Abstract
The TORC1 signaling pathway plays a major role in the control of cell growth and response to stress. Here we demonstrate that the SEA complex physically interacts with TORC1 and is an important regulator of its activity. During nitrogen starvation, deletions of SEA complex components lead to Tor1 kinase delocalization, defects in autophagy, and vacuolar fragmentation. TORC1 inactivation, via nitrogen deprivation or rapamycin treatment, changes cellular levels of SEA complex members. We used affinity purification and chemical cross-linking to generate the data for an integrative structure modeling approach, which produced a well-defined molecular architecture of the SEA complex and showed that the SEA complex comprises two regions that are structurally and functionally distinct. The SEA complex emerges as a platform that can coordinate both structural and enzymatic activities necessary for the effective functioning of the TORC1 pathway.
The highly conserved Target of Rapamycin Complex 1 (TORC1)1 controls eukaryotic cell growth and cellular responses to a variety of signals, including nutrients, hormones, and stresses (1, 2). In a nutrient-rich environment, TORC1 promotes anabolic processes including ribosome biogenesis and translation. Nutrient limitation or treatment with rapamycin inhibits the Tor1 kinase and initiates autophagy, a catabolic process that mediates the degradation and recycling of cytoplasmic components. However, the nutrient-sensing function of TORC1 is not fully understood, and the mechanisms of TORC1 modulation by amino acid and nitrogen availability are not yet clear.
In the yeast Saccharomyces cerevisiae, the TOR1 complex is composed of four subunits (Tor1, Kog1, Tco89, and Lst8) and is localized to the vacuole membrane. Amino acid levels are signaled to TORC1 (at least partially) via the EGO complex (Ragulator-Rag in mammals), which consists of Ego1, Ego3, Gtr1 (RagA/RagB), and Gtr2 (RagC/RagD) (3–6). The small GTPases Gtr1 and Gtr2 function as heterodimers and in their active form exist as the Gtr1-GTP/Gtr2-GDP complex. Amino acid sensing via the EGO complex involves the conserved vacuolar membrane protein Vam6, a member of the HOPS tethering complex. Vam6 is a GDP exchange factor that regulates the nucleotide-binding status of Gtr1 (6). At the same time, the GTP-bound state of Gtr1 is controlled by a leucyl t-RNA synthetase (7). In mammals, amino acids promote interaction of Ragulator-Rag with mTORC1 and its translocation to the lysosomal membrane (3, 4). Ragulator interacts with the v-ATPase complex at the lysosomal membrane (8), and leucyl t-RNA synthetase binds to RagD to activate mTORC1 (9).
A genome-wide screen for TORC1 regulators in yeast identified two proteins, Npr2 and Npr3, as proteins that mediate amino acid starvation signal to TORC1 (10). Npr2 and Npr3 are both members of the SEA complex that we discovered recently (11–13). Besides Npr2 and Npr3, the SEA complex also contains four previously uncharacterized proteins (Sea1–Sea4) and two proteins also found in the nuclear pore complex, Seh1 and Sec13, the latter of which is additionally a component of the endoplasmic-reticulum-associated COPII coated vesicle. However, the SEA complex localizes to the vacuole membrane, and not to the nuclear pore complex or endoplasmic reticulum.
The Sea proteins contain numerous structural elements present in intracellular structural trafficking complexes (11). For example, proteins Sea2–Sea4 are predicted to possess β-propeller/α-solenoid folds and contain RING domains, architectural combinations characteristic to protein complexes that form coats around membranes (e.g. coated vesicles, nuclear pore complexes) or participate in membrane tethering (e.g. HOPS, CORVET complexes). Npr2 and Npr3 possess a longin domain, found in many guanine nucleotide exchange factors (GEFs) (14–16), and Sea1/Iml1 is a GTPase activating protein (GAP) for Gtr1 (17). These structural characteristics, taken together with functional data, indicate a role for the SEA complex in intracellular trafficking, amino acid biogenesis, regulation of the TORC1 pathway, and autophagy (11–13, 17–20). A mammalian analog of the SEA complex, termed GATOR1/GATOR2, has recently been identified (21). GATORS are localized at the lysosome membrane and serve as upstream regulators of mammalian TORC1 via GATOR1 GAP activity toward RagA and RagB (21).
In this study, we characterized the structural and functional organization of the yeast SEA complex. We present here a well-defined molecular architecture of the SEA complex obtained via an integrative modeling approach based on a variety of biochemical data. The structure reveals the relative positions and orientations of two SEA subcomplexes, Sea1/Npr2/Npr3 (or SEACIT (19)) and Sea2/Sea3/Sea4/Sec13/Seh1 (or SEACAT (19)), and identifies the Sea3/Sec13 dimer as a major interacting hub within the complex. We describe how the SEA complex interacts physically with TORC1 and the vacuole and is required for the relocalization of Tor1, and how every member of the Sea1/Npr2/Npr3 subcomplex is required for general autophagy.
EXPERIMENTAL PROCEDURES
Materials
The following materials were used in this study: Dynabeads M-270 Epoxy (Invitrogen/LifeTechnologies, 143.02D), rabbit IgG (Sigma, 15006), protease inhibitor mixture (Sigma, P-8340), disuccinimidyl suberate (Creative Molecules, 001S), HRP-mouse IgG (Jackson ImmunoResearch Laboratories West Grove, PA), anti-GFP antibody (Roche, 11814460001), anti-PGK1 antibody (Sigma, 459250), and concanavalin A (Sigma, C7275).
Yeast Strains and Growth Conditions
Yeast strains used in this study are listed in supplemental Table S1. Yeast were grown to mid-log phase in Wickerham media for immunoprecipitation experiments (0.3% Bacto malt extract, 0.3% yeast extract, 0.5% Bacto Peptone, and 1% glucose), in yeast synthetic complete media for imaging (0.67% yeast nitrogen base without amino acids and carbohydrates, 0.2% complete drop-out mix, and 2% glucose), and in YPD (2% Bacto-Peptone, 1% yeast extract, and 2% glucose) or an appropriate drop-out media for all other purposes. Starvation experiments were conducted in synthetic media lacking nitrogen (0,17% yeast nitrogen base without ammonium and amino acids, 2% glucose).
Immunoprecipitation of the SEA Complex Components
Three types of SEA members were used for immunopurifications: (i) PrA tagged proteins expressed in the wild-type background; (ii) PrA tagged proteins expressed in cells where a gene of another component of the SEA complex was deleted; and (iii) PrA tagged C-terminal truncations. Points of C-terminal truncations for SEA proteins were selected based on the secondary structure prediction and PAL data (11). The C-terminal deletions carried a human rhinovirus 3C protease site (GLEVLFQGPS) between a SEA protein and PrA tag and were constructed essentially as described in Ref. 22. Affinity purifications of SEA complex protein complexes from whole cell lysates using magnetic beads were performed as described previously (11). The extraction and washing buffers used for immunoprecipitations are listed in supplemental Table S2.
Mass Spectrometry Analysis of Immunoprecipitations
Protein bands appearing after Coomassie staining were cut from the gel. Gel bands were washed first with 100 μl of 25 mm ammonium bicarbonate (Sigma, 11204)/acetonitrile (Sigma, 34967) 50/50 v/v over 10 min at room temperature and then with 100 μl of 100% acetonitrile for 10 min at room temperature. These washes were repeated twice. Samples were dried in a SpeedVac for 2 min; then 20 μl of 11.55 ng/μl trypsin (Calbiochem, 650279) was added to each gel piece, and gels were incubated at room temperature for 15 min. Samples were further incubated overnight at 37 °C with 20 μl of 50 mm ammonium bicarbonate. Supernatants were separated from gel pieces and transferred to analysis vials. 20 μl of 5% formic acid (Sigma, 33015)/acetonitrile 30/70 v/v was added to each piece of a gel to extract remaining peptides. Supernatants were combined together and dried in a SpeedVac. 10 μl of 3% acetonitrile, 0.1% formic acid solution in water was added to solubilize peptides. The peptide mixture obtained from tryptic digestion of gel bands was analyzed via nano-HPLC (Agilent Technologies 1200, Santa Clara, CA) directly coupled to an ion trap mass spectrometer (Bruker 6300) equipped with a nano-electrospray source. 4 μl of peptide mixture were separated on a ProtID-Chip-43 II 300A C18 43-mm column (Agilent Technologies, G4240–62005) with a 3% to 97% acetonitrile gradient over 30 min. The acquisition was performed as follows: one full MS scan over the range of 200–2200 m/z, followed by three data-dependent MS/MS scans on the three most abundant ions in the full scan. The data were analyzed using Spectrum Mill MS Proteomics Workbench Rev A.03.03.084 SR4, with the following settings: Data Extractor, MH+ 200 to 4400 Da; scan range, 0 to 30 min; MS/MS search, Swiss-Prot database; S. cerevisiae; trypsin; two missed cleavages; oxidized methionine (M), phosphorylated S, T, Y: monoisotopic masses; cutoff score/expectation value for accepting individual MS/MS spectra, 17; precursor mass tolerance, ±2 Da; and product mass tolerance, ±0.8 Da. The lists of putative proteins were obtained by searching against the Swiss-Prot protein database (updated weekly; last version used for the analysis was from October 15, 2013), and the number of protein entries was 526,969.
