Abstract
Background:
The diagnosis of the follicular variant of papillary thyroid carcinoma (FVPTC) is increasingly common. Recent studies have suggested that FVPTC is heterogeneous and comprises multiple tumor types with distinct biological behaviors and underlying genetics.
Objectives:
The purpose of this work was to identify the prevalence of mutations and gene fusions in known oncogenes in a panel representative of the common spectrum of FVPTC diagnosed at an academic medical center and correlate the clinical and pathological features obtained at the initial diagnosis with the tumor genotype.
Materials and Methods:
We performed SNaPshot genotyping on a panel of 129 FVPTCs of ≥1 cm for 90 point mutations or small deletions in known oncogenes and tumor suppressors and identified gene fusions using an anchored multiplex PCR assay targeting a panel of rearranged oncogenes.
Results:
We identified a mutation or gene fusion in 70% (89 of 127) of cases. Mutations targeting the RAS family of oncogenes were the most frequently observed class of alterations, present in 36% (46 of 127) of cases, followed by BRAF mutation, present in 30% (38 of 127). We also detected oncogenic rearrangements not previously associated with FVPTC, including TFG-ALK and CREB3L2-PPARγ. BRAF mutation was significantly associated with unencapsulated tumor status.
Conclusions:
These data support the hypothesis that FVPTC is composed of distinct biological entities, with one class being identified by BRAF mutation and support the use of clinical genotyping assays that detect a diverse array of rearrangements involving ALK and PPARγ. Additional studies are necessary to identify genetic drivers in the 30% of FVPTCs with no known oncogenic alteration and to better predict behavior in tumors with known genotypes.
Thyroid cancer, the most common endocrine malignancy, is increasing in frequency (1). Although most thyroid cancers exhibit an indolent clinical course, an increasing number of patients require therapies targeted to oncogenic alterations present in tumor cells (2). At the same time, many patients with indolent forms of thyroid cancer receive unnecessary treatments. Therefore, additional studies are needed to improve individualized therapy in subclasses of thyroid cancer.
The follicular variant of papillary thyroid carcinoma (FVPTC) exhibits many of the nuclear features of papillary thyroid cancer (PTC) with a predominantly microfollicular histologic growth pattern. The incidence of FVPTC increased 3-fold between 1973 and 2003 and currently comprises >25% of all PTCs (3, 4). The pathological criteria for FVPTC are well defined, but subjective, leading to inter- and intraobserver variability (5). Recent studies suggest that a subset of FVPTCs exhibit infiltrative growth within the thyroid parenchyma, whereas other FVPTCs are circumscribed by a fibrous tumor capsule (6, 7). The infiltrative (unencapsulated) tumors are more likely to harbor BRAFT1799A (BRAFV600E) mutations and have a higher prevalence of lymph node metastases and local recurrences (8). Encapsulated FVPTCs are more likely to harbor mutations in the RAS family of oncogenes and exhibit a low recurrence rate in the absence of capsular or vascular invasion (9). Therefore, encapsulated FVPTCs without capsular or vascular invasion are predicted to behave in a benign fashion, akin to follicular adenoma, whereas those with vascular or capsular invasion may behave similarly to follicular thyroid carcinoma. Each of these studies required pathological review for study entry; therefore, it is not known whether this classification will apply to patients with FVPTCs routinely encountered in practice.
Here, we characterize a large panel of FVPTC specimens for mutations and translocations in a broad panel of oncogenes and tumor suppressors and associate the clinical and pathological features obtained at the initial diagnosis with mutation status.
Subjects and Methods
Patients
We identified patients with a diagnosis of FVPTC of ≥1 cm by the final surgical pathology report who underwent surgery at Massachusetts General Hospital between 2000 and 2011. Formalin-fixed paraffin-embedded tumor specimens were available for 129 cases. Patients harboring FVPTCs with synchronous thyroid cancers that affected staging were excluded. We specifically requested that the pathological diagnosis not be rereviewed as a requirement for study entry, as we designed the study to closely reflect the diversity of FVPTC diagnoses made in clinical practice, rather than a more narrowly defined population of tumors. Patient data, including demographic, clinical, and pathological, were abstracted from electronic medical records. Evidence of persistent or recurrent disease after the initial treatment with surgery and/or radioiodine was defined by a suppressed thyroglobulin level of >1 or a stimulated thyroglobulin level of >2 or imaging evidence of structural or radioactive iodine-avid disease or by biopsy-proven disease. No evidence of disease was defined by the absence of all of these criteria. Recurrence was defined by a period of no evidence of disease followed by evidence of disease. Protocol approval was obtained from the Partners Human Research Committee (institutional review board).
