Abstract
Light-harvesting complex II (LHCII) contains three highly homologous chlorophyll-a/b-binding proteins (Lhcb1, Lhcb2 and Lhcb3), which can be assembled into both homo- and heterotrimers. Lhcb1 and Lhcb2 are reversibly phosphorylated by the action of STN7 kinase and PPH1/TAP38 phosphatase in the so-called state-transition process. We have developed antibodies that are specific for the phosphorylated forms of Lhcb1 and Lhcb2. We found that Lhcb2 is more rapidly phosphorylated than Lhcb1: 10 sec of ‘state 2 light’ results in Lhcb2 phosphorylation to 30% of the maximum level. Phosphorylated and non-phosphorylated forms of the proteins showed no difference in electrophoretic mobility and dephosphorylation kinetics did not differ between the two proteins. In state 2, most of the phosphorylated forms of Lhcb1 and Lhcb2 were present in super- and mega-complexes that comprised both photosystem (PS)I and PSII, and the state 2-specific PSI–LHCII complex was highly enriched in the phosphorylated forms of Lhcb2. Our results imply distinct and specific roles for Lhcb1 and Lhcb2 in the regulation of photosynthetic light harvesting.
Keywords: Arabidopsis, Lhcb2, LHCII, phosphorylation, state transitions
Introduction
Protein phosphorylation regulates many important biological processes in plants. One of the first plant processes found to be regulated by protein phosphorylation was the so-called ‘state transition’. State transitions, which were first described by Bonaventura and Myers (1969) and Murata and Sugahara (1969), and subsequently by, for example, Bennett (1977) and Allen (1992), alleviate the imbalance in the photosynthetic machinery during unequal excitation of two photosystems, PSI and PSII. This imbalance is sensed by changes in the redox state of the plastoquinone (PQ) pool that activate a specific kinase, which phosphorylates an amino acid close to the N-termini of the Lhcb1 and Lhcb2 polypeptides of LHCII (light-harvesting complex of PSII; see (Jansson, 1994) for review). As a result, part of LHCII becomes functionally detached from PSII and acts as an external antenna of PSI. The name ‘state transition’ originates from the technique used to detect the changes; the fluorescence properties of intact leaves or algal cells can be reversibly shifted from ‘state 1’ to ‘state 2’ simply by changing the spectrum of incident light. The kinetics of the process are typically in the order of minutes. Ten minutes are often long enough to reach a steady state of fluorescence and the process follows first-order kinetics (Islam, 1987; Larsson et al., 1987). In higher plants, only some of the Lhcb proteins, ca. 20–25%, are thought to participate in state transitions (Vener, 2007). Although the two photosystems have different excitation spectra – this is the reason why changes in light quality can induce excitation imbalance – the vast majority of photons is absorbed equally well by the two photosystems. Nevertheless, photons with wavelengths >680 nm are preferentially used by PSI, although recently it has been suggested that PSII electron transfer can also be driven by far-red light of wavelengths up to 800 nm (Thapper et al., 2009). State transitions are important for plants that live under a canopy, where the majority of the visible light has already been absorbed by the photosynthetic apparatus of other leaves, but where a large proportion of the photons with wavelengths around 700 nm are transmitted. In addition to changes in light quality, variations in light quantity can also provoke reversible LHCII protein phosphorylation (Rintamaki et al., 2000), and it is likely that, under natural conditions, state transitions may be more important for acclimation of the photosynthetic apparatus to fluctuations in light quantity than for adjustments to changes in light quality (Tikkanen and Aro, 2011; Tikkanen et al., 2012).
In the last few years, the molecular details of state transitions have been elucidated. A crucial breakthrough was the identification of a kinase responsible for the phosphorylation of LHCII, first in Chlamydomonas (where it is named Stt7; Depege et al., 2003) and later in Arabidopsis, in which it is named STN7 (Bellafiore et al., 2005). Subsequent studies, using for example mutants that lacked STN7, have produce much important information regarding the function and regulation of the kinase. A homologous kinase, STN8, primarily phosphorylates PSII core subunits (Bonardi et al., 2005; Vainonen et al., 2005). PPH1/TAP38 phosphatase, which catalyzing the reverse reaction, has also been identified (Pribil et al., 2010; Shapiguzov et al., 2010). The elucidation of the three-dimensional structures of different supercomplexes of PSI and PSII with LHCII has also been important for a better understanding of the phosphorylation and subsequent lateral migration of LHCII in the membrane. There is, however, one area in which progress has been limited: the importance of substrate specificity in the regulatory phosphorylation of Lhcb proteins. The major LHCII complex, which consists of the highly homologous Lhcb1, Lhcb2 and Lhcb3 gene products, has long been known to occur as trimers in the thylakoid membrane, although monomeric forms may also exist. Lhcb1 is typically found in large excess compared with the other subunits, so Lhcb13 trimers are abundant; most of the Lhcb2 and Lhcb3 subunits are probably in Lhcb12/Lhcb2 and Lhcb12/Lhcb3 trimers but Lhcb1/Lhcb22 trimers are also found (Standfuss and Kuhlbrandt, 2004), and there are also reports of Lhcb1/Lhcb2/Lhcb3 heterotrimers (Caffarri et al., 2004; Jackowski, 2004). It is conceivable that additional kinds of LHCII trimers exist and that the relative abundance of each trimer is a simple consequence of the relative stoichiometries of the Lhcb1, Lhcb2 and Lhcb3 polypeptides. Different LHCII subpopulations have different phosphorylation properties (Bassi et al., 1988a,b). It should be noted that Lhcb1 and Lhcb2 are also phosphorylated at other sites, but this phosphorylation does not seem to be catalyzed by STN7 or STN8 (Ingelsson and Vener, 2012).
Lhcb3 differs from Lhcb1 and Lhcb2 as it lacks the ‘state-transition phosphorylation site’, a Thr (or, in some species, a Ser) residue very close to the N-terminus of the protein. Lhcb12/Lhcb3 trimers appear also to have a specific binding pocket on PSII (Damkjaer et al., 2009) and not to participate in state transitions. In Arabidopsis that lack Lhcb3, the binding pocket is instead occupied by trimers that lack Lhcb3, and state transitions in such plants are slightly faster than in wild-type plants, presumably because there are more substrates available for the STN7 kinase, as all remaining LHCII trimer subunits have three potential phosphorylation sites. However, to date, it has not been possible to investigate any putative differences in the function between the Lhcb1 and Lhcb2 subunits using molecular genetics. The Lhcb1 and Lhcb2 gene products are extremely similar; only 14 amino acids consistently distinguish them from each other (Jansson and Gustafsson, 1990). Yet both protein sequences have been conserved for over 300 million years (MY) and they are thus likely to possess specific functions (Jansson and Gustafsson, 1990). Arabidopsis Lhcb1 is encoded by five genes and Lhcb2 by three genes. Some of these genes lie in close proximity on the chromosomes and it has not been possible to generate quintuple Lhcb1 or triple Lhcb2 knockout plants, only antisense plants that lack both Lhcb1 and Lhcb2 (Andersson et al., 2003; Ruban et al., 2003).
