observed by quantitative PCR are of the same order of magnitude as the number of retrieved elements from the database. Note that we retrieved only nearly intact elements by MAK, thus the number of retrieved elements may be under-estimated.
Figure 2.
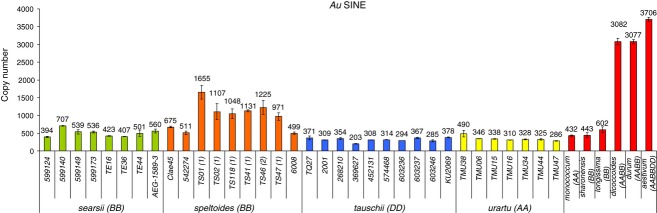
Copy numbers for Au SINE in ten wheat species, including 40 accessions, based on estimation of element copy number in a 454 database for Chinese Spring, and relative quantification of the element by quantitative PCR on genomic DNA. Values are means ± standard deviations based on three technical replicates.
Copy number variation of Au SINE in newly formed allopolyploids
We then studied whether allopolyploidization affects the copy number of Au SINE in three newly formed allopolyploid combinations using quantitative PCR, and found that the relative Au SINE quantities in newly formed allohexaploids (from parental species T. turgidum ssp. durum and Ae. tauschii) and allotetraploids (from parental lines Triticum monococcum and Ae. sharonensis) showed no significant increase in SINE copy number (varying between 78 and 106% of the expected values). In contrast, newly formed allotetraploids (from parental lines T. urartu and Ae. longissima) showed a significant increase in the first generation to 187% from the expected values [1666 ÷ (499 + 392)] the RQ of the tetraploid divided by the additive RQs of its parental lines, returning to expected levels in the second generation (Figure 3). This indicates that, in some newly formed allopolyploid combinations in wheat, there may be massive dynamics of SINEs.
Figure 3.
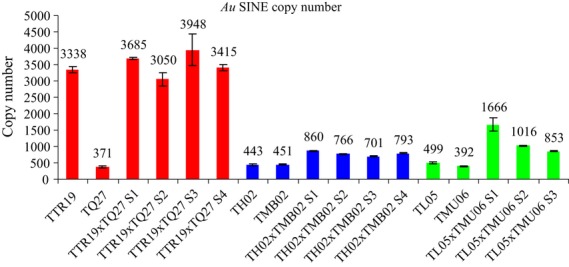
Copy numbers for Au SINE in the newly formed polyploid systems, calculated from quantitative PCR and the estimated copy number in Chinese Spring. The polyploid systems include: T. turgidum ssp. durum (accession TTR19) and Ae. tauschii (accession TQ27) (colored red), T. monococcum ssp. aegilopoides (accession TMB02) and Ae. sharonensis (accession TH02) (colored blue), T. urartu (accession TMU06) and Ae. longissima (accession TL05) (colored green), and three or four generations of the newly formed polyploids (S1–S4) for each pair mentioned. Values are means ± standard deviations based on three technical replicates.
Epigenetic regulation of Au SINE in newly formed allopolyploids
In order to study the epigenetic regulation of SINEs, we tested the methylation status of Au SINEs in the newly formed allohexaploid versus its parental lines, using transposon methylation display (Kashkush and Khasdan, 2007; Kraitshtein et al., 2010; Yaakov and Kashkush, 2011a). Transposon methylation display analysis allows an assessment of the methylation status of CCGG sites flanking TEs, using HpaII and MspI restriction enzymes, which recognize CCGG sites but have different sensitivity to cytosine methylation. Thus, the methylation status of CCGG sites may be measured based on the number of sites that are polymorphic between HpaII and MspI digestions (Kashkush and Khasdan, 2007; Kraitshtein et al., 2010; Yaakov and Kashkush, 2011a). Analysis of approximately 90 CCGG sites revealed high overall cytosine methylation surrounding Au SINE (78.38%) in natural T. aestivum (accession TAA01) and a significant increase (hypermethylation) in a newly formed allohexaploid, from 37.74 ± 3.22% in the parental species and two generations (S1 and S2) of the newly formed allohexaploid, to 75.28% in the third generation. Interestingly, the level of methylation in the third generation of the newly formed allohexaploid was similar to that for the natural allohexaploid. Recent studies in wheat have investigated the methylation status near several TE families in detail (Kraitshtein et al., 2010; Yaakov and Kashkush, 2011a,b; Zhao et al., 2011). In one study, similar to the results reported here, hypomethylation of Veju-flanking CCGG sites was seen in the S1 and S2 generations of the newly formed allohexaploid, and hypermethylation was seen in the third generation of the synthetic allohexaploid (Kraitshtein et al., 2010).
