Abstract
Context:
To date, few systems genetics studies in the bone field have been performed. We designed our study from a systems-level perspective by integrating genome-wide association studies (GWASs), human protein-protein interaction (PPI) network, and gene expression to identify gene modules contributing to osteoporosis risk.
Methods:
First we searched for modules significantly enriched with bone mineral density (BMD)-associated genes in human PPI network by using 2 large meta-analysis GWAS datasets through a dense module search algorithm. One included 7 individual GWAS samples (Meta7). The other was from the Genetic Factors for Osteoporosis Consortium (GEFOS2). One was assigned as a discovery dataset and the other as an evaluation dataset, and vice versa.
Results:
In total, 42 modules and 129 modules were identified significantly in both Meta7 and GEFOS2 datasets for femoral neck and spine BMD, respectively. There were 3340 modules identified for hip BMD only in Meta7. As candidate modules, they were assessed for the biological relevance to BMD by gene set enrichment analysis in 2 expression profiles generated from circulating monocytes in subjects with low versus high BMD values. Interestingly, there were 2 modules significantly enriched in monocytes from the low BMD group in both gene expression datasets (nominal P value <.05). Two modules had 16 nonredundant genes. Functional enrichment analysis revealed that both modules were enriched for genes involved in Wnt receptor signaling and osteoblast differentiation.
Conclusion:
We highlighted 2 modules and novel genes playing important roles in the regulation of bone mass, providing important clues for therapeutic approaches for osteoporosis.
Osteoporosis is a skeletal disease characterized by a reduction in bone mass and diagnosed through measurement of bone mineral density (BMD). Low BMD is associated with increased risk of osteoporotic fracture (1). BMD variation is under strong genetic control with heritability (h2) estimates ranging from 0.5 to 0.9 (2). In recent years, more than 15 genome-wide association studies GWASs have identified more than 60 genes/loci associated with BMD (3). However, such conventional GWASs focus on disease-associated single-nucleotide polymorphisms (SNPs) at genome-wide significance level (eg, P < 5 × 10−8), which explains only a small proportion of genetic risks. To search for missing heritability and enhance our understanding of biological mechanisms, several advanced approaches have been developed.
The network-assisted GWAS has been developed by incorporating protein-protein interaction (PPI) networks into GWASs to search for groups of functional related genes whose protein products interact with one another. The rationale behind this approach is the principle of “guilt by association” (4), which states that genes (or gene products) interconnected in the network are more likely to share the same or similar function (5). Inspirationally, the network-assisted method has been successfully applied to multiple complex diseases, including autoimmune and neurological diseases (6–9).
The advantages of network-assisted GWAS are as follows. First, it can improve power, because the annotations of PPI data cover a much larger proportion of human proteins than predefined gene sets. For example, the human PPI network may recruit >10 000 proteins and >60 000 protein interaction pairs with experimental evidence (10). However, the popular KEGG database has >200 pathways, covering only 5000 to 5500 genes, accounting for less than 30% of genes in GWAS datasets (6). Therefore, the association signals from GWAS may converge on only part of the pathway, resulting in loss of power. Second, network-assisted methods can define de novo gene sets, which may collectively contribute to disease risk, by dynamically searching for subnetworks in the whole interactome (9). Predefined gene sets covered by current annotations limit the analysis by their fixed sizes and may be too general for their roles in disease-related biological functions (11).
As an emerging and novel approach, systems genetics has been applied to complex diseases from a systems-level perspective to determine how genetic variations perturb cellular systems and ultimately disease (12). In a systems genetics study, data from genome, transcriptome, epigenome, proteome, metabolome, and interactome will be analyzed using a suite of analytical approaches including GWAS and network analysis (13). The network analysis may represent the first statistical step in systems genetics to identify multiple genetic perturbations, which alter the states of molecular networks and therefore form systems into disease states (14). By integrating high dimensional biological data from multiple sources, systems genetics provides a global view of the molecular architecture of complex traits and greatly enhances the identification of genes, pathways, and networks as key drivers of complex diseases (15).
Inspired by recent pioneering systems genetics studies in the bone field (16, 17), the present study applied systems genetics approach in its own way by integrating GWASs, human PPI network, and gene expression to identify gene modules contributing to osteoporosis risk. We highlighted 2 modules as well as novel genes in modules playing important roles in the regulation of BMDs.
