Abstract
The alteration of photoperiod sensitivity has let breeders diversify flowering time in Oryza sativa (rice) and develop cultivars adjusted to a range of growing season periods. Map-based cloning revealed that the rice flowering-time quantitative trait locus (QTL) Heading date 16 (Hd16) encodes a casein kinase-I protein. One non-synonymous substitution in Hd16 resulted in decreased photoperiod sensitivity in rice, and this substitution occurred naturally in an old rice cultivar. By using near-isogenic lines with functional or deficient alleles of several rice flowering-time genes, we observed significant digenetic interactions between Hd16 and four other flowering-time genes (Ghd7, Hd1, DTH8 and Hd2). In a near-isogenic line with the weak-photoperiod-sensitivity allele of Hd16, transcription levels of Ehd1, Hd3a, and RFT1 increased under long-day conditions, and transcription levels of Hd3a and RFT1 decreased under short-day conditions. Expression analysis under continuous light and dark conditions showed that Hd16 was not likely to be associated with circadian clock regulation. Biochemical characterization indicated that the functional Hd16 recombinant protein specifically phosphorylated Ghd7. These results demonstrate that Hd16 acts as an inhibitor in the rice flowering pathway by enhancing the photoperiod response as a result of the phosphorylation of Ghd7.
Keywords: Oryza sativa L., flowering time, photoperiod sensitivity, natural variation, casein kinase I
Introduction
Many plant species have the ability to initiate flowering at a time that is best suited to their reproduction. This ability depends mainly on the accurate measurement of seasonal changes in day length and temperature (Thomas and Vince-Pure, 1997; Hayama et al., 2003). Photoperiod sensitivity is therefore an important factor for measuring the critical day length and for determining flowering time in plants. An alteration of photoperiod sensitivity changes the flowering time and can enhance the adaptability to local environmental conditions in many plant species (Jung and Muller, 2009). Many researchers have reported that photoperiod insensitivity has played an important role in expanding the range of cultivation to high latitudes in Zea mays (maize; Gouesnard et al., 2002), Hordeum vulgare (barley; Turner et al., 2005; Zakhrabekova et al., 2012), Sorghum (Murphy et al., 2011) and Oryza sativa (rice; Fujino and Sekiguchi, 2005; Izawa, 2007; Xue et al., 2008). This is because of the requirement for early heading and maturity to coincide with a period of optimal climatic conditions at high latitudes.
During the last 15 years, significant progress has been made in our understanding of the molecular regulation of flowering time in plants. Many genes that control flowering time have been identified, and their molecular genetic pathways have been well characterized in Arabidopsis thaliana and rice (Simpson and Dean, 2002; Hayama and Coupland, 2004; Tsuji et al., 2011). Rice photoperiodic flowering is controlled by two independent gene pathways. The OsGI-Hd1-Hd3a (rice GIGANTEA, Heading date 1 and Heading date 3a) signaling pathway has been evolutionarily conserved in rice, as was the GI-CO-FT (GIGANTEA, CONSTANS, and FLOWERING LOCUS T) pathway in Arabidopsis. The OsGI-Hd1-Hd3a pathway is regulated by light perception and the circadian clock (Searle and Coupland, 2004; Imaizumi and Kay, 2006). Regulation of Hd3a expression comes from the functional conversion of Hd1. Hd1 promotes Hd3a expression under short-day (SD) conditions, but inhibits Hd3a expression under long-day (LD) conditions. This day length-dependent conversion of Hd1 activity is caused by phytochrome-mediated signaling (Hayama and Coupland, 2004; Izawa, 2007).
The other pathway includes Ghd7 (Grain number, plant height and heading date 7) and Ehd1 (Early heading date 1), which control flowering time by regulating the transcription of Hd3a, RFT1 (Rice flowering locus T1), or both genes (Xue et al., 2008). Ghd7 and Ehd1 have no orthologs in the Arabidopsis genome, indicating that these genes are associated with a rice-specific flowering pathway. The expressions of Ghd7 and Ehd1 are regulated by circadian gating of light responses through phytochromes (red light) and cryptochromes (blue light) (Itoh et al., 2010). Photoperiodic signals are perceived in the leaves, from which a long-distance signal (‘florigen’), consisting of Hd3a and RFT1 proteins, is transmitted to the shoot apical meristem, where it stimulates reproductive development (Tamaki et al., 2007; Komiya et al., 2009). Photoperiod pathways and the circadian clock are controlled through an intricate network of feedback regulation at the level of gene expression and at a post-transcriptional level (Izawa, 2007; Itoh et al., 2010; Tsuji et al., 2011). For example, phosphorylation of clock proteins plays a critical role in generating a proper circadian rhythm in Arabidopsis (Sugano et al., 1999). A central clock component protein CCA1 (CIRCADIAN CLOCK ASSOCIATED 1) is phosphorylated by the kinase encoded by CK2 (CASEIN KINASE 2) to acquire its DNA-binding activity in Arabidopsis (Daniel et al., 2004). The protein encoded by Hd6 (Heading date 6), which is a rice homolog of an alpha subunit of CK2, enhances the Hd1 repressor function under LD conditions through the intermediary of phosphorylation of the protein encoded by an unknown gene (Ogiso et al., 2010).
There is a wide range of variation in flowering time in rice cultivars (Izawa, 2007). Sequence analysis of the flowering-time genes has indicated that allelic differences have contributed greatly to this variation (Yano et al., 2000; Xue et al., 2008; Takahashi et al., 2009; Wei et al., 2010; Ebana et al., 2011). Functional and non-functional alleles of Hd1 are associated with late and early flowering, respectively, and Hd1 is one of the major determinants of natural variation in flowering time in cultivated rice (Yano et al., 2000; Takahashi et al., 2009). Similarly, the geographic distribution of Ghd7 alleles suggests that favorable alleles were selected by breeders to enhance rice productivity and adaptability for each cultivation region (Xue et al., 2008). The japonica group of rice cultivars is cultivated from tropical regions to temperate regions at the northern limit of rice cultivation. Previous studies revealed that japonica cultivars contain allelic variation in flowering-time genes, including Hd1, Ghd7, Hd6, DTH8 (Days to heading 8), Hd2 (Heading date 2) and Hd17 (Heading date 17) (Fujino and Sekiguchi, 2005; Ebana et al., 2011; Shibaya et al., 2011; Matsubara et al., 2012).
In the present study, we performed map-based cloning of a flowering-time quantitative trait locus (QTL), Heading date 16 (Hd16), and revealed that Hd16 encodes a casein kinase-I protein. One non-synonymous substitution in Hd16 changes the photoperiod sensitivity. The functional Hd16 recombinant protein phosphorylated Ghd7 in vitro. Our results suggested that Hd16 is involved in the photoperiodic flowering pathway by means of its phosphorylation of Ghd7. Although Hd16 is identical to Early flowering 1 (EL1), a gene previously reported to control the gibberellin signaling pathway (Dai and Xue, 2010), our findings provided evidence for a new phosphorylation-related function of Hd16/EL1 (i.e. the phosphorylation of Ghd7) in the photoperiod response pathway that controls rice flowering.