Conclusions
TEs are implicated in creating genetic variation, giving rise to diversification of related species and diploidization of polyploid species. This genetic variation may arise as a result of evolutionary changes (following the divergence of two species) or revolutionary changes (over short periods of stress) (Feldman and Levy, 2005). The genetic and epigenetic involvement of TEs in genomic diversification has been studied extensively (Feschotte and Pritham, 2007; Mansour, 2007; Slotkin and Martienssen, 2007; Yaakov and Kashkush, 2011b). Transcriptional activation of retrotransposons was shown previously in wheat (Kashkush et al., 2003) and in other polyploidy systems such as Arabidopsis (Madlung et al., 2005), and the transcriptional activation of LTR retrotransposons correlated with their methylation status (Kashkush and Khasdan, 2007). However, in all studies, research has been restricted mainly to DNA transposons and LTR retrotransposons and epigenetic regulation of the element was not associated with its transpositional activity.
In this study, we focused on the genome-wide impact of a SINE family in various wheat species and newly formed allopolyploid wheat. Au SINE copy number was observed to be most variable in accessions of Ae. speltoides, which suggests that Au SINE has transposed in the B-genome of this species. This evidence suggests activity of this transposon family during the evolution of wheat.
Here we present compelling evidence for retrotransposition of Au SINE elements following allopolyploidization of wheat. Interestingly, we observed target-site deletion produced by Au SINE insertions, as well as target-site duplication. The similar numbers of novel and absent bands from the TD of Au SINE suggest a combination of amplification and removal of the element (probably by retrotransposition and recombinational mechanisms, respectively) following allopolyploidization, which may have led to a rapid increase in epigenetic regulation of these elements by cytosine methylation, as indicated by the hypermethylation in the S3 generation. Taken together, we conclude that the Au SINE family has retained its capacity for activity throughout the evolution of wheat, and has probably been epigenetically de-regulated following polyploidization events in wheat. Thus, we infer that Au SINE has contributed to the diversification of wheat species and differentiation of polyploid species by inducing genetic and epigenetic changes, and possibly by modifying gene regulation. However, the underlying mechanism by which independent retrotransposition events may occur at the same position following independent allohexaploidization events, as seen in various natural hexaploid wheat species (Figure S8) and in a newly formed allohexaploid (Figure 1 and Figure S5), remains to be determined. Epigenetic mechanisms, such as DNA methylation and structural modification of heterochromatin accompanying the creation of the nascent allopolyploid species, may play a prominent role in restricting the sites available for DNA recombination and transposition.
Experimental procedures
Plant material
In this study, 40 accessions of ten wheat species (Table S1) were used. In addition, three combinations of newly formed allopolypoids were used: (i) four generations (S1–S4) of a newly formed allotetraploid and its parental lines, T. monococcum ssp. aegilopoides (accession TMB02) and Ae. sharonensis (accession TH02), (ii) three generations (S1–S3) of a newly formed allotetraploid and its parental lines, T. urartu (accession TMU06) and Ae. longissima (accession TL05), and (iii) four generations (S1–S4) of a newly formed allohexaploid and its parental lines, T. turgidum ssp. durum (accession TTR19) and Ae. tauschii (accession TQ27). Seed material was kindly provided by Moshe Feldman, Plant Sciences Department, the Weizmann Institute of Science, Rehovot, Israel and the US Department of Agriculture (http://www.ars-grin.gov/npgs/acc/acc_queries.html). Genomic DNA was isolated using a DNeasy plant kit (Qiagen, Hilden, Germany) from green leaves of plants that were approximately 4 weeks old. Total RNA was isolated under the same conditions, using an Aurum total RNA mini kit (Bio-Rad, Hercules, CA, USA), and converted to cDNA using a Verso® cDNA kit (Thermo Scientific, Waltham, MA, USA).