Materials and Methods
GWAS dataset
The first dataset was from an imputation-based meta-analysis (referred to as Meta7), including 7 individual GWAS samples, which consisted of a total of 11 140 individuals with BMDs at lumbar spine (SPN), hip, and femoral neck (FN) measured by dual-energy x-ray absorptiometry (DXA) scanners. Details of this dataset are provided in Ref. 18. Briefly, each GWAS sample was genotyped by high-throughput SNP genotyping array. Each individual GWAS sample was imputed by the relevant population's reference haplotypes in the 1000 genomes project (1000G). SNPs with high accuracy in at least 2 samples and minor allele frequency >0.05 in at least 1 sample were included in the association analysis. In unrelated GWAS samples, the linear regression model was used in MACH2QTL to examine the association between BMDs and allele dosages as the predictor (19). In familial GWAS samples, a mixed linear model was used in which the effect of genetic relatedness within each pedigree was also taken into account. Genomic control inflation factor was estimated for each individual GWAS. Weighted fixed-effect meta-analyses were performed in METAL to combine summary statistics of associations from each GWAS. Cochran's Q statistic and I2 were calculated by METAL as measures of between-study heterogeneity (20). Random-effects meta-analyses were performed on those SNPs with a Q P value less than .05 or I2 value higher than 50%. In total 5 842 825 SNPs were qualified in the meta-analyses and overall genomic control inflation factors for 3 BMD traits ranged from 0.99 to 1.04.
The second GWAS dataset was the meta-analysis from the Genetic Factors for Osteoporosis Consortium (GEFOS2), the largest meta-analysis to date in the bone field, including 17 GWASs and 32 961 individuals of European and East Asian ancestry with the study detailed in an earlier publication (21). Each SNP and its association to SPN and FN BMD in the meta-analysis were downloaded from http://www.gefos.org/?q=content/data-release.
To enrich for potentially functional variants, SNPs with nominal evidence of association in meta-analyses (P < .05) were taken into account for subsequent network-assisted GWASs. A SNP was mapped to a human protein-coding gene downloaded from the NCBI ftp site (Build 36), if it was located within or 20 kb upstream/downstream from the gene (22).
Human PPI network
A comprehensive human PPI dataset was obtained from Goh et al (10). This dataset consisted of 2 high-quality systematic yeast 2-hybrid experiments and PPIs obtained from the literature by manual curation (23, 24). The PPI network included 10 174 nodes (genes) and 61 070 interactions.
Dense module search analysis
Dense module search (DMS) algorithm was applied to GWAS datasets for module searching and construction. Details of the DMS algorithm are provided in Ref. 25. The procedures were briefly summarized as follows. First, for each gene, a gene-wise P value was calculated by Simes method using multiple SNPs mapped to the gene (26, 27). For L SNPs ranked by their P value, P(1),…, P(L), the Simes P value was calculated as min(Lp(i)/i) where 1 ≤ i ≤ L. As a conservative approach, the Simes method provided an overall P value for the entire collection of L hypotheses. Gene-wise P values were overlaid to the PPI network to generate a node-weighted network with each node weighted by z=Φ−1(1 − p), where Φ−1 is the inverse normal cumulative density function and P is the gene-wise P value. Second, each of the nodes in the network was taken as a seed gene and obtain a best scored module by searching for the node with the highest score in the neighborhood within a distance d (d = 2). Each module was scored by Zm = ∑zi/, where k is the number of nodes (genes) in the module. Nodes will be added if the increment is greater than Zm × r, where r, the rate of proportion increment, equals 0.1 as suggested in the study (25). That is, the expanded module had a score Zm + 1 greater than Zm × (1 + r). To adjust for module size and make modules comparable to each other, the module score was further normalized by ZN = (Zm − mean(Zm(π)))/SD(Zm(π)), where Zm(π) was generated by randomly selecting the same number of genes in a module from the whole network 100 000 times. For each module generated in the discovery dataset, the corresponding P(Zm(eval)) based on the gene weights of the same genes in the same module will be also computed in evaluation dataset. Modules were selected if they have P(Zm(eval)) < .05.