So far, all quantitative studies on LHCII phosphorylation have been performed using either radioactively labelled phosphate, or antibodies against phosphothreonine (P-Thr) that are unable to distinguish between phosphorylated forms of Lhcb1 and Lhcb2, hereafter named P-Lhcb1 and P-Lhcb2, at least not in Arabidopsis in which Lhcb1 and Lhcb2 have the same electrophoretic mobility. Moreover, the antibodies also naturally react with the other ca. 25 thylakoid proteins that can be phosphorylated (Aro et al., 2004; Pesaresi et al., 2009). In addition, different brands – even different batches – of P-Thr antibodies show significant differences in specificity towards the different thylakoid phosphoproteins (see e.g. Martinsuo et al., 2003; Ingelsson and Vener, 2012). The ProQ phospho-staining method combined with the Sypro total protein stain has also been used to quantify thylakoid protein phosphorylation (e.g. Tikkanen et al., 2006; Fristedt et al., 2010), but the difficulty in separating the Lhcb1 and Lhcb2 proteins remains a problem. Mass spectrometry methods, on the other hand, have limited quantitative power (Vainonen et al., 2005). Overall, carrying out quantitative studies on the localization of the P-Lhcb1 and P-Lhcb2 in different supermolecular complexes and thylakoid domains is not a trivial undertaking.
To overcome these limitations we have generated antibodies that specifically recognize Arabidopsis P-Lhcb1 or P-Lhcb2 and describe their use for investigating LHCII phosphorylation kinetics and dynamics in wild-type and mutant plants.
Results
Development of specific antibodies for the phosphorylated forms of Lhcb1 and Lhcb2
We designed and synthesized oligopeptides that contained the phosphorylated Thr residue and the conserved regions that surround the phosphorylation sites of Arabidopsis Lhcb1 and Lhcb2 (Figure 1) and used them to generate antibodies against P-Lhcb1 and P-Lhcb2. It should be noted that one of the five Lhcb1 gene products, Lhcb1.4, lacks the Thr residue at position 3 in the mature protein that corresponds to the canonical phosphorylation site. The possibility cannot be ruled out that this protein is instead phosphorylated at the Ser residue at position 4 in the protein; if so, phosphorylation at that position would not be picked up by our antibodies. The antisera were subjected to immunopurification to increase the activity and specificity of the antibodies. The antibodies (P-Lhcb1 and P-Lhcb2 antibodies) detected proteins of the expected size in wild-type Arabidopsis plants. To confirm that the antibodies recognized only the phosphorylated forms of Lhcb1 and Lhcb2, we used wild-type plants and knockout mutants that lacked Stn7 expression, the gene coding for LHCII kinase protein and therefore LHCII phosphorylation (SALK_073254; Bellafiore et al., 2005), or Stn8 (SALK_060869; Vainonen et al., 2005), the homologous kinase mainly phosphorylating core proteins of photosystem II, as well as the stn7stn8 double mutant. We also included psal, a knockout mutant (SALK_010554), as antisense psal line has been shown to have higher phosphorylation of LHCII in state 2 (Lunde et al., 2000). We exposed these lines to state 1 and state 2 conditions and assayed thylakoid proteins by immunoblot analysis (Figure 2). The signal from both P-Lhcb1 antibody and P-Lhcb2 antibody was much stronger in state 2 than in state 1 in wild-type as well as in stn8, and koPsaL plants (Figure 2). Only a minute signal was detected in state 2 in the stn7 line. Lhcb1 and Lhcb2 phosphorylation levels were only marginally affected in stn8, but was completely missing in the stn7xstn8 double mutant. Both Lhcb1 and Lhcb2 phosphorylation was elevated in the psal mutant in state 1, but not in state 2, and the phosphorylation level of Lhcb1 in stn8 in state 1 was slightly elevated.
Figure 1.

Alignment of N-terminal regions of premature Arabidopsis thaliana Lhcb1 and Lhcb2 protein isoforms.
Mature proteins are indicated in black and transit peptides in grey. The peptides used for obtaining P-Lhcb1 and P-Lhcb2 antibodies are indicated in bold and threonine residues phosphorylated by STN7 kinase are boxed.
Figure 2.
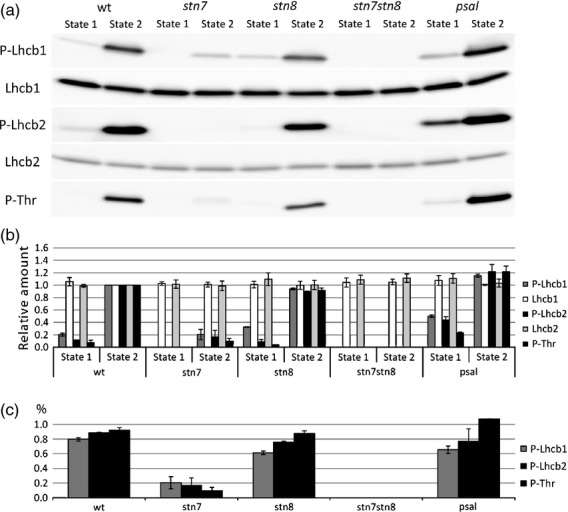
Specificity of P-Lhcb1 and P-Lhcb2 antibodies.
Arabidopsis wild-type (wt), stn7 (SALK_073254) and psal (SALK_010554), stn8 (SALK_060869), stn7stn8 plants were subjected to state 1 and state 2 light conditions, and then proteins were extracted and immunoblotted with antibodies specific against phosphorylated and non-phosphorylated Lhcb1 and Lhcb2 forms.
(a) Immunoblots. The specificity of antibodies is indicated on the left side.
(b) Densitometric analysis of immunoblots. Intensity of signals has been normalized respect to the value in wild-type state 2.
(c) Difference in phosphorylation between state 1 and state 2.
We also confirmed these data by analysing the samples using an antibody against P-Thr. The signals given by this antibody in the region of the gel corresponding to Lhcb1 and Lhcb2 (i.e. 25–30 kDa) matched fairly well the signals detected by the P-Lhcb1 and P-Lhcb2 antibodies, but the signal obtained with P-Thr antibody was lower in state 1 for all examined lines, compared with P-Lhcb1 and P-Lhcb2 antibodies. Taken together, these observations show that both P-Lhcb1 and P-Lhcb2 antibodies only recognized the phosphorylated forms of the proteins, and that STN8, in addition to STN7, could phosphorylate both proteins.
We compared the electrophoretic mobility of phosphorylated and non-phosphorylated forms of Lhcb1 and Lhcb2 by immunoblot analysis of the same electrophoretic lane cut in half and incubated in one case with either P-Lhcb1 antibody or Lhcb1 antibody, in the other case with P-Lhcb2 antibody or Lhcb2 antibody (Figure 3a). No difference in mobility between phosphorylated and non-phosphorylated forms of the proteins could be detected. In the same way, we also compared the mobilities of the phosphorylated forms of Lhcb1 and Lhcb2. A comparison of immunoblot patterns in Figure 3(b) using P-Lhcb1 antibody or P-Lhcb2 antibody and the P-Thr antibody indicated that the phosphorylated form of Lhcb1 may migrate slightly slower than the phosphorylated form of Lhcb2 (Figure 3b). However, the difference was small and irreproducible even when we performed electrophoretic separation on a different and longer gels allowing a better resolution and illustrating the need for specific antibodies against phosphorylated and non-phosphorylated forms of the proteins if the relative phosphorylation levels Lhcb1 and Lhcb2 are to be analysed.
Figure 3.