Computer-assisted analysis
The sequence for Au SINE was retrieved from Repbase (http://www.girinst.org/repbase/). Access to the 454 pyrosequencing database for Chinese Spring, which was used to estimate Au SINE copy number, was kindly provided by members of the Chinese Spring sequencing consortium (http://www.cerealsdb.uk.net/). For validation of the quantitative PCR and bioinformatics analyses, the publicly available 454 pyrosequencing database of Ae. tauschii (National Center for Biotechnology Information Sequence Read Archive submission SRA052214) was used. SINE elements were retrieved from these databases using MITE analysis kit (mak) software (Yang and Hall, 2003) with an E-value of 10−3. MAK was originally designed for downstream analysis of MITE, but some functions, such as sequence retrieval from databases, are applicable to other types of TEs as well (Janicki et al., 2011). Each hit was retrieved with 100 bp of flanking sequence, and redundant reads were excluded by comparing the flanking sequences with to the databases. Searching of the publicly available Triticum and Aegilops genomic, EST and mRNA sequences from the National Center for Biotechnology Information was performed using BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) with an E-value threshold of 10−10. The multiple sequence alignment of Au SINE retrotranspositions was performed with ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The multiple sequence alignments for the Au SINE elements and target-site duplications retrieved from the database were performed using MAFFT (Katoh et al., 2009), and all logos were produced using WebLogo 3.3 (Crooks et al., 2004). Nucleotide density calculations for Au SINE insertion sites were performed using the ‘Density’ program from EMBOSS (http://bips.u-strasbg.fr/EMBOSS/).
Site-specific PCR
Primer design for the flanking regions and insertion analysis was performed using primer3 version 0.4.0 (http://frodo.wi.mit.edu/primer3/). The reaction consisted of 12 μl ultrapure water (Biological Industries, Beit Haemek, Israel), 2 μl of 10 x Taq DNA polymerase buffer (EURX, Gdansk, Poland), 2 μl of 25 mm MgCl2 (EURX), 0.8 μl of 2.5 mm dNTPs, 0.2 μl Taq DNA polymerase (5 U μl−1, EURX), 1 μl of each site-specific primer (50 ng μl−1), and 1 μl of template genomic DNA (approximately 50 ng μl−1). The PCR conditions for these reactions were 94°C for 3 min, 3o cycles of 94°C for 1 min, 60°C for 1 min and 72°C for 1 min, and 72°C for 3 min. The PCR products were purified using an Invisorb® Spin PCRapid kit (Invitek, STRATEC Molecular GmbH, Berlin, Germany) or extracted from an agarose gel using an Invisorb® Spin DNA extraction kit (Invitek), ligated into the pGEM-T easy vector (Promega, Madison, WI, USA), used to transform Escherichia coli DH5α and sequenced using a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA) at Ben-Gurion University, Israel. Primer sequences are listed in Table S2.
Transposon display and transposon methylation display
Transposon display was performed by restricting 0.5 μg of genomic DNA with MseI (recognition site TTAA), ligating the fragments to adaptors, and PCR amplifying using primers specific to the adaptor and to the transposon. The PCR adaptor primer included three selective bases (CTG), the products were run on 5% polyacrylamide gel, and equally-sized bands were counted as one site. A change in the band pattern was counted when a band appeared or disappeared in a certain polyploid generation (S1–S4) and all subsequent generations. Transposon methylation display was performed in the same manner, but using HpaII and MspI (recognition site CCGG), which cleave DNA differently, depending on its cytosine methylation, and with four selective nucleotides in the adaptor primer (TCAG). In this case, the overall methylation was calculated by counting all bands present in the lane corresponding to restriction by one enzyme but not the other, and dividing by all amplified sites in each sample. More details on TD and transposon methylation display are provided in Kashkush and Khasdan (2007), Kraitshtein et al. (2010) and Yaakov and Kashkush (2011a).