Gene expression data processing
Expression data were obtained from 2 in-house gene expression samples. Both samples were recruited for the purpose of systemically searching for differentially expression genes underlying BMD variations. Both datasets consisted of expression profiles generated from circulating monocytes isolated and purified in subjects with low vs high BMD values. The first sample (Chinese sample) consisted of 12 unrelated healthy young Chinese women with low hip BMD and 14 matched unrelated young Chinese women with high hip BMD (28). The second sample (Caucasian sample) contained 40 unrelated white women with low hip BMD and 40 matched unrelated white women with high hip BMD (29). The detailed characteristics of the 2 gene expression samples are shown in Supplemental Table 1. Both samples were recruited by adopting the same strict exclusion criteria. Subjects with chronic diseases and conditions that potentially may affect bone mass have been excluded from the study. These diseases/conditions included chronic disorders involving vital organs, serious metabolic diseases, skeletal diseases, chronic use of drugs affecting bone metabolism, and malnutrition conditions. Briefly, a monocyte-negative isolation kit (Dynal Biotech, Inc) was used to isolate circulating monocytes from 50 mL whole blood, and then total RNA was extracted from monocytes using a QIAGEN kit (QIAGEN, Inc). The Chinese sample and Caucasian sample used the Affymetrix U-133 Plus 2.0 Gene Chips and the Affymetrix U-133 A Gene Chips for profiling, respectively.
First, the raw cell intensity files from both datasets were processed by the Affy package in R (30). Second, the robust multiarray algorithm was used to normalize and generate probe-level expression data (31). Third, a clustering and principal components analysis (PCA) was performed to identify potential outliers in each dataset by using functions hclust and prcomp in R. For the Caucasian sample, the first principal component (PC1) explained 98.45% of the overall variance. After clustering samples based on global expression values and principal component 1, 3 samples were identified as outliers and therefore removed from subsequent analysis (Supplemental Figure 1). For the Chinese sample, 2 outliers were identified and removed. Details are shown in a previous study (17).
Biological significance of network modules
The biological significance of modules identified by DMS was evaluated by the gene set enrichment analysis (GSEA) algorithm in gene expression datasets to determine whether the modules were associated with BMDs. The GSEA has been a popular tool for interpreting gene expression data at the gene set level (32). Modules that showed significant results in both gene expression datasets (nominal P value < .05) were considered as final candidate modules. To gain further insight into the functional significance of these final candidate modules, genes in modules were submitted for gene ontology (GO) term enrichment analysis based on GO level 4 annotations. A hypergeometric test was implemented in the WebGestalt website (http://bioinfo.vanderbilt.edu/webgestalt/) (33) to compute the enrichment P value for each GO term, adjusted by the Bonferroni method.
Results
An overview of the integrative analysis framework for network-assisted GWAS and gene expression
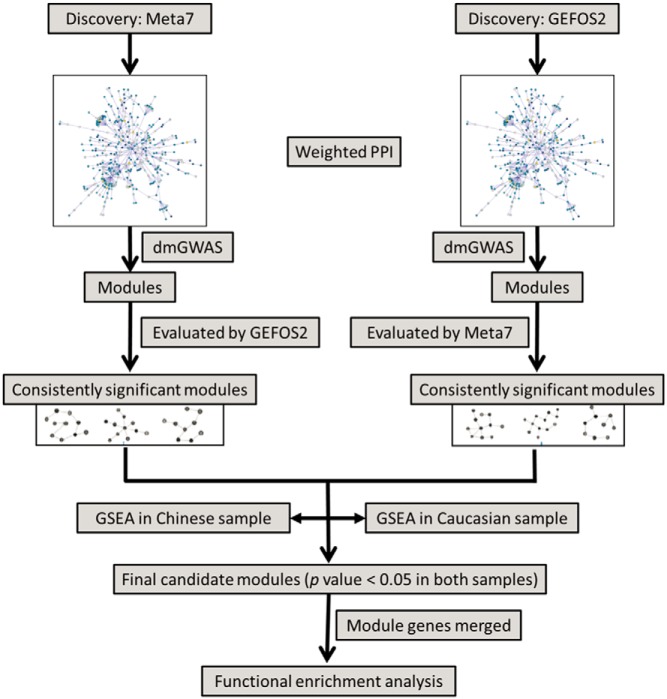
The integrative analysis workflow is shown in Figure 1. Module discovery was started from Meta7 dataset, followed by module evaluation using GEFOS2 dataset. In the parallel thread, GEFOS2 was used for module discovery and Meta7 dataset for evaluation. The modules, which passed the significance criteria in both datasets, were selected as de novo gene sets associated with BMD. The roles of selected gene sets in the regulation of bone mass were evaluated by GSEA in 2 independent microarray expression datasets. For hip BMD, which was not available in the GEFOS2 dataset, the module discovery was based only on the Meta7 dataset, and then the modules were evaluated in both gene expression datasets. Modules significantly enriched in both gene expression datasets were considered as final candidate modules. Finally we performed GO enrichment and pathway analysis using genes merged in final modules for the characterization of final modules content and function.
