Abstract
Mutations in JAK2, MPL and CALR are highly relevant to the Philadelphia chromosome (Ph)-negative myeloproliferative neoplasms (MPNs). We performed high resolution melting analysis and Sanger sequencing together with T-A cloning to elucidate the unique mutation profile of these genes, in Chinese patients with MPNs. Peripheral blood DNA samples were obtained from 80 patients with polycythemia vera (PV), 80 patients with essential thrombocytosis (ET) and 50 patients with primary myelofibrosis (PMF). Ten PV patients were identified with diverse JAK2 exon 12 mutations. Five novel JAK2 Exon 12 mutation patterns (M532V/E543G, N533D, M535I/H538Y/K549I, E543G and D544N) were described. JAK2 V617F was detected in 140 samples (66 PV, 45 ET and 29 PMF). JAK2 Exon 12 mutations were prevalent (13%) and variable in the Chinese patients. Compared with PV patients with JAK2 V617F mutations, PV patients with JAK2 exon 12 mutations had an earlier median onset of disease (P = 0.0013). MPL W515L/K mutations were discerned in 4 ET and 3 PMF patients. Two kinds of CALR mutation, c. 1179_1230del and c. 1234_1235insTTGTC were detected in 20 ET and 16 PMF patients. A novel CALR mutation pattern (c. 1173_1223del/c. 1179_1230del) was identified in 2 PMF samples. In addition, 17 scattered point mutations in CALR c.1153 to c.1255 were also detected in 13 cases with CALR frame-shifting variations and 2 cases without CALR frame-shifting variations. Female patients showed a predisposition to CALR mutations (P = 0.0035). Chinese Ph-negative MPN patients have a unique mutation landscape in the common molecular markers of MPN diagnosis. Validation of the molecular diagnostic pipeline should be emphasized since there is a considerable ethnical diversity in the molecular profiles of Ph-negative MPNs.
Introduction
Myeloproliferative neoplasms (MPNs), the malignant conditions characterized by myeloid cell excessive proliferation, were first proposed by the American hematologist Dr. William Dameshek in 1951 [[1]]. Apart from chronic myelogenous leukemia (CML) characterized by the Philadelphia chromosome (Ph) [[2]], the classical Ph-negative MPNs comprise polycythemia vera (PV), essential thrombocytosis (ET) and primary myelofibrosis (PMF), according to the 2008 World Health Organization (WHO) classification criteria [[3]].
Routine diagnostic tests for MPNs include red cell mass determination, bone marrow aspirate and trephine biopsy, arterial oxygen saturation and carboxyhaemoglobin level, neutrophil akaline phosphatase level, vitamin B12 and serum urate [[4]]. Although these tests strongly support the diagnosis of MPNs, the disease could still not be well discriminated from the reactive hyperplasia. Moreover, misdiagnosis could occur when the symptoms of disease are atypical.
In 2005, somatic mutation in JAK2 exon 14 (JAK2 V617F) was first reported to occur in over 95% of PV and approximately 50% of ET and PMF patients, resulting in the auto-activation of this tyrosine kinase and enhanced signaling of the down-stream JAK-STAT pathway [[5],[6]]. Soon after this remarkable discovery, mutations in JAK2 exon 12 were reported to prevail in JAK2 V617F-negative MPNs patients, specifically those with PV in most cases [[7]]. Apart from the mutations in JAK2 gene, genetic alteration in MPL, mainly manifested as MPL W515L and W515K variation in the 10th exon, also plays an important role in the pathogenesis of ET and PMF [[8],[9]]. With a view to promote the molecular diagnosis of MPNs, these mutations were consequently integrated into the WHO diagnostic criteria in 2008 [[10]].
Recently, another milestone in the molecular diagnosis of MPNs, somatic mutations in the CALR gene, was reported [[11],[12]]. In terms of frame shifting insertion and deletion, these JAK2- and MPL-exclusive mutations were found to present in 30% to 40% of ET and PMF patients and were considered highly likely to be integrated into the next version of MPN diagnostic criteria [[13],[14]].
Although the molecular variation in Ph-negative MPNs patients is increasingly a concern for both scientific researchers and clinical professionals, the panoramagram of major genetic alternations, which could be highly variable between the different ethnicities [[15]], has not been depicted in Chinese Han patients with MPNs.
In this study, we assessed the major mutations in the JAK2, MPL and CALR genes in 210 Chinese Han MPNs patients, employing high resolution melting curve analysis (HRMA) for preliminary screening and Sanger sequencing for mutation validation, in order to unveil the MPN-specific mutations in the Chinese Han population.
Materials and methods
Patients
Peripheral blood samples were obtained from 80 PV, 80 ET and 50 PMF patients in the Department of Hematology in Huashan Hospital and Department of Hematology of Shanghai Tenth People’s Hospital. All these Ph-negative patients were diagnosed according to the WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (2008) [[3]]. Written informed consent was received from all the participants. DNA was extracted from the blood samples collected in ethylenediaminetetraacetic acid anticoagulant with QiaAmp DNA Blood Mini kit, and diluted with ddH2O to a final concentration of 15–25 ng/μl. In compliance with Helsinki Declaration of 1975 as revised in 1996, this study was approved by the Institutional Review Board of Huashan Hospital. The patients’ clinical features, including age, gender and hematological test results were illustrated in Table 1.
Table 1.