Purification of Native SEA Complex and Analysis of Its Relative Stoichiometry
5 to 20 g of cryo-grindate obtained from ySD227 strain (Sea1-ppx-PrA, supplemental Table S1) were used for immunopurification of native SEA complex. 20 mm K/HEPES, pH 7.4, 110 mm KOAc, 300 mm NaCl, 0.1% CHAPS, 2 mm MgCl2, 1 mm DTT, 1/500 protease inhibitors was used as extraction and washing buffer. The complex was released from magnetic beads by protease digestion through incubation with 1 μg of protease per 1 μg of complex in extraction buffer (without protease inhibitors) for 1 h at 4 °C. The recovered sample was centrifuged at 20,000g for 10 min. 100 to 150 μl of supernatant was loaded on top of a 5%–20% sucrose gradient in a buffer containing 20 mm K/HEPES, pH 7.4, 110 mm KOAc, 150 mm NaCl, 0.01% CHAPS, 0.2 mm MgCl2, 0.1 mm DTT, 1/1000 protease inhibitors. Gradients were centrifuged on an SW 55 Ti rotor (Beckman Coulter) at 35,000 rpm and 5 °C for 6 h. Gradients were manually unloaded from the top in 12 fractions of 410 μl. Fractions were precipitated using 90% methanol. Pellets were resuspended in protein loading buffer, and the proteins were separated in 4–12% Bis-Tris gels (Novex/LifeTechnologies, Grand Island, NY) and visualized with Coomassie stain. For stoichiometry, quantification gels were stained with SYPRO Ruby (Molecular Probes/Life Technologies, Grand Island, NY) and visualized on an LAS-3000 system (linear detection range; Fujifilm). The SEA complex protein band intensities were measured using ImageJ software (National Institutes of Health), with values normalized for protein molecular weight. The copy number of SEA members was calculated as relative to Sea1 (the handle used for the affinity purification). As a control, the same procedure was applied to affinity-purified Nup84 complex samples showing the expected 1:1 stoichiometry for all Nup84 complex members (22).
Chemical Cross-linking of the Purified SEA Complex
∼10 to 20 μg of the SEA complex purified from ySD227 strain were cleaved off from the affinity beads via protease treatment (see above) and eluted in 250 μl of elution buffer. The complex was cross-linked by incubation with 0.1 mm disuccinimidyl suberate at room temperature for 30 min with constant agitation at 750 rpm and quenched by the addition of ammonium bicarbonate at a final concentration of 50 mm. The cross-linked complex was subsequently reduced with 5 mm tris(2-carboxyethyl)phosphine and alkylated in the dark with 20 mm iodoacetamide for 20 min.
Mass Spectrometric Analysis of Cross-linked Peptides
After cross-linking with disuccinimidyl suberate, the SEA complex was digested either in-solution or in-gel with trypsin to identify the cross-linked peptides. For in-solution digestion, ∼20 μg of purified complex was digested with 1 μg of trypsin (Promega) in 1 m urea and ∼2% acetonitrile at 37 °C. After 12 to 16 h of incubation, an additional 0.5 μg of trypsin was added to the digest, which was then incubated for an additional 4 h. The resulting proteolytic peptide mixture was purified using a C18 cartridge (Sep-Pak, Waters, Milford, MA), lyophilized, and fractionated via peptide size exclusion chromatography (23). For in-gel digestion, ∼10 μg of purified complex was precipitated by methanol, resuspended, and heated at 95 °C in 1× SDS loading buffer. The sample was cooled to room temperature for cysteine alkylation and separated via electrophoresis in a 4–12% SDS-PAGE gel. The gel region above ∼160 kDa was sliced, crushed into small pieces, and digested in-gel by trypsin. After extraction and purification, the resulting proteolytic peptide mixture was dissolved in 20 μl of a solution containing 30% acetonitrile and 0.2% formic acid and fractionated via peptide size exclusion chromatography (Superdex Peptide PC 3.2/30, GE Healthcare) using off-line HPLC separation with an autosampler (Agilent Technologies). Three size exclusion chromatography fractions in the molecular mass range of ∼2.5 kDa to 8 kDa were collected and analyzed via LC/MS.
Purified peptides were dissolved in the sample loading buffer (5% MeOH, 0.2% formic acid) and loaded onto a self-packed PicoFrit® column with an integrated electrospray ionization emitter tip (360 outer diameter, 75 inner diameter, 15-μm tip; New Objective, Woburn, MA). The column was packed with 5 cm of reverse-phase C18 material (3-μm porous silica, 200-Å pore size, Dr. Maisch GmbH, Ammerbuch-Entrigen, Germany). Mobile phase A consisted of 0.5% acetic acid, and mobile phase B of 70% acetonitrile with 0.5% acetic acid. The peptides were eluted in a 150-min LC gradient (8% B to 46% B, 0–118 min; 46% B to 100% B, 118–139 min; equilibrated with 100% A until 150 min) using an HPLC system (Agilent Technologies) and analyzed with an LTQ Velos Orbitrap Pro mass spectrometer (Thermo Fisher). The flow rate was ∼200 nl/min. The electron-spray voltage was set at 1.7–2.2 kV. The capillary temperature was 275 °C, and ion transmission on Velos S lenses was set at 35%. The instrument was operated in the data-dependent mode, where the top eight most abundant ions were fragmented by means of higher energy collisional dissociation (24) (dissociation energy 27–33, 0.1-ms activation time) and analyzed in the Orbitrap mass analyzer. The target resolution for MS1 was 60,000, and that for MS2 was 7500. Ions (370–1700 m/z) with charge states of ≥3 were selected for fragmentation. A dynamic exclusion of 15/2/55 s was used. Other instrumental parameters included the following: lock mass at 371.1012 Da, monoisotopic mass selection off, minimal threshold of 5000 to trigger an MS/MS event, and ion trap accumulation limits of 105 and 106, respectively, for the linear ion trap and Orbitrap. The maximum ion injection times for the LTQ and Orbitrap were, respectively, 100 ms and 500 ms.
The raw data were transformed to Mascot generic format and searched by pLink software (25) with a database containing sequences of the eight protein subunits of SEA complex together with BSA. Other search parameters included mass accuracy of MS1 ≤ 10 ppm and MS2 ≤ 20 ppm for the initial database search, cysteine carboxymethylation as a fixed modification, methionine oxidation as a variable modification, and a maximum of two trypsin miscleavages allowed. The results were filtered at a 5% false discovery rate, which led to a total of 295 unique cross-linked peptides. We noticed that false discovery rate estimation of cross-linking might not have been accurate (even with a high mass accuracy measurement), presumably because of the greatly expanded database search space and the fact that many cross-linked peptides are of low abundance, leading their identification to be complicated by other low-abundant peptide species resulting from, for example, trypsin miscleavages, nonspecific cleavages, chemically modified peptides, or combinations of these. We treated the 5% false discovery rate (a default parameter of the pLink software) as a rough initial filter of the raw data (albeit quite permissive). Next, we applied additional stringent filters (including high mass accuracy, large enough individual chain lengths, and extensive fragmentation information) to remove potential false positive identifications from our dataset. We applied the following criteria for verification of cross-linked peptides: (i) only identifications with mass accuracy ≤ 5 ppm for MS1 and ≤ 10 ppm at MS/MS were considered, as 94% of the identifications have an MS1 mass accuracy of ≤±2 ppm; and (ii) for positive identifications, both peptide chains had to contain at least four amino acids (in 97% of the identifications, each peptide chain contained at least five amino acids). For both peptide chains, the major MS/MS fragmentation peaks had to be assigned and follow a pattern that contained a continuous stretch of fragmentations. The appearance of dominant fragment ions N-terminal to proline and C-terminal to aspartic acid and glutamic acid for arginine-containing peptides was generally expected (26, 27). 188 of the high-confidence cross-linked peptides (selected from 295 of 5% false discovery rate–filtered cross-links) passed these criteria and were used as restraints for determination of the SEA complex architecture. Thus, we did not allow uncertainty of the cross-linking data for our integrative modeling approach.
Determination of the SEA Complex Architecture through Integrative Modeling
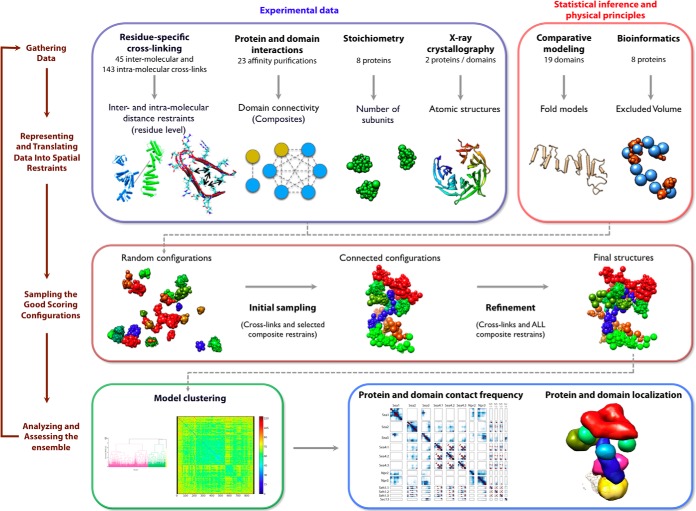
Our integrative approach to determining the SEA complex structure proceeds through four stages (22, 28–31) (Figs. 2 and 3): (i) gathering of data, (ii) representation of subunits and translation of the data into spatial restraints, (iii) configurational sampling to produce an ensemble of models that satisfies the restraints, and (iv) analysis of the ensemble. The modeling protocol (i.e. Stages 2, 3, and 4) was scripted using the Python Modeling Interface (https://github.com/salilab/pmi), version 47dafcc, a library to model macromolecular complexes based on our open source Integrative Modeling Platform package (http://salilab.org/imp/), version 65734ec (32).
Fig. 2.