Quantitative PCR
Primers for Au SINE (forward: 5′-AGCTGCTGCCTTGTGACCAT-3′; reverse: 5′-GGGAAGGGTCCGACCACTT-3′) were designed from a conserved region (based on multiple sequence alignment) using primer express version 2.0 (Applied Biosystems). All quantitative PCR reactions included 7.5 μl KAPA SYBR® FAST Universal Master Mix (KAPA Biosystems, Boston, MA, USA), 5 μl DNA template (0.24 ng μl−1) or a 50 x dilution of cDNA template, 1 μl Au SINE forward primer (10 μm), 1 μl Au SINE reverse primer (10 μm), 0.3 μl ROX (a reference dye used to normalize changes in background fluorescence) and 0.2 μl ultrapure water (Biological Industries). For reactions on genomic DNA, the VRN1 gene was used to normalize for variations in input template concentration, and Ae. tauschii (accession TQ27) was used a reference sample (thus its value is always 1). All samples were corrected for ploidy level, as the VRN1 gene exists as two copies in diploids, four copies in tetraploids and six copies in hexaploids (Kraitshtein et al., 2010). In addition, the primers were used to amplify serial dilutions of DNA or cDNA, and their efficiency was calculated as (10−1/y – 1) × 100, where y is the slope of the linear regression. Thus, the relative quantity (RQ) for genomic DNA samples was calculated as (ploidy level) × (2 × primer efficiency)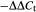 , where ΔΔCt = (Ct(sample) –Ct(VRN1)) – (Ct(TQ27) – Ct(VRN1)) (Livak and Schmittgen, 2001). In order to calculate the copy number of Au SINE in each sample, we multiplied the copy number for T. aestivum (3706 copies, as determined from the 454 database) with the ratio of the RQ of each sample to the RQ of T. aestivum. For example, the RQ of T. monococcum was 1.163358 and the RQ of T. aestivum was 9.976081, thus the copy number of Au SINE in T. monococcum is 3706 × 1.163358 ÷ 9.976081 = 432 (rounded to the nearest integer). It is important to note that to ensure that the quantitative PCR amplification was accurate, we used two different primer pairs for three different transposons, one MITE (Yaakov et al., 2013) and two LTR retrotransposons, and received similar results. In addition, a previous study successfully predicted the copy number of the MITE mPing in various rice species using the same method (Baruch and Kashkush, 2012).
, where ΔΔCt = (Ct(sample) –Ct(VRN1)) – (Ct(TQ27) – Ct(VRN1)) (Livak and Schmittgen, 2001). In order to calculate the copy number of Au SINE in each sample, we multiplied the copy number for T. aestivum (3706 copies, as determined from the 454 database) with the ratio of the RQ of each sample to the RQ of T. aestivum. For example, the RQ of T. monococcum was 1.163358 and the RQ of T. aestivum was 9.976081, thus the copy number of Au SINE in T. monococcum is 3706 × 1.163358 ÷ 9.976081 = 432 (rounded to the nearest integer). It is important to note that to ensure that the quantitative PCR amplification was accurate, we used two different primer pairs for three different transposons, one MITE (Yaakov et al., 2013) and two LTR retrotransposons, and received similar results. In addition, a previous study successfully predicted the copy number of the MITE mPing in various rice species using the same method (Baruch and Kashkush, 2012).
Acknowledgments
We would like to thank Guojun Yang (Toronto University, Canada) for providing the updated stand-alone mak software, Moshe Feldman (The Weismann Institute of Science, Israel) and Hakan Ozkan (Sütçü İmam University, Turkey) for providing the seed material, and Mike Bevan (John Innes Center, Norwich, UK), Neil Hall (Liverpool University, UK) and Keith Edwards (Bristol University, UK) for providing access to the 454 database, and for their permission to publish the data. This work was supported by a grant from the Israel Science Foundation (number 142/08) to K.K.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Figure S1. Sequence logos of retrieved elements and their TSDs.
Figure S2. Nucleotide density of novel insertion sites.
Figure S3. Sequence logos from all 3′ and 5′ flanking sequences surrounding Au SINE.
Figure S4. Association of Au SINE with annotated genes.
Figure S5. Retrotransposition of Au SINE in newly formed allohexaploid wheat.