Clinical and laboratory features of 210 patients with myeloproliferative neoplasms, stratified by the clinical diagnosis of polycythemia vera (PV), essential thrombocytosis (ET) and primary myelofibrosis (PMF)
| Variables | PV (n = 80) | ET (n = 80) | PMF (n = 50) | All patients (n = 210) |
|---|---|---|---|---|
| Age in years; median (range) |
61 (20–89) |
59 (19–94) |
60 (36–89) |
60 (19–94) |
| Age > 60 years; n (%) |
51 (63%) |
45 (56%) |
31 (62%) |
127 (60%) |
| Leukocytes, × 109/l; median (range) |
10.94 (1–42.41) |
9.94 (2.83–22.71) |
10.38 (0.69–92.67) |
10.43 (0.69–92.67) |
| Erythrocytes, × 1012/l; median (range) |
6.36 (2.22–9.93) |
4.53 (1.51–6.84) |
4.11 (1.97–9.36) |
5.15 (1.51–9.93) |
| Hemoglobin, g/dl; median (range) |
186.25 (73–245) |
132.94 (48–182) |
116.95 (44–278) |
150.22 (44–278) |
| Platelets, × 109/l; median (range) | 292.61 (56–715) | 774.89 (328–2887) | 416.30 (16–2890) | 509.47 (16–2890) |
Mutation screening of JAK2 Exon 12 with high resolution melting analysis
High-resolution melting analysis (HRMA) was used to screen for genetic alternations in JAK2 Exon 12. The whole Exon 12 was amplified by PCR with the mutation screen primers (sequences described in Table 2), amplifying a 127 bp small amplicon in order to achieve a high mutation detecting sensitivity [[16]]. Each PCR reaction was performed in a 50 μl of reaction volume. The master mix contained: 2× Premix ExTaq Hotstart (TaKaRa BIO, Shiga, Japan), 0.25 μM E12HRM-F primer, 0.25 μM E12HRM-R primer, 1.5 μM SYTO-9 DNA dye (Invitrogen, Carlsbad, CA) and 15-25 ng DNA template. The mix was subjected to an ABI 9700 thermal cycler (Applied Biosystems, Foster City, CA) for the gene amplification. The PCR protocol included an initial denaturation of 95°C for 10 min, followed by 35 cycles of 95°C for 30 sec, 58°C for 30 sec, and 72°C for 30 sec. The products were then brought into the HRMA assay on a Rotor-Gene Q HRMA platform (Qiagen, Germany). During HRMA, these amplicons were heated to 98°C for 2 min and then cooled to 40°C in order to form the abundant heteroduplex; then they were melted at a ramping rate of 0.1°C/s from 65°C to 85°C. The melting fluorescent data were collected using Rotor-Gene Q 1.7 software. For each assay, 2 peripheral blood DNA samples obtained from the healthy people were also amplified and melted as the wild-type control. Any sample identified with a melting curve not fitting that of the wild-type control was considered as a mutation suspect.
Table 2.
Primers for JAK2 Exon 12, JAK2 V617F and CALR mutation screening and sequencing
| Primer | Sequence (5’-3’) | Amplicon length (bp) | |
|---|---|---|---|
| JAK2 exon 12 mutation screening |
E12HRM-F |
AATGGTGTTTCTGATGTACC |
127 |
| |
E12HRM-R |
AGACAGTAATGAGTATCTAATGAC |
|
| JAK2 exon 12 sequencing |
E12SEQ-F |
CTCCTCTTTGGAGCAATTCA |
496 |
| |
E12SEQ-R |
GAGAACTTGGGAGTTGCGATA |
|
| JAK2 V617F |
V617F-F |
AGCTTTCTCACAAGCATTTGG |
150 |
| |
V617F-R |
TGACACCTAGCTGTGATCCTG |
|
| |
V617F-P |
AAATTATGGAGTATGTTTCTGTGGAGACGAGA |
|
| CALR exon sequencing |
CALRExon1-F |
CGGGTGGGTATAAAAGTG |
348 |
| |
CALRExon1-R |
GGGACGCAGAAGAGAAAT |
|
| |
CALRExon2-F |
GTTGGAATGGGGAGTGTC |
337 |
| |
CALRExon2-R |
CTTCCTCCACCTGTCCTC |
|
| |
CALRExon3-F |
CGGTGACGAGGAGAAAGATA |
493 |
| |
CALRExon3-R |
TAAGAAAGTCAATGGGGTCT |
|
| |
CALRExon4.5-F |
GTTTCTCTTCTCAGCCTTG |
678 |
| |
CALRExon4.5-R |
TCTCTTCATCCCAGTCCTC |
|
| |
CALRExon6.7-R |
ATCCCACAGACTCCAAGC |
611 |
| |
CALRExon6.7-R |
GGACTTCACAGGGACAGAC |
|
| |
CALRExon8.9-F |
CGGTGTTCCTTGTCTTCTC |
584 |
| CALRExon8.9-R | ACAGAGACATTATTTGGCG |
Genotyping of JAK2 Exon 12 mutations
Sanger sequencing was further applied to validate the suspected JAK2 Exon 12 mutations. The sequencing primers flanked the intron 11, exon 12 and intron 12 of the JAK2 gene (sequences described in Table 2). Gene amplification was carried out in a 20 μl PCR mix, which contained 2× Premix ExTaq Hotstart, 0.25 μM E12SEQ-F primer, 0.25 μM E12SEQ-R primer and 15-25 ng DNA template with the thermal cycling procedure described in the Mutation Screening of JAK2 Exon 12 with High Resolution Melting section. After the amplification, all the products were separated by agarose electrophoresis and delivered to the Technical Support Department of BGI genomics institution (Shanghai, China) for the subsequent bi-directional sequencing procedure on an ABI 3730 XL genetic analyzer (Applied Biosystems).