The four-stage scheme for integrative structure determination of the SEA complex. Our integrative approach proceeds through four stages (22, 28–31): (i) gathering of data, (ii) representation of subunits and translation of data into spatial restraints, (iii) configurational sampling to produce an ensemble of models that satisfy the restraints, and (iv) analysis and assessment of the ensemble. The modeling protocol (i.e. Stages 2, 3, and 4) was scripted using the Python Modeling Interface, version 47dafcc, a library to model macromolecular complexes based on our open source Integrative Modeling Platform package, version 65734ec (32).
Fig. 3.
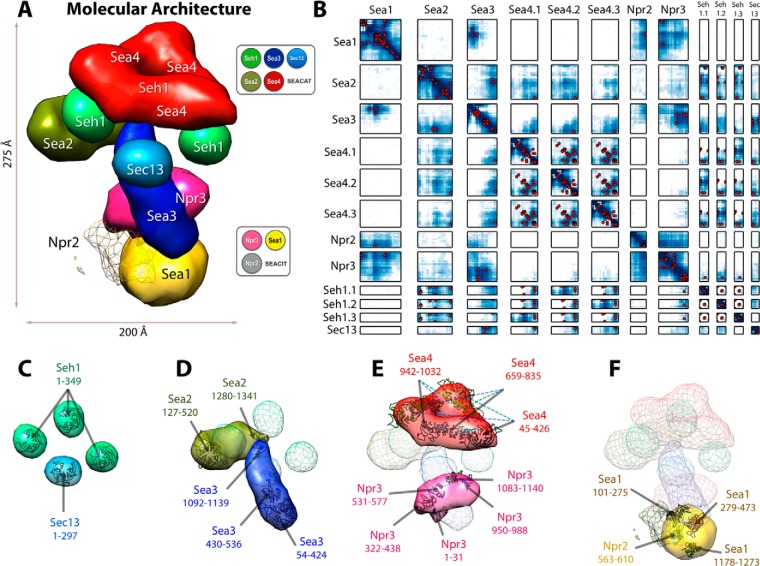
Molecular architecture and contact frequency of the SEA complex. A, the molecular architecture of the SEA complex was obtained through integrative modeling based on various biochemical data (Fig. 2). The localization of each SEA complex protein is defined by a density map, contoured here at the threshold that results in 1.5 times its volume estimated from sequence (supplemental Table S6). Three copies of Seh1 and Sea4 were included in the complex, based on the stoichiometry data, with a symmetry constraint applied to increase the model ensemble precision. Npr2 was localized with relatively low precision, indicated by a mesh. The approximate dimensions of the SEA complex are 200 Å × 200 Å × 275 Å. B, the proximities of any two residues in the molecular architecture were measured by their relative contact frequency. A contact between a pair of residues was defined as an instance when their corresponding bead surfaces were less than 30 Å from each other. Cross-links are plotted as red dots, and the residue contact frequency is indicated by a color ranging from white (0) to dark blue (1). Each box contains the contact frequency between the corresponding pair of SEA complex proteins. C–F, Although the x-ray structures of Seh1 and Sec13 (C) and comparative models of Sea2 and Sea3 (D), Sea4 and Npr3 (E), and Sea1 and Npr2 (F) are placed inside the density map, their orientations are arbitrary; for contrast, other SEA complex proteins are shown as faint meshes.
Stage 1: Gathering of Data
The stoichiometry was determined via biochemical quantitation of the density-gradient purified SEA complex (supplemental Fig. S1). 45 intermolecular and 143 intramolecular cross-links were identified via mass spectrometry (Fig. 1C, supplemental Table S4). Six composites with wild-type proteins, one composite with a mutant, and 16 composites with domain deletion constructs were determined via affinity purification (Figs. 1A and 1B, supplemental Fig. S2, supplemental Tables S2 and S3). A composite is a single subcomplex of physically interacting proteins or a mixture of such complexes overlapping at least at the tagged protein (28). The atomic structures of Seh1, Sec13, and a homolog of the longin domains in Npr2 and Npr3 have been previously determined via x-ray crystallography (PDB codes 3F3F, 2PM7, and 3TW8, respectively) (14, 15, 33, 34). In addition, putative homologs of known structures were detected for domains in other SEA components using HHpred (35) (supplemental Table S5). Domain boundaries, secondary structures, and disordered regions were predicted by DomPRED (36), PSIPRED (37), and DISOPRED (38), respectively. A single unit of a 7-blade β-propeller was predicted by SMURF (39) for each N-terminal domain of Sea2, Sea3, and Sea4.
Fig. 1.
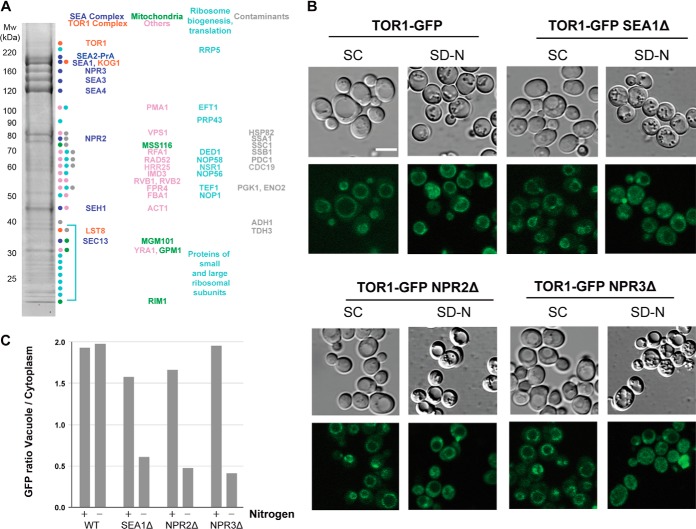
Identification of SEA complex interconnectivity and domain interaction by immunoprecipitation and chemical cross-linking. A, immunoprecipitation of Protein A (PrA) tagged proteins (indicated and underlined on the top of the gel lanes) was performed as described under “Experimental Procedures.” SEA complex proteins and their partners were resolved via SDS-PAGE and visualized with Coomassie Blue staining. Proteins identified via mass spectrometry are marked by solid circles at the right of the gel lane and are listed in order below (S1 = Sea1, S2 = Sea2, S3 = Sea3, S4 = Sea4, N3 = Npr3). PrA tagged proteins are indicated in red, co-purifying proteins in black, and IgG contaminant in gray. Shown are only members of the SEA complex. For the complete set of co-purifying proteins in lanes 6, 8, 12, 18, and 20, see supplemental Fig. S2 and supplemental Table S3. The identity of a truncated protein (in amino acid residues) or deleted SEA member is indicated on the top of the gel lane. WT, wild type; Sea1G (lane 2) is a fraction from the sucrose gradient gel (supplemental Fig. S1). Each individual gel image was differentially scaled along its length so that its molecular mass standards aligned to a single reference set of molecular mass standards. Contrast was adjusted to improve visibility. All original gel figures are available upon request. B, co-purification profile of different SEA deletion and truncation strains. Horizontal gray lines represent the number of amino acid residues in each protein; amino acid residue positions are shown on top of the lines. Co-purifying SEA complex proteins are indicated by “+,” and missing proteins by “−.” The Sea1, Npr2, and Npr3 proteins are colored in blue; others are in yellow. C, summary of identified interprotein cross-links of the SEA complex, generated using AUTOCAD (Autodesk INC., educational version). A representative high-resolution MS/MS spectrum of a cross-linked peptide connecting two different proteins (inter-cross-link) of the SEA complex is shown on the right. An example MS spectra is shown in which the cross-linking site Sea3(1072)-Sea4(885) is unambiguously identified.
Stage 2: Representation of Subunits and Translation of the Data into Spatial Restraints
The domains of the SEA complex components were represented by beads of varying sizes, arranged into either a rigid body or a flexible string of beads, based on the available crystallographic structures and comparative models (supplemental Fig. S3, supplemental Table S5). The bead radii were determined using the statistical relationship between the volume and the number of residues (28, 40).
To balance computational efficiency with scoring precision in light of varying data precision, we represented the structures in a multi-scale fashion. For the cross-link, excluded volume, and composite restraints, the crystallographic structures of Seh1 and Sec13 were coarse-grained by representing each consecutive segment of 1, 5, and 100 residues with a single bead, centered on the center of mass, respectively. Sequence fragments missing in the crystal structures were substituted by a single bead or multiple beads of the corresponding size.
For predicted non-disordered domains of the remaining sequences, comparative models were built with MODELLER 9.12 (41) based on the closest known structure detected by HHPred, SMURF (for the β-propellers), and the literature (for the longin domains in Npr2 and Npr3) (supplemental Fig. S3, supplemental Table S5). 35% of the residues in the SEA complex were in at least one model. Similarly to the x-ray structures, the modeled regions were represented in a multi-scale fashion. To reflect the uncertainties in comparative modeling, including those in the target-template alignment, 5 residues per bead were used for the cross-link and excluded volume restraints, and 100 residues per bead were used for the composite restraints.
For each protein, the beads representing a structured region were kept rigid with respect to each other during configurational sampling.
Regions without a crystallographic structure or a comparative model (i.e. regions predicted to be disordered or structured without a known putative homolog) were represented by a flexible string of large beads corresponding to 100 residues each (supplemental Fig. S3 and supplemental Table S5), applied to the cross-link, excluded volume, and composite restraints.