Results
The flowering-time response of Hd16 near-isogenic lines
The japonica rice cultivars Nipponbare and Koshihikari differed in their flowering time and in the response of their flowering to the photoperiod (Figure 1a,b). Nipponbare flowered after 45.0 days under SD conditions, 89.2 days under LD conditions and 115.0 days under natural day-length (ND) conditions. Koshihikari flowered after 50.3 days under SD conditions, 72.5 days under LD conditions and 106.6 days under ND conditions. The number of days to flowering (DTF) of Nipponbare increased more than that of Koshihikari under LD conditions, indicating that the photoperiod sensitivity of Nipponbare was greater than that of Koshihikari. We previously found a QTL, designated Hd16, that is associated with the flowering-time difference between Nipponbare and Koshihikari (Matsubara et al., 2008). Two near-isogenic lines (NILs) were developed to verify the genetic effects of Hd16: one carrying the Koshihikari Hd16 allele in the genetic background of Nipponbare [N-NIL(Hd16)], and the other carrying the Nipponbare Hd16 allele in the genetic background of Koshihikari [K-NIL(Hd16)] (Figure 1c). DTF for N-NIL(Hd16) was longer (by 3 days) under SD conditions, and was shorter (by 20 days) under LD conditions, compared with the parent Nipponbare. DTF for K-NIL(Hd16) was shorter (by 2 days) under SD conditions, and longer (by 19 days) under LD conditions, compared with the parent Koshihikari. These results indicated that an allelic difference in Hd16 mainly explained the difference in photoperiod response between Nipponbare and Koshihikari.
Figure 1.
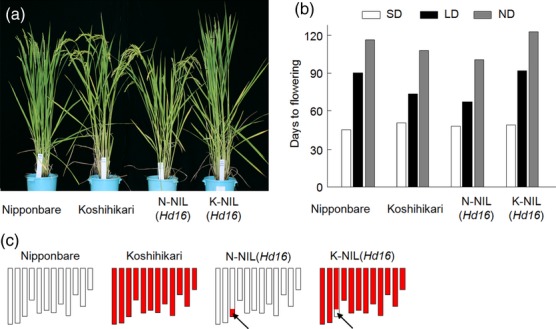
Phenotypes and genotypes of Nipponbare, Koshihikari, a Nipponbare near-isogenic line (NIL), N-NIL(Hd16), and a Koshihikari NIL, K-NIL(Hd16).
(a) Typical plants of Nipponbare, Koshihikari, N-NIL(Hd16) and K-NIL(Hd16), grown under natural day-length (ND) conditions after the flowering of Nipponbare.
(b) Days to flowering under different photoperiod conditions. Values are means ± standard deviation (n = 10). SD, short-day conditions (10 h of light/14 h of dark); LD, long-day conditions (14.5 h of light/9.5 h of dark).
(c) Graphical representations of the genotypes of the parent cultivars and the NILs. The 12 vertical bars represent the rice chromosomes. White and red bars indicate the Nipponbare and Koshihikari chromosome regions, respectively. Arrows indicate the positions of the Hd16 locus.
Hd16 encodes casein kinase I
Hd16 was previously mapped on the long arm of rice chromosome 3 (Matsubara et al., 2008). We performed high-resolution linkage analysis using 2918 BC4F3 plants. We delimited the candidate genomic region of Hd16 within a 29.4-kb genomic region between two molecular markers, SNP-1 (single nucleotide polymorphism 1) and SSR-2 (simple sequence repeat 2) (Figure 2a; Table S1). In this candidate region, four putative open reading frames were predicted in the Rice Annotation Project Database (http://rapdb.dna.affrc.go.jp). We isolated and sequenced a bacterial artificial chromosome clone containing a Koshihikari genomic sequence encompassing the candidate genomic region of Hd16. Comparison of the genomic sequences of the candidate region between Nipponbare and Koshihikari revealed the existence of a single non-synonymous single nucleotide polymorphism (SNP) within the tenth exon of Os03g0793500, which is annotated as encoding a casein kinase I (CKI) protein (Figure 2b). A guanine nucleotide in the Nipponbare allele changed to adenine in the Koshihikari allele; consequently, an alanine amino acid in Nipponbare changed to a threonine amino acid in Koshihikari. No other nucleotide polymorphisms were observed in the 29.4-kb genome sequence (Figure 2a,b).
Figure 2.
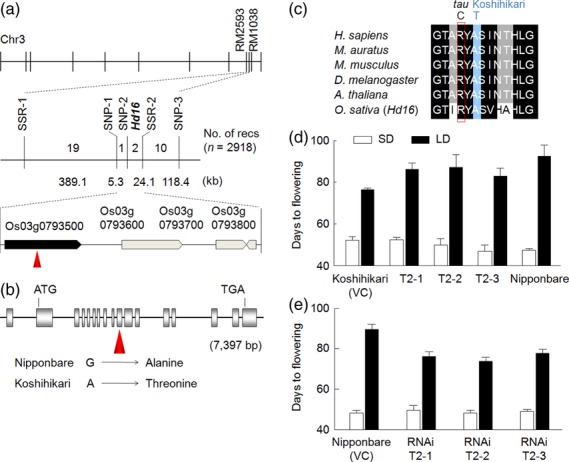
Map-based cloning of Hd16.
(a) Chromosomal location and high-resolution linkage map of Hd16 on rice chromosome 3. The pentagons indicate candidate open reading frames within the 29.4-kbp candidate quantitative trait locus (QTL) region. The red arrowheads indicate the position of a single non-synonymous nucleotide substitution.
(b) Structure of Hd16, which consists of 17 exons (gray boxes) and 16 introns.
(c) Partial alignment of non-synonymous nucleotide substitutions in casein kinase isoforms. The red box indicates the tau mutation site, and the aqua box indicates the Koshihikari mutation site. The accession numbers and entire alignments are described in Figure S1.
(d) Days to flowering (DTF) of three independent T2 lines homozygous for the Hd16 transgene from Nipponbare under short-day (SD) and long-day (LD) conditions. VC, vector control.
(e) Mean values and standard deviations for DTF of three independent T2 lines for knock-down constructs of Hd16 under SD and LD conditions.
The predicted protein sequence of CKI is highly conserved among plants and animals (Figures S1 and S2). The non-synonymous substitution was located near a catalytic activation domain of CKI. Only the Koshihikari allele encodes a threonine amino acid at the nucleotide position of the non-synonymous substitution; all other genes encode an alanine amino acid at this position (Figure 2c). Based on these observations, we hypothesize that Os03g0793500 is Hd16, and that the Nipponbare allele is the ancestral type and the Koshihikari allele is a mutant type. Os03g0793500 is reported as Early flowering 1 (EL1), which is associated with the gibberellin-mediated flowering pathway (Dai and Xue, 2010). The genome sequence of Hd16 in Nipponbare is the same as that of the wild-type allele of EL1.