Mutational analysis
Samples were reviewed to identify FVPTC regions for coring from paraffin blocks, and nucleic acid was extracted and analyzed for mutations using the Clinical Laboratory Improvement Amendments–approved SNaPshot multiplexed targeted sequencing platform (10). This sequencing platform interrogated 90 genetic loci frequently mutated in 21 cancer genes (Supplemental Table 1). Technical failure precluded molecular analysis of 2 samples, and SNaPshot analysis was successfully performed on 127 specimens.
Gene fusion discovery
The anchored multiplex PCR assay was based on multiplex PCR technology used for fusion transcript detection using targeted next generation sequencing. Total nucleic acid was isolated from formalin-fixed paraffin-embedded tumor specimens and reverse transcribed with random hexamers, followed by second-strand synthesis to create double-stranded cDNA. The double-stranded cDNA was end-repaired, adenylated, and ligated with a half-functional adapter. Two hemi-nested PCRs were applied to create a fully functional sequencing library that targets ALK exons 19 to 22, ROS1 exons 31 to 37, RET exons 8 to 13, PPARγ exons 3 to 8, NTRK1 exons 8 to 13 and 15 and 16, and controls, B2M exon 2, CTBP1 exon 6, and GAPDH exon 6. Illumina MiSeq 2 × 151 bp paired-end sequencing results were aligned to the hg19 reference genome using bwa and BLAT (11, 12). A laboratory-developed algorithm was used for fusion transcript detection and annotation. The integrity of the input nucleic acid and the technical performance of the assay were assessed with control sequences from the B2M, CTBP1, and GAPDH targets. We observed 4 technical failures in our panel of FVPTCs using this assay. All observed fusions were validated by fluorescence in situ hybridization (FISH). In addition, we used immunohistochemistry to detect ALK expression in ALK-rearranged samples.
Statistical analysis
Clinical variables were chosen based on established risk factors for thyroid cancer survival, including American Joint Committee on Cancer (AJCC) stage and MACIS score (13). These variables included age at diagnosis, sex, family history, history of radiation exposure, and primary tumor characteristics (size, multifocality, capsular invasion, and vascular invasion). Dichotomous pathological variables were based solely on the original surgical pathology report. Univariate comparisons of categorical variables (eg, mutation status) were analyzed using a Fisher exact test. Multivariate analysis to assess the correlation between mutation status and pathological variables (eg, encapsulated tumor status), adjusting for those variables significant by univariate analysis, was performed using logistic regression. A P value of <.05 was considered significant.
Results
Point mutation detection
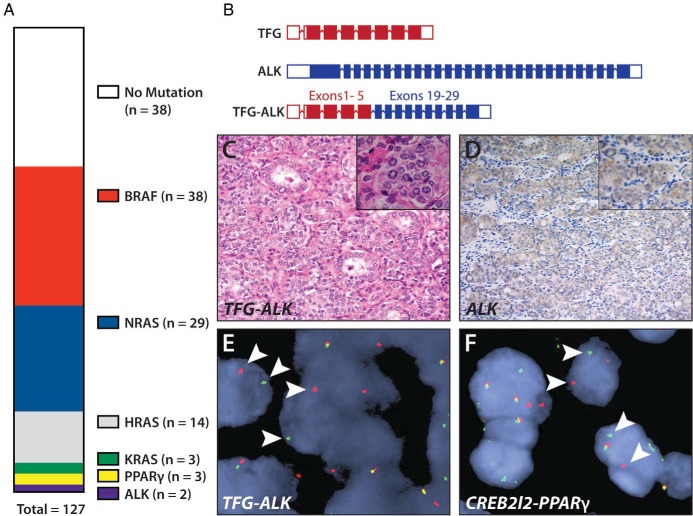
We performed SNaPshot genotyping on 127 FVPTC cases and identified point mutations in 84 of 127 (66%) specimens: BRAF in 38 of 127 (30%), NRAS in 29 of 127 (23%), HRAS in 14 of 127 (11%), and KRAS in 3 of 127 (2%) (Figure 1A). BRAF and RAS mutations were mutually exclusive in all cases. We identified one TP53 mutation that occurred at low allelic frequency in a BRAF-mutant tumor. This may suggest that the TP53 mutation occurred in a fraction of tumor cells in an emerging subclone. No other point mutations were identified in this cohort.