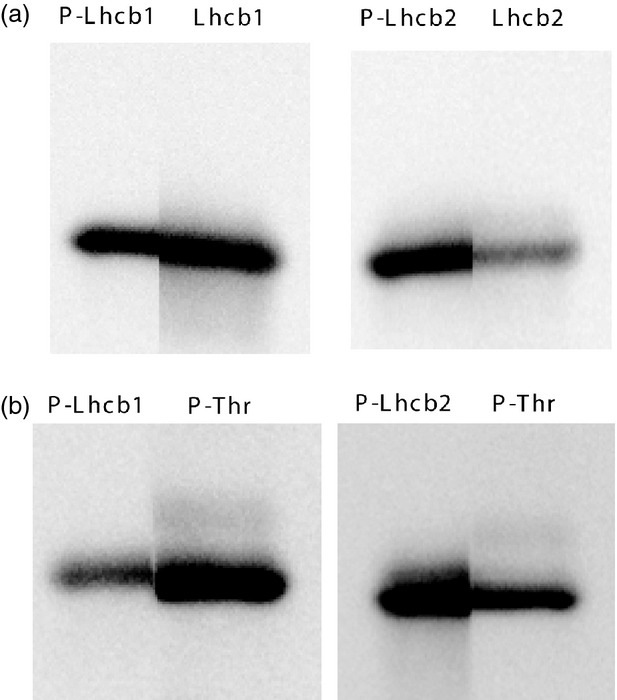
Comparison of gel mobilities of Lhcb1 and Lhcb2 proteins.
(a) Immunoblot analysis with P-Lhcb1/Lhcb1 and P-Lhcb2/Lhcb2 antibodies obtained by cutting the membrane through the middle of the electrophoretic lane.
(b) Immunoblot analysis of P-Lhcb1/P-Thr antibodies and PLhcb2/P-Thr antibodies.
Lhcb2 showed very rapid phosphorylation kinetics
To study the phosphorylation kinetics of Lhcb1 and Lhcb2, wild-type plants were first treated for 60 min with red light, supplemented with far-red light, to induce state 1 and then exposed to red light alone for different time intervals, to bring plants into state 2. The kinetics of phosphorylation was very different for the two proteins: Lhcb2 was more rapidly phosphorylated than Lhcb1 (Figure 4a). When expressed on a scale where 0% corresponds to the phosphorylation level in state 1 and 100% is the maximum phosphorylation level in state 2 of the wild-type (measured after 60 min when the reaction has reached steady state), within 10 sec after the light shift, Lhcb2 phosphorylation reached about 30% of the maximum level and after 10 min it was approximately 85% of the maximum. In contrast, Lhcb1 was phosphorylated to only 15% of the maximum level after 30 sec, and to only slightly more than 60% after 10 min. Again, P-Thr antibody was used for confirmation. In this case, two bands in the Lhcb1/2 region were recognized by the P-Thr antibody. The phosphorylation kinetics of both Lhcb1 and Lhcb2 during the transition from state 1 to state 2 were dependent only on the substrate specificity of the LHCII kinase; the kinetics in the KO mutant that lacked the phosphatase pph1/tap38 (SALK_025713C, (Pribil et al., 2010; Shapiguzov et al., 2010)) were similar to that described in wild-type plants (ca. 60–80% phosphorylation after 10 min). We also compared phosphorylation/dephosphorylation kinetics when state transitions were induced by shifts from dark to low light and vice versa. Although the changes in phosphorylation levels were less pronounced (Figure S2) Lhcb2 phosphorylation appeared also under these conditions to have faster kinetics.
Figure 4.
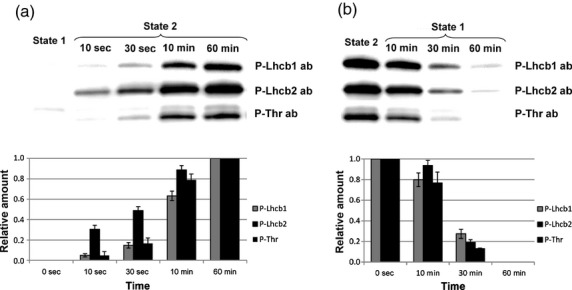
Phosphorylation (a) and dephosphorylation (b) kinetics of Arabidopsis Lhcb1 and Lhcb2 during state transitions, analysed using P-Lhcb1 and P-Lhcb2 antibodies.
Immunoblot analysis of phosphorylated proteins during State 1 and State 2 are shown at the top, while the lower graphs show quantification of the immunoblot data.
In order to study dephosphorylation kinetics, plants were treated with red light for 40 min to induce state 2 and then exposed to additional far-red light for different time intervals to induce dephosphorylation and return the plants to state 1. The dephosphorylation of both Lhcb1 and Lhcb2 was much slower than phosphorylation; after 10 min only 10–20% of both Lhcb1 and Lhcb2 was dephosphorylated (Figure 4b).
Lhcb1 and Lhcb2 phosphorylation kinetics were influenced by PSII antenna protein composition
We then studied the phosphorylation and dephosphorylation kinetics of Lhcb1 and Lhcb2 proteins in mutant and transgenic lines with modified PSII antenna composition. To this end, we compared wild-type Arabidopsis with knockout mutants that lacked Lhcb3 (Damkjaer et al., 2009), Lhcb5 (SALK_014869C) or Lhcb6 (SALK_035761C, (Kovacs et al., 2006)) and the npq4-1 mutant that lacked PsbS (Li et al., 2000). We also included a Lhcb2 antisense line (asLhcb2-12) that has been shown to be devoid of both Lhcb1 and Lhcb2 proteins (Andersson et al., 2003). However, some of the plants generated from seeds harvested from the asLhcb2-12 did not express such a pale phenotype as we expected. When several asLhcb2-12 plants were analysed for chlorophyll, Lhcb1 and Lhcb2 levels (Figure S1), there was a significant variation in the amount of, in particular, Lhcb1 between the individual plants, and this correlated with the chlorophyll content. Apparently, although the asLhcb2 construct consistently repressed the accumulation of Lhcb2 proteins, repression of Lhcb1 was variable. Only those asLhcb2-12 plants with a pale phenotype, as described before (Andersson et al., 2003) showed strong suppression of Lhcb1 and only these were used for further experiments.
Most mutants did not show significant differences from wild-type in the phosphorylation levels of either Lhcb1 or Lhcb2 after either 30 sec, 10 min or 60 min (Figure 5a,b). Similar results were also obtained using P-Thr antibodies (Figure 5c). We have previously shown that LHCII phosphorylation is more rapid in koLhcb3 mutants (Damkjaer et al., 2009). However, in order to pick up this subtle difference, more time points, and possibly more replicates, would be needed. The aim of this experiment was to carry out a broader screen to see whether absence of any antenna protein had a particularly large effect on the phosphorylation kinetics. The koLhcb6 and npq4 lines showed kinetics similar to those of wild-type plants, a finding that indicated that the absence of CP24 and PsbS did not significantly influence the rate of phosphorylation and, hence, state transitions. Phosphorylation of Lhcb1 was faster in koLhcb5 mutants, which showed significantly higher Lhcb1 phosphorylation after 30 sec than WT, and reached maximum phosphorylation within 10 min. This situation may indicate that the lack of CP26 either increases activation of the kinase or allows for better access to the substrate (LHCII).
Figure 5.