Complementation analysis was performed to test whether the non-synonymous SNP was responsible for the flowering-time difference. A 10.7-kb Nipponbare genomic fragment (including a 2.6-kb region upstream from the transcription start site, the approximately 7.3-kb putative coding region for the gene encoding CKI and a 0.8-kb downstream sequence) was transformed into Koshihikari. T2 plants homozygous for the transgene showed earlier flowering (by 5 days) under SD conditions, and later flowering (by 11 days) under LD conditions, compared with the vector control line (Figure 2d). In addition, RNA interference (RNAi) with Hd16 expression significantly affected flowering time (Figures 2e and S3). T2 plants of the RNAi construct in the Nipponbare genetic background showed later flowering (by 2 days) under SD conditions, and earlier flowering (by 15 days) under LD conditions, compared with the vector control line. RNAi plants in the N-NIL(Hd16) genetic background showed no significant differences in flowering time under SD and LD conditions compared with the vector control (Figure S4). These results clearly suggest that Hd16 corresponds to Os03g0793500 and encodes a CKI protein, and that it is involved in photoperiodic flowering.
Transcript levels of Hd16 were investigated in several rice tissues (Figure S5). Transcription of Hd16 was observed in all tissues and at all developmental stages, and tended to increase in the leaf blade during later developmental stages. These expression patterns were coincident with those reported in the Rice Expression Profile Database (http://ricexpro.dna.affrc.go.jp). We also performed an Hd16 promoter::GUS (β-glucuronidase) reporter gene analysis to profile Hd16 expression. GUS expression in T1 transformants was detected in the vascular bundles of leaves and in proximal regions of the shoot and roots (Figure S6).
Origin of the non-synonymous substitution in Hd16
We investigated the origin of the non-synonymous substitution SNP in the Koshihikari Hd16 allele using 306 accessions of international rice cultivars (both indica and japonica subspecies) and 50 accessions of wild rice (Oryza rufipogon). Tables S1 and S2 list the SNP detection primers and the accessions used, respectively. The SNP of the Koshihikari Hd16 allele was only observed in Japanese japonica cultivars, and was not found in any of the wild rice, indica accessions or japonica accessions from outside of Japan (Figure S7). Based on the pedigree of Koshihikari, it is likely that the non-synonymous substitution originated from an old japonica cultivar: Moritawase.
Genetic interactions between Hd16 and other flowering-time genes
To clarify the role of Hd16 in the flowering-time gene network, which is mainly involved in photoperiodic flowering control, we developed a series of NILs with functional or defective alleles of four flowering-time genes (Hd1, Ghd7, DTH8 and Hd2), and each of the Koshihikari and Nipponbare Hd16 alleles (Figure 3a–d). In the combinations with Hd1 and Hd16 under LD conditions, DTF increased greatly with the Nipponbare Hd16 allele in a genetic background with a functional Hd1, but not in the background with a defective Hd1 (Figure 3a). In the combinations with Ghd7 and Hd16, the Nipponbare Hd16 allele also increased DTF in a genetic background with a functional Ghd7, but not with a defective Ghd7 (Figure 3b). Similarly, the Nipponbare Hd16 allele significantly increased DTF in a genetic background with a functional DTH8, but not with a defective DTH8 (Figure 3c). The defective Hd2 allele prevented the Nipponbare Hd16 allele from increasing DTF under LD conditions, whereas in the presence of a functional Hd2 allele, the Nipponbare Hd16 allele greatly increased DTF (Figure 3d). These observations clearly demonstrated that Hd16 is involved in a genetic control pathway for photoperiodic flowering under LD conditions that includes Hd1, Ghd7, DTH8 and Hd2.
Figure 3.
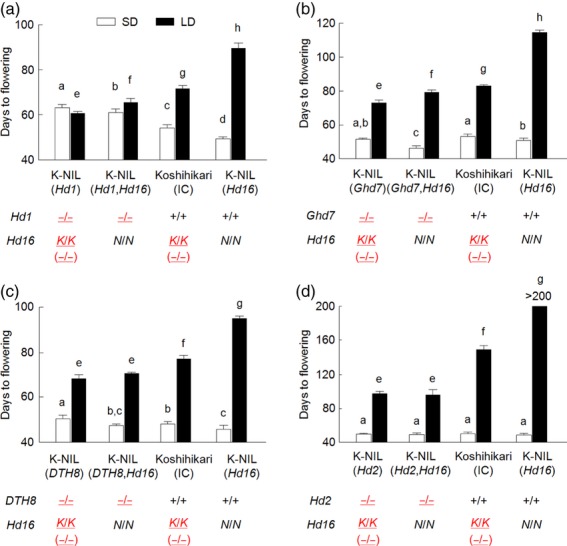
Effects of the Hd16 allele on DTF in backgrounds with functional and defective alleles of (a) Hd1, (b) Ghd7, (c) DTH8 and (d) Hd2.
IC, isogenic control. The phenotypes of each line are presented under short-day (SD) and long-day (LD) conditions. + and − represent functional and defective alleles, respectively. N and K are the Nipponbare and Koshihikari alleles of the Hd16 gene, respectively. Values are means ± standard deviations (n = 10). Means followed by different letters are significantly different among Koshihikari and near-isogenic lines (NILs) at P < 0.05 by Tukey’s honestly significant difference test (a–d, SD conditions; e–h, LD condition). Red and underlined characters indicate defective alleles of each gene.
Expression patterns of flowering-time genes
To reveal the molecular function of Hd16 within the photoperiodic flowering pathway, we examined mRNA levels of flowering-time genes (Hd16, OsLHY, OsPRR1, OsGI, DTH8, Ghd7, Hd1, Ehd1, Hd3a and RFT1) using TaqMan quantitative real-time PCR (qRT-PCR; Figure 4). Gene expression patterns were investigated using 14- and 30-day-old plants grown under SD and LD conditions, respectively. The developmental stages of these plants were roughly 35 days before flowering (the floral induction period) for Nipponbare under SD conditions, and N-NIL(Hd16) under LD conditions. The mRNA level of Hd16 itself showed no clear difference between Nipponbare and N-NIL(Hd16), and little or no diurnal change under either SD or LD conditions. Similarly, we observed no clear differences in mRNA levels between Nipponbare and N-NIL(Hd16) for OsLHY, OsGI, OsPRR1, DTH8, Ghd7 and Hd1 under SD and LD conditions (Figure 4). In contrast, the mRNA level of Ehd1 was significantly higher in N-NIL(Hd16) than in Nipponbare under LD conditions. Likewise, the mRNA levels of Hd3a and RFT1 were also significantly higher in N-NIL(Hd16) under LD conditions, whereas the reverse was true under SD conditions. Ehd1 acts as a promoter of Hd3a and RFT1 expression (Doi et al., 2004). Therefore, these results suggest that Hd16 negatively regulated the transcription of Ehd1, Hd3a and RFT1 under LD conditions.
Figure 4.
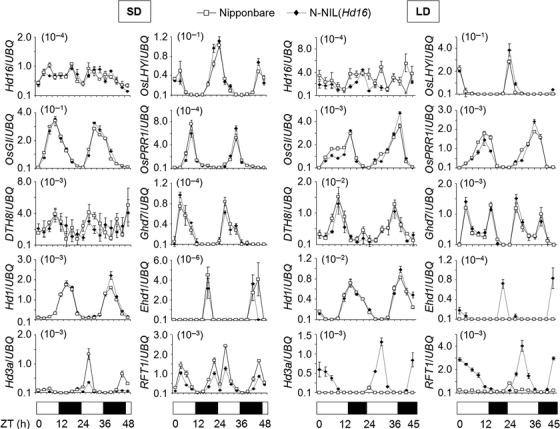
Diurnal changes in transcript levels of Hd16 and nine other genes related to rice flowering under short-day (SD) and long-day (LD) conditions in Nipponbare and N-NIL(Hd16).