Figure S6. Spikes from parental species and their newly formed polyploid offspring.
Figure S7. Examples of site-specific PCR for Au SINE retrotransposition sites on natural diploid and tetraploid species.
Figure S8. Site-specific PCR for Au SINE retrotransposition sites on natural hexaploid species.
Figure S9. Site-specific PCR for novel retrotranspositions in natural T. aestivum.
Figure S10. Au SINE retrotransposition events in two independent crosses of the newly formed polyploid.
Figure S11. Transposon display of Au SINE in natural and synthetic polyploids.
Table S1. Wheat accessions used in the study.
Table S2. Primer sequences and expected product sizes for all Au SINE sites used in the study.
References
- Baruch O, Kashkush K. Analysis of copy-number variation, insertional polymorphism, and methylation status of the tiniest class I (TRIM) and class II (MITE) transposable element families in various rice strains. Plant Cell Rep. 2012;31:885–893. doi: 10.1007/s00299-011-1209-5. [DOI] [PubMed] [Google Scholar]
- Borodulina OR, Kramerov DA. Wide distribution of short interspersed elements among eukaryotic genomes. FEBS Lett. 1999;457:409–413. doi: 10.1016/s0014-5793(99)01059-5. [DOI] [PubMed] [Google Scholar]
- Brenchley R, Spannagl M, Pfeifer M, et al. Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature. 2012;491:705–710. doi: 10.1038/nature11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churakov G, Sadasivuni MK, Rosenbloom KR, Huchon D, Brosius J, Schmitz J. Rodent evolution: back to the root. Mol. Biol. Evol. 2010;27:1315–1326. doi: 10.1093/molbev/msq019. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deragon JM, Zhang XY. Short interspersed elements (SINEs) in plants: origin, classification, and use as phylogenetic markers. Syst. Biol. 2006;55:949–956. doi: 10.1080/10635150601047843. [DOI] [PubMed] [Google Scholar]
- Deragon JM, Landry BS, Pelissier T, Tutois S, Tourmente S, Picard G. An analysis of retroposition in plants based on a family of SINEs from Brassica napus. J. Mol. Evol. 1994;39:378–386. doi: 10.1007/BF00160270. [DOI] [PubMed] [Google Scholar]
- Fawcett JA, Kawahara T, Watanabe H, Yasui Y. A SINE family widely distributed in the plant kingdom and its evolutionary history. Plant Mol. Biol. 2006;61:505–514. doi: 10.1007/s11103-006-0026-7. [DOI] [PubMed] [Google Scholar]
- Feldman M, Levy AA. Allopolyploidy – a shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005;109:250–258. doi: 10.1159/000082407. [DOI] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han-yu G, Jian-bo W. Au elements and their evolution in some allopolyploid genomes of Aegilops. Agric. Sci. China. 2006;5:895–904. [Google Scholar]
- Hedges DJ, Callinan PA, Cordaux R, Xing J, Barnes E, Batzer MA. Differential Alu mobilization and polymorphism among the human and chimpanzee lineages. Genome Res. 2004;14:1068–1075. doi: 10.1101/gr.2530404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki M, Rooke R, Yang G. Bioinformatics and genomic analysis of transposable elements in eukaryotic genomes. Chromosome Res. 2011;19:787–808. doi: 10.1007/s10577-011-9230-7. [DOI] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl Acad. Sci. USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajikawa M, Okada N. LINEs mobilize SINEs in the eel through a shared 3’ sequence. Cell. 2002;111:433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. A novel class of SINE elements derived from 5S rRNA. Mol. Biol. Evol. 2003;20:694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- Kashkush K, Khasdan V. Large-scale survey of cytosine methylation of retrotransposons and the impact of readout transcription from long terminal repeats on expression of adjacent rice genes. Genetics. 2007;177:1975–1985. doi: 10.1534/genetics.107.080234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Gene loss, silencing and activation in a newly synthesized wheat allotetraploid. Genetics. 