Figure 1.
Identification of oncogenic alterations in the FVPTC. A, Vertical parts-of-whole diagram of observed mutations in the panel of 127 FVPTC cases. B, Diagram of the TFG-ALK rearrangement observed in one tumor. C, Hematoxylin and eosin histology of the TFG-ALK rearranged tumor. D, Immunohistochemical detection of increased ALK expression in TFG-ALK rearranged tumor. E, FISH showing break-apart probes for ALK being separated in tumor cells (white arrowheads) in a TFG-ALK-positive tumor, indicating genomic translocation of ALK. F, FISH capture showing break-apart probes (white arrowheads) for PPARγ in a CREB3L2-PPARγ-positive tumor.
Identification of gene fusions in FVPTC
We used an anchored multiplex PCR assay to identify expressed fusion transcripts involving the ALK, ROS, RET, PPARγ, and NTRK oncogenes. We identified expressed gene fusions in 5 of 126 (4%) specimens; all were validated by FISH. Three gene fusions involved the PPARγ transcriptional regulator. In 2 cases, the translocation partner was CREB3L2, whereas the other case harbored a PAX8-PPARγ fusion. We also detected 2 gene fusions involving the ALK gene, one in which EML4 was the partner and one harboring a TFG-ALK fusion (Figure 1B). ALK-rearranged tumors also exhibited high levels of ALK expression, as observed previously (Figure 1, C–D) (14–16). All gene fusions were mutually exclusive with mutations in the SNaPshot panel, and we did not detect RET-PTC fusions in this cohort. No mutation or rearrangement was found in 38 of 127 (30%) patients with FVPTCs.
Clinical correlations
Clinical features of the patient cohort are summarized in Table 1. We found a strong correlation between BRAF mutation status and unencapsulated tumor status at the initial pathological diagnosis, which remained significant after adjustment for age and AJCC stage. BRAF mutations were associated with older age at diagnosis, and RAS mutations were associated with younger age. Probably as a result of a younger age at diagnosis, we observed an association of RAS mutation and lower MACIS score at the initial diagnosis (mean MACIS for RAS-mutant tumors = 4.7 vs 5.1 for RAS wild-type tumors).
Table 1.
Clinical and Pathological Characteristics of Patients With FVPTCs
| All Patients (n = 126) |
BRAF Status |
RAS Status |
|||||
|---|---|---|---|---|---|---|---|
| Negative (n = 89) | Positive (n = 37) | P | Negative (n = 81) | Positive (n = 45) | P | ||
| Demographics | |||||||
| Age, y, mean ± SD | 49 ± 16 | 47 ± 16 | 54 ± 13 | .04 | 51 ± 17 | 45 ± 13 | .04 |
| Female, n (%) | 101 (80) | 70 (79) | 31 (84) | .62 | 66 (81) | 35 (78) | .65 |
| Radiation exposure, n (%) | 5 (4) | 4 (5) | 1 (3) | .62 | 4 (5) | 1 (2) | .66 |
| Family history, n (%) | 5 (4) | 2 (2) | 3 (8) | .16 | 4 (5) | 1 (2) | .66 |
| Primary tumor characteristics | |||||||
| Tumor diameter, cm, median (IQR) | 2.0 (1.6) | 2.1 (1.5) | 1.6 (1.5) | .13 | 1.8 (1.7) | 2.1 (1.5) | .67 |
| Multifocality, n (%) | 31 (25) | 23 (26) | 8 (22) | .65 | 19 (23) | 12 (27) | .83 |
| LVI, n (%) | 14 (11) | 8 (9) | 6 (16) | .35 | 12 (15) | 2 (4) | .08 |
| Encapsulated, n (%) | 40 (32) | 35 (40) | 5 (14) | <.01 | 23 (29) | 17 (38) | .32 |
| Capsular invasion, n (%) | 5 (4) | 5 (6) | 0 (0) | .32 | 4 (5) | 1 (2) | .65 |
| Vascular invasion, n (%) | 3 (2) | 3 (3) | 0 (0) | .55 | 2 (3) | 1 (2) | .92 |
| Extrathyroidal extension, n (%) | 1 (1) | 0 (0) | 1 (6) | .23 | 1 (2) | 0 (0) | .40 |