Transcription levels were investigated every 3 h from 14-day-old plants grown under SD conditions and from 30-day-old plants grown under LD conditions. Values are means ± standard deviations of three biological replicates. UBQ, ubiqutin used to normalize the values; ZT, Zeitgeber time.
Expression patterns of clock-related and clock-regulated genes
The genes that encode CKI have a pivotal function in the mammalian circadian clock, where they phosphorylate core components of the circadian oscillator (Tuazon and Traugh, 1991; Gross and Anderson, 1998). To reveal whether Hd16 is involved in the similar regulation of the circadian clock in rice, we analyzed the expression patterns of OsLHY and OsPRR1 under constant light (LL) conditions. The expression patterns of these genes were very similar between Nipponbare and N-NIL(Hd16) (Figure S8). Furthermore, we investigated the expression of a luciferase gene driven by the CHLOROPHYLL a/b-BINDING PROTEIN promoter (Cab1R:LUC) under a 12-h dark/12-h light cycle (DL), constant light (LL) and constant dark (DD) conditions. Cab1R:LUC expression was quickly induced by light at dawn under DL conditions (Figure S9). The periods of the Cab1R:LUC expression rhythms were very similar between Nipponbare and N-NIL(Hd16) under DL, LL and DD conditions. These results indicate that Hd16 was not required for clock function in either the presence or the absence of light signals.
Hd16 phosphorylates Ghd7
We observed differences in the transcript levels of Ehd1, Hd3a and RFT1 between Nipponbare and N-NIL(Hd16), but no clear difference in upstream genes such as Ghd7 and Hd1 (Figure 4). To clarify the role of Hd16 in the transcriptional regulation of Ehd1, we investigated the function of the protein encoded by Hd16. Hd16 recombinant proteins encoded by the Nipponbare allele [rHd16(Ni)] and the Koshihikari allele [rHd16(Ko)] were expressed as glutathione-S-transferase (GST) fusion proteins. Purified rHd16(Ni) exhibited significant in vitro kinase activity, leading to the phosphorylation of the common CKI substrate phosvitin protein (Figure 5a). rHd16(Ko) had weak phosvitin phosphorylation activity, indicating decreased kinase activity. rHd16(Ko) also showed decreased activity in auto-phosphorylation. To investigate whether Hd1 and Ghd7 were direct targets of Hd16, we purified GST fusions with Hd1 and Ghd7, and characterized their potentials as Hd16 substrates by means of an in vitro phosphorylation assay. Ghd7 was strongly phosphorylated by rHd16(Ni), but not by rHd16(Ko) (Figure 5b). Hd1 was not phosphorylated by either rHd16(Ni) or rHd16(Ko) (Figure S10). An amino acid residue of serine in the 68th position (Ser-68) of Ghd7 was predicted as a CKI phosphorylation site by KinasePhos, a kinase-specific phosphorylation site prediction tool (Huang et al., 2005). Although we looked for potential phosphorylation sites in the Ghd7 protein using another prediction tool, NetPhosK, only KinasePhos predicted a phosphorylation site. This observation suggests that Hd16 plays a role in the phosphorylation of Ghd7, thereby enhancing the function of Ghd7, downregulating the transcription of Ehd1 and of other downstream genes, such as Hd3a and RFT1, and obviously delaying flowering.
Figure 5.
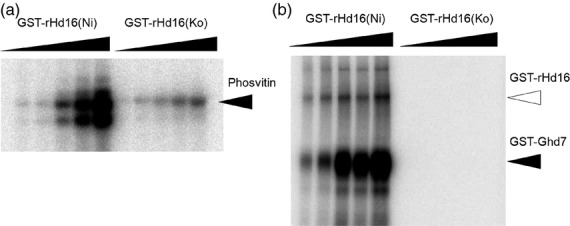
Autoradiographs of the SDS-PAGE analyses of (a) phosvitin and (b) rGhd7 after in vitro rHd16 phosphorylation assays.
The black triangles on the autoradiograph represent concentrations of substrates and rHd16s.
We performed a yeast two-hybrid assay using β-galactosidase to examine whether there was a direct protein–protein interaction between Hd16 and Ghd7, and whether the level of the protein–protein interaction with Ghd7 differed between the Nipponbare Hd16 [Hd16(Ni)] and the Koshihikari Hd16 [Hd16(Ko)]. Hd16(Ni) interacted with the full-length Ghd7 [1–257 amino acids (aa)], with the N-terminal region of Ghd7 (1–128 aa) and with the C-terminal region of Ghd7 (70–257 and 117–257 aa) (Figure 6a,b). Hd16(Ko) also interacted with these Ghd7 sequences, but the interaction between Hd16(Ko) and Ghd7 was weaker than the interaction with Hd16(Ni). Taken together, these results suggest that Hd16(Ko) exhibits greatly decreased kinase activity, but does not significantly alter its ability to interact with Ghd7. These results showed that Hd16 is involved in a Ghd7-mediated photoperiodic flowering pathway in rice.
Figure 6.
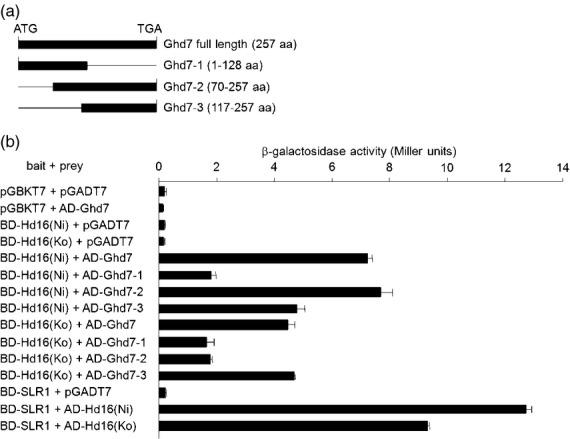
Yeast two-hybrid interaction between Hd16 and Ghd7.
(a) Schematic diagrams of the Ghd7 fragments used in the yeast two-hybrid interaction assay. Numbers in parentheses indicate the number of amino acid residues (aa).
(b) Results of the yeast two-hybrid interaction assay between Hd16 and Ghd7. Yeast strain AH109 was transformed with the indicated plasmid combinations of the bait and the prey, respectively. Values are means ± standard deviations of β-galactosidase activities from three individual colonies. pGBKT7, bait vector; pGADT7, prey vector; Hd16(Ni), Nipponbare Hd16 protein; Hd16(Ko), Koshihikari Hd16 protein.