With this representation in hand, we next encoded the spatial restraints based on the information gathered in Stage 1. First, for each cross-link, we applied an upper-bound harmonic restraint of 17 Å on the distance between the surfaces of the beads containing the cross-linked residues. Second, for the cross-links involving Sea4 and Seh1, which occur in three copies each, we used a conditional restraint (28) that required that at least one copy of the restrained subunit satisfy the observed cross-link. Third, for each composite, we applied a conditional connectivity restraint that ensured the proximity between the protein types defined by the composite (22). Fourth, the sequence connectivity restraints enforced proximity between beads representing consecutive sequence segments; we used a harmonic upper-bound restraint between consecutive beads to enforce sequence connectivity. The upper-bound distance D = [√(5/3)]Rg was determined using the average radius of gyration, Rg = 1.93n0.6, of a random coil polypeptide with n = (n1 + n2)/2 residues (42), where n1 and n2 are the numbers of residues in the two beads, respectively. Fifth, we applied the excluded volume restraints to all pairs of beads. Finally, the three-fold C3 symmetry constraint was imposed separately on the three copies of Seh1 and Sea4 (see Stage 4). The scoring function is defined as the sum of all spatial restraints enumerated above.
Stage 3: Sampling the Good Scoring Configurations
Models of the SEA complex that satisfied all the spatial restraints were obtained through independent applications of two consecutive stochastic optimization runs (initial sampling and refinement), with each application starting from a different random initial configuration. Metropolis Monte Carlo enhanced by simulated annealing was used to sample configurations in both runs. Each Monte Carlo step consisted of sequential random rotations and random translations of all rigid bodies and all beads. During the initial optimization run, 50,000 models were produced by saving a configuration every 100 Monte Carlo steps (supplemental Movie S1); no computationally expensive composite restraints were used in the scoring function. The best-scoring model from the initial sampling was then refined with the complete scoring function (including all composite restraints) using the same sampling algorithm. A final refined model was chosen as the best-scoring model among the 20,000 refined snapshot models. The entire two-run procedure was repeated to obtain an ensemble of 885 refined models.
Stage 4: Analysis of the Ensemble
The ensemble of 885 refined models was analyzed to find a cluster of 340 models that best satisfied the restraints in terms of subunit configurations, contacts, and positions.
The ensemble of 885 refined models was first superposed on a randomly selected model using a rigid body least-squares superposition. Hierarchical clustering based on the root-mean-square deviation distance matrix (43) identified a single dominant cluster containing 340 models (supplemental Fig. S4). The average root-mean-square deviations of cluster models from the center of the cluster and all pairs of models were 61.7 and 67.9 Å, respectively. Random subsets of 10% of the 885 refined models also form a single dominant cluster with the average root-mean-square deviations from the center of the cluster of 60 to 67 Å. Thus, the precision of the ensemble did not significantly change even if only a small fraction of the good-scoring solutions was used, demonstrating that our optimization procedure is likely to have exhaustively sampled the set of possible solutions, given the data. The variability in the ensemble of good-scoring structures reflects the heterogeneity (e.g. flexibility) of the sample, as well as the lack of information from which to determine a highly precise structure.
Next, the proximities of any two residues in the structure were measured based on their relative contact frequency, which was defined by how often the two residues contacted each other in the cluster of the 340 models (28, 29). A contact between a pair of residues was defined as an instance when their corresponding bead surfaces were less than 30 Å apart (Fig. 3B). The biochemical data, including the cross-links (red dots in Fig. 3B) and protein and domain interactions, were well satisfied by the 340 models, as demonstrated by the match of these datasets and the contact frequency map in Fig. 3B.
The superposed structures in the cluster of the 340 models were then converted into the probability of any volume element being occupied by a given protein (that is, the “localization probability”) (22, 28, 29) (Fig. 3A). The spread around the maximum localization probability of each protein describes how precisely its position was defined by the input data. The positions that have a single narrow maximum in their probability distribution in the ensemble were determined most precisely. When multiple maxima are present in the distribution at the precision of interest, the input restraints are insufficient to define the single native state of that protein (or there are multiple native states) (28, 29). The localization of each SEA complex protein is defined by a density map, contoured at the threshold that results in 1.5 times its volume estimated from sequence (supplemental Table S6).
The stoichiometry used in the construction of the molecular architecture is further supported by our inability to find structures that satisfy all the data assuming the other stoichiometries, including a single copy of Sea4 and Seh1 (supplemental Fig. S5). For example, three (off-diagonal intramolecular) cross-link pairs of Sea4 (441)-Sea4 (859), Sea4 (606)-Sea4 (864), and Sea4 (606)-Sea4 (922) could only be satisfied when assigned to different copies of Sea4, resulting in proximities between Sea4.1 and Sea4.2, Sea4.1 and Sea4.3, and Sea4.2 and Sea4.3 (Fig. 3B and supplemental Fig. S5). We did not computationally explore either 1:2 or 2:3 stoichiometries for Sea3. The molecular weights of the SEA complex calculated by assuming these stoichiometries (supplemental Table S7) were not in agreement with the molecular weight of ∼1 MDa estimated experimentally (11).
The C3 symmetry constraints on the three copies of Sea4 and Seh1 each were supported by their approximate C3 symmetries in a molecular architecture computed without any symmetry constraints (data not shown). As a result, we imposed symmetry constraints in an effort to increase the precision of the final molecular architecture. We note that similar proteins in evolutionarily related COPI and COPII complexes are also arranged in various symmetric configurations (44, 45).
Autophagy Analysis
To test autophagy, wild-type and various deletion strains of the SEA complex members were transformed with a plasmid coding for GFP-ATG8, grown in drop-out media without uracil until mid-log phase and shifted to synthetic media lacking nitrogen. Samples were taken at between 45 min and 20 h of starvation, and whole cell lysates were prepared and used for Western blotting with anti-GFP and anti-PGK1 antibodies. Microscope observations were performed after 20 h of starvation. To prevent cell movement, yeast were placed on a slide covered with 1 mg/ml concanavalin A and visualized at room temperature. Steady-state images were obtained on a custom confocal microscope comprising a Leica TCS SpE with a ×63 ON 1,3 oil objective and a 491-nm solid-state laser. All components were driven by Leica software (LAS AF).
RESULTS
Structural Analyses and Molecular Architecture of the SEA Complex
To further test our initial observation that the octameric SEA complex exists as a discrete assembly (11), we performed sucrose gradient fractionation of the affinity-purified complex. Our results demonstrate that all the proteins of the SEA complex migrated primarily as a single entity of ∼1 MDa (Fig. 1, lane 2; supplemental Fig. S1). We also analyzed the relative stoichiometry of the isolated complex through quantification of SDS-PAGE separated protein bands. Our results were consistent with the complex being formed by one copy of each component except for Sea4 and Seh1, which were each present in approximately three copies per complex (supplemental Fig. S1).
Next, we used several approaches to obtain detailed information about the organization of the SEA complex (Fig. 1, supplemental Figs. S1 and S2, supplemental Tables S1–S4): (a) fractionation of affinity-purified complexes by sucrose velocity gradients (Fig. 1A (lane 2), supplemental Fig. S1); (b) isolation of components from strains in which another protein of the SEA complex was deleted (Fig. 1A, lanes 3, 4, 12, 13, and 18–20); (c) purification of C-terminal truncated versions of a given SEA component (Fig. 1A, lanes 5, 7, 10, 14–16, 21, and 22); (d) application of extraction buffers of increasing stringency to purify only the most tightly connected proteins (Fig. 1A; compare lanes 14 and 15, 18 and 19); and (e) chemical cross-linking in combination with mass spectrometry to identify adjacent lysine residues from different components (Fig. 1C, supplemental Table S4) (23, 46–48). For Sea1, Sea2, Sea3, and Sea4, we also performed affinity purifications from diploid (in addition to haploid) strains to detect an untagged copy, which can indicate a potential homotypic interaction. For both Sea1 and Sea4, an untagged copy was indeed identified (supplemental Fig. S2, lanes 1 and 6).
We then computed the molecular architecture of the SEA complex (Figs. 2 and 3, supplemental Movie S1) by fitting a structural representation of the SEA complex to the various biochemical and proteomic data described above (Fig. 1, supplemental Fig. S1, supplemental Tables S3 and S4). The resulting configuration of SEA proteins is shown as a density map that is sufficiently precise to pinpoint the locations, but generally not orientations, of the component proteins (Fig. 3A and supplemental Movie S2). This configuration satisfies the data used to compute it, including stoichiometry, chemical cross-links (red dots in Fig. 3B and supplemental Fig. S6) within a distance of 30 Å (supplemental Table S8), and protein and domain interactions from affinity purification (see the contact frequency pattern in Fig. 3B). No alternative solutions satisfying all the data were found.
The model reveals that the SEA complex consists of two structurally distinct and physically connected subcomplexes. The first subcomplex is composed of Sea1, Npr3, and Npr2 and corresponds to SEACIT (Figs. 1A (lane 14), 3A, and 3B). Npr2 is proximal to Sea1, and both Npr3 and Sea1 are proximal to the N-terminal domain of Sea3 (Figs. 1A (lanes 4, 14–16), 1C, 3A, and 3B). The position of Npr2 within the SEA complex was determined at relatively low precision, mainly because of the lack of proximity and contact data for Npr2. Indeed, Npr2 was restrained by a single intermolecular cross-link of Sea1 (562)–Npr2 (562) (supplemental Table S4) and a domain interaction of Npr3–Npr2 (1–496) (Fig. 1A, lane 7). The uncertainty in Npr2, however, does not affect the uncertainty in the remaining parts of the structure.