Since the JAK2 exon 12 mutation enjoys a predisposition to rare allele load [[17]], T-A cloning was performed to isolate the pure mutant subclones for accurate mutation genotyping before the final conformation. For each JAK2 exon 12 mutation candidate, the PCR amplicon was ligated to the PMD19-T simple vector (Takara), and then transformed into DH5α competent E. coli cells. The bacteria were proliferated in Luria-Bertani broth and spread onto the IPTG-Xgel (Invitrogen) coated ampicillin-LB araga dishes for blue/white selection, and plasmids were extracted from the enriched white isolations with Plasmid Mini Kit (Qiagen). For each sample, 20 bacteria clones were isolated for the subsequent DNA sequencing.
Detection of JAK2 V617F mutation with unlabeled probe melting assay
As we previously described, the unlabeled probe melting technology is a robust tool for identifying the JAK V617F mutation [[18]]. This unlabeled probe HRMA was again performed on the Rotor-Gene Q platform to identify the JAK2 V617F mutation in the peripheral DNA of 210 MPNs patients.
Mutation scanning of MPL W515L and W515K mutations with Taqman qPCR
All 130 ET and PMF samples were subjected to a Taqman probe based qPCR assay [[19]] to detect the MPL W515L and MPL W515K mutations. The qPCR procedure was carried out with the Quantitect probe qPCR kit (Qiagen) on the Rotor-Gene Q instrument according to the manufacturer’s instructions, and the data analysis was finished with the Rotor-Gene Q 1.7 software.
Sanger sequencing of CALR gene
Five different DNA poolings were prepared for the sequencing of all 9 exons in CALR. Each pooling comprised 3 equivalent portions of peripheral blood DNA from diverse JAK2/MPL mutation-free ET or PMF patients, to ensure the final sequencing data embodied the genetic information in CALR exons from 15 patients. These poolings were amplified with the sequencing primers (sequences listed in Table 2) and gene amplification procedure described in the Genotyping of JAK2 Exon 12 Mutations section. Agarose electrophoresis purified amplicons were then subjected to bi-directional Sanger sequencing. Moreover, all 210 peripheral blood DNA samples were amplified with sequencing primer targeting the CALR Exon 9 (CALRExon8.9-F and CALRExon8.9-R). Purified amplicons were also bi-directionally sequenced. All DNA sequencing experiments were carried out on an ABI XL3730 genetic analyzer in the BGI Shanghai laboratory. For each mutation-susceptive sample, T-A cloning was also used to confirm the mutation with pure mutant subclones.
Mutation calling in JAK2 Exon 12 and CALR
All the sequencing results were integrated into the Mutation Surveyor V4.0.6 software (SoftGenetics, State College, PA) and then aligned with National Center for Biotechnology Information (NCBI) reference gene contigs (NM_004972.3 for JAK2 and NM_004343.3 for CALR), respectively. To eliminate the false positives from regions of low data quality, we set the mutation trimming score to 20 for the final mutation calling.
Statistical analysis
The clinical parameters were statistically analyzed with Mann–Whitney U test (for measurement data) or Fisher’s exact test (for enumeration data).
Results
Mutation screening of JAK2 Exon 12 by HRMA and genotyping
The HRMA method discerned 10 JAK2 Exon 12 mutation positive samples from 200 MPN samples with wild-type Exon 12. All these mutation-susceptive samples came from patients diagnosed with PV or showed a distinctive erythremia-related feature. The mutation genotyping and conformation were carried out with further Sanger sequencing and T-A cloning. Among these 10 samples, we identified five novel genetic alternation patterns, M532V/E543G (c. 2048 T > C/c. 2088A > G), N533D (c. 2091A > G), M535I/H538Y/K549I (c. 2099G > C/c. 2106C > T/c. 2110A > T), E543G (c. 2122A > G) and D544N (c. 2124G > A) (Figure 1), besides the previously reported mutation models such as F537L (c. 2103 T > C) [http://www.cancer.sanger.ac.uk/cosmic/mutation/overview?id=1462560], F537-I546dup10/F547L [[20]], K539L, N542-E543del and H538K539delinsL [[7]]. The clinical parameters and the JAK2 Exon 12 mutation profiles of each patient are listed in Table 3.
Figure 1.
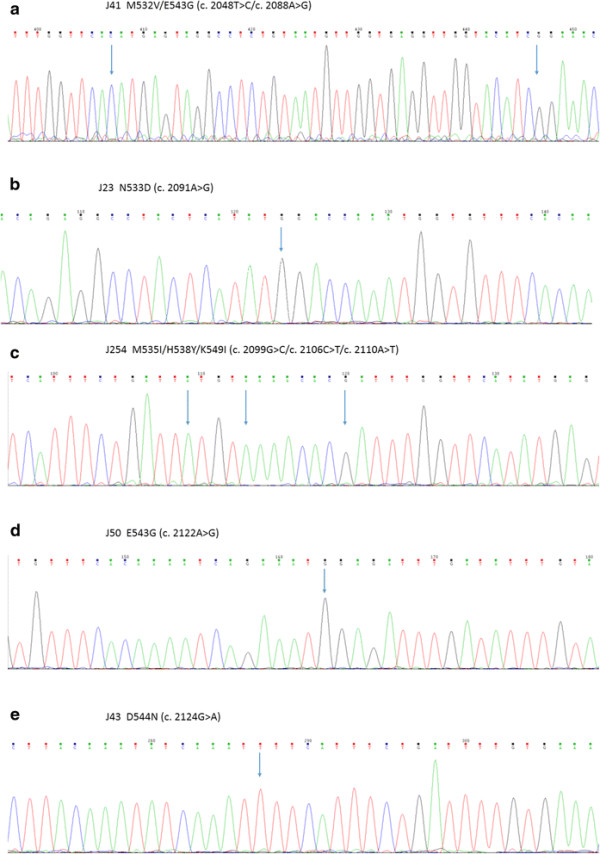
Five novel mutation profiles in JAK2 exon 12. Arrows indicate the mutation site. a) M532V/E543G (c. 2048 T > C/c. 2088A > G). b) N533D (c. 2091A > G). c) M535I/H538Y/K549I (c. 2099G > C/c. 2106C > T/c. 2110A > T). d) E543G (c. 2122A > G). e) D544N (c. 2124G > A). a), c), e): with reverse sequencing primer. b), d): with forward sequencing primer.