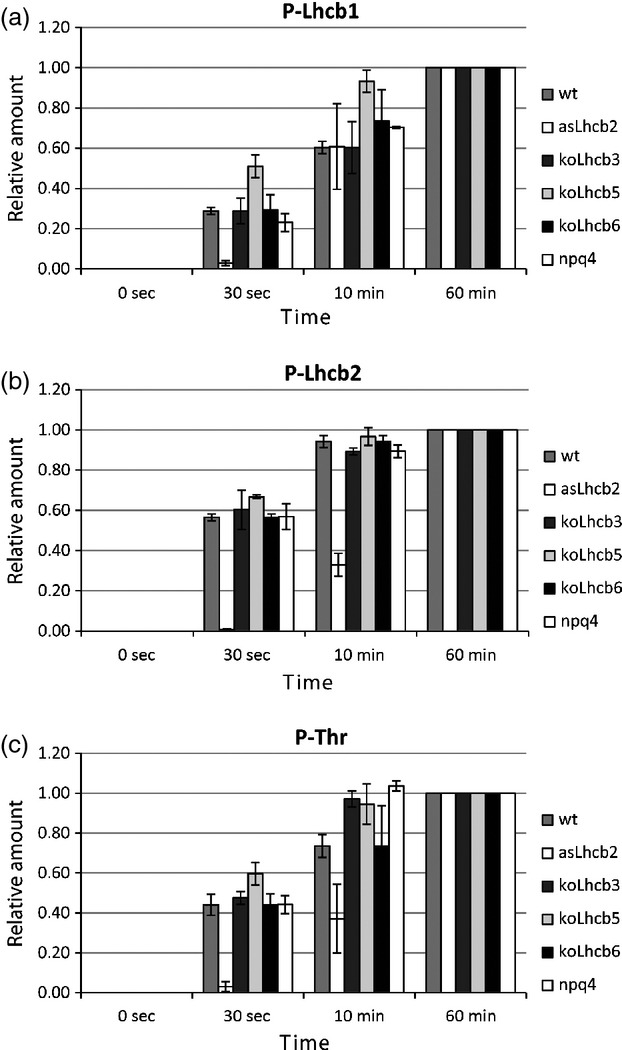
Phosphorylation kinetics for wild-type (wt) and asLhcb2, koLhcb3, koLhcb5, koLhcb6 and npq4-1 mutants with altered PSII antenna composition.
Quantification of immunoblot data for samples incubated with P-Lhcb1 (a), P-Lhcb2 (b) and P-Thr (c) antibodies. Values for wild-type (wt) have not been included, and letters A, B, and C are not present in the figure.
The residual amount of Lhcb1 in asLhcb2 plants had a lower phosphorylation level after 30 sec compared with that in wild-type, but after 10 min no difference was recorded (Figure 5a).
P-Lhcb1 and P-Lhcb2 are located in both LHCII trimers and large complexes
In order to study the distribution within supermolecular complexes of P-Lhcb1 and P-Lhcb2 in state 1 and 2, we subjected wild-type and psal thylakoids to large pore Blue Native (lpBN) electrophoresis and subsequent immunoblotting. Before thylakoid preparation, the plants were subjected for 60 min to far-red light or red light conditions in order to induce state 1 and 2, respectively. We used digitonin to solubilize the thylakoid membrane. Digitonin is a mild detergent and is known to preferably solubilize the stroma-exposed lamellae and grana margins, allowing preservation of weak protein–protein interactions (Järvi et al., 2011). This method thus allows for better visualization of, for example, less abundant state 2-specific complexes such as the LHCII–PSI complexes (Zhang and Scheller, 2004; Galka et al., 2012) and the PSI–PSII–LHCII mega-complexes (Järvi et al., 2011), which are found in these parts of the thylakoid membranes. To separate these high molecular mass complexes, the membranes solubilized with 1% digitonin were subjected to lpBN electrophoresis (Figure 6). The polypeptide composition of each complex was determined by 2D-electrophoresis (Figure S3).
Figure 6.
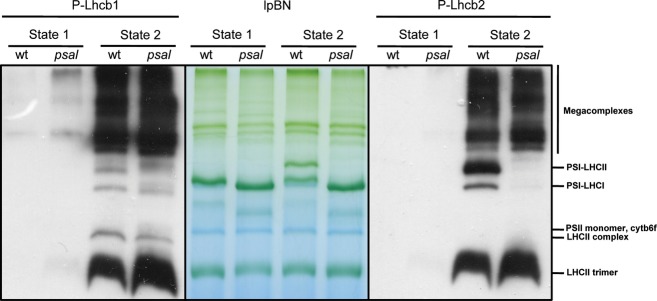
Immunolocalization of phosphorylated and non-phosphorylated forms of Lhcb1 and Lhcb2 in thylakoid membrane complexes of Arabidopsis wild-type and psaL knockout (KO) mutant.
Thylakoids from plants in state 1 and state 2 conditions were separated by large pore Blue Native (lpBN) gel electrophoresis. From left: Immunoblot with P-Lhcb1 antibody; lpBN; immunoblot with P-Lhcb2 antibody.
Most of both the phosphorylated and the non-phosphorylated forms of the Lhcb1 and Lhcb2 proteins were found in the high molecular mass mega-complexes. However, free LHCII trimers also contained P-Lhcb1 and P-Lhcb2. In state 1, Lhcb1 and Lhcb2 phosphorylation was very low. Only in psal mutants some phosphorylation was observed in large complexes. In state 1, the PSI–LHCII complexes were, as expected, absent. Induction of state 2 by turning off the far-red light resulted in an increase in the PSI–LHCII complex. Interestingly, this complex (absent in psaL mutants, as described by Pesaresi et al. (2009)) was highly enriched in P-Lhcb2 but virtually lacked P-Lhcb1.
A yet undescribed complex, denoted ‘LHCII complex’ in Figure 6, and that migrated between cytb6f and LHCII trimer, contained only P-Lhcb1, not P-Lhcb2. Protein complexes from psal plants in state 2 were hyperphosphorylated (Figures 2 and 6) as observed by Lunde et al. (2000). The overall amount of super- and mega-complexes was higher in state 2; this is visible both in the stained gels and in the immunoblots using antibodies against Lhcb1 and Lhcb2 (Figure S3). As digitonin preferentially solubilizes stroma-exposed thylakoids and grana margins, this result shows that these complexes accumulate in these areas of the chloroplast in state 2, or alternatively that grana stacks in state 2 are easier to solubilize. The majority of P-Lhcb1 or P-Lhcb2 was associated with mega-complexes. Moreover, it seemed that P-Lhcb1 was more enriched than P-Lhcb2 in these mega-complexes.
Discussion
Although the primary photosynthetic process is very similar in all plants, including green algae, there are significant differences in the way it is regulated. Different proteins have been employed to regulate the so-called qE quenching: PsbS is used in higher plants (Li et al., 2000), LI818/LHCSR in Chlamydomonas (Peers et al., 2009) and diatoms (Depauw et al., 2012), while both systems are operational in the moss Physcomitrella patens (Alboresi et al., 2010). The kinases that regulate state transitions are conserved between Chlamydomonas and Arabidopsis (Depege et al., 2003; Bellafiore et al., 2005), but their substrate polypeptides differ; the split between the genes coding for Lhcb1, Lhcb2 and Lhcb3 polypeptides occurred after the radiation of green algae and higher plants so there are no direct counterparts of Lhcb1, Lhcb2 and Lhcb3 in green algae (Elrad and Grossman, 2004). Therefore, results from Chlamydomonas (Ferrante et al., 2012) cannot be directly used to inform about the situation in higher plants. Lhcb3 does not carry a phosphorylation site and therefore probably does not play a direct role in state transitions, but as plants respond to the lack of Lhcb3 by increasing the amount of (phosphorylatable) Lhcb1 and Lhcb2 subunits, Lhcb3 KO mutants have slightly faster state transitions (Damkjaer et al., 2009).