Figure 1.
The workflow of integration analysis to identify gene modules for BMD.
Module identification by integrating GWAS data and PPI
Using Meta7 as the discovery dataset, a total of 3238 modules were identified for FN BMD. Among these 3238 modules, 37 modules were significant in the GEFOS2 evaluation dataset (p(Zm(eval)) < .05). For SPN BMD, there were 3257 modules identified in Meta7 dataset, 111 of which passed the criteria (p(Zm(eval)) < .05) in the GEFOS2 dataset. For hip BMD, there were 3340 modules identified in Meta7. The module size for 3 traits all ranged between 5 and 15, with a median value of 11 for FN BMD and of 10 for both SPN and hip BMD.
Similarly, using GEFOS2 as the discovery dataset, there were 2693 modules and 2635 modules identified for FN and SPN BMD, respectively. Among these modules, 5 modules and 18 modules were significant in the Meta7 evaluation dataset for FN and SPN BMD, respectively.
In total, 42 modules and 129 modules were identified significantly in both the Meta7 and GEFOS2 dataset for FN and SPN BMD, respectively, and 3340 modules were identified for hip BMD only in Meta7. The results are shown in Supplemental Table 2. As candidate modules, they were used in the gene expression dataset for further analysis.
GSEA of modules in gene expression datasets
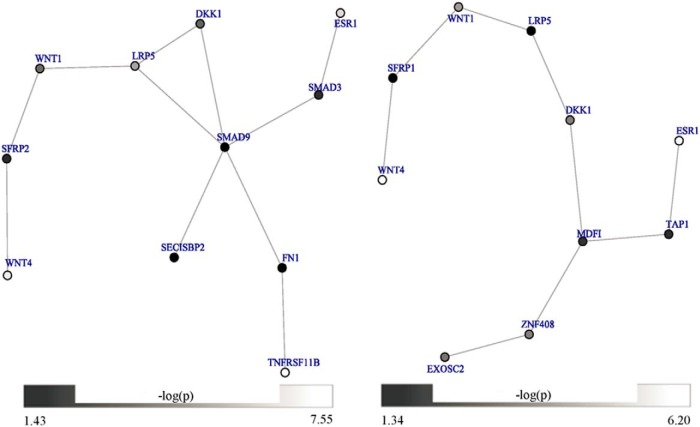
GSEA was performed in 2 gene expression datasets to investigate the roles of modules mentioned above in the regulation of BMD. For FN BMD, 1 of the 42 modules showed significant results only in the Chinese sample's low-BMD group. For SPN BMD, 22 of 129 modules were significantly enriched in the low-BMD group in the Chinese sample even after multiple correction (false discovery rate q-value <0.05). Interestingly, 1 of these 22 modules also had significant results in low-BMD group in the Caucasian sample with nominal P value <.05. Similarly, for hip BMD, there was 1 module of those 3340 modules enriched in monocytes from the low-BMD group in both gene expression datasets (nominal P value <.05). The GSEA results of the 3 modules above are shown in Table 1. Supplemental Table 3 shows the statistics for each module in Meta7 and GEFOS2. Especially, the 2 modules enriched in monocytes of both low-BMD groups were considered as final candidate modules (Figure 2), which overlapped in their gene content and had 16 nonredundant genes.
Table 1.
The 3 Modules Selected by GSEA Results in Gene Expression Datasets
| Trait | Module | Genes in the Module | Gene Expression Sample | ESa | NESa | NOM P Valuea |
|---|---|---|---|---|---|---|
| FN | 1 | SP1, BCL6, ESR1, WNT16, ESRRA, MEF2C | Caucasian | – | – | – |
| Chinese | −0.779 | −1.457 | .043 | |||
| SPN | 2b | SMAD3, ESR1, DKK1, FN1, SFRP2, TNFRSF11B, SMAD9, LRP5, WNT1, SECISBP2, WNT4 | Caucasian | −0.583 | −1.500 | .049 |
| Chinese | −0.621 | −1.722 | .024 | |||
| Hip | 3b | MDFI, ESR1, TAP1, DKK1, LRP5, ZNF408, SFRP1, EXOSC2, WNT1, WNT4 | Caucasian | −0.685 | −1.618 | .023 |
| Chinese | −0.657 | −1.577 | .021 |
Abbreviations: ES, enrichment score; NES, normalized enrichment score; NOM, nominal. Dashes, not significant.