Table 3.
Clinical features of 10 myeloproliferative neoplasm patients with JAK2 Exon 12 mutations
| Patient no. | Clinical diagnosis | Age | Gender | JAK2 Exon 12 mutation | WBC (10E9/L) | RBC (10E12/L) | Hb (g/L) | PLT (10E9/L) |
|---|---|---|---|---|---|---|---|---|
| J2 |
PV |
44 |
Male |
F537-I546dup10/F547L |
14.25 |
3.94 |
161 |
438 |
| J23 |
PV |
60 |
Male |
N533D |
9.93 |
4.45 |
153 |
136 |
| J27 |
PV with splenomegaly |
44 |
Female |
F537L |
N/A* |
|||
| J32 |
PV |
43 |
Male |
K539L |
7.57 |
5.83 |
182 |
159 |
| J41 |
PV |
35 |
Male |
M532V/E543G |
8.33 |
5.72 |
180 |
192 |
| J43 |
PV with recurrent brain stem hemorrhage |
47 |
Male |
D544N |
9.11 |
3.86 |
112 |
494 |
| J50 |
PV |
44 |
Male |
E543G |
14.46 |
5.1 |
166 |
257 |
| J71 |
PV |
41 |
Female |
N542-E543del |
6.46 |
4.14 |
143 |
293 |
| J207 |
PV |
43 |
Female |
H538K539delinsL |
5.85 |
5.81 |
178 |
85 |
| J254 | PV | 23 | Female | M535I/H538Y/K539I | 7.08 | 7.46 | 216 | 151 |
*Complete blood count was not available as the patient was in an impaired hematopoiesis condition
JAK2 V617F Mutation Screening with Unlabeled Probe Melting Assay
The unlabeled probe melting JAK2 V617F detecting system indicated there were 66 (82%) PV, 45 (56%) ET and 29 (58%) PMF cases in the MPN patient cohort. No concurrent JAK2 Exon 12/Exon14 (V617F) mutation was identified in any of these samples.
Compared with those PV patients afflicted with JAK2 exon 12 mutations, V617F-positive PV patients showed a delayed median disease onset (P = 0.0013). Among all 140 JAK2 V617F-positive MPNs patients, 124 (88%) were over 60 years old. However, there was no statistical difference according to gender and complete blood cell count (CBC) in the PV patients.
Identification of MPL W515L and W515K mutations with Taqman Probe qPCR
The qPCR method identified 3 MPL W515L mutant and 1 MPL W515K mutant ET samples, and 2 W515L and 1 W515K mutant PMF cases. All these 7 samples were with the wild-type JAK2 exon 12 and exon 14.
CALR mutations in the MPN patients
In the 5 poolings of MPN patients’ DNA, neither novel frame shifting nor recurrent point mutation was detected within all the 9 CALR exons. The mutations in CALR exon 9 were further screened and confirmed by the bi-directional Sanger sequencing and T-A cloning in all 210 samples. A total of 36 patients were found to harbor the CALR mutations. Twenty of these patients were diagnosed with ET, while the remainder were characterized with the PMF diagnosis. The previously reported 52 bp deletion (c. 1179_1230del) and 5 bp insertion (c. 1234_1235insTTGTC) were detected in 17 and 19 patients, respectively. Moreover, we also identified one new kind of CALR genetic variation, c. 1173_1223del in the PCR amplicon subclones, which was derived from two independent PMF samples with the c. 1179_1230del mutation (Figure 2).
Figure 2.
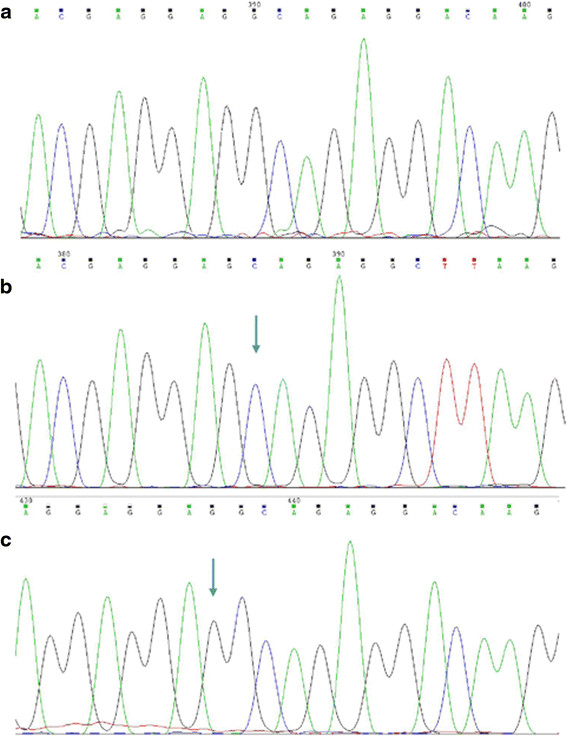
CALR c. 1173_1223del mutation. The 51 bp deletion was detected by Sanger sequencing using forward sequencing primer. a) CALR c. 1173_1223del sequence. b) wild-type sequence. Arrow indicates c. 1173 truncating site. c) wild-type sequence. Arrow indicates c. 1223 truncating site.