Discussion
We demonstrated that Hd16 acts as an activator of Ghd7, which is encoded by one of the key genes in the photoperiodic control of rice flowering (Figure 7). Ghd7 is a major floral repressor under LD conditions in rice: it strongly represses the expression of Ehd1 and the genes downstream of Ehd1, such as Hd3a and RFT1 (Xue et al., 2008; Itoh et al., 2010). In the present study, the transcript levels of three flowering-time genes (Ehd1, Hd3a and RFT1) were higher in N-NIL(Hd16), which has the Koshihikari Hd16 allele in a Nipponbare background, than in Nipponbare under LD conditions, but Hd16 did not affect the level of Ghd7 mRNA under either SD or LD conditions (Figure 4). Hd16 and Ghd7 showed a significant protein–protein interaction, and Ghd7 was phosphorylated by Hd16 in vitro (Figures 5 and 6). Although an in vivo kinase assay would be necessary to confirm the phosphorylation of Ghd7 by Hd16, our observations suggest that Hd16 directly regulates the function of Ghd7 by means of phosphorylation, and thereby affects the expression of downstream genes (Figure 7).
Figure 7.
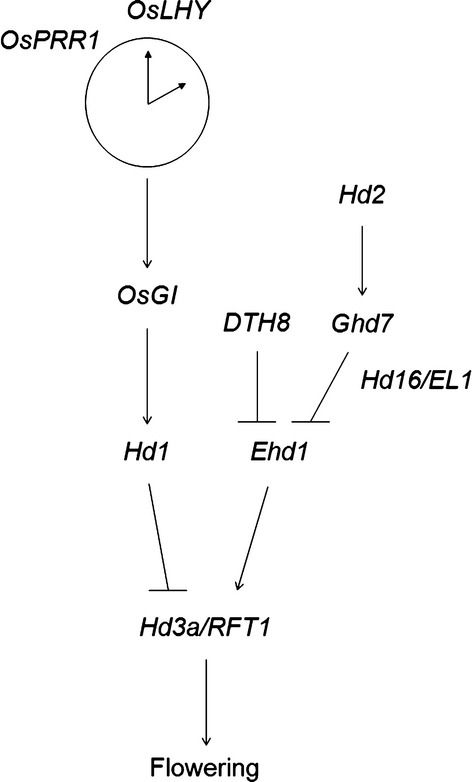
Schematic representation of the proposed model of Hd16/EL1 in the part of the gene regulatory network that controls photoperiodic flowering under long-day conditions in rice.
It is likely that Hd16/EL1 does not control the circadian clock. Phosphorylation of Ghd7 by Hd16/EL1 controls the transcriptional level of Ehd1. Pointed arrows indicate the upregulation of a gene; blunt-ended arrows indicate the downregulation of a gene.
Genetic interactions were observed between Hd16 and four flowering-time genes (Hd1, Ghd7, DTH8 and Hd2) under LD conditions (Figure 3). Previous studies have suggested that DTH8 is a member of a Ghd7-mediated photoperiodic flowering pathway (Wei et al., 2010; Tsuji et al., 2011). The position of Hd2 in the photoperiodic flowering pathway is not clear, but Hd2 and Ghd7 have been suggested to control rice flowering time within the same pathway (Shibaya et al., 2011). Therefore, the genetic interactions we observed in our analysis of the NILs could also be explained by differences in the activation state of this Ghd7 pathway. Arabidopsis CO forms a trimeric complex of HAP2/HAP3/HAP5 with high homology to the yeast HAP complex (Wenkel et al., 2006). Interestingly, Hd1 and Ghd7 contain the CCT domain. The CCT domain of these proteins is similar to a domain of HAP2 (Ishikawa et al., 2011). DTH8 encodes a putative HAP3 subunit (Wei et al., 2010). Therefore, Hd16 might be associated with the formation of Hd1/DTH8/HAP5 or Ghd7/DTH8/HAP5 trimetric complexes to suppress flowering and to downregulate Ehd1 under LD conditions. Additional protein–protein interaction assays with Hd1 and DTH8 might confirm the formation of these trimeric HAP complexes. Under SD conditions, Hd16 promotes flowering, but Ghd7 and DTH8 have no significant effects on flowering time (Xue et al., 2008; Wei et al., 2010). There are two possible explanations for this observation. One is that the Hd16 control of flowering time is mediated by the formation of the Hd1/DTH8/HAP5 complex under SD conditions. Alternatively, Hd16 may regulate flowering time by interacting with other unknown genes under SD conditions. Further analysis will be necessary to identify the genes that interact with Hd16 under SD conditions and to clarify this control pathway. Gene expression analysis of Hd16 and Ehd1 in an Hd1-deficient genetic background would corroborate the role of Hd16 in the photoperiodic flowering pathway of rice.
Hd16 was previously identified as Early flowering 1 (EL1) (Dai and Xue, 2010). EL1 controls rice flowering time by downregulating the gibberellin signaling pathway mediated by the phosphorylation of SLR1 (Slender rice 1). We confirmed the in vitro phosphorylation of SLR1 by Hd16 (Figure S11). N-NIL(Hd16) and RNAi transformants of Hd16 showed taller culms and an enhanced gibberellin response (longer leaf sheaths) compared with wild-type Nipponbare (Figure S12). These phenotypes were similar to those in the previous study of the EL1 mutant (Dai and Xue, 2010). Our results also revealed that N-NIL(Hd16/EL1) exhibited decreased photoperiod sensitivity (Figure 1) and that Hd16/EL1 phosphorylated Ghd7 (Figure 5). Phosphorylation of SLR1 by Hd16 suppresses the gibberellin response, and phosphorylation of Ghd7 by Hd16 suppresses the production of Ehd1 mRNA. Both phosphorylations lead to delayed flowering under LD conditions. Therefore, Hd16/EL1 appears to be associated with both photoperiod and gibberellin responses in the rice flowering pathway. In the NILs with defective Ghd7 alleles, there is a small flowering-time difference between the Nipponbare and Koshihikari Hd16 alleles under LD conditions (Figure 3b). Although we cannot confirm whether the gibberellin signaling pathway promotes flowering time either directly or indirectly, our results suggest that Hd16/EL1 mainly controls flowering time in rice by mediation of the photoperiodic flowering pathway. It would be interesting to evaluate the effects of Hd16 on plant height and flowering time in the slr1 mutant genetic background. Recently, Asano et al. (2009) described the Slr1-d series of dominant dwarf mutants, caused by a gain-of-function mutation in the N-terminal region of SLR1. Future studies using these mutant lines could reveal how Hd16 is involved in the control of plant height and flowering time in rice.