The second subcomplex is composed of Sea2, Sea3, Sea4, Seh1, and Sec13 and corresponds to SEACAT. Immunoprecipitations, cross-linking analysis, and the molecular architecture all confirm that Sec13, Sea3, Seh1, Sea4, and Sea2 are in close proximity to one another (Figs. 1A (lanes 13 and 20), 1C, and 3A). Sec13 requires the presence of Sea3 to interact with the rest of the complex, suggesting that these two proteins can form a dimer (Figs. 1A, 3A, and 3B), similar to the Sea4/Seh1 dimer (11). Moreover, the Sea3/Sec13 and Sea4/Seh1 dimers interact with each other (Figs. 1A (lane 12), 3A, and 3B). The interactions among Sea2, Sea3/Sec13, and Sea4/Seh1 strongly depend on the C-terminal RING domains of Sea2, Sea3, and Sea4 (Figs. 3D and 3E), because RING domain deletion constructs of these proteins do not interact with each other (Figs. 1A (lanes 10, 16, and 22) and 3B). Sea3 can be cross-linked with all other members of the SEA complex except Npr2 and appears to be an interacting hub within the complex (Figs. 1C and 3A). Npr2 and Sea2 can easily dissociate from the complex, especially under stringent extraction conditions (Figs. 1A (lanes 15 and 19) and supplemental Fig. S1), consistent with a more peripheral localization in the complex. Seh1, Sec13, and the N termini of Sea4 and Sea2 seem to form a large cluster of β-propeller domains in the molecular architecture (Figs. 3C–3E). Similar arrangements of β-propeller domains have been described at the vertex of the evolutionarily related complexes COPI and COPII (44).
In summary, we have described here the molecular architecture of the yeast SEA complex. The structure was determined via an integrative approach based on interactions between proteins and domains obtained via affinity purification and residue-specific cross-links from chemical cross-linking and mass spectrometry. Most of the affinity purification data are consistent with the cross-linking data. However, these data are not redundant but, rather, complementary: first, a composite of multiple subunits implies a longer-range spatial restraint than a cross-link, which always restrains only two subunits; second, a cross-link may be observed when a co-purification is not, and vice versa—for example, although Npr2 and Npr3 co-purified, we could not obtain any high-confidence cross-links between these two subunits. The available data were sufficient to determine the molecular architecture of SEA, even in the absence of electron microscopy and/or small-angle X-ray scattering (SAXS) data for the entire complex or its individual components. However, if this additional information were available, it would undoubtedly further improve the precision and accuracy of our structure. Finally, our integrative modeling approach could be used to study any conformational or compositional changes larger than the precision of the solution ensemble; importantly, however, chemical cross-linking experiments need to be feasible under the conditions that trigger the change.
The SEA Complex Interacts with Mitochondria, the v-ATPase, and TORC1
We also explored a wide variety of affinity capture conditions to survey the interactome of the SEA complex beyond its immediate core components (Fig. 4A, supplemental Fig. S2, supplemental Tables S2 and S3). We noticed that a number of mitochondrial proteins co-purified with the SEA complex (Fig. 4A and supplemental Fig. S2). This was especially evident for Sea1-PrA when purified with a new variant of our standard extraction buffer (supplemental Fig. S2, lane 2). In this case, mitochondrial membrane components represented the majority of precipitated proteins, including members of the inner membrane of F1F0 ATPase (Atp1, Atp2), the 1.2-MDa prohibitin ring (Phb1, Phb2), the cytochrome bc1 complex (Cor1, Qcr2, Cyt1, Rip1, Qcr7), and the cytochrome c oxidase complex (Cox4, Cox5a), Pet9 and a major protein of the mitochondrial outer membrane, Por1. In other affinity purifications we also found mitochondrial proteins Mss116, Mgm1, Mir1, Rim1, and Gpm1 (supplemental Fig. S2). Mitochondrial proteins are often considered as contaminants during immunoprecipitation experiments. Over the years we have performed a large number of affinity purifications with all necessary controls, which has allowed us (as well as other groups doing similar studies) to establish a common list of contaminants typical for such experiments (28, 49–52). Many mitochondrial proteins mentioned above do not belong to the list of common contaminants (e.g. prohibitins)., and porin1, which is indeed often considered as a contaminant, can be seen in only a few immunoprecipitations (see, for example, supplemental Fig. S2, lane 2).
Fig. 4.
SEA complex is involved in the regulation of the TORC1 pathway. A, the yeast SEA complex physically interacts with the TORC1 complex. Sea2-PrA was immunoprecipitated as described under “Experimental Procedures.” Co-precipitating proteins were resolved via SDS-PAGE, visualized with Coomassie Blue stain, and identified via mass spectrometry (supplemental Tables S2 and S3). SEA complex members are marked in blue, TORC1 members in orange, mitochondria proteins in green, proteins involved in ribosome biogenesis and translation in blue, contaminants in gray, and others in pink. B, deletions of Sea1, Npr2, and Npr3 provoked Tor1 relocalization to the cytoplasm during nitrogen starvation. The localization of Tor1-GFP was followed by light fluorescence microscopy in wild-type and deletion strains of indicated SEA complex members, either in synthetic complete (SC) medium or in synthetic media lacking nitrogen SD-N, (nitrogen starvation). Observations were made for the strains grown in YPD or subjected to nitrogen starvation. C, the “vacuole-to-cytoplasm” GFP signal ratio was calculated for 25 cells in each strain shown in B.
Among co-purifying proteins, we found proteins belonging to two complexes associated with the vacuole membrane. The first were members of the V-ATPase complex: Vma1, Vma2, Vma6, and Vph1 (Fig. 4A, supplemental Fig. S2, supplemental Table S3). Interestingly, under certain conditions, the amount of Vph1 appeared to be almost stoichiometric with amounts of other members of the SEA complex (Fig. 4A, supplemental Fig. S2, supplemental Table S3). We also found the dynamin-like GTPase Vps1, involved in trafficking from the late endosome to the vacuole and required for vacuole fragmentation (53, 54). The components of the second complex were members of the TORC1: Tor1, Kog1, and Lst8 (Fig. 4A, supplemental Fig. S2, supplemental Table S3). Interestingly, TORC1 was not present when the purifications were performed with Npr2-PrA or Npr3-PrA (supplemental Fig. S2, supplemental Table S3). In contrast, TORC1 was almost always detected when Sea2 was present in the purifications (Fig. 4A, supplemental Fig. S2, supplemental Table S3).
The SEA Complex Functionally Interacts with TORC1
To confirm the physiological relevance of the detected interactions, we next examined whether the localization and activity of Tor1 kinase were affected in deletion mutants of the SEA complex (Figs. 4B and 4C). We followed the localization of Tor1-GFP in wild-type cells and in deletion mutants of SEA1, NPR2, and NPR3. In agreement with previous observations (6, 55, 56), Tor1-GFP was localized to the vacuole membrane in wild-type cells, in both rich (YPD) and nitrogen-free media. Although Tor1-GFP also localized to the vacuole in the deletion strains when grown in YPD (Figs. 4B and 4C), in all three mutant strains, after 20 h of nitrogen starvation, Tor1-GFP was no longer found at the vacuole membrane; instead it was dispersed in the cytoplasm, where it concentrated in distinct foci (this was especially obvious upon NPR2 deletion).
Sea1 Is Required for General Autophagy and Maintenance of Vacuole Integrity during Nitrogen Deprivation
One of the consequences of TORC1 inhibition is the induction of autophagy (1). We and others have previously shown that deletions of Npr2 and Npr3 cause defects in both general and specific forms of autophagy, whereas deletions of Sea2–Sea4 do not have significant effects (11, 18, 20). However, the role of Sea1 in autophagy was not explored in detail. To determine whether Sea1 is required for general autophagy, we followed maturation and localization of the autophagy marker GFP-ATG8 in various SEA1 deletion strains (57), with GFP localization monitored via fluorescence microscopy (Fig. 5A). Autophagic induction (indicated by an increase of total GFP signal over time) and flux (indicated by a change in the ratio of free GFP to total GFP signal over time) were monitored with Western blots (Figs. 5B and 5C). As has been previously observed in Npr2 and Npr3 mutants (11, 18), both induction and flux were decreased in all strains carrying deletions of the SEA1 gene (Fig. 5). However, the SEA1 deletions appeared to have consequences beyond defects in autophagic induction and flux, because we also observed significant vacuolar fragmentation (Fig. 5A) in SEA1 mutants subjected to nitrogen starvation. Vacuole fission in these strains is specifically stimulated by nitrogen deprivation, as deletion mutants grown in YPD have non-fragmented vacuoles (11).
Fig. 5.
Sea1 is involved in the regulation of general autophagy. A, wild-type and indicated deletion strains transformed with a plasmid expressing GFP-ATG8 were subjected to nitrogen starvation as described under “Experimental Procedures” and examined under a fluorescent microscope after 20 h of starvation. Scale bar = 5 μm. B, strains were grown as in A. Samples were taken at the indicated time points and analyzed via Western blotting with anti-GFP or anti-PGK1 antibodies. C, autophagic flux was calculated as a ratio (percentage) of free GFP to total GFP signal (combined free GFP and GFP-ATG8) in the corresponding Western blots from B. D, to estimate autophagic induction, the total GFP signal was normalized to the PGK1 signal. Normalized values at time point 0 were set as 1.
Yeast vacuoles adjust their morphology in response to environmental stresses through fusion and fission. When TORC1 is inactivated by rapamycin or during nutrition restriction, vacuoles fuse into a single organelle to facilitate the degradation of materials delivered via autophagy (58). Because Sea1, Npr2, and Npr3 are involved in TORC1 inhibition upon nitrogen starvation (10, 11, 17), one might expect that their deletion would inhibit vacuolar fusion. Indeed, in sea1Δ and npr2Δ/npr3Δ cells, vacuolar fusion was strongly inhibited upon nitrogen starvation (Fig. 5A). Interestingly, in sea1Δ/sea3Δ and sea1Δ/sea4Δ strains, we observed two cell populations: one showing fragmented vacuoles with GFP signal concentrated in the vacuoles, and a second one showing single-lobed vacuoles with GFP signal concentrated in the cytoplasm. Nevertheless, taken together these data remain consistent with the idea that Sea1, Npr2, and Npr3 work in synergy to maintain effective autophagy and vacuole integrity.