Twenty seven (75%) of the 36 patients identified with CALR insertion or deletion were female, suggesting the female patients showed a vulnerability to the CALR mutation (P = 0.0035). No further statistical difference was observed between the clinical parameters of CALR mutation-positive cases and the CALR mutation-free ones.
It is worth noting that we found 17 non-recurrent point mutations accumulated in the CALR Exon 9 in 1 ET and 2 PMF cases free of CALR exon 9 insertion or deletion and in 6 ET and 6 PMF cases with the frame shifting mutations (Figure 3 and Additional file 1: Figure S1). The detailed mutation profile of each patient is listed in Table 4.
Figure 3.
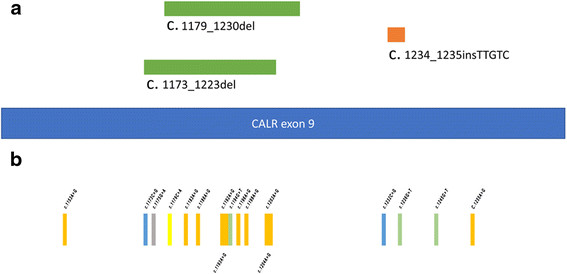
Mutations in CALR exon 9. a) Three types of CALR exon 9 frame-shifting mutations (c. 1173_1223del, c. 1179_1230del, 1234_1235insTTGTC). b) 17 scattered point mutations in c.1153 to c.1255.
Table 4.
CALR exon 9 point mutation profile in 15 MPN patients
| Patient no. | Clinical diagnosis | CALR exon 9 frame-shifting mutations | CALR exon 9 point mutations |
|---|---|---|---|
| J31 |
PMF |
c. 1173_1223del/c. 1179_1230del |
c. 1255A > G |
| J91 |
PMF |
c. 1234_1235insTTGTC |
c. 1232C > G |
| J94 |
PMF |
c. 1179_1230del |
c. 1173C > G |
| J101 |
PMF |
c. 1234_1235insTTGTC |
c. 1203A > G |
| J167 |
PMF |
Undetected |
c. 1175G > A |
| J172 |
PMF |
c. 1234_1235insTTGTC |
c. 1153A > G |
| J183 |
ET |
c. 1179_1230del |
c. 1196A > G/c. 1204A > G |
| J211 |
PMF |
Undetected |
c.1186A > G |
| J233 |
ET |
c. 1234_1235insTTGTC |
c. 1236G > T |
| J243 |
PMF |
c. 1173_1223del/c. 1179_1230del |
c. 1198A > G |
| J259 |
ET |
c. 1179_1230del |
c. 1179C > A/c. 1193A > G |
| J265 |
ET |
c. 1234_1235insTTGTC |
c. 1194G > T |
| J296 |
ET |
Undetected |
c. 1183A > G |
| J307 |
ET |
c. 1179_1230del |
c. 1245G > T |
| J312 | ET | c. 1234_1235insTTGTC | c. 1192A > G |
Compared with those ET and PMF patients with JAK2 V617F or MPL mutations, the CALR mutant patients showed a lower white blood cell count (P = 0.0061), elevated platelet (P = 0.0168) and younger age (P = 0.0002).
Discussion
Since the first description of myeloproliferative diseases by Dr. William Dameshek in 1951 [[1]], there has been a consecutive progression in the understanding of these disease conditions characterized by abnormal bone marrow hyperplasia. Apart from the characterization of the Philadelphia chromosome in the CML, the discovery of the JAK2 V617F mutation in 2005 [[5],[6]] is the most thrilling development in the molecular diagnosis of Ph-negative MPNs. The subsequently reported somatic mutation in JAK2 exon 12 [[7]], though much less prevalent in the patients, is considered as another robust molecular marker for Ph-negative MPNs, especially for PV patients.
Apart from these established pathological markers in JAK2, the genetic mutations in MPL, especially the W515L and W515K substitution in exon 10 could also play a role in the diagnosis of ET and PMF [[21],[22]].
Recently, a total of 36 types of frame-shifting insertions and deletions were detected in the CALR gene, which encodes a Ca2+ binding protein in endoplasmic reticulum called calreticulin. These mutations are located in the 9th exon of the gene, leading to the imperilment of the protein’s C-terminal structure, and were reported to have a incidence of over 60% to 80% in JAK2 and MPL mutation-negative ET and PMF patients [[11],[12]].
In this study, we used the preliminary HRMA and Sanger sequencing method, together with the T-A cloning strategy to reveal the unique genetic variation background of JAK2, MPL and CALR in Chinese Ph-negative MPN patients.
JAK2 V617F is the most prevalent genetic alternation in MPN patients, which could be detected in approximately 95% of patients with PV [[23],[24]]. However, we noticed the Chinese PV patients have a relatively low mutation frequency in the JAK2 V617F mutation (82%), while the mutations in JAK2 exon 12 were much more pervasive (13%) than in the Westerns [[25],[26]], and other East Asians [[27],[28]]. This observation is compatible with the previous small-scale investigation from Taiwan [[29]]. Meanwhile, all the 10 JAK2 Exon 12-mutant patients showed a unique genetic variation pattern, half of which has not been previously reported, indicating the mutation patterns in the JAK2 Exon 12 were highly diverse in the Chinese PV patients.
In our patient cohort, the JAK2 exon 12 mutations were inclined to affect young and middle-aged patients, while the V617F mutation was predominantly found in patients over age 60, which was consistent with a previous report [[30]]. In addition, the high prevalence of exon12 highlights its detection value in Chinese PV patients.