CKI is a protein serine/threonine kinase that is highly conserved among plant and animal species (Tuazon and Traugh, 1991; Gross and Anderson, 1998). CKI has various functions in both the cytoplasm and the nucleus, such as DNA repair, regulation of the cell cycle, vesicular trafficking, morphogenesis, and circadian rhythm (Liu et al., 2003; Rumpf et al., 2010). Another function of CKI is to maintain the circadian rhythm. Phosphorylation of clock components by CKI is the key step that both initiates and regulates the circadian oscillation (Rumpf et al., 2010). The tau, doubletime (dbt), and familial advanced sleep phase syndrome (FASPS) genes encode CKIε in Mesocricetus auratus, Drosophila melanogaster and Homo sapiens, respectively. The tau mutant allele drastically reduces the period of the circadian rhythms (Ralph and Menaker, 1988). The dbt and FASPS mutants also show an altered circadian period (Kloss et al., 1998; Xu et al., 2005). The non-synonymous substitution in the Koshihikari Hd16 allele was localized near the tau mutation site (Figure S1). RNAi plants of N-NIL(Hd16), which contain the Koshihikari Hd16 allele, showed no significant difference in flowering time compared with the control plants (Figure S4). The Hd16 protein encoded by the Koshihikari allele showed no phosphorylation of Ghd7 in our in vitro phosphorylation assay (Figure 5b). These results suggest that the non-synonymous substitution in the Koshihikari Hd16 allele influences the ability of the protein to phosphorylate Ghd7, which, in turn, decreases the photoperiod sensitivity of the plant. However, no alterations of the period of the circadian rhythm were observed in the Cab:LUC experiments that compared the Nipponbare and Koshihikari Hd16 alleles (Figure S9). Hd16 was classified in a different clade than the clade containing rice orthologs of CKIε in the phylogenetic tree of CKI (Figure S2). Therefore, Hd16 might have different functions from CKI genes that control the circadian rhythm, and might regulate flowering time mediated by the photoperiod sensitivity pathway without affecting the regulation of the circadian rhythm. Cab:LUC experiments with RNAi plants and T-DNA tagged lines that presumably produce little or no Hd16 protein would be necessary to determine whether Hd16 functions differently from the other CKI genes that control the circadian rhythm. In rice, there is evidence of the involvement of CKI genes in the regulation of plant root development, hormone-related functions and hybrid breakdown (Liu et al., 2003; Dai and Xue, 2010; Yamamoto et al., 2010). The present study elucidated the involvement of rice CKI in flowering regulation and as a physiological substrate for flowering-time genes.
During the process of introducing rice to higher latitudes, rice breeders have selected lines with weaker sensitivity to photoperiod to produce an early heading date and to ensure maturity during the period of optimal climatic conditions (Izawa, 2007; Xue et al., 2008; Takahashi et al., 2009; Wei et al., 2010; Ebana et al., 2011). Photoperiod insensitivity resulting from the deficient functioning of flowering-time genes has been used extensively across many crop species to facilitate seasonal adaptation and extension of the geographic range of a species. For example, Ma1 in Sorghum has been used to develop early-flowering cultivars that are suitable for grain production in temperate regions of the world (Murphy et al., 2011). Mat-a in barley has extended barley cultivation into Scandinavia and Colombia (Zakhrabekova et al., 2012). Because the non-synonymous substitution in the Koshihikari Hd16 allele resulted in weak photoperiod sensitivity, this SNP may have permitted the extension of the cultivation area for rice cultivars into more northern temperate regions. Agronomic traits differ significantly between Koshihikari and K-NIL(Hd16) under SD, LD and ND conditions (Figure S13). A recent large-scale QTL analysis for agronomic traits of importance in rice breeding programs clearly demonstrated that Hd16 had pleiotropic effects on many other agronomic traits (Hori et al., 2012). The proportion of modern rice cultivars that contain the Koshihikari Hd16 allele has been increased by breeders (Figure S7). Therefore, the Koshihikari Hd16 allele might have been selected to provide a more appropriate flowering time for temperate environments, thereby allowing cultivation in a wider range of areas. For the practical application of our results in breeding programs, it would be possible to use PCR primers capable of amplifying the non-synonymous substitution of Hd16 (Table S1) in marker-assisted selection. Information on epistatic interactions between Hd16 and other flowering-time genes (Figure 3) would also be helpful to predict the flowering time of future rice cultivars. It will also be important to identify epistatic interactions for plant height between Hd16 and other flowering-time genes to support breeding efforts. Hd16, Ghd7 and DTH8 affect plant height (Figures S12b and S13; Xue et al., 2008; Wei et al., 2010; Dai and Xue, 2010). Predicting such gene interactions will be a valuable contribution to future rice breeding efforts.
Taken together, our results suggest that Hd16 regulates the rice flowering pathway by enhancing the photoperiod response caused by the phosphorylation of Ghd7. Allele combinations among various flowering-time genes determine the natural variation in rice flowering time. Understanding this natural variation and genetic bases of flowering time more completely will be necessary before breeders can develop crop cultivars that are better adapted to specific geographic regions.
Experimental Procedures
Plant materials and growth conditions
We developed NILs for Hd16, Hd1, Ghd7, DTH8 and Hd2 by means of four backcrosses with the rice cultivars Nipponbare and Koshihikari, as the recurrent parents. Donor alleles of each gene were introduced from chromosome segment substitution lines of Koshihikari/Kasalath, Nipponbare/Koshihikari or Koshihikari/Hayamasari (Ebitani et al., 2005; Hori et al., 2010; Shibaya et al., 2011). Plants were grown in a controlled-environment cabinet under SD conditions (10 h of light/14 h of dark, at 28°C for 12 h and 24°C for 12 h, respectively) or LD conditions (14.5 h of light/9.5 h of dark, at 28°C for 12 h and 24°C for 12 h, respectively; in the Hd2 background, 18 h of light/6 h of dark, at 28°C for 12 h and 24°C for 12 h, respectively). The relative humidity was maintained at 60% under a photosynthetic photon flux density of 500 μmol m−2 s−1 provided by metal halide lamps that covered the spectrum from 300 to 1000 nm. For evaluation under ND conditions, plants were grown from mid-April until the end of September in a paddy field at the National Institute of Agrobiological Sciences (36.03°N, 140.11°E). DTF under SD, LD and ND conditions was scored as the number of days required from germination to emergence of the first panicle from a leaf sheath.
Quantitative real-time PCR analysis of gene expression
Total RNA (2.5 μg) was extracted from leaves using the sodium dodecyl sulfate phenol method and primed with the oligo(dT)12–18 primer using a SuperScript II reverse transcriptase (Invitrogen, http://www.invitrogen.com). cDNA corresponding to 50 ng of total RNA was used as the template for each TaqMan PCR reaction using an ABI PRISM 7900 Sequence Detection System (Applied Biosystems, http://www.appliedbiosystems.com). Transcription levels of Hd16 and DTH8 were quantified using gene-specific primers and probes (Table S1). To measure the transcription levels of OsLHY, OsPRR1, OsGI, Hd1, Ghd7, Ehd1, Hd3a, RFT1 and UBQ2, we quantified their abundance in accordance with the methods in previously published reports (Ogiso et al., 2010; Matsubara et al., 2012). The results represent the means of at least three biological replicates, with three technical repeats for each biological replicate.
Protein purification and in vitro kinase assays
Plasmids used for the expression of four glutathione-S-transferase (GST) fusion proteins were constructed by means of PCR amplification using appropriate primers (Table S1), and were subcloned into pCR-Blunt II-TOPO vectors (Invitrogen). DNA fragments that included the Hd16, Ghd7 and SLR1 coding sequences were digested from the TOPO vectors with SalI/NotI restriction enzymes, and then ligated into the pGEX-4T-1 bacterial expression vector (GE Healthcare, http://www.gehealthcare.com). GST fusion proteins were expressed in Escherichia coli strain BL21 and purified using GST Sepharose 4B beads (GE Healthcare). Kinase assays were performed using 150 ng of the recombinant Hd16 proteins after cleavage of GST by thrombin. An exogenous substrate, 15 μg phosvitin (Sigma-Aldrich, http://www.sigmaaldrich.com), and recombinant Ghd7 protein were added to the assays. The phosphorylation assay was performed by adding [γ-P32]ATP (20 μm, 10 μCi; PerkinElmer, http://www.perkinelmer.com). Phosphorylation of the exogenous substrates was analyzed by means of autoradiography.