We have been able to generate antibodies specific for the phosphorylated forms of Lhcb1 and Lhcb2, which we believe will be important tools for gaining a better understanding of the biochemistry of state transitions and, in the longer term, of the true physiological role of LHCII phosphorylation in natural environments. By using these antibodies, we have obtained information that sheds new light on the state-transition process in Arabidopsis. Our key findings are: (i) Lhcb2 is more rapidly phosphorylated than Lhcb1 after a shift from ‘state 1 light’ to ‘state 2 light’; within only 10 sec, Lhcb2 phosphorylation reached ca 1/3 of the maximum level, (ii) STN8 shows, at least in stn7 mutants, some activity towards both Lhcb1 and Lhcb2, (iii) there is no difference in gel mobility between the phosphorylated and non-phosphorylated forms of the proteins, (iv) there is no difference in the kinetics of dephosphorylation between Lhcb1 and Lhcb2, in either the presence or the absence of the PPH1/TAP38 phosphatase, in other words the PPH1/TAP38 phosphatase is not selective, (v) in neither case is phosphorylation of the protein strongly dependent on the presence of Lhcb3, Lhcb5, Lhcb6 or PsbS, (vi) in plants with very little Lhcb1 – and no Lhcb2 – Lhcb1 phosphorylation is slower, (vii) some, but by no means all, P-Lhcb1 and P-Lhcb2 associate with PSI complexes, (viii) the PSI–LHCII complex formed in state 2 is highly enriched in the phosphorylated forms of Lhcb2, (ix) a large proportion of the Lhcb1 and P-Lhcb1 proteins is associated with mega-complexes comprising both PSI and PSII, and finally (x) we visualized a new LHCII complex enriched in P-Lhcb1.
Interestingly, early studies using isolated spinach chloroplasts or thylakoids identified the so-called 25 kDa peptide (later found to correspond to Lhcb2, Jansson et al., 1990) as having three times faster kinetics of phosphorylation than the so-called 27 kDa peptide (Ljungberg et al., 1986), which was later found to be encoded by Lhcb1 genes (Jansson et al., 1990). At this time the third LHCII protein, Lhcb3, had not yet been discovered, which complicates interpretation of the early data, but it appears that in both Arabidopsis and spinach Lhcb2 has more rapid phosphorylation kinetics than Lhcb1. To induce state transitions we mainly employed light conditions that cause a rapid and drastic shift in light quality as an effective mean of studying the underlying mechanistic details of the state-transition process. Our results suggest that the STN7 kinase is localized in close proximity to its substrate. Whether or not it is the structure of the substrate itself, i.e. the difference in the primary sequences of Lhcb1 and Lhcb2, that makes the Lhcb2 protein a better substrate for STN7, or whether Lhcb2 has a location in the thylakoid that makes it more accessible to the kinase, is a question that cannot yet be answered.
It has been suggested that 20–25% of LHCII is engaged in state transitions in higher plants (Allen, 1992) while in Chlamydomonas up to 80% is engaged (Delosme et al., 1996). There are very few data on absolute phosphorylation levels of LHCII in plants, but a few reports (e. g. Islam, 1987) have estimated that about one-third of LHC could be phosphorylated. It is evident that not all P-Lhcb1 and P-Lhcb2 are associated with PSI under state 2, as revealed by our lpBN gel electrophoresis, with much of the P-Lhcb1 and P-Lhcb2 also being found in the LHCII trimer fraction. It is of course possible that most of the phosphorylated LHCII trimers are functionally attached to PSI in vivo, but dissociated during solubilization. Alternatively, the amount of phosphorylated LHCII may not be directly correlated with the extent of state transition, as suggested by studies of Arabidopsis plants that lacked Lhcb3 (Damkjaer et al., 2009) and analyses of purified PSI–LHCII complexes isolated with digitonin (Zhang and Scheller, 2004). P-Lhcb2 was enriched in the PSI–LHCII complex that appears rapidly after a shift to state 2 light, suggesting that Lhcb2 may have a major role in the initial stage of state transitions; interestingly, the non-phosphorylated form of Lhcb2 was also enriched in that complex (Figure S3). This finding is in accordance with earlier observations on the purified PSI–LHCII complex, which also contained non-phosphorylated LHCII (Zhang and Scheller, 2004). It should also be mentioned that we saw no evidence for any phosphorylation-induced monomerisation of LHCII trimers. Moreover, the distribution of the Lhcb2 protein responded to the state transition, as more Lhcb2 was found in the PSI–LHCII supercomplexes in state 2 as compared with state 1. This situation is consistent with a remodelling of the PSII–LHCII supercomplexes in the context of state transitions, as recently documented by Dietzel et al. (2011). It must, however, be borne in mind that our solubilization protocol has, like other methods, a bias for different thylakoid compartments. The thylakoid membrane is remodelled during state transitions; during state 1 membranes are tightly connected forming grana stacks, while upon shift to state 2 membranes undergo unstacking (Chuartzman et al., 2008). This solubilization bias complicates comparisons of data generated by different methods, but we believe that the P-Lhcb1 and P-Lhcb2 antibodies will become useful tools for analysing the composition of LHCII using different techniques.
As mentioned above, if the Lhcb1.4 gene product, which lacks the STN7-phosphorylatable Thr residue, is instead phosphorylated at the Ser residue close to its N-terminus we would not be able to quantify it with our antibodies, but phosphoproteomics have failed to detect phosphorylation at this residue (Ingelsson and Vener, 2012). Even if Lhcb1.4 were phosphorylated by STN7, we think it is highly unlikely that this could affect our conclusions, as Lhcb1.4 is not the major Lhcb1 gene product in Arabidopsis (Jansson, 1999). Lhcb1.4 is phosphorylated (Wang et al., 2012), but apparently only at the Ser48 residue – marked with an asterisk in the sequence GPSGS*PWYGSDR (Ingelsson and Vener, 2012) where the other four Arabidopsis Lhcb1 gene products are also phosphorylated. To our knowledge, there are no data linking the STN7- and STN8-independent phosphorylations of this residue (Ingelsson and Vener, 2012) to state transitions, so we believe that Lhcb1.4 has no direct role in state transitions. It should be pointed out that some Lhcb1 gene products from other species may also lack the phosphorylatable Thr residue. However, there is no subgroup among Lhcb1 gene sequences that would suggest the existence of two functionally distinct types of Lhcb1, one phosphorylated and participating in state transitions and one not, so it appears that it is not necessary for plants with multiple Lhcb1 gene copies to have all of their products subject to phosphorylation by STN7.
In this work, we have mainly used illumination of intact plants with red/far-red light to induce state transitions in order to reveal the mechanistic consequences of differential phosphorylation kinetics of the Lhcb1 and Lhcb2 proteins. This approach does not mean that we regard LHCII phosphorylation as a way of regulating PSI/PSII imbalances in excitation due to fluctuations in light intensity as being less important, merely that light quality fluctuations have certain experimental advantages. However, after a light intensity shift, Lhcb2 seemed also to have more rapid phosphorylation kinetics.
We have made no attempt to quantify exactly the differences in relative stoichiometries of Lhcb1, P-Lhcb1, Lhcb2 and P-Lhcb2 in the different super- and mega-complexes of PSI and/or PSII, but we have provided indications that most, if not all, of the large complexes that could be separated by lpBN-PAGE contain phosphorylated LHCII subunits. This situation indicates that the mega-complexes are prone to dynamically regulated reversible LHCII phosphorylation, and are thus functionally involved in the regulation of photosynthesis. Our antibodies provide a useful tool for studying protein complexes isolated under different conditions in order to better understand the relationships between state transitions and the variety of different thylakoid membrane super- and mega-complexes.