Compared with the JAK2 mutations, genetic abnormalities in MPL exon 10, namely the MPL W515L and W515K mutations, are rare but specific markers for ET and PMF patients. In the Chinese MPN patients, these two types of MPL mutations could be found in approximately 5% of ET and 6% of PMF patients.
For the CALR gene, we identified a novel type of exon 9 long-fragment deletion, c. 1173_1223del, which was isolated in the subclones from patients with the previously reported c. 1179_1230del deletion in exon 9. Meanwhile, the overwhelming majority (94%) of the CALR mutation pattern still lies in the c. 1179_1230del and c. 1234_1235insTTGTC. Our data also indicated that female patients were predisposed to the CALR insertion/deletion. Moreover, we also found a number of point mutations accumulated in c.1153 to c.1255 of the CALR gene, in both the patients with and without the exon 9 insertion/deletion. This phenomenon implied that these scattered point mutations could be the predecessors of the frame-shifting alternation and indicated that the genetic alternations in CALR exon 9 could be an acquired clonal evolution event [[31],[32]]. Although we extracted the data from a moderately-sized patient cohort, this is the first time the landscape of CALR mutations in Chinese MPN patients was revealed.
As reported in the previous literature, a number of gene loci are frequently affected by complicated mutations in MPN patients, such as those in JAK2, CALR, TET2, DNMT3A, and ASXL1 [[33]–[36]]. However, the mechanism behind the pathogenesis and development of these MPNs relevant to genetic imperilment is still unexplored. Given the high variability of Ph-negative MPN relevant gene mutations, we hypothesize that the Ph-negative MPN-related gene variation could be a sequel to the structural instability of nucleotide sequence, and the DNA repair pathway impairment may play a role in the early pathogenesis of MPNs [[37]].
Mutations in JAK2 exon 12, exon 14, MPL exon 10 and CALR exon 9 were presented in each individual exclusively. Patients with genetic alternations in these three genes accounted for up to 90% of all the recruited patients (Figure 4). The highlighted prevalence of these mutations provides us with a multi-gene scanning strategy that has favorable sensitivity for the diagnosis of the Ph-negative MPNs.
Figure 4.
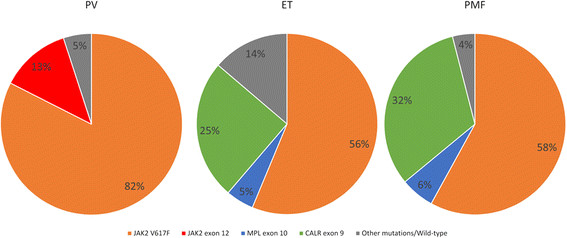
Variation frequency of JAK2 exon 12, JAK2 V617F, MPL W515L/K and CALR exon 9 frame-shifting mutations.
JAK2 V617F and CALR mutations lead to disparate clinical features in the ET and PMF patients. Several previous publications reported the the higher platelet count and lower leukocyte and hemoglobin values in CALR mutant ET patients compared with those with JAK2 V617F or MPL mutations [[38]–[40]]. Our findings confirmed part of those findings (elevated platelet, decreased hemoglobin and earlier disease onset) in the Chinese Han population. However, the different gender inclination in CALR mutation prevalence may be due to the ethnic dissimilarity and sample size.
Despite the progress in the development of MPN molecular markers, the diagnosis of individual patients with MPNs has been a complicated issue demanding intensive collaboration between the clinic and laboratory [[41]]. Our results shed light on a combined molecular diagnostic model, which utilizes the most prevalent DNA variation signatures in three MPN pathogenesis related genes. Moreover, we also suggest that the validation of the molecular diagnostic pipeline should be heeded since there is a considerable ethnical diversity in the molecular profiles of Ph-negative MPNs.
Abbreviations
Ph: Philadelphia chromosome
MPNs: Myeloproliferative neoplasms
CML: Chronic myelogenous leukemia
PV: Polycythemia vera
ET: Essential thrombocytosis
PMF: Primary myelofibrosis
HRMA: High resolution melting curve analysis
CBC: Complete blood cell count
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
WZ, ZX, and XX carried out the molecular genetic studies and participated in the sequence alignment. WZ drafted the manuscript. CY and HT performed the statistical analysis. KZ, LS, WH helped to draft the manuscript. WZ, LW, MX and GM participated in the design of the study. All the authors read and approved the final manuscript.
Additional file
Supplementary Material
Seventeen Scattered Point Mutations in CALR Exon 9.
Contributor Information
Zhiyuan Wu, Email: wuzhiyuan111@hotmail.com.
Xinju Zhang, Email: pp_dou@126.com.
Xiao Xu, Email: nausicaa.dx@live.cn.
Yuming Chen, Email: chenyuming192@126.com.
Tingting Hu, Email: hutingting124@126.com.
Zhihua Kang, Email: kangban@163.com.
Shibao Li, Email: lishibao19851103@163.com.
Hua Wang, Email: ntdx05yjswh@126.com.
Weiwei Liu, Email: liuweiwei221@126.com.
Xiaochao Ma, Email: MXIAOCHA@pitt.edu.
Ming Guan, Email: guanming88@yahoo.com.
Acknowledgements
We thank Dr. Martha Montello from University of Kansas Medical Center for editing the manuscript.
This study is supported by the Key Basic Research Project of Shanghai Science and Technology Commission [11JC1401800], Shanghai Municipal Health Bureau Hundred Talents Program [XBR2011044] and the Scientific Research Project supported by Huashan hospital, Fudan University [2013QD22].