Yeast two-hybrid analysis and β-galactosidase assay
We used full-length and partial sequences of Hd16, Ghd7 and SLR1 as baits and prey, and performed PCR amplifications from the full-length cDNA clones of Hd16, Ghd7 and SLR1. We divided the Ghd7 sequence into three parts: from 1 to 128 amino acids, from 70 to 257 amino acids and from 117 to 257 amino acids. Amplified cDNA fragments were cloned into pGBKT7 DNA-BD and pGADT7 AD vectors from the Matchmaker Two-Hybrid System 3 (Clontech, http://www.clontech.com). The resultant constructs were transformed into Saccharomyces cerevisiae strain AH109. Yeast two-hybrid assays were performed in the absence of adenine, leucine, histidine and tryptophan in solid media (BD Biosciences, http://www.bdbiosciences.com). We performed the β-galactosidase assay in accordance with the manufacturer’s instructions (Clontech).
Supporting experimental procedures
Experimental protocols for the fine mapping of Hd16, Agrobacterium-mediated transformation, phylogenetic analysis, histochemical analysis of GUS expression, bioluminescence assays for circadian rhythm and the measurement of shoot elongation in response to gibberellins are described in Appendix S1.
Acknowledgments
This work was supported by the Ministry of Agriculture, Forestry and Fisheries of Japan (Integrated Research Project for Plant, Insect, and Animal using Genome Technology, IP1001; Genomics for Agricultural Innovation, GPN0001), and by the Ministry of Education, Culture, Sports, Science and Technology of Japan through a Grant-in-Aid For Young Scientists (B) (20780008). We thank Ms H.J. Zhu and Ms K. Ono at the National Institute of Agrobiological Sciences for their excellent technical support.
Author Contributions
K.H., E.O.T., K.M., and M.Y. designed the research; K.H., E.O.T., K.M., K.E., and U.Y. performed the research; and K.H., E.O.T., and M.Y. wrote the paper.
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Figure S1. Alignment of casein kinase I isoforms from various species.
Figure S2. Phylogenetic relationships among the casein kinase-I genes.
Figure S3. RNA interference with Hd16, showing mRNA production in T2 lines.
Figure S4. Phenotypes of RNA interference in N-NIL(Hd16).
Figure S5. Levels of Hd16 mRNA in different tissues during development.
Figure S6. Localization of GUS expression at the seedling stage.
Figure S7. Allelic distributions of the Hd16 gene in cultivated and wild rice.
Figure S8. Diurnal changes in transcript levels of OsLHY and OsPRR1 under constant light conditions.
Figure S9. Bioluminescence analysis of Cab1R:LUC expression.
Figure S10. Autoradiograph from the SDS-PAGE analyses of Hd1 and Ghd7 after in vitro Hd16 phosphorylation assays.
Figure S11. Autoradiograph from the SDS-PAGE analyses of SLR1 after in vitro Hd16 phosphorylation assays.
Figure S12. Gibberellin (GA3) and paclobutrazol (PBZ) responses in Nipponbare, N-NIL(Hd16) and the RNAi transformant.
Figure S13. Agronomic traits in Koshihikari and K-NIL(Hd16).
Table S1. Primer and probe sequences used in this study.
Table S2. Rice accessions used in this study.
Appendix S1. Supporting experimental procedures.
References
- Asano K, Hirano K, Ueguchi-Tanaka M, Angeles-Shim RB, Komura T, Satoh H, Kitano H, Matsuoka M, Ashikari M. Isolation and characterization of dominant dwarf mutants, Slr1-d, in rice. Mol. Genet. Genomics. 2009;281:223–231. doi: 10.1007/s00438-008-0406-6. [DOI] [PubMed] [Google Scholar]
- Dai C, Xue HW. Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 2010;29:1916–1927. doi: 10.1038/emboj.2010.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel X, Sugano S, Tobin EM. CK2 phosphorylation of CCA1 is necessary for its circadian oscillator function in Arabidopsis. Proc. Natl Acad. Sci. USA. 2004;101:3292–3297. doi: 10.1073/pnas.0400163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi K, Izawa T, Fuse T, Yamanouchi U, Kubo T, Shimatani Z, Yano M, Yoshimura A. Ehd1, a B-type response regulator in rice, confers short-day promotion of flowering and controls FT-like gene expression independently of Hd1. Genes Dev. 2004;18:926–936. doi: 10.1101/gad.1189604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebana K, Shibaya T, Wu J, et al. Uncovering of major genetic factors generating naturally occurring variation in heading date among Asian rice cultivars. Theor. Appl. Genet. 2011;122:1199–1210. doi: 10.1007/s00122-010-1524-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebitani T, Takeuchi Y, Nonoue Y, Yamamoto T, Takeuchi K, Yano M. Construction and evaluation of chromosome segment substitution lines carrying overlapping chromosome segments of indica rice cultivar Kasalath in a genetic background of japonica elite cultivar Koshihikari. Breed. Sci. 2005;55:65–73. [Google Scholar]
- Fujino K, Sekiguchi H. Mapping of QTLs conferring extremely early heading in rice (Oryza sativa L.) Theor. Appl. Genet. 2005;111:393–398. doi: 10.1007/s00122-005-2035-3. [DOI] [PubMed] [Google Scholar]
- Gouesnard B, Rebourg C, Welcker C, Charcosset A. Analysis of photoperiod sensitivity within a collection of tropical maize populations. Genet. Resour. Crop Evol. 2002;49:471–481. [Google Scholar]
- Gross SD, Anderson RA. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell. Signal. 1998;10:699–711. doi: 10.1016/s0898-6568(98)00042-4. [DOI] [PubMed] [Google Scholar]
- Hayama R, Coupland G. The molecular basis of diversity in the photoperiodic flowering responses of Arabidopsis and rice. Plant Physiol. 2004;135:677–684. doi: 10.1104/pp.104.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama R, Yokoi S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- Hori K, Sugimoto K, Nonoue Y, Ono N, Matsubara K, Yamanouchi U, Abe A, Takeuchi Y, Yano M. Detection of quantitative trait loci controlling pre-harvest sprouting resistance by using backcrossed populations of japonica rice cultivars. Theor. Appl. Genet. 2010;120:1547–1557. doi: 10.1007/s00122-010-1275-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K, Kataoka T, Miura K, Yamaguchi M, Saka N, Nakahara T, Sunohara Y, Ebana K, Yano M. Variation in heading date conceals quantitative trait loci for other traits of importance in breeding selection of rice. Breed. Sci. 2012;62:223–234. doi: 10.1270/jsbbs.62.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HD, Lee TY, Tseng SW, Horng JT. KinasePhos: a web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 2005;33:W226–W229. doi: 10.1093/nar/gki471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11:550–558. doi: 10.1016/j.tplants.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Ishikawa R, Aoki M, Kurotani K, Yokoi S, Shinomura T, Takano M, Shimamoto K. Phytochrome B regulates Heading date 1Hd1)-mediated expression of rice florigen Hd3 and critical day length in rice. Mol. Genet. Genomics. 2011;285:461–470. doi: 10.1007/s00438-011-0621-4. [DOI] [PubMed] [Google Scholar]