2002;160:1651–1659. doi: 10.1093/genetics/160.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat. Genet. 2003;33:102–106. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with MAFFT. Methods Mol. Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- Koval AP, Veniaminova NA, Kramerov DA. Additional box B of RNA polymerase III promoter in SINE B1 can be functional. Gene. 2011;487:113–117. doi: 10.1016/j.gene.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Kraitshtein Z, Yaakov B, Khasdan V, Kashkush K. Genetic and epigenetic dynamics of a retrotransposon after allopolyploidization of wheat. Genetics. 2010;186:801–812. doi: 10.1534/genetics.110.120790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramerov DA, Vassetzky NS. SINEs. Wiley Interdiscip. Rev. RNA. 2011;2:772–786. doi: 10.1002/wrna.91. [DOI] [PubMed] [Google Scholar]
- Kriegs J, Matzke A, Churakov G, Kuritzin A, Mayr G, Brosius J, Schmitz J. Waves of genomic hitchhikers shed light on the evolution of gamebirds (Aves: Galliformes) BMC Evol. Biol. 2007;7:190. doi: 10.1186/1471-2148-7-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenoir A, Lavie L, Prieto JL, Goubely C, Cote JC, Pélissier T, Deragon JM. The evolutionary origin and genomic organization of SINEs in Arabidopsis thaliana. Mol. Biol. Evol. 2001;18:2315–2322. doi: 10.1093/oxfordjournals.molbev.a003778. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Madlung A, Tyagi AP, Watson B, Jiang HM, Kagochi T, Doerge RW, Martienssen R, Comai L. Genomic changes in synthetic Arabidopsis polyploids. Plant J. 2005;41:221–230. doi: 10.1111/j.1365-313X.2004.02297.x. [DOI] [PubMed] [Google Scholar]
- Mansour A. Epigenetic activation of genomic retrotransposons. J. Cell Mol. Biol. 2007;6:99–107. [Google Scholar]
- Murata S, Takasaki N, Saitoh M, Okada N. Determination of the phylogenetic relationships among Pacific salmonids by using short interspersed elements (SINES) as temporal landmarks of evolution. Proc. Natl Acad. Sci. USA. 1993;90:6995–6999. doi: 10.1073/pnas.90.15.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigumann P, Redik K, Matlik K, Speek M. Many human genes are transcribed from the antisense promoter of L1 retrotransposon. Genomics. 2002;79:628–634. doi: 10.1006/geno.2002.6758. [DOI] [PubMed] [Google Scholar]
- Nikaido M, Rooney AP, Okada N. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interpersed elements: hippopotamuses are the closest extant relatives of whales. Proc. Natl Acad. Sci. USA. 1999;96:10261–10266. doi: 10.1073/pnas.96.18.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna NT, Batzer MA, Deininger PL, Stoneking M. Alu insertion polymorphism – a new type of marker for human-population studies. Hum. Biol. 1992;64:641–648. [PubMed] [Google Scholar]
- Piskurek O, Austin C, Okada N. Sauria SINEs: novel short interspersed retroposable elements that are widespread in reptile genomes. J. Mol. Evol. 2006;62:630–644. doi: 10.1007/s00239-005-0201-5. [DOI] [PubMed] [Google Scholar]
- Roy-Engel AM, Carroll ML, Vogel E, Garber RK, Nguyen SV, Salem AH, Batzer MA, Deininger PL. Alu insertion polymorphisms for the study of human genomic diversity. Genetics. 2001;159:279–290. doi: 10.1093/genetics/159.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem AH, Ray DA, Xing J, Callinan PA, Myers JS, Hedges DJ, Garber RK, Witherspoon DJ, Jorde LB, Batzer MA. Alu elements and hominid phylogenetics. Proc. Natl Acad. Sci. USA. 2003;100:12787–12791. doi: 10.1073/pnas.2133766100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Tatout C, Lavie L, Deragon JM. Similar target site selection occurs in integration of plant and mammalian retroposons. J. Mol. Evol. 1998;47:463–470. doi: 10.1007/pl00006403. [DOI] [PubMed] [Google Scholar]
- Umeda M, Ohtsubo H, Ohtsubo E. Diversification of the rice waxy gene by insertion of mobile DNA elements into introns. Jpn. J. Genet. 1991;66:569–586. doi: 10.1266/jjg.66.569. [DOI] [PubMed] [Google Scholar]