To conclude, we have shown that in Arabidopsis the two major LHCII proteins Lhcb1 and Lhcb2 differ substantially in the kinetics of STN7-dependent phosphorylation. The implication of this finding is that the two proteins probably have unique roles in the state-transition process, despite their very high degree of sequence conservation.
Experimental procedures
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) and the mutant lines asLhcb2-12 (Andersson et al., 2003), koLhcb3 (SALK_02314C) (Damkjaer et al., 2009), koLhcb5 (SALK_014869C), koLhcb6 (SALK_035761C) (Kovacs et al., 2006), npq4-1 (SALK_095156), stn7 (SALK_073254) (Bellafiore et al., 2005), stn8 (SALK_060869) (Vainonen et al., 2005), stn7stn8 (Bonardi et al., 2005), and psal (SALK_010554) were grown for 6 weeks under 250 μmol m−2 s−1 (or, for lpBN, 120 μmol m−2 s−1) light irradiance and an 8 h light (23°C)/16 h dark (18°C) photoperiod. The humidity was maintained at 75%.
State-transition induction
To induce state 1 and state 2 artificially, Arabidopsis plants were treated with a customized light-emitting diode (LED) Light Source (SL 3500-R-D) from Photon System Instruments using the Light Studio program. To trigger phosphorylation, plants were subjected for 60 min to 100% far-red light supplemented with 15% red light to develop state 1, and then the far-red light was switched off for another 60 min to induce a shift to state 2. The reverse light regime was used to study dephosphorylation kinetics. Leaves for thylakoid isolation were sampled at different time intervals. To induce state 1 and state 2 by changes in light intensity wild-type plants were transferred from the growth light into either the dark (D) or low light (LL) to induce state 1 and state 2 respectively, and then shifted to LL or D for phosphorylation and dephosphorylation induction. Leaves samples were collected at the same time intervals as for LED lights treatment.
Thylakoid extraction and immunoblot analysis
Thylakoids were extracted as previously described by Järvi et al. (2011). For the (de)phosphorylation assays, 6-week-old leaves were homogenized together with ice-cold grinding medium [0.45 m sorbitol, 20 mm tricine pH 8.4, 10 mm EDTA, 10 mm NaHCO3, 0.1% (w/v) BSA] and, after filtration through two layers of muslin, centrifuged at 4000 g for 10 min. The pellet was resuspended in 15 ml of hypotonic buffer (20 mm tricine pH 7.8, 5 mm MgCl2), followed by the addition of 15 ml of osmoticum medium (0.66 m sorbitol, 20 mm tricine pH 7.8, 5 mm MgCl2), and re-centrifuged for 10 min at 4000 g. The pelleted thylakoid proteins were then resuspended in resuspension buffer (0.33 m sorbitol, 20 mm tricine pH 7.8, 5 mm MgCl2). All the buffers contained 10 mm NaF to inhibit the activity of phosphatases and maintain the in vivo phosphorylation state (Bellafiore et al., 2005). All the stages in thylakoid protein preparation were performed on ice or in a cold room (4°C). Protein extracts were used for chlorophyll quantification according to Porra et al. (1989).
To generate antibodies specific for phosphorylated Lhcb1 and Lhcb2, synthetic peptides with the sequences RKT*VAKPKGP and RRT*VKSTPQS, where T* indicates phospho-Thr, were synthesized by standard Fmoc peptide synthesis by Innovagen (Lund, Sweden) and injected into rabbits. For immunopurification, 5–6 mg of each peptide was coupled to 2 ml Ultralink iodoacetyl gel (Pierce). Antibodies from 20 ml of serum was first immuno-purified with the phosphopeptides used for immunization bound to the gel essentially according to the Vendors manual. After elution, a second step was performed where instead the Ultralink with corresponding non-phosphorylated peptide coupled was used, to remove all antibodies recognizing the non-phosphorylated form. The flowthrough from this step consisted of the purified antibodies. Immunoblotting analysis was performed, using 1 μg of thylakoid proteins, essentially as described by Ganeteg et al. (2001), with slight modifications according to the instructions provided by the antibody manufacturer. Chemiluminescence was detected after 1 and 5 min using a LAS-3000 cooled CCD camera. Images were recorded, processed and quantified according to Damkjaer et al. (2009).
Three biological replicates for each state-transition condition and genotype were used for detection of Lhcb1 and Lhcb2 phosphorylated proteins. Statistical analysis was performed using the R application package and Student’s t-test and data normality test (Ihaka and Gentleman, 1996).
Large pore Blue Native (lpBN) gel electrophoresis and 2-D immunoblot analysis
Samples were prepared and run as described previously (Sirpiö et al., 2011). Thylakoid membranes were resuspended in ice-cold 25BTH20G buffer [25 mm BisTris–HCl, pH 7.0, 20% (v/v) glycerol and 0.25 mg ml-1 Pefabloc] with the addition of 10 mm NaF, to a chlorophyll concentration of 1 μg μl-1, and an equal volume of digitonin solution was added to give a final concentration of 1%. Samples were solubilized for 8 min in darkness with gentle mixing. Insoluble material was removed by centrifugation at 18000 g at 4°C for 20 min. 1/10 volume of Serva Blue G buffer (100 mm BisTris–HCl, pH 7.0, 0.5 m amino-n-caproic acid, 30% sucrose and 50 mg ml-1 Serva Blue G) was added to each sample prior to loading to introduce a negative charge and to increase the solubility of the sample. lpBN gels were run as described in Sirpiö et al. (2011) and Järvi et al. (2011).
For immunoblotting, the whole lpBN gels were electroblotted on to a polyvinylidene fluoride (PVDF) membrane (Millipore, Watford Herts, UK). For two-dimensional analyses, the gel strips from the lpBN were excised and incubated with gentle shaking in Laemmli buffer [138 mm Tris–HCl, pH 6.8, 6 m urea, 22.2% (v/v) glycerol, 4.3% (w/v) sodium dodecyl sulphate (SDS), 5% (v/v) 2-mercaptoethanol] for 1 h, Thereafter, the gel strips were transferred to the top of an SDS-PAGE gel [15% (w/v) polyacrylamide, 6 m urea] and sealed with 0.5% agarose in SDS-PAGE running buffer [25 mm Tris base, 190 mm glycine, 0.1% (w/v) SDS]. After electrophoretic separation, the 2D gels were stained with silver as described in Blum et al. (1987).
Acknowledgments
This work was supported by the European Union project FP7-PEOPLE-ITN-2008 ‘HARVEST: Control of Light Use Efficiency in Plants and Algae From Light to Harvest’, the Swedish Research Council (VR), the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (Formas), the Academy of Finland (project Nos 118637 and 138703) and C. Leoni by a 1-year fellowship from the University of Bari (Rectoral Decree No. 1598). The authors declare no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Immunoblot analysis of Lhcb1 and Lhcb2 in individual asLhcb2 plants.
Phosphorylation (a) and dephosphorylation (b) kinetics of Arabidopsis Lhcb1 and Lhcb2 during state transitions in induced by dark to LL shift (and back) analyzed using P-Lhcb1 and P-Lhcb2 antibodies.
Polypeptide composition of thylakoid protein complexes from leaves in state 1 and state 2 separated by large pore Blue Native (lpBN) gel electrophoresis (top).