- Itoh H, Nonoue Y, Yano M, Izawa T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 2010;42:635–638. doi: 10.1038/ng.606. [DOI] [PubMed] [Google Scholar]
- Izawa T. Adaptation of flowering-time by natural and artificial selection in Arabidopsis and rice. J. Exp. Bot. 2007;58:3091–3097. doi: 10.1093/jxb/erm159. [DOI] [PubMed] [Google Scholar]
- Jung C, Muller AE. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009;14:563–573. doi: 10.1016/j.tplants.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell. 1998;94:97–107. doi: 10.1016/s0092-8674(00)81225-8. [DOI] [PubMed] [Google Scholar]
- Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136:3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- Liu W, Xu ZH, Luo D, Xue HW. Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 2003;36:189–202. doi: 10.1046/j.1365-313x.2003.01866.x. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Kono I, Hori K, et al. Novel QTLs for photoperiodic flowering revealed by using reciprocal backcross inbred lines from crosses between japonica rice cultivars. Theor. Appl. Genet. 2008;117:935–945. doi: 10.1007/s00122-008-0833-0. [DOI] [PubMed] [Google Scholar]
- Matsubara K, Ogiso-Tanaka E, Hori K, Ebana K, Ando T, Yano M. Natural variation in Hd17, a homolog of Arabidopsis ELF3 that is involved in rice photoperiodic flowering. Plant Cell Physiol. 2012;53:709–716. doi: 10.1093/pcp/pcs028. [DOI] [PubMed] [Google Scholar]
- Murphy RL, Klein RR, Morishige DT, Brady JA, Rooney WL, Miller FR, Dugas DV, Klein PE, Mullet JE. Coincident light and clock regulation of pseudoresponse regulator protein 37PRR37) controls photoperiodic flowering in sorghum. Proc. Natl Acad. Sci. USA. 2011;109:10281–10286. doi: 10.1073/pnas.1106212108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogiso E, Takahashi Y, Sasaki T, Yano M, Izawa T. The role of casein kinase II in flowering time regulation has diversified during evolution. Plant Physiol. 2010;152:808–820. doi: 10.1104/pp.109.148908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph MR, Menaker M. A mutation of the circadian system in golden hamsters. Science. 1988;241:1225–1227. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- Rumpf C, Cipak L, Dudas A, Benko Z, Pozgajova M, Riedel CG, Ammerer G, Mechtler K, Gregan J. Casein kinase 1 is required for efficient removal of Rec8 during meiosis I. Cell Cycle. 2010;9:2655–2660. doi: 10.4161/cc.9.13.12146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle I, Coupland G. Induction of flowering by seasonal changes in photoperiod. EMBO J. 2004;23:1217–1222. doi: 10.1038/sj.emboj.7600117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibaya T, Nonoue Y, Ono N, Yamanouchi U, Hori K, Yano M. Genetic interactions involved in the inhibition of heading by heading date QTL, Hd2 in rice under long-day conditions. Theor. Appl. Genet. 2011;123:1133–1143. doi: 10.1007/s00122-011-1654-0. [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dean C. Arabidopsis, the Rosetta stone of flowering time? Science. 2002;296:285–289. doi: 10.1126/science.296.5566.285. [DOI] [PubMed] [Google Scholar]
- Sugano S, Andronis C, Ong MS, Green RM, Tobin EM. The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc. Natl Acad. Sci. USA. 1999;96:12362–12366. doi: 10.1073/pnas.96.22.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Teshima KM, Yokoi S, Innan H, Shimamoto K. Variations in Hd1 proteins, Hd3a promoters, and Ehd1 expression levels contribute to diversity of flowering time in cultivated rice. Proc. Natl Acad. Sci. USA. 2009;106:4555–4560. doi: 10.1073/pnas.0812092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316:1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- Thomas B, Vince-Pure D. Photoperiodium in Plants. 2nd edn. San Diego, California: Academic Press; 1997. [Google Scholar]
- Tsuji H, Taoka K, Shimamoto K. Regulation of flowering in rice: two florigen genes, a complex gene network, and natural variation. Curr. Opin. Plant Biol. 2011;14:45–52. doi: 10.1016/j.pbi.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Tuazon P, Traugh J. Casein kinase I and II-multipotential serine protein kinases: structure, function, and regulation. Adv. Second Messenger Phosphoprotein Res. 1991;23:123–164. [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–1034. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou Z, Hu P, Zhai H, Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissota L, Le Gourriereca J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18:2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptacek LJ, Fu YH. Functional consequences of a CKIδ mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- Xue W, Xing Y, Weng X, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 2008;40:761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- Yamamoto E, Takashi T, Morinaka Y, Lin S, Wu J, Matsumoto T, Kitano H, Matsuoka M, Ashikari M. Gain of deleterious function causes an autoimmune response and Bateson–Dobzhansky–Muller incompatibility in rice. Mol. Genet. Genomics. 2010;283:305–315. doi: 10.1007/s00438-010-0514-y. [DOI] [PubMed] [Google Scholar]
- Yano M, Katayose Y, Ashikari M, et al. Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell. 2000;12:2473–2484. doi: 10.1105/tpc.12.12.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakhrabekova S, Gough SP, Braumann I, et al. Induced mutations in circadian clock regulator Mat-a facilitated short-season adaptation and range extension in cultivated barley. Proc. Natl Acad. Sci. USA. 2012;109:4326–4331. doi: 10.1073/pnas.1113009109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Alignment of casein kinase I isoforms from various species.
Figure S2. Phylogenetic relationships among the casein kinase-I genes.
Figure S3. RNA interference with Hd16, showing mRNA production in T2 lines.
Figure S4. Phenotypes of RNA interference in N-NIL(Hd16).
Figure S5. Levels of Hd16 mRNA in different tissues during development.
Figure S6. Localization of GUS expression at the seedling stage.
Figure S7. Allelic distributions of the Hd16 gene in cultivated and wild rice.
Figure S8. Diurnal changes in transcript levels of OsLHY and OsPRR1 under constant light conditions.
Figure S9. Bioluminescence analysis of Cab1R:LUC expression.
Figure S10. Autoradiograph from the SDS-PAGE analyses of Hd1 and Ghd7 after in vitro Hd16 phosphorylation assays.
Figure S11. Autoradiograph from the SDS-PAGE analyses of SLR1 after in vitro Hd16 phosphorylation assays.
Figure S12. Gibberellin (GA3) and paclobutrazol (PBZ) responses in Nipponbare, N-NIL(Hd16) and the RNAi transformant.
Figure S13. Agronomic traits in Koshihikari and K-NIL(Hd16).
Table S1. Primer and probe sequences used in this study.
Table S2. Rice accessions used in this study.
Appendix S1. Supporting experimental procedures.