References
- Dameshek W. Editorial: some speculations on the myeloproliferative syndromes. Blood. 1951;6:372–375. [PubMed] [Google Scholar]
- Nowell PC. A minute chromosome in human chronic granulocytic leukemia. Science. 1960;132:1497. [Google Scholar]
- Swerdllow S, Campo E, Harris NL. Who Classification Of Tumours Of Haematopoietic And Lymphoid Tissues. IARC Press, France; 2008. [Google Scholar]
- Lewis SM, Bain BJ, Bates I, Dacie JV. Dacie And Lewis Practical Haematology. Elsevier Health Sciences, Philadelphia; 2006. [Google Scholar]
- James C, Ugo V, Le Couédic J-P, Staerk J, Delhommeau F, Lacout C, Garçon L, Raslova H, Berger R, Bennaceur-Griscelli A. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, Vassiliou GS, Bench AJ, Boyd EM, Curtin N. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, Futreal PA, Erber WN, McMullin MF, Harrison CN. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–468. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, Cuker A, Wernig G, Moore S, Galinsky I. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3:e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardanani AD, Levine RL, Lasho T, Pikman Y, Mesa RA, Wadleigh M, Steensma DP, Elliott MA, Wolanskyj AP, Hogan WJ. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood. 2006;108:3472–3476. doi: 10.1182/blood-2006-04-018879. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Vardiman J. Classification and diagnosis of myeloproliferative neoplasms: the 2008 World Health Organization criteria and point-of-care diagnostic algorithms. Leukemia. 2008;22:14–22. doi: 10.1038/sj.leu.2404955. [DOI] [PubMed] [Google Scholar]
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, Them NC, Berg T, Gisslinger B, Pietra D. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369:2379–2390. doi: 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369:2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Thiele J, Vannucchi A, Barbui T. An overview on CALR and CSF3R mutations and a proposal for revision of WHO diagnostic criteria for myeloproliferative neoplasms. Leukemia. 2014;ᅟ:ᅟ. doi: 10.1038/leu.2014.35. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Tefferi A, Pardanani A. Genetics: CALR mutations and a new diagnostic algorithm for MPN. Nat Rev Clin Oncol. 2014;11:125–126. doi: 10.1038/nrclinonc.2014.16. [DOI] [PubMed] [Google Scholar]
- Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- Jones AV, Cross NC, White HE, Green AR, Scott LM. Rapid identification of JAK2 exon 12 mutations using high resolution melting analysis. Haematologica. 2008;93:1560–1564. doi: 10.3324/haematol.12883. [DOI] [PubMed] [Google Scholar]
- Theocharides A, Passweg JR, Medinger M, Looser R, Li S, Hao-Shen H, Buser AS, Gratwohl A, Tichelli A, Skoda RC. The allele burden of JAK2 mutations remains stable over several years in patients with myeloproliferative disorders. Haematologica. 2008;93:1890–1893. doi: 10.3324/haematol.13074. [DOI] [PubMed] [Google Scholar]
- Wu Z, Yuan H, Zhang X, Liu W, Xu J, Zhang W, Guan M. Development and inter-laboratory validation of unlabeled probe melting curve analysis for detection of JAK2 V617F mutation in polycythemia vera. PLoS One. 2011;6:e26534. doi: 10.1371/journal.pone.0026534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancrazzi A, Guglielmelli P, Ponziani V, Bergamaschi G, Bosi A, Barosi G, Vannucchi AM. A sensitive detection method for MPL W515L or MPL W515K mutation in chronic myeloproliferative disorders with locked nucleic acid-modified probes and real-time polymerase chain reaction. J Mol Diagn. 2008;10:435–441. doi: 10.2353/jmoldx.2008.080015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra D, Li S, Brisci A, Passamonti F, Rumi E, Theocharides A, Ferrari M, Gisslinger H, Kralovics R, Cremonesi L. Somatic mutations of JAK2 exon 12 in patients with JAK2 (V617F)-negative myeloproliferative disorders. Blood. 2008;111:1686–1689. doi: 10.1182/blood-2007-07-101576. [DOI] [PubMed] [Google Scholar]
- Vannucchi AM, Antonioli E, Guglielmelli P, Pancrazzi A, Guerini V, Barosi G, Ruggeri M, Specchia G, Lo-Coco F, Delaini F. Characteristics and clinical correlates of MPL 515W> L/K mutation in essential thrombocythemia. Blood. 2008;112:844–847. doi: 10.1182/blood-2008-01-135897. [DOI] [PubMed] [Google Scholar]
- Beer PA, Campbell PJ, Scott LM, Bench AJ, Erber WN, Bareford D, Wilkins BS, Reilly JT, Hasselbalch HC, Bowman R. MPL mutations in myeloproliferative disorders: analysis of the PT-1 cohort. Blood. 2008;112:141–149. doi: 10.1182/blood-2008-01-131664. [DOI] [PubMed] [Google Scholar]
- Lippert E, Boissinot M, Kralovics R, Girodon F, Dobo I, Praloran V, Boiret-Dupré N, Skoda RC, Hermouet S. The JAK2-V617F mutation is frequently present at diagnosis in patients with essential thrombocythemia and polycythemia vera. Blood. 2006;108:1865–1867. doi: 10.1182/blood-2006-01-013540. [DOI] [PubMed] [Google Scholar]