TORC1 Inhibition Changes the Stability of SEA Complex Members
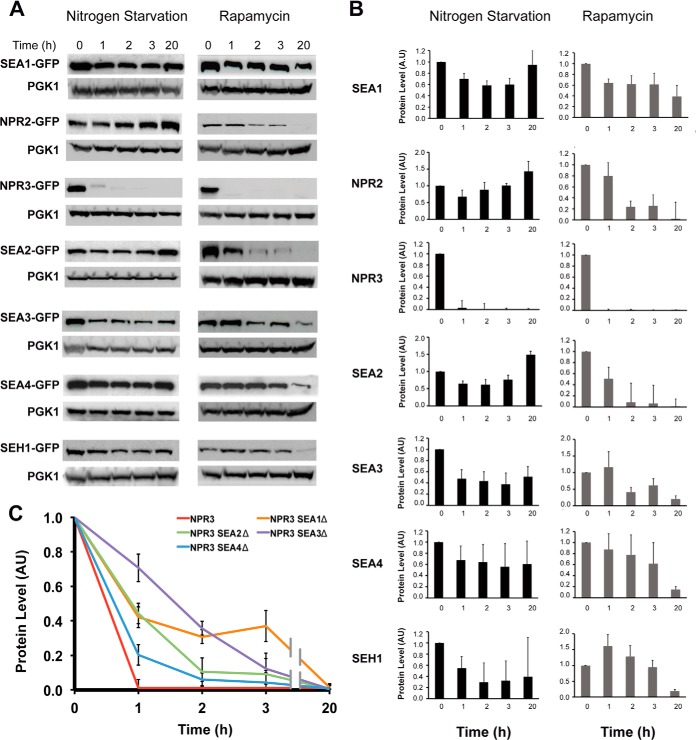
Because the SEA complex regulates TORC1 activity and localization, it is necessary to follow the behavior of the SEA complex when cells are subjected to nitrogen starvation or treated with rapamycin, two conditions that inhibit Tor1 kinase and induce autophagy (Fig. 6). The stability of SEA components is altered upon TORC1 inactivation, in a manner dependent upon the mode of TORC1 inhibition. In general, after 20 h of incubation the majority of SEA complex members were still detectable in the cells subjected to nitrogen starvation, whereas their amount was largely decreased in the rapamycin-treated cells (Figs. 6A and 6B). For example, Sea2 and Npr2 were completely degraded after 20 h of rapamycin treatment, whereas the amounts of these proteins were stable or marginally increased after 20 h of nitrogen deprivation. Likewise, Sea4 and Seh1 levels were constant during the whole period of nitrogen starvation but were significantly decreased after 20 h of rapamycin treatment.
Fig. 6.
Stability of SEA complex components during nitrogen starvation and rapamycin treatment. A, yeast cells carrying a SEA member tagged with GFP were subjected to nitrogen starvation or rapamycin treatment (20 nm final concentration). Samples were collected at indicated time points, and whole cell extracts were prepared and analyzed via Western blotting with anti-GFP or anti-PGK1 antibodies. Error bars represent the standard deviation in three independent experiments. B, the protein level in arbitrary units (AU) was calculated by normalizing the GFP signal to the corresponding PGK1 signal from blots shown in A. The signal at time 0 was set at 1. C, the protein level of Npr3-GFP in indicated deletion strains subjected to rapamycin treatment at different time points was calculated as in B and is represented as a graph. Error bars represent the standard deviation in three independent experiments.
The level of Npr3 was already significantly decreased after 1 h of either treatment. The degradation of Npr3 during nitrogen starvation is probably induced by some posttranslational modifications, because after 1 h of starvation we could detect a higher migrating band on a Western blot that disappeared after prolonged starvation. Importantly, deletions of SEA1, SEA2, SEA3, or SEA4 increased Npr3 stability (Fig. 6C), indicating that these SEA members might participate in Npr3 degradation.
DISCUSSION
Molecular Architecture of the SEA Complex
It has been suggested that the SEA proteins are divided between two distinct complexes (termed GATOR1 and GATOR2 in vertebrates and SEACIT and SEACAT in yeast) (17, 19, 21). We show here that the yeast SEA proteins form a single complex, the SEA complex, albeit composed of two structurally and functionally distinct subcomplexes that are intimately connected to each other and perform complementary functions (Figs. 1, 3, and 7). Thus, one end of the SEA complex is made of the Sea1/Npr2/Npr3 trimer (SEACIT), involved in TORC1 inhibition, and the other end, composed of Sea2/Sea3/Sea4/Seh1/Sec13 (SEACAT), is involved in TORC1 activation.
Fig. 7.
An overview of the proposed SEA complex activities and interactions. The SEA complex is situated at the vacuole membrane and interacts with V-ATPase, mitochondria, and TORC1 (straight blue arrows). The SEA complex possesses GAP activity (curved blue arrow) toward another TORC1 regulator, the EGO complex. Both SEA and EGO act upstream of the TORC1 (curved red arrows).
The members of the first subcomplex contain motifs found in GAPs and GEFs, which regulate GTPases. Sea1 has been shown to be a GAP for Gtr1 (17), and both Npr2 and Npr3 possess longin domains, found in various GEFs, though GEF activity has not yet been demonstrated for these two proteins (14–16). In mammals the homolog of SEACIT exhibits GAP activity toward RagA (21), though in this case an enzymatic activity for individual proteins was not determined. In addition, it is also unclear whether yeast SEACIT can have GEF or GAP activity, or both (17). The GAP (Sea1) and the GEF (Vam6) for Gtr1 have been identified (6, 17), but there is no information about equivalent factors for Gtr2. Intriguingly, the Vam6 protein, also called Vps39, is a member of the HOPS tethering complex and is structurally very similar to Sea4. It remains to be determined whether Sea4 has GEF activity.
The members of the second subcomplex (Seh1, Sec13, and the N termini of Sea4 and Sea2) form a large β-propeller cluster (13) that may serve as a structural platform for the multiple functions of the SEA complex. At the same time the N-terminal β-propeller domain of Sea3 connects the SEACAT to the Sea1/Npr2/Npr3 subcomplex. The C-terminal part of Sea3, which contains a RING domain, interacts with the C-terminal RING domains of Sea2 and Sea4. This RING domain interaction seems to be crucial to maintain the contacts among Sea2, Sea3, and Sea4 and the rest of the SEA complex, because in the absence of these domains, Sea2 is no longer connected to the complex and Sea4 interacts only with Seh1. In contrast, in the absence of the Sea3 RING domain, the N-terminal part of the protein still interacts with Sea1/Npr2/Npr3. Given the overall conservation of components of the complex in humans (11, 21), we expect that the structure of the human SEA complex will closely resemble that of the yeast SEA complex.
The structural organization of the TORC1 pathway components indicates that the TORC1 network is another example of an evolutionary connection among the different complexes involved in the control of intracellular membrane trafficking systems (59). The TORC1 interactome at the yeast vacuolar membrane (Fig. 7) includes EGO, HOPS, and SEA complexes. All these assemblies are enriched in proteins containing β-propellers and α-solenoid-like folds, characteristic folds of membrane-coating systems such as clathrin, COPI, COPII, and the nuclear pore complex (11, 60–63). In addition, RING, DEP, and longin domains appear as novel structural elements essential for TORC1 signaling. In this context the SEA complex emerges as a platform that can coordinate both structural and enzymatic activities necessary for the effective functioning of the TORC1. We suggest, based on its structural composition (similar to those of known membrane coating complexes), molecular architecture, dynamic localization, and functional associations, that the SEACAT subcomplex forms a membrane-associated scaffold for TORC1 activation. In contrast, SEACIT apparently represents a regulatory subcomplex that participates in TORC1 inhibition. GAP activity was demonstrated for the SEACIT component Sea1 (17), and a putative GEF activity was suggested for Npr2 and Npr3 (16). Thus, remarkably, the two connected subcomplexes of the SEA complex have opposite activities: one activates TORC1, whereas the other one suppresses it (17, 19, 21). The physical interaction between these subcomplexes might provide an opportunity for additional regulation of this major cellular signaling pathway.
The SEA Complex Is a Major Regulator of the TORC1 Pathway
The role of the SEA complex in TORC1 regulation seems to be quite elaborate; it is possible that it assesses various cellular and metabolic states and integrates them into concerted signals regulating TORC1 (Fig. 7). We show here that the SEA complex physically interacts with TORC1 (Fig. 4). Deletions of SEA1, NPR2, or NPR3 during nitrogen starvation caused dramatic relocalization of the Tor1 kinase to the cytoplasm, where it accumulated in distinct foci. Recently, TORC1 sequestration in cytoplasmic stress granules was demonstrated for yeast cells exposed to heat stress (64). Therefore, TORC1 response to stresses involves not only changes in its enzymatic activity, but also alterations in its location. Given the structural and evolutionary relationships of the SEA complex with the coating and tethering assemblies (11), it is conceivable that the SEA complex helps maintain TORC1 at the vacuole membrane during nitrogen starvation. In turn, the SEA complex also seems to be subjected to regulation by the TORC1 signaling pathway, as both nitrogen starvation and rapamycin treatment influenced the stability of particular SEA complex components (Fig. 6).