Results in low-BMD group.
Final candidate module.
Figure 2.
The 2 final candidate modules for SPN (left) and Hip (right), respectively. The gray color of a node is based on its P value.
GO enrichment analysis
Table 2 summarized the results of enrichment analysis of the 16 module genes. It shows that even after Bonferroni correction, a number of GO terms still remained significantly enriched. Some of these enriched GO terms were particularly interesting, including the Wnt receptor signaling pathway, regulation of the Wnt receptor signaling pathway, and osteoblast differentiation.
Table 2.
GO Enrichment Analysis of Genes in 2 Final Candidate Modules
| GO Term | Biological Process | Overlapped Genes | P Valuea | Adjusted P Valueb |
|---|---|---|---|---|
| GO:0061053 | Somite development | SMAD3, WNT1, SFRP1, WNT4, SFRP2, DKK1 | 7.43 × 10−11 | 1.99 × 10−8 |
| GO:0016055 | Wnt receptor signaling pathway | SMAD3, WNT1, SFRP1, SFRP2, LRP5, MDFI, WNT4, DKK1 | 3.53 × 10−10 | 9.46 × 10−8 |
| GO:0003002 | Regionalization | SMAD3, WNT1, SFRP1, SFRP2, LRP5, MDFI, WNT4, DKK1 | 4.02 × 10−10 | 1.08 × 10−7 |
| GO:0060070 | Canonical Wnt receptor signaling pathway | SMAD3, WNT1, SFRP2, LRP5, MDFI, WNT4, DKK1 | 4.11 × 10−10 | 1.10 × 10−7 |
| GO:0030111 | Regulation of Wnt receptor signaling pathway | SMAD3, SFRP1, SFRP2, LRP5, MDFI, WNT4, DKK1 | 4.44 × 10−10 | 1.19 × 10−7 |
| GO:0009952 | Anterior/posterior pattern specification | SMAD3, WNT1, SFRP1, LRP5, WNT4, SFRP2, DKK1 | 1.09 × 10−9 | 2.92 × 10−7 |
| GO:0009887 | Organ morphogenesis | SMAD3, ESR1, TNFRSF1B, SFRP1, SFRP2, LRP5, MDFI, WNT1, WNT4, DKK1 | 1.37 × 10−9 | 3.67 × 10−7 |
| GO:0060828 | Regulation of canonical Wnt receptor signaling pathway | SMAD3, SFRP1, SFRP2, LRP5, WNT4, DKK1 | 2.70 × 10−9 | 7.24 × 10−7 |
| GO:0060562 | Epithelial tube morphogenesis | SMAD3, SFRP1, SFRP2, LRP5, WNT4, WNT1, ESR1 | 2.96 × 10−9 | 7.93 × 10−7 |
| GO:0001649 | Osteoblast differentiation | SMAD3, SFRP1, SFRP2, LRP5, WNT4, ESR1 | 4.68 × 10−9 | 1.25 × 10−6 |
Hypergeometric test P value.
Bonferroni correction-adjusted P value.
Discussion
In the present study, we performed a systematic analysis and identified 2 modules underlying BMD by incorporating GWASs, human PPI network, and gene expression. We first adopted a DMS algorithm onto the human PPI network weighted by association signals separately from 2 large meta-analyses of BMD GWASs. Through a discovery-evaluation strategy, a number of modules were consistent in both meta-analysis datasets for 1 specific site of BMD. Importantly, we were able to validate the biological relevance of these modules as de novo gene sets in 2 independent gene expression datasets with high- and low-BMD subjects. Two final candidate modules were identified as they were both significantly enriched in both low-BMD groups of the Chinese and Caucasian samples, suggesting that they played important roles in bone regulation. Functional enrichment analysis further revealed that both modules were enriched for genes involved in Wnt receptor signaling and osteoblast differentiation. From a system-level analysis above, we highlighted 2 modules as well as novel genes in the modules playing important roles in the regulation of bone mass.