| Microscopic ETE, n (%) | 2 (2) | 1 (2) | 0 (0) | .58 | 1 (2) | 0 (0) | .40 |
| Incomplete resection, n (%) | 7 (6) | 5 (6) | 2 (6) | .97 | 5 (6) | 2 (5) | .71 |
| Cervical LN status | |||||||
| Central LN+, n (%) | 9 (7) | 9 (9) | 0 (0) | .60 | 6 (8) | 3 (7) | .42 |
| Lateral LN+, n (%) | 4 (3) | 4 (5) | 0 (0) | .63 | 2 (3) | 2 (4) | .65 |
| PTmicroC, n (%) | 34 (27) | 24 (27) | 10 (27) | .99 | 23 (28) | 11 (24) | .68 |
| AJCC TNM stage | |||||||
| I, n (%) | 117 (94) | 83 (94) | 34 (92) | .59 | 73 (91) | 44 (98) | .52 |
| II, n (%) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | ||
| III, n (%) | 7 (6) | 4 (5) | 3 (8) | 6 (8) | 1 (2) | ||
| IV, n (%) | 1 (1) | 1 (1) | 0 (0) | 1 (1) | 0 (0) | ||
| MACIS score, mean ± SD | 4.9 ± 1.0 | 4.9 ± 1.1 | 5.1 ± 1.2 | .23 | 5.1 ± 1.2 | 4.7 ± 1.0 | .05 |
Abbreviations: ETE, extrathyroidal extension; IQR, interquartile range; LN, lymph node; LVI, lymphovascular invasion; PTmicroC, Papillary thyroid microcarcinoma; TNM, tumor, node, metastasis. There were no tumors with T4a/b extrathyroidal extension or extranodal extension.
We manually examined clinical data for patients harboring oncogenic translocations. No patients had a history of radiation exposure. All tumors harboring PPARγ fusions were encapsulated, and no lymph node or distant metastases were identified in these patients (0 of 3). No recurrences were observed in these patients. This result is consistent with the identification of PPARγ fusions in follicular thyroid carcinoma and encapsulated FVPTC and suggests that these tumors are more likely to behave like follicular tumors (8, 17). In contrast, both tumors harboring ALK rearrangements exhibited features consistent with classic PTC, including multifocality and lymph node metastases (1 of 2), although one of these tumors was encapsulated (with multifocal capsular and vascular invasion).
Finally, it is noteworthy that we observed only 2 recurrences in our patient cohort over a median follow-up period of 2.3 years. This result is consistent with prior studies and suggests that as a group FVPTCs are indolent tumors, particularly in the absence of aggressive pathological features at the initial diagnosis.
Discussion
FVPTC is increasing in incidence and remains a diagnostic and clinical challenge. We found that almost two thirds of the FVPTCs in this study harbored oncogenic mutations, either BRAF (30%) or RAS (36%). Other investigators also reported predominantly BRAF or RAS mutations, but most studies included a smaller number of patients, underwent consensus pathological review, and found a lower percentage of BRAF mutations, generally 10% to 20% (8, 17–20). All of our patients with BRAF mutations harbored the V600E variant (T1779A mutation), and none had other BRAF mutations that have infrequently been reported in FVPTC (17, 19). It is likely that the differences reflect local or regional differences in the pathological diagnosis of FVPTC.
Our data support the hypothesis that FVPTC is composed of distinct biological entities: PTC-like unencapsulated FVPTC harboring BRAF mutations and encapsulated follicular-like tumors (malignant or benign) with or without capsular or vascular invasion. This association was confirmed in our large panel of unselected FVPTCs that may be representative of the spectrum of diagnoses observed in clinical practice.