TORC1 signaling regulates a variety of cellular processes, including autophagy. Because SEACIT inhibits TORC1 signaling, deletion of SEACIT components results in hyperactive TORC1, and therefore suppresses autophagy (Fig. 5) (11, 12, 18, 20). Deletion of SEA1 or double deletion of NPR2 and NPR3 also results in inhibition of vacuolar fusion upon nitrogen starvation. Autophagic defects are not commonly associated with such inhibition; however, a recent study reported that inactivation of TORC1 during nitrogen deprivation, and therefore induction of autophagy, promotes vacuolar coalescence (58). These data are in agreement with our results showing that deletions of Sea1/Npr2/Npr3 maintained TORC1 activity during starvation and in turn led to increased vacuolar fragmentation and defects in autophagy. Thus, we have demonstrated that upstream TORC1 signaling controls vacuolar fusion and fission events. Apart from TORC1, vacuolar fission requires the activity of various factors, including the dynamin-like GTPase Vps1 (53) and the vacuole protein pump V-ATPase (65). Remarkably, our proteomic survey revealed that the SEA complex interacted not only with TORC1, but also with Vps1 and V-ATPase (supplemental Fig. S2). Thus, our results identify the SEA complex as a new component involved in the maintenance of vacuole integrity.
The characterization of the SEA complex as a novel major regulator of the TORC1 pathway might open new avenues in the study of the role of TORC1 signaling in disease and development. Various mutations of Nprl2 have been detected in different tumors, including lung cancer (66) and hepatocellular carcinoma (67). Nprl2 and Depdc5 (the human homolog of Sea1) are also deregulated in ovarian cancers and glioblastomas (21, 68). In addition, loss-of-function mutation in DEPDC5 has been detected in individuals with familial local epilepsy (69, 70). Mice in which a promoter of the NPRL3 gene has been deleted die in late gestation, often with severe cardiac defects (71). Finally, Drosophila homologs of Seh1 and Sea4 (Mio) are required for fly oogenesis (72), and Nprl2 and Nprl3 are essential for female fertility during nitrogen starvation (73). We anticipate that our structural and functional characterization of the SEA complex will offer a valuable framework for understanding the defects leading to these various diseases.
Supplementary Material
Acknowledgments
We thank Rosemary Williams, Sophie Desnoulez, and the Proteomic Platform of Institut Gustave Roussy for the help with experiments. We are grateful to C. De Virgilio for the Tor1-GFP strain.
Footnotes
Author contributions: J.F., Y.S., S.K., R.P., and S.D. designed research; R.A., J.F., Y.S., S.K., R.P., P.C., E.C., and S.D. performed research; R.A., J.F., Y.S., S.K., R.P., P.C., A.S., B.T.C., M.P.R., and S.D. analyzed data; A.S., B.T.C., M.P.R., and S.D. wrote the paper.
* S.D. is grateful for financial support from Fondation Gustave Roussy, Fondation de France, Fondation pour la Recherche Médicale, La Ligue National contre le Cancer (Comité de Paris/Ile-de-France and Comité de l'Oise); S.D. and M.P.R. acknowledge support from CNRS (grant PICS); R.A. is grateful for the support from Université Paris-Sud and Fondation Gustave Roussy. This work was supported in part by National Institutes of Health Grant No. U54 GM103511 to M.P.R., A.S., and B.T.C.; Grant No. R01 GM083960 to A.S.; Grant No. P41 GM103314 to B.T.C.; and Grant No. U54 GM094662 to S.K. P.C. is supported by a Howard Hughes Medical Institute Predoctoral Fellowship, and R.P. is supported by Swiss National Science Foundation Grants PA00P3_139727 and PBZHP3–133388.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- TORC1
- target of rapamycin complex 1
- EGO
- escape from rapamycin growth arrest (complex)
- GAP
- GTPase activating protein
- GEF
- guanine nucleotide exchange factor
- SEA
- Seh1 associated complex
- SEACAT
- SEA subcomplex activating TORC1
- SEACIT
- SEA subcomplex inhibiting TORC1.
REFERENCES
- 1. Loewith R., Hall M. N. (2011) Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189, 1177–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Laplante M., Sabatini D. M. (2012) mTOR signaling in growth control and disease. Cell 149, 274–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bar-Peled L., Schweitzer L. D., Zoncu R., Sabatini D. M. (2012) Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Binda M., Peli-Gulli M. P., Bonfils G., Panchaud N., Urban J., Sturgill T. W., Loewith R., De Virgilio C. (2009) The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 35, 563–573 [DOI] [PubMed] [Google Scholar]
- 7. Bonfils G., Jaquenoud M., Bontron S., Ostrowicz C., Ungermann C., De Virgilio C. (2012) Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell 46, 105–110 [DOI] [PubMed] [Google Scholar]
- 8. Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D. M. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 334, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han J. M., Jeong S. J., Park M. C., Kim G., Kwon N. H., Kim H. K., Ha S. H., Ryu S. H., Kim S. (2012) Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149, 410–424 [DOI] [PubMed] [Google Scholar]
- 10. Neklesa T. K., Davis R. W. (2009) A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 5, e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dokudovskaya S., Waharte F., Schlessinger A., Pieper U., Devos D. P., Cristea I. M., Williams R., Salamero J., Chait B. T., Sali A., Field M. C., Rout M. P., Dargemont C. (2011) A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol. Cell. Proteomics 10, M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dokudovskaya S., Rout M. P. (2011) A novel coatomer-related SEA complex dynamically associates with the vacuole in yeast and is implicated in the response to nitrogen starvation. Autophagy 7, 1392–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Algret R., Dokudovskaya S. (2012) The SEA complex—the beginning. Biopolymers Cell 28, 281–284 [Google Scholar]
- 14. Nookala R. K., Langemeyer L., Pacitto A., Ochoa-Montano B., Donaldson J. C., Blaszczyk B. K., Chirgadze D. Y., Barr F. A., Bazan J. F., Blundell T. L. (2012) Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2, 120071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang D., Iyer L. M., He F., Aravind L. (2012) Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front. Genet. 3, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Levine T. P., Daniels R. D., Wong L. H., Gatta A. T., Gerondopoulos A., Barr F. A. (2013) Discovery of new Longin and Roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases 4, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Panchaud N., Peli-Gulli M. P., De Virgilio C. (2013) Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6, ra42. [DOI] [PubMed] [Google Scholar]
- 18. Graef M., Nunnari J. (2011) Mitochondria regulate autophagy by conserved signalling pathways. EMBO J. 30, 2101–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Panchaud N., Peli-Gulli M. P., De Virgilio C. (2013) SEACing the GAP that nEGOCiates TORC1 activation: evolutionary conservation of Rag GTPase regulation. Cell Cycle 12, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu X., Tu B. P. (2011) Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol. Biol. Cell 22, 4124–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bar-Peled L., Chantranupong L., Cherniack A. D., Chen W. W., Ottina K. A., Grabiner B. C., Spear E. D., Carter S. L., Meyerson M., Sabatini D. M. (2013) A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fernandez-Martinez J., Phillips J., Sekedat M. D., Diaz-Avalos R., Velazquez-Muriel J., Franke J. D., Williams R., Stokes D. L., Chait B. T., Sali A., Rout M. P. (2012) Structure-function mapping of a heptameric module in the nuclear pore complex. J. Cell Biol. 196, 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Leitner A., Reischl R., Walzthoeni T., Herzog F., Bohn S., Forster F., Aebersold R. (2012) Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics 11, M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 25. Yang B., Wu Y. J., Zhu M., Fan S. B., Lin J., Zhang K., Li S., Chi H., Li Y. X., Chen H. F., Luo S. K., Ding Y. H., Wang L. H., Hao Z., Xiu L. Y., Chen S., Ye K., He S. M., Dong M. Q. (2012) Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 [DOI] [PubMed] [Google Scholar]
- 26. Qin J., Chait B. T. (1995) Preferential fragmentation of protonated gas-phase peptide ions adjacent to acidic amino-acid-residues. J. Am. Chem. Soc. 117, 5411–5412 [Google Scholar]
- 27. Michalski A., Neuhauser N., Cox J., Mann M. (2012) A systematic investigation into the nature of tryptic HCD spectra. J. Proteome Res. 11, 5479–5491 [DOI] [PubMed] [Google Scholar]
- 28. Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Rout M. P., Sali A. (2007) Determining the architectures of macromolecular assemblies. Nature 450, 683–694 [DOI] [PubMed] [Google Scholar]
- 29. Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 30. Lasker K., Phillips J. L., Russel D., Velazquez-Muriel J., Schneidman-Duhovny D., Tjioe E., Webb B., Schlessinger A., Sali A. (2010) Integrative structure modeling of macromolecular assemblies from proteomics data. Mol. Cell. Proteomics 9, 1689–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lasker K., Sali A., Wolfson H. J. (2010) Determining macromolecular assembly structures by molecular docking and fitting into an electron density map. Proteins 78, 3205–3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Russel D., Lasker K., Webb B., Velazquez-Muriel J., Tjioe E., Schneidman-Duhovny D., Peterson B., Sali A. (2012) Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 10, e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fath S., Mancias J. D., Bi X., Goldberg J. (2007) Structure and organization of coat proteins in the COPII cage. Cell 129, 1325–1336 [DOI] [PubMed] [Google Scholar]