References
- Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T. Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc. Natl Acad. Sci. USA. 2010;107:11128–11133. doi: 10.1073/pnas.1002873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen JF. Protein phosphorylation in regulation of photosynthesis. Biochim. Biophys. Acta. 1992;1098:275–335. doi: 10.1016/s0005-2728(09)91014-3. [DOI] [PubMed] [Google Scholar]
- Andersson J, Wentworth M, Walters RG, Howard CA, Ruban AV, Horton P, Jansson S. Absence of the Lhcb1 and Lhcb2 proteins of the light-harvesting complex of photosystem II – effects on photosynthesis, grana stacking and fitness. Plant J. 2003;35:350–361. doi: 10.1046/j.1365-313x.2003.01811.x. [DOI] [PubMed] [Google Scholar]
- Aro EM, Rokka A, Vener AV. Determination of phosphoproteins in higher plant thylakoids. Methods Mol. Biol. 2004;274:271–285. doi: 10.1385/1-59259-799-8:271. [DOI] [PubMed] [Google Scholar]
- Bassi G, Rigoni F, Barbato R, Giacometti GM. Light-harvesting chlorophyll a/b protein (LHCII) populations in phosphorylated membranes. Biochim. Biophys. Acta. 1988a;936:29–38. [Google Scholar]
- Bassi R, Giacometti GM, Simpson DJ. Changes in the organization of stroma membranes induced by in vivo state1 – state2 transition. Biochim. Biophys. Acta. 1988b;935:152–165. [Google Scholar]
- Bellafiore S, Barneche F, Peltier G, Rochaix J-D. State transitions and light adaptation require chloroplast thylakoid protein kinase STN7. Nature. 2005;433:892–895. doi: 10.1038/nature03286. [DOI] [PubMed] [Google Scholar]
- Bennett J. Phosphorylation of chloroplast membrane polypeptides. Nature. 1977;269:344–346. [Google Scholar]
- Blum H, Beier H, Gross HJ. Improved silver staining of plant proteins, RNA and DNA in polyacrylamide gels. Electrophoresis. 1987;8:93–99. [Google Scholar]
- Bonardi V, Pesaresi P, Becker T, Schleiff E, Wagner R, Pfannschmidt T, Jahns P, Leister D. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature. 2005;437:1179–1182. doi: 10.1038/nature04016. [DOI] [PubMed] [Google Scholar]
- Bonaventura C, Myers J. Fluorescence and oxygen evolution from Chlorella pyrenoidosa. Biochim. Biophys. Acta. 1969;189:366–383. doi: 10.1016/0005-2728(69)90168-6. [DOI] [PubMed] [Google Scholar]
- Caffarri S, Croce R, Cattivelli L, Bassi R. A look within LHCII: differential analysis of the Lhcb1-3 complexes building the major trimeric antenna complex of higher-plant photosynthesis. Biochemistry. 2004;43:9467–9476. doi: 10.1021/bi036265i. [DOI] [PubMed] [Google Scholar]
- Chuartzman SG, Nevo R, Shimoni E, Charuvi D, Kiss V, Ohad I, Brumfeld V, Reich Z. Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant Cell. 2008;20:1029–1039. doi: 10.1105/tpc.107.055830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damkjaer JT, Kereiche S, Johnson MP, Kovacs L, Kiss AZ, Boekema EJ, Ruban AV, Horton P, Jansson S. The photosystem II light-harvesting protein Lhcb3 affects the macrostructure of photosystem II and the rate of state transitions in Arabidopsis. Plant Cell. 2009;21:3245–3256. doi: 10.1105/tpc.108.064006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delosme R, Olive J, Wollman F-A. Change in light energy distribution upon state transitions: an in vivo photoacoustic study of the wild-type and photosynthesis mutants from Chlamydomonas reinhardtii. Biochim. Biophys. Acta. 1996;1273:150–158. [Google Scholar]
- Depauw FA, Rogato A, Ribera d’Alcala M, Falciatore A. Exploring the molecular basis of responses to light in marine diatoms. J. Exp. Bot. 2012;63:1575–1591. doi: 10.1093/jxb/ers005. [DOI] [PubMed] [Google Scholar]
- Depege N, Bellafiore S, Rochaix J-D. Role of chloroplast protein kinase Stt7 in LHCII phosphorylation and state transition in Chlamydomonas. Science. 2003;299:1572–1575. doi: 10.1126/science.1081397. [DOI] [PubMed] [Google Scholar]
- Dietzel L, Brautigam K, Steiner S, Schuffler K, Lepetit B, Grimm B, Schottler MA, Pfannschmidt T. Photosystem II supercomplex remodeling serves as an entry mechanism for state transitions in Arabidopsis. Plant Cell. 2011;23:2964–2977. doi: 10.1105/tpc.111.087049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrad D, Grossman AR. A genome’s-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii. Curr. Genet. 2004;45:61–75. doi: 10.1007/s00294-003-0460-x. [DOI] [PubMed] [Google Scholar]
- Ferrante P, Ballottari M, Bonente G, Giuliano G, Bassi R. LHCBM1 and LHCBM2/7 polypeptides, components of major LHCII complex, have distinct functional roles in photosynthetic antenna system of Chlamydomonas reinhardtii. J. Biol. Chem. 2012;287:16276–16288. doi: 10.1074/jbc.M111.316729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fristedt R, Granath P, Vener AV. A protein phosphorylation threshold for functional stacking of plant photosynthetic membranes. PLoS One. 2010;5:e10963. doi: 10.1371/journal.pone.0010963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galka P, Santabarbara S, Khuong TT, Degand H, Morsomme P, Jennings RC, Boekema EJ, Caffarri S. Functional analyses of the plant photosystem I-light-harvesting complex II supercomplex reveal that light-harvesting complex II loosely bound to photosystem II is a very efficient antenna for photosystem I in state II. Plant Cell. 2012;24:2963–2978. doi: 10.1105/tpc.112.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganeteg U, Strand A, Gustafsson P, Jansson S. The properties of the chlorophyll a/b-binding proteins Lhca2 and Lhca3 studied in vivo using antisense inhibition. Plant Physiol. 2001;127:150–158. doi: 10.1104/pp.127.1.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman D. A language for data analysis and graphics. J. Comput. Graph. Stat. 1996;5:299–314. [Google Scholar]
- Ingelsson B, Vener AV. Phosphoproteomics of Arabidopsis chloroplasts reveals involvement of the STN7 kinase in phosphorylation of nucleoid protein pTAC16. FEBS Lett. 2012;586:1265–1271. doi: 10.1016/j.febslet.2012.03.061. [DOI] [PubMed] [Google Scholar]
- Islam K. The rate and extent of phosphorylation of the two light-harvesting chlorophyll a/b-binding protein complex (LHCII) polypeptides in isolated spinach thylakoids. Biochem. Biophys. Acta. 1987;893:333–341. [Google Scholar]
- Jackowski G. Separation, purification, and characterization of polypeptide composition of subcomplexes of the main light-harvesting chlorophyll a/b-protein complex of photosystem II. Methods Mol. Biol. 2004;274:115–128. doi: 10.1385/1-59259-799-8:115. [DOI] [PubMed] [Google Scholar]
- Jansson S. The light-harvesting chlorophyll a/b-binding proteins. Biochim. Biophys. Acta. 1994;1184:1–19. doi: 10.1016/0005-2728(94)90148-1. [DOI] [PubMed] [Google Scholar]
- Jansson S. A guide to the Lhc genes and their relatives in Arabidopsis/IT>. Trends Plant Sci. 1999;4:236–240. doi: 10.1016/s1360-1385(99)01419-3. [DOI] [PubMed] [Google Scholar]
- Jansson S, Gustafsson P. Type I and type II genes for the chlorophyll a/b-binding protein in the gymnosperm Pinus sylvestris (Scots pine): cDNA cloning and sequence analysis. Plant Mol. Biol. 1990;14:287–296. doi: 10.1007/BF00028766. [DOI] [PubMed] [Google Scholar]