Our study also used a gene fusion detection assay to identify diverse translocation partners of known oncogenes. PAX8-PPARγ rearrangements have been described in FVPTCs and follicular thyroid tumors, and one of our patients harbored a PAX8-PPARγ fusion. We also identified 2 patients with CREB3L2-PPARγ fusions. CREB3L2 is a cAMP response element–binding protein previously identified as a fusion partner of PPARγ in thyroid cancer (21, 22). ALK rearrangements involving EML4 and STRN partners have recently been described in anaplastic thyroid cancer and thyroid cancers with follicular features (14–16, 23). We identified 2 patients harboring ALK fusions with previously described partners: (1) EML4, a common translocation partner of ALK in lung cancer (24) and (2) TFG, a translocation partner of ALK in anaplastic lymphoma not previously described in thyroid cancer (25). Notably, TFG was first discovered as an oncogenic translocation partner of NTRK1 in PTC (26). It is interesting to note that the tumor harboring the EML4-ALK rearrangement exhibited no features associated with clinically aggressive disease at the initial surgical pathology examination, and the patient has exhibited no tumor recurrence to date. Therefore, it is possible that additional cooperating genetic alterations, including events not detected by the focused SNaPshot panel, may be necessary for progression to clinically aggressive disease in the context of EML4-ALK. Additional studies, however, will be necessary to define the clinical features associated with ALK rearrangement in thyroid cancer.
Our gene fusion assay makes no assumption regarding the specific translocation partner of the target oncogene. This permitted the identification of 2 oncogenic rearrangements (CREB3L2-PPARγ and TFG-ALK) that would not have been identified by many clinical genotyping assays. The identification of ALK rearrangements in a small number of thyroid cancers and the approval of small molecule ALK inhibitors for clinical use highlight the importance of using clinical genotyping assays that detect the diversity of rearrangements targeting ALK in patients harboring clinically aggressive thyroid cancers. Finally, the absence of a known oncogene or tumor suppressor alteration in 30% of our FVPTC cohort suggests that additional studies, such as the ongoing The Cancer Genome Atlas (TCGA) study of PTC, are needed to more completely define the spectrum of genetic drivers of thyroid cancer.
Acknowledgments
This work was supported by a grant award to G.H.D. from the Ellison Foundation. D.G.M. is supported by the National Cancer Institute (Career Development Award K08-CA160658). C.C.L. is supported by the National Cancer Institute program in Cancer Outcomes (Research Training Grant (R25 CA092203), Massachusetts General Hospital Department of Surgery and a Massachusetts General Hospital American Cancer Society Institutional Research Grant.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AJCC
- American Joint Committee on Cancer
- FISH
- fluorescence in situ hybridization
- FVPTC
- follicular variant of papillary thyroid carcinoma
- PTC
- papillary thyroid cancer.
References
- 1. Davies L, Welch HG. Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg. 2014;140:317–322. [DOI] [PubMed] [Google Scholar]
- 2. Haugen BR, Sherman SI. Evolving approaches to patients with advanced differentiated thyroid cancer. Endocr Rev. 2013;34:439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Albores-Saavedra J, Henson DE, Glazer E, Schwartz AM. Changing patterns in the incidence and survival of thyroid cancer with follicular phenotype–papillary, follicular, and anaplastic: a morphological and epidemiological study. Endocr Pathol. 2007;18:1–7. [DOI] [PubMed] [Google Scholar]
- 4. Lin HW, Bhattacharyya N. Clinical behavior of follicular variant of papillary thyroid carcinoma: presentation and survival. Laryngoscope. 2010;120:712–716. [DOI] [PubMed] [Google Scholar]
- 5. Daniels GH. What if many follicular variant papillary thyroid carcinomas are not malignant? A review of follicular variant papillary thyroid carcinoma and a proposal for a new classification. Endocr Pract. 2011;17:768–787. [DOI] [PubMed] [Google Scholar]