The module genes merged from 2 final modules included several well-known candidate genes for osteoporosis, such as ESR1, LRP5, and TNFRSF11B (also known as osteoprotegerin), and it was notable that 8 of the 16 module genes are involved in the Wnt receptor signaling pathway, including SMAD3, SFRP1, SFRP2, MDFI, LRP5, WNT1, WNT4, and DKK1. Especially, the last 4 genes were shared by both final candidate modules. Wnt signaling induces differentiation of bone-forming cells (osteoblasts) and suppresses the development of bone-resorbing cells (osteoclasts). Wnt receptor signaling can regulate osteogenesis by repressing adipocyte differentiation and promoting the proliferation, expansion, survival, and mineralization activity of osteoblasts while blocking osteoblast apoptosis (34). Wnt signaling represses bone resorption by an osteoprotegerin-independent mechanism acting directly on osteoclast precursors such as monocytes (35). Wnt signaling requires the interaction of the LRP5 and frizzled receptors and can be inhibited by Dickkopf (DKK; an inhibitor of LRP5) and secreted frizzled-related protein (SFRP), because Dkks and Sfrps antagonize Wnt signaling in osteoblasts and may downregulate the pathway in mature cells to induce terminal differentiation (36). Interestingly, both final candidate modules, which identified though a systems genetics approach, contained DKK1, LRP5, and SFRP, and interactions among them regulated the Wnt signaling pathway. It is important to fully understand the biological relevance of the Wnt signaling pathway with osteoporosis through further mechanistic study and therefore find novel therapeutic approaches to enhance bone formation.
The biggest advantage of the present study was that it investigated BMD traits from a systems-level perspective by incorporating a protein-protein interactome with 2 biological components, genome and transcriptome. The PPI map, crucial for all biological processes, provided a better understanding of the functional organization of the proteome. From a systems biology point of view, it helped uncover the genetic organization principles of the functional cellular network (24). The modules identified in network-assisted GWASs basically do not have as specific and explicit biological function as canonical pathways have. So an important issue is how to interpret the modules identified and find their biological evidence associated with diseases. Here, importantly, the component transcriptome was used to evaluate the biological roles of the modules. We used monocyte microarray expression profiles from 2 datasets having subjects with extremely low or high BMD. One dataset had 26 Chinese subjects, and the other had 80 Caucasian subjects. To our knowledge, the present study had the largest samples with expression data on individuals having discordant BMD values. We used somehow strict criteria in the GSEA results of 2 gene expression datasets to confirm the biological roles of modules, because these modules had been already validated in both meta-analyses (for FN and SPN BMD traits). Benefiting from such an approach, the present study uncovered interactions among DKK1, LRP5, and SFRP in the final candidate modules. These genes and their interactions had a great impact on the Wnt signaling pathway, which may result in loss of bone mass. The integrative approach provided more insights into biological mechanisms and important findings for further study into the etiology of osteoporosis as well as intervention points for its treatment.
The DMS algorithm that the present study adopted was successfully applied to complex diseases such as schizophrenia (9) and alcohol dependence (8). It allowed for a quantitative global search using the association signals. Other advantages of the present study were as follows. First, unlike previous studies (8, 9), the present study used association signals from large-scale meta-analyses, which can be much more powerful for the detection of more significant loci. Especially, signals of the genome came from the 2 largest meta-analyses of BMD for spine, FN, and hip. One is the largest meta-analysis to date in the bone field, including 17 GWASs and 32 961 individuals, and the other is a newly published meta-analysis of 7 GWAS samples including 11 140 individuals. We made full use of both datasets in a bidirectional framework, which provided robust results with validation. Second, one challenge was how to compute a gene-based statistical value from multiple SNPs in a gene to represent its overall association signal, which weighted the PPI network. The most popular method is to directly use the smallest P value among all SNPs mapped to the gene. However, this easy implementation will introduce biases because larger genes are likely to have more significant P values (5). To overcome this disadvantage, we applied the Simes method (27), which is a global test applied to multiple SNPs in a gene jointly, rather than any individual SNP. Because we had already taken into account the potential biases in the gene-based P value calculation, a permutation-based test would not be necessary in the downstream analysis to adjust for potential biases resulting from SNP density and/or gene size (5). Third, network-assisted analysis uncovered several susceptibility genes that may be missed by conventional approaches. Only LRP5 and TNFRSF11B in final candidate modules reached genome-wide significance level (P = 5 × 10−8). Although other genes, including SMAD3, SFRP1, SFRP2, MDFI, WNT1, WNT4, and DKK1 would not be highlighted by SNP-based GWASs, they may collectively contribute to BMD susceptibility in the context of the Wnt signaling pathway as discussed above. Furthermore, the present study provided several novel candidate genes in the final modules such as FN1, SECISBP2, ZNF408, and EXOSC2. For example, study showed that FN1 is required for osteoblast survival and immunodetection of FN1 showed its upregulation in differentiated osteoblasts compared with their nondifferentiated osteoblasts (37). And ZNF408 was reported in a meta-analysis of gene-based GWASs to be associated with hip BMD with a P value of 7.9 × 10−5 (38). Further investigations are needed to illustrate their roles in the regulation of BMD.