- Jansson S, Selstam E, Gustafsson P. The rapidly phosphorylated 25 kDa polypeptide of the light- harvesting complex of photosystem II is encoded by the type 2 cab-II genes. Biochim. Biophys. Acta. 1990;1019:110–114. doi: 10.1016/0005-2728(90)90130-v. [DOI] [PubMed] [Google Scholar]
- Järvi S, Suorsa M, Paakkarinen V, Aro EM. Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem. J. 2011;439:207–214. doi: 10.1042/BJ20102155. [DOI] [PubMed] [Google Scholar]
- Kovacs L, Damkjaer J, Kereiche S, Ilioaia C, Ruban AV, Boekema EJ, Jansson S, Horton P. Lack of the light-harvesting complex CP24 affects the structure and function of the grana membranes of higher plant chloroplasts. Plant Cell. 2006;18:3106–3120. doi: 10.1105/tpc.106.045641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson UK, Anderson JM, Andersson B. Variations in the relative content of the peripheral and inner light-harvesting chlorophyll a/b-binding complex (LHCII) subpopulations during thylakoid light adaptation and development. Biochim. Biophys. Acta. 1987;894:69–75. [Google Scholar]
- Li XP, Bjorkman O, Shih C, Grossman AR, Rosenquist M, Jansson S, Niyogi KK. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature. 2000;403:391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- Ljungberg U, Akerlund HE, Andersson B. Isolation and characterization of the 10-kDa and 22-kDa polypeptides of higher plant photosystem 2. Eur. J. Biochem. 1986;158:477–482. doi: 10.1111/j.1432-1033.1986.tb09779.x. [DOI] [PubMed] [Google Scholar]
- Lunde C, Jensen PE, Haldrup A, Knoetzel J, Scheller HV. The PSI-H subunit of photosystem I is essential for state transitions in plant photosynthesis. Nature. 2000;408:613–615. doi: 10.1038/35046121. [DOI] [PubMed] [Google Scholar]
- Martinsuo P, Pursiheimo S, Aro EM, Rintamaki E. Dithiol oxidant and disulfide reductant dynamically regulate the phosphorylation of light-harvesting complex II proteins in thylakoid membranes. Plant Physiol. 2003;133:37–46. doi: 10.1104/pp.103.027268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata N, Sugahara K. Control of excitation transfer in photosynthesis. 3. Light-induced decrease of chlorophyll a fluorescence related to photophosphorylation system in spinach chloroplasts. Biochim. Biophys. Acta. 1969;189:182–192. doi: 10.1016/0005-2728(69)90046-2. [DOI] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462:518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- Pesaresi P, Hertle A, Pribil M, Kleine T, Wagner R, Strissel H, Ihnatowicz A, Bonardi V, Scharfenberg M, Schneider A, Pfannschmidt T, Leister D. Arabidopsis STN7 kinase provides a link between short and long-term photosynthetic acclimation. Plant Cell. 2009;21:2402–2423. doi: 10.1105/tpc.108.064964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra RJ, Thompson WA, Kriedmann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochem. Biophys. Acta. 1989;975:384–394. [Google Scholar]
- Pribil M, Pesaresi P, Hertle A, Barbato R, Leister D. Role of plastid protein phosphatase TAP38 in LHCII dephosphorylation and thylakoid electron flow. PLoS Biol. 2010;8:e1000288. doi: 10.1371/journal.pbio.1000288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rintamaki E, Martinsuo P, Pursiheimo S, Aro EM. Cooperative regulation of light-harvesting complex II phosphorylation via the plastoquinol and ferredoxin-thioredoxin system in chloroplasts. Proc. Natl Acad. Sci. USA. 2000;97:11644–11649. doi: 10.1073/pnas.180054297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruban AV, Wentworth M, Yakushevska AE, Andersson J, Lee PJ, Keegstra W, Dekker JP, Boekema EJ, Jansson S, Horton P. Plants lacking the main light-harvesting complex retain photosystem II macro-organization. Nature. 2003;421:648–652. doi: 10.1038/nature01344. [DOI] [PubMed] [Google Scholar]
- Shapiguzov A, Ingelsson B, Samol I, Andres C, Kessler F, Rochaix JD, Vener AV, Goldschmidt-Clermont M. The PPH1 phosphatase is specifically involved in LHCII dephosphorylation and state transitions in Arabidopsis. Proc. Natl Acad. Sci. USA. 2010;107:4782–4787. doi: 10.1073/pnas.0913810107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirpiö S, Suorsa M, Aro EM. Analysis of thylakoid protein complexes by two-dimensional electrophoretic systems. Methods Mol. Biol. 2011;775:19–30. doi: 10.1007/978-1-61779-237-3_2. [DOI] [PubMed] [Google Scholar]
- Standfuss J, Kuhlbrandt W. The three isoforms of the light-harvesting complex II: spectroscopic features, trimer formation, and functional roles. J. Biol. Chem. 2004;279:36884–36891. doi: 10.1074/jbc.M402348200. [DOI] [PubMed] [Google Scholar]
- Thapper A, Mamedov F, Mokvist F, Hammarstrom L, Styring S. Defining the far-red limit of photosystem II in spinach. Plant Cell. 2009;21:2391–2401. doi: 10.1105/tpc.108.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikkanen M, Aro EM. Thylakoid protein phosphorylation in dynamic regulation of photosystem II in higher plants. Biochim. Biophys. Acta. 2011;1817:232–238. doi: 10.1016/j.bbabio.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Piippo M, Suorsa M, Sirpio S, Mulo P, Vainonen J, Vener AV, Allahverdiyeva Y, Aro EM. State transitions revisited-a buffering system for dynamic low light acclimation of Arabidopsis. Plant Mol. Biol. 2006;62:779–793. doi: 10.1007/s11103-006-9044-8. [DOI] [PubMed] [Google Scholar]
- Tikkanen M, Suorsa M, Gollan PJ, Aro EM. Post-genomic insight into thylakoid membrane lateral heterogeneity and redox balance. FEBS Lett. 2012;586:2911–2916. doi: 10.1016/j.febslet.2012.07.035. [DOI] [PubMed] [Google Scholar]
- Vainonen JP, Hansson M, Vener AV. STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of photosystem II core proteins. J. Biol. Chem. 2005;280:33679–33686. doi: 10.1074/jbc.M505729200. [DOI] [PubMed] [Google Scholar]
- Vener AV. Environmentally modulated phosphorylation and dynamics of proteins in photosynthetic membranes. Biochim. Biophys. Acta. 2007;1767:449–457. doi: 10.1016/j.bbabio.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Zou H, Sun SS, He JX. A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. J. Proteome Res. 2012;11:2301–2315. doi: 10.1021/pr2010764. [DOI] [PubMed] [Google Scholar]
- Zhang S, Scheller HV. Light-harvesting complex II binds to several small subunits of photosystem I. J. Biol. Chem. 2004;279:3180–3187. doi: 10.1074/jbc.M311640200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Immunoblot analysis of Lhcb1 and Lhcb2 in individual asLhcb2 plants.
Phosphorylation (a) and dephosphorylation (b) kinetics of Arabidopsis Lhcb1 and Lhcb2 during state transitions in induced by dark to LL shift (and back) analyzed using P-Lhcb1 and P-Lhcb2 antibodies.
Polypeptide composition of thylakoid protein complexes from leaves in state 1 and state 2 separated by large pore Blue Native (lpBN) gel electrophoresis (top).