- 6. Rivera M, Tuttle RM, Patel S, Shaha A, Shah JP, Ghossein RA. Encapsulated papillary thyroid carcinoma: a clinico-pathologic study of 106 cases with emphasis on its morphologic subtypes (histologic growth pattern). Thyroid. 2009;19:119–127. [DOI] [PubMed] [Google Scholar]
- 7. Vivero M, Kraft S, Barletta JA. Risk stratification of follicular variant of papillary thyroid carcinoma. Thyroid. 2013;23:273–279. [DOI] [PubMed] [Google Scholar]
- 8. Rivera M, Ricarte-Filho J, Knauf J, et al. Molecular genotyping of papillary thyroid carcinoma follicular variant according to its histological subtypes (encapsulated vs infiltrative) reveals distinct BRAF and RAS mutation patterns. Mod Pathol. 2010;23:1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howitt BE, Jia Y, Sholl LM, Barletta JA. Molecular alterations in partially-encapsulated or well-circumscribed follicular variant of papillary thyroid carcinoma. Thyroid. 2013;23:1256–1262. [DOI] [PubMed] [Google Scholar]
- 10. Dias-Santagata D, Akhavanfard S, David SS, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hay ID, Bergstralh EJ, Goellner JR, Ebersold JR, Grant CS. Predicting outcome in papillary thyroid carcinoma: development of a reliable prognostic scoring system in a cohort of 1779 patients surgically treated at one institution during 1940 through 1989. Surgery. 1993;114:1050–1057; discussion 1057–10588. [PubMed] [Google Scholar]
- 14. Demeure MJ, Aziz M, Rosenberg R, Gurley SD, Bussey KJ, Carpten JD. Whole-genome sequencing of an aggressive BRAF wild-type papillary thyroid cancer identified EML4-ALK translocation as a therapeutic target. World J Surg. 2014;38:1296–1305. [DOI] [PubMed] [Google Scholar]
- 15. Kelly LM, Barila G, Liu P, et al. Identification of the transforming STRN-ALK fusion as a potential therapeutic target in the aggressive forms of thyroid cancer. Proc Natl Acad Sci USA. 2014;111:4233–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Perot G, Soubeyran I, Ribeiro A, et al. Identification of a recurrent STRN/ALK fusion in thyroid carcinomas. PLoS One. 2014;9:e87170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santarpia L, Myers JN, Sherman SI, Trimarchi F, Clayman GL, El-Naggar AK. Genetic alterations in the RAS/RAF/mitogen-activated protein kinase and phosphatidylinositol 3-kinase/Akt signaling pathways in the follicular variant of papillary thyroid carcinoma. Cancer. 2010;116:2974–2983. [DOI] [PubMed] [Google Scholar]
- 18. Lee SR, Jung CK, Kim TE, et al. Molecular genotyping of follicular variant of papillary thyroid carcinoma correlates with diagnostic category of fine-needle aspiration cytology: values of RAS mutation testing. Thyroid. 2013;23:1416–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park JY, Kim WY, Hwang TS, et al. BRAF and RAS mutations in follicular variants of papillary thyroid carcinoma. Endocr Pathol. 2013;24:69–76. [DOI] [PubMed] [Google Scholar]
- 20. Proietti A, Giannini R, Ugolini C, et al. BRAF status of follicular variant of papillary thyroid carcinoma and its relationship to its clinical and cytological features. Thyroid. 2010;20:1263–1270. [DOI] [PubMed] [Google Scholar]
- 21. Lui WO, Zeng L, Rehrmann V, et al. CREB3L2-PPARγ fusion mutation identifies a thyroid signaling pathway regulated by intramembrane proteolysis. Cancer Res. 2008;68:7156–7164. [DOI] [PubMed] [Google Scholar]
- 22. Ricarte-Filho JC, Li S, Garcia-Rendueles ME, et al. Identification of kinase fusion oncogenes in post-Chernobyl radiation-induced thyroid cancers. J Clin Invest. 2013;123:4935–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamatani K, Mukai M, Takahashi K, Hayashi Y, Nakachi K, Kusunoki Y. Rearranged anaplastic lymphoma kinase (ALK) gene in adult-onset papillary thyroid cancer amongst atomic bomb survivors. Thyroid. 2012;22:1153–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol. 2013;31:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hernández L, Pinyol M, Hernández S, et al. TRK-fused gene (TFG) is a new partner of ALK in anaplastic large cell lymphoma producing two structurally different TFG-ALK translocations. Blood. 1999;94:3265–3268. [PubMed] [Google Scholar]
- 26. Greco A, Mariani C, Miranda C, et al. The DNA rearrangement that generates the TRK-T3 oncogene involves a novel gene on chromosome 3 whose product has a potential coiled-coil domain. Mol Cell Biol. 1995;15:6118–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]