On the other hand, the present study still had some limitations. First, as a gene-centered analysis, the network-assisted method may still neglect genes that are not represented in the PPI network. Second, it is difficult and impractical to examine all possible modules because of the heavy computation burden. The modules identified by different methods or algorithms may greatly vary (9). Nevertheless, the DMS algorithm used in present study allowed for validation by using another GWAS, making our findings more convincing. Third, we acknowledge that there was a lack of evaluation for hip BMD.
In summary, through an integrative analysis of GWASs, PPI, and gene expression, 2 important modules were identified to be significantly associated with BMD. Interestingly, the module genes not only supported previously reported associations with BMD but also implicated functional components such as the Wnt receptor signaling pathway, regulation of the Wnt receptor signaling pathway, and osteoblast differentiation. Therefore, it provided an important clue for further mechanistic studies to find novel therapeutic approaches for treatment of osteoporosis.
Acknowledgments
We thank all staff who provided the clinical expertise and collected and managed the data.
This work was partially supported by grants from the National Institutes of Health (NIH) (P50AR055081, R01AG026564, R01AR050496, RC2DE020756, R01AR057049, and R03TW008221) and startup funds from Tulane University, University of Missouri–Kansas City.
The Framingham Heart Study (FHS) is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract N01-HC-25195). Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding support for the Framingham Bone Mineral Density datasets was provided by NIH Grant R01AR/AG 41398. Funding support for the Genetic Determinants of Bone Fragility (the Indiana fragility study [IFS]) was provided through the National Institute on Aging (NIA) Division of Geriatrics and Clinical Gerontology. Genetic Determinants of Bone Fragility is a GWAS funded as part of the NIA Division of Geriatrics and Clinical Gerontology. Support for the collection of datasets and samples was provided by the parent grant, Genetic Determinants of Bone Fragility (P01-AG018397). Funding support for the genotyping that was performed at the Johns Hopkins University Center for Inherited Diseases Research was provided by the NIH NIA. The Women's Health Initiative (WHI) program is funded by the NHLBI, NIH, U.S. Department of Health and Human Services through Contracts N01WH22110, 24152, 32100–2, 32105–6, 32108–9, 32111–13, 32115, 32118, 32119, 32122, 42107–26, 42129–32, and 44221. WHI PAGE is funded through the National Human Genome Research Institute Population Architecture Using Genomics and Epidemiology (PAGE) network (Grant U01HG004790).
This manuscript was not prepared in collaboration with investigators of the FHS and does not necessarily reflect the opinions or views of the FHS, Boston University, or NHLBI. The FHS datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000007.v14.p6. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, of Genetic Determinants of Bone Fragility study was provided by the NIA Division of Geriatrics and Clinical Gerontology and the NIA Division of Aging Biology. The IFS datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000138.v2.p1. This manuscript was not prepared in collaboration with investigators of the WHI, has not been reviewed and/or approved by the WHI and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. Assistance with phenotype harmonization, SNP selection, data cleaning, meta-analyses, data management and dissemination, and general study coordination was provided by the PAGE Coordinating Center (U01HG004801-01). The WHI datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000200.v6.p2.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BMD
- bone mineral density
- DMS
- dense module search
- FN
- femoral neck
- GEFOS2
- Genetic Factors for Osteoporosis Consortium
- GO
- gene ontology
- GSEA
- gene set enrichment analysis
- PPI
- protein-protein interaction
- SNP
- single-nucleotide polymorphism
- SPN
- spine
- WAS
- genome-wide association study.