- 34. Debler E. W., Ma Y., Seo H. S., Hsia K. C., Noriega T. R., Blobel G., Hoelz A. (2008) A fence-like coat for the nuclear pore membrane. Mol. Cell 32, 815–826 [DOI] [PubMed] [Google Scholar]
- 35. Soding J., Biegert A., Lupas A. N. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bryson K., Cozzetto D., Jones D. T. (2007) Computer-assisted protein domain boundary prediction using the Dom-Pred server. Curr. Protein Peptide Sci. 8, 181–188 [DOI] [PubMed] [Google Scholar]
- 37. McGuffin L. J., Bryson K., Jones D. T. (2000) The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405 [DOI] [PubMed] [Google Scholar]
- 38. Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 39. Menke M., Berger B., Cowen L. (2010) Markov random fields reveal an N-terminal double beta-propeller motif as part of a bacterial hybrid two-component sensor system. Proc. Natl. Acad. Sci. U.S.A. 107, 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen M. Y., Sali A. (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci. 15, 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sali A., Blundell T. L. (1993) Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 42. Kohn J. E., Millett I. S., Jacob J., Zagrovic B., Dillon T. M., Cingel N., Dothager R. S., Seifert S., Thiyagarajan P., Sosnick T. R., Hasan M. Z., Pande V. S., Ruczinski I., Doniach S., Plaxco K. W. (2004) Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12491–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oliphant T. E. (2007) Python for scientific computing. Comput. Sci. Eng. 9, 10–20 [Google Scholar]
- 44. Lee C., Goldberg J. (2010) Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell 142, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gurkan C., Stagg S. M., Lapointe P., Balch W. E. (2006) The COPII cage: unifying principles of vesicle coat assembly. Nat. Rev. Mol. Cell Biol. 7, 727–738 [DOI] [PubMed] [Google Scholar]
- 46. Seebacher J., Mallick P., Zhang N., Eddes J. S., Aebersold R., Gelb M. H. (2006) Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J. Proteome Res. 5, 2270–2282 [DOI] [PubMed] [Google Scholar]
- 47. Herzog F., Kahraman A., Boehringer D., Mak R., Bracher A., Walzthoeni T., Leitner A., Beck M., Hartl F. U., Ban N., Malmstrom L., Aebersold R. (2012) Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 [DOI] [PubMed] [Google Scholar]
- 48. Chen Z. A., Jawhari A., Fischer L., Buchen C., Tahir S., Kamenski T., Rasmussen M., Lariviere L., Bukowski-Wills J. C., Nilges M., Cramer P., Rappsilber J. (2010) Architecture of the RNA polymerase II-TFIIF complex revealed by cross-linking and mass spectrometry. EMBO J. 29, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Archambault V., Chang E. J., Drapkin B. J., Cross F. R., Chait B. T., Rout M. P. (2004) Targeted proteomic study of the cyclin-Cdk module. Mol. Cell 14, 699–711 [DOI] [PubMed] [Google Scholar]
- 50. Tackett A. J., DeGrasse J. A., Sekedat M. D., Oeffinger M., Rout M. P., Chait B. T. (2005) I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res. 4, 1752–1756 [DOI] [PubMed] [Google Scholar]
- 51. Niepel M., Strambio-de-Castillia C., Fasolo J., Chait B. T., Rout M. P. (2005) The nuclear pore complex-associated protein, Mlp2p, binds to the yeast spindle pole body and promotes its efficient assembly. J. Cell Biol. 170, 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Oeffinger M., Wei K. E., Rogers R., DeGrasse J. A., Chait B. T., Aitchison J. D., Rout M. P. (2007) Comprehensive analysis of diverse ribonucleoprotein complexes. Nat. Methods 4, 951–956 [DOI] [PubMed] [Google Scholar]
- 53. Peters C., Baars T. L., Buhler S., Mayer A. (2004) Mutual control of membrane fission and fusion proteins. Cell 119, 667–678 [DOI] [PubMed] [Google Scholar]
- 54. Hayden J., Williams M., Granich A., Ahn H., Tenay B., Lukehart J., Highfill C., Dobard S., Kim K. (2013) Vps1 in the late endosome-to-vacuole traffic. J. Biosci. 38, 73–83 [DOI] [PubMed] [Google Scholar]
- 55. Urban J., Soulard A., Huber A., Lippman S., Mukhopadhyay D., Deloche O., Wanke V., Anrather D., Ammerer G., Riezman H., Broach J. R., De Virgilio C., Hall M. N., Loewith R. (2007) Sch9 is a major target of TORC1 in Saccharomyces cerevisiae. Mol. Cell 26, 663–674 [DOI] [PubMed] [Google Scholar]
- 56. Sturgill T. W., Cohen A., Diefenbacher M., Trautwein M., Martin D. E., Hall M. N. (2008) TOR1 and TOR2 have distinct locations in live cells. Eukaryot. Cell 7, 1819–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Klionsky D. J., Cuervo A. M., Seglen P. O. (2007) Methods for monitoring autophagy from yeast to human. Autophagy 3, 181–206 [DOI] [PubMed] [Google Scholar]
- 58. Michaillat L., Baars T. L., Mayer A. (2012) Cell-free reconstitution of vacuole membrane fragmentation reveals regulation of vacuole size and number by TORC1. Mol. Biol. Cell 23, 881–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Field M. C., Sali A., Rout M. P. (2011) Evolution: on a bender—BARs, ESCRTs, COPs, and finally getting your coat. J. Cell Biol. 193, 963–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Devos D., Dokudovskaya S., Alber F., Williams R., Chait B. T., Sali A., Rout M. P. (2004) Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2, e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Devos D., Dokudovskaya S., Williams R., Alber F., Eswar N., Chait B. T., Rout M. P., Sali A. (2006) Simple fold composition and modular architecture of the nuclear pore complex. Proc. Natl. Acad. Sci. U.S.A. 103, 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dokudovskaya S., Williams R., Devos D., Sali A., Chait B. T., Rout M. P. (2006) Protease accessibility laddering: a proteomic tool for probing protein structure. Structure 14, 653–660 [DOI] [PubMed] [Google Scholar]
- 63. DeGrasse J. A., DuBois K. N., Devos D., Siegel T. N., Sali A., Field M. C., Rout M. P., Chait B. T. (2009) Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics 8, 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Takahara T., Maeda T. (2012) Transient sequestration of TORC1 into stress granules during heat stress. Mol. Cell 47, 242–252 [DOI] [PubMed] [Google Scholar]
- 65. Baars T. L., Petri S., Peters C., Mayer A. (2007) Role of the V-ATPase in regulation of the vacuolar fission-fusion equilibrium. Mol. Biol. Cell 18, 3873–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ueda K., Kawashima H., Ohtani S., Deng W. G., Ravoori M., Bankson J., Gao B., Girard L., Minna J. D., Roth J. A., Kundra V., Ji L. (2006) The 3p21.3 tumor suppressor NPRL2 plays an important role in cisplatin-induced resistance in human non-small-cell lung cancer cells. Cancer Res. 66, 9682–9690 [DOI] [PubMed] [Google Scholar]
- 67. Otani S., Takeda S., Yamada S., Sakakima Y., Sugimoto H., Nomoto S., Kasuya H., Kanazumi N., Nagasaka T., Nakao A. (2009) The tumor suppressor NPRL2 in hepatocellular carcinoma plays an important role in progression and can be served as an independent prognostic factor. J. Surg. Oncol. 100, 358–363 [DOI] [PubMed] [Google Scholar]
- 68. Seng T. J., Ichimura K., Liu L., Tingby O., Pearson D. M., Collins V. P. (2005) Complex chromosome 22 rearrangements in astrocytic tumors identified using microsatellite and chromosome 22 tile path array analysis. Genes Chrom. Cancer 43, 181–193 [DOI] [PubMed] [Google Scholar]
- 69. Dibbens L. M., de Vries B., Donatello S., Heron S. E., Hodgson B. L., Chintawar S., Crompton D. E., Hughes J. N., Bellows S. T., Klein K. M., Callenbach P. M., Corbett M. A., Gardner A. E., Kivity S., Iona X., Regan B. M., Weller C. M., Crimmins D., O'Brien T. J., Guerrero-Lopez R., Mulley J. C., Dubeau F., Licchetta L., Bisulli F., Cossette P., Thomas P. Q., Gecz J., Serratosa J., Brouwer O. F., Andermann F., Andermann E., van den Maagdenberg A. M., Pandolfo M., Berkovic S. F., Scheffer I. E. (2013) Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat. Genet. 45, 546–551 [DOI] [PubMed] [Google Scholar]
- 70. Ishida S., Picard F., Rudolf G., Noe E., Achaz G., Thomas P., Genton P., Mundwiller E., Wolff M., Marescaux C., Miles R., Baulac M., Hirsch E., Leguern E., Baulac S. (2013) Mutations of DEPDC5 cause autosomal dominant focal epilepsies. Nat. Genet. 45, 552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kowalczyk M. S., Hughes J. R., Babbs C., Sanchez-Pulido L., Szumska D., Sharpe J. A., Sloane-Stanley J. A., Morriss-Kay G. M., Smoot L. B., Roberts A. E., Watkins H., Bhattacharya S., Gibbons R. J., Ponting C. P., Wood W. G., Higgs D. R. (2012) Nprl3 is required for normal development of the cardiovascular system. Mamm. Genome 23, 404–415 [DOI] [PubMed] [Google Scholar]
- 72. Senger S., Csokmay J., Akbar T., Jones T. I., Sengupta P., Lilly M. A. (2011) The nucleoporin Seh1 forms a complex with Mio and serves an essential tissue-specific function in Drosophila oogenesis. Development 138, 2133–2142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wei Y., Lilly M. A. (2014) The TORC1 inhibitors Nprl2 and Nprl3 mediate an adaptive response to amino-acid starvation in Drosophila. Cell Death Differ. 21(9), 1460–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.