- Vicient CM, Jaaskelainen MJ, Kalendar R, Schulman AH. Active retrotransposons are a common feature of grass genomes. Plant Physiol. 2001;125:1283–1292. doi: 10.1104/pp.125.3.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Nikaido M, Tsuda TT, Inoko H, Mindell DP, Murata K, Okada N. The rise and fall of the CR1 subfamily in the lineage leading to penguins. Gene. 2006;365:57–66. doi: 10.1016/j.gene.2005.09.042. [DOI] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, et al. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Xing J, Witherspoon DJ, Ray DA, Batzer MA, Jorde LB. Mobile DNA elements in primate and human evolution. Am. J. Phys. Anthropol. 2007;134(Suppl. 45):2–19. doi: 10.1002/ajpa.20722. [DOI] [PubMed] [Google Scholar]
- Xing JC, Zhang YH, Han K, et al. Mobile elements create structural variation: analysis of a complete human genome. Genome Res. 2009;19:1516–1526. doi: 10.1101/gr.091827.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaakov B, Kashkush K. Massive alterations of the methylation patterns around DNA transposons in the first four generations of a newly formed wheat allohexaploid. Genome. 2011a;54:42–49. doi: 10.1139/G10-091. [DOI] [PubMed] [Google Scholar]
- Yaakov B, Kashkush K. Methylation, transcription, and rearrangements of transposable elements in synthetic allopolyploids. Int. J. Plant Genomics. 2011b;2011:569826. doi: 10.1155/2011/569826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaakov B, Kashkush K. Mobilization of Stowaway-like MITEs in newly formed allohexaploid wheat species. Plant Mol. Biol. 2012;80:419–427. doi: 10.1007/s11103-012-9957-3. [DOI] [PubMed] [Google Scholar]
- Yaakov B, Ben-David S, Kashkush K. Genome-wide analysis of Stowaway-like MITEs in wheat reveals high sequence conservation, gene association, and genomic diversification. Plant Physiol. 2013;161:486–496. doi: 10.1104/pp.112.204404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GJ, Hall TC. MAK, a computational tool kit for automated MITE analysis. Nucleic Acids Res. 2003;31:3659–3665. doi: 10.1093/nar/gkg531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui Y, Nasuda S, Matsuoka Y, Kawahara T. The Au family, a novel short interspersed element (SINE) from Aegilops umbellulata. Theor. Appl. Genet. 2001;102:463–470. [Google Scholar]
- Yoshioka Y, Matsumoto S, Kojima S, Ohshima K, Okada N, Machida Y. Molecular characterization of a short interspersed repetitive element from tobacco that exhibits sequence homology to specific transfer-RNAs. Proc. Natl Acad. Sci. USA. 1993;90:6562–6566. doi: 10.1073/pnas.90.14.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N, Zhu B, Li M, Wang L, Xu L, Zhang H, Zheng S, Qi B, Han F, Liu B. Extensive and heritable epigenetic remodeling and genetic stability accompany allohexaploidization of wheat. Genetics. 2011;188:499–510. doi: 10.1534/genetics.111.127688. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequence logos of retrieved elements and their TSDs.
Figure S2. Nucleotide density of novel insertion sites.
Figure S3. Sequence logos from all 3′ and 5′ flanking sequences surrounding Au SINE.
Figure S4. Association of Au SINE with annotated genes.
Figure S5. Retrotransposition of Au SINE in newly formed allohexaploid wheat.
Figure S6. Spikes from parental species and their newly formed polyploid offspring.
Figure S7. Examples of site-specific PCR for Au SINE retrotransposition sites on natural diploid and tetraploid species.
Figure S8. Site-specific PCR for Au SINE retrotransposition sites on natural hexaploid species.
Figure S9. Site-specific PCR for novel retrotranspositions in natural T. aestivum.
Figure S10. Au SINE retrotransposition events in two independent crosses of the newly formed polyploid.
Figure S11. Transposon display of Au SINE in natural and synthetic polyploids.
Table S1. Wheat accessions used in the study.
Table S2. Primer sequences and expected product sizes for all Au SINE sites used in the study.