References
- 1. Johnell O, Kanis JA, Oden A, et al. Predictive value of BMD for hip and other fractures. J Bone Miner Res. 2005;20:1185–1194. [DOI] [PubMed] [Google Scholar]
- 2. Ralston SH, de Crombrugghe B. Genetic regulation of bone mass and susceptibility to osteoporosis. Genes Dev. 2006;20:2492–2506. [DOI] [PubMed] [Google Scholar]
- 3. Richards JB, Zheng HF, Spector TD. Genetics of osteoporosis from genome-wide association studies: advances and challenges. Nat Rev Genetics. 2012;13:576–588. [DOI] [PubMed] [Google Scholar]
- 4. Lee I, Blom UM, Wang PI, Shim JE, Marcotte EM. Prioritizing candidate disease genes by network-based boosting of genome-wide association data. Genome Res. 2011;21:1109–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jia P, Zhao Z. Network.assisted analysis to prioritize GWAS results: principles, methods and perspectives. Hum Genet. 2014;133:125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. International Multiple Sclerosis Genetics Consortium. Network-based multiple sclerosis pathway analysis with GWAS data from 15,000 cases and 30,000 controls. Am J Hum Genet. 2013;92:854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee YH, Bae SC, Choi SJ, Ji JD, Song GG. Genome-wide pathway analysis of genome-wide association studies on systemic lupus erythematosus and rheumatoid arthritis. Mol Biol Rep. 2012;39:10627–10635. [DOI] [PubMed] [Google Scholar]
- 8. Han S, Yang BZ, Kranzler HR, et al. Integrating GWASs and human protein interaction networks identifies a gene subnetwork underlying alcohol dependence. Am J Hum Genet. 2013;93:1027–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jia P, Wang L, Fanous AH, Pato CN, Edwards TL, Zhao Z. Network-assisted investigation of combined causal signals from genome-wide association studies in schizophrenia. PLoS Comput Biol. 2012;8:e1002587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun YV. Integration of biological networks and pathways with genetic association studies. Hum Genet. 2012;131:1677–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farber CR. Systems genetics: a novel approach to dissect the genetic basis of osteoporosis. Curr Osteoporos Rep. 2012;10:228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Farber CR, Lusis AJ. Integrating global gene expression analysis and genetics. Adv Genet. 2008;60:571–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sieberts SK, Schadt EE. Moving toward a system genetics view of disease. Mamm Genome. 2007;18:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Farber CR. Systems-level analysis of genome-wide association data. G3 (Bethesda). 2013;3:119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farber CR. Identification of a gene module associated with BMD through the integration of network analysis and genome-wide association data. J Bone Miner Res. 2010;25:2359–2367. [DOI] [PubMed] [Google Scholar]
- 18. Zhang L, Choi HJ, Estrada K, et al. Multi-stage genome-wide association meta-analyses identified two new loci for bone mineral density. Hum Mol Genet. 2014;23:1923–1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Estrada K, Styrkarsdottir U, Evangelou E, et al. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44:491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jia P, Wang L, Meltzer HY, Zhao Z. Pathway-based analysis of GWAS datasets: effective but caution required. Int J Neuropsychopharmacol. 2011;14:567–572. [DOI] [PubMed] [Google Scholar]
- 23. Rual JF, Venkatesan K, Hao T, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. [DOI] [PubMed] [Google Scholar]
- 24. Stelzl U, Worm U, Lalowski M, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. [DOI] [PubMed] [Google Scholar]
- 25. Jia P, Zheng S, Long J, Zheng W, Zhao Z. dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics. 2011;27:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 2007;81:1278–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. SIMES RJ. An improved Bonferroni procedure for multiple tests of significance. Biometrika. 1986;73:751–754. [Google Scholar]
- 28. Lei SF, Wu S, Li LM, et al. An in vivo genome wide gene expression study of circulating monocytes suggested GBP1, STAT1 and CXCL10 as novel risk genes for the differentiation of peak bone mass. Bone. 2009;44:1010–1014. [DOI] [PubMed] [Google Scholar]
- 29. Lei SF, Papasian CJ, Deng HW. Polymorphisms in predicted miRNA binding sites and osteoporosis. J Bone Miner Res. 2011;26:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. [DOI] [PubMed] [Google Scholar]
- 31. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. [DOI] [PubMed] [Google Scholar]
- 32. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Krishnan V, Bryant HU, Macdougald OA. Regulation of bone mass by Wnt signaling. J Clin Invest. 2006;116:1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Canalis E. Wnt signalling in osteoporosis: mechanisms and novel therapeutic approaches. Nat Rev Endocrinol. 2013;9:575–583. [DOI] [PubMed] [Google Scholar]
- 36. Westendorf JJ, Kahler RA, Schroeder TM. Wnt signaling in osteoblasts and bone diseases. Gene. 2004;341:19–39. [DOI] [PubMed] [Google Scholar]
- 37. Komori T. Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. 2010;339:189–195. [DOI] [PubMed] [Google Scholar]
- 38. Cheung CL, Sham PC, Xiao SM, Bow CH, Kung AW. Meta-analysis of gene-based genome-wide association studies of bone mineral density in Chinese and European subjects. Osteoporos Int. 2012;23:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]