- Passamonti F, Rumi E, Pietra D, Elena C, Boveri E, Arcaini L, Roncoroni E, Astori C, Merli M, Boggi S. A prospective study of 338 patients with polycythemia vera: the impact of JAK2 (V617F) allele burden and leukocytosis on fibrotic or leukemic disease transformation and vascular complications. Leukemia. 2010;24:1574–1579. doi: 10.1038/leu.2010.148. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Lasho T, Finke C, Hanson C, Tefferi A. Prevalence and clinicopathologic correlates of JAK2 exon 12 mutations in JAK2V617F-negative polycythemia vera. Leukemia. 2007;21:1960–1963. doi: 10.1038/sj.leu.2404810. [DOI] [PubMed] [Google Scholar]
- Passamonti F, Elena C, Schnittger S, Skoda RC, Green AR, Girodon F, Kiladjian J-J, McMullin MF, Ruggeri M, Besses C. Molecular and clinical features of the myeloproliferative neoplasm associated with JAK2 exon 12 mutations. Blood. 2011;117:2813–2816. doi: 10.1182/blood-2010-11-316810. [DOI] [PubMed] [Google Scholar]
- Kim JT, Cho YG, Choi SI, Lee YJ, Kim HR, Jang SJ, Moon DS, Park YJ, Park G. JAK2 V617F and Exon 12 Genetic Variations in Korean Patients with BCR/ABL1-negative Myeloproliferative Neoplasms. Korean J Lab Med. 2010;30:567–574. doi: 10.3343/kjlm.2010.30.6.567. [DOI] [PubMed] [Google Scholar]
- Ohyashiki JH, Hisatomi H, Shimizu S, Sugaya M, Ohyashiki K. Detection of low allele burden of JAK2 exon 12 mutations using TA-cloning in patients with erythrocytosis. Jpn J Clin Oncol. 2009;39:509–513. doi: 10.1093/jjco/hyp048. [DOI] [PubMed] [Google Scholar]
- Yeh Y-M, Chen Y-L, Cheng H-Y, Su W-C, Chow N-H, Chen T-Y, Ho C-L. High percentage of JAK2 exon 12 mutation in Asian patients with polycythemia vera. Am J Clin Pathol. 2010;134:266–270. doi: 10.1309/AJCPK7KGOWPHYWM0. [DOI] [PubMed] [Google Scholar]
- Schnittger S, Bacher U, Haferlach C, Geer T, Müller P, Mittermüller J, Petrides P, Schlag R, Sandner R, Selbach J. Detection of JAK2 exon 12 mutations in 15 patients with JAK2V617F negative polycythemia vera. Haematologica. 2009;94:414–418. doi: 10.3324/haematol.13223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumi E, Harutyunyan AS, Pietra D, Milosevic JD, Casetti IC, Bellini M, Them NC, Cavalloni C, Ferretti VV, Milanesi C. CALR exon 9 mutations are somatically acquired events in familial cases of essential thrombocythemia or primary myelofibrosis. Blood. 2014;123:2416–2419. doi: 10.1182/blood-2014-01-550434. [DOI] [PubMed] [Google Scholar]
- Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I, Girsberger S, Lehmann T, Passweg J, Stern M. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123:2220–2228. doi: 10.1182/blood-2013-11-537167. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Pardanani A, Lim K, Abdel-Wahab O, Lasho T, Patel J, Gangat N, Finke C, Schwager S, Mullally A. TET2 mutations and their clinical correlates in polycythemia vera, essential thrombocythemia and myelofibrosis. Leukemia. 2009;23:905–911. doi: 10.1038/leu.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24:1128–1138. doi: 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegelmann F, Bullinger L, Schlenk R, Paschka P, Griesshammer M, Blersch C, Kuhn S, Schauer S, Döhner H, Döhner K. DNMT3A mutations in myeloproliferative neoplasms. Leukemia. 2011;25:1217–1219. doi: 10.1038/leu.2011.77. [DOI] [PubMed] [Google Scholar]
- Brecqueville M, Rey J, Bertucci F, Coppin E, Finetti P, Carbuccia N, Cervera N, Gelsi‐Boyer V, Arnoulet C, Gisserot O. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer. 2012;51:743–755. doi: 10.1002/gcc.21960. [DOI] [PubMed] [Google Scholar]
- Vorechovsky I, Jones A, Cross N. Why do we see JAK2 exon 12 mutations in myeloproliferative neoplasms&quest. Leukemia. 2013;27:1930–1932. doi: 10.1038/leu.2013.85. [DOI] [PubMed] [Google Scholar]
- Rotunno G, Mannarelli C, Guglielmelli P, Pacilli A, Pancrazzi A, Pieri L, Fanelli T, Bosi A, Vannucchi AM. Impact of calreticulin mutations on clinical and hematological phenotype and outcome in essential thrombocythemia. Blood. 2014;123:1552–1555. doi: 10.1182/blood-2013-11-538983. [DOI] [PubMed] [Google Scholar]
- Rumi E, Pietra D, Ferretti V, Klampfl T, Harutyunyan AS, Milosevic JD, Them NC, Berg T, Elena C, Casetti IC. JAK2 or CALR mutation status defines subtypes of essential thrombocythemia with substantially different clinical course and outcomes. Blood. 2014;123:1544–1551. doi: 10.1182/blood-2014-01-550434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A, Wassie E, Lasho T, Finke C, Belachew A, Ketterling R, Hanson C, Pardanani A, Gangat N, Wolanskyj A. Calreticulin mutations and long-term survival in essential thrombocythemia. Leukemia. 2014;ᅟ:ᅟ. doi: 10.1038/leu.2014.148. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Gianelli U, Iurlo A, Cattaneo D, Lambertenghi-Deliliers G. Cooperation between pathologists and clinicians allows a better diagnosis of Philadelphia chromosome-negative myeloproliferative neoplasms. Expert Rev Hematol. 2014;7:255–264. doi: 10.1586/17474086.2014.876898. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Seventeen Scattered Point Mutations in CALR Exon 9.


