Abstract
Post-translational modifications of histones, such as acetylation and methylation, are differentially positioned in chromatin with respect to gene organization. For example, although histone H3 is often trimethylated on lysine 4 (H3K4me3) and acetylated on lysine 14 (H3K14ac) at active promoter regions, histone H3 lysine 36 trimethylation (H3K36me3) occurs throughout the open reading frames of transcriptionally active genes. The conserved yeast histone acetyltransferase complex, NuA3, specifically binds H3K4me3 through a plant homeodomain (PHD) finger in the Yng1 subunit, and subsequently catalyzes the acetylation of H3K14 through the histone acetyltransferase domain of Sas3, leading to transcription initiation at a subset of genes. We previously found that Ylr455w (Pdp3), an uncharacterized proline-tryptophan-tryptophan-proline (PWWP) domain-containing protein, copurifies with stable members of NuA3. Here, we employ mass-spectrometric analysis of affinity purified Pdp3, biophysical binding assays, and genetic analyses to classify NuA3 into two functionally distinct forms: NuA3a and NuA3b. Although NuA3a uses the PHD finger of Yng1 to interact with H3K4me3 at the 5′-end of open reading frames, NuA3b contains the unique member, Pdp3, which regulates an interaction between NuA3b and H3K36me3 at the transcribed regions of genes through its PWWP domain. We find that deletion of PDP3 decreases NuA3-directed transcription and results in growth defects when combined with transcription elongation mutants, suggesting NuA3b acts as a positive elongation factor. Finally, we determine that NuA3a, but not NuA3b, is synthetically lethal in combination with a deletion of the histone acetyltransferase GCN5, indicating NuA3b has a specialized role at coding regions that is independent of Gcn5 activity. Collectively, these studies define a new form of the NuA3 complex that associates with H3K36me3 to effect transcriptional elongation. MS data are available via ProteomeXchange with identifier PXD001156.
Eukaryotic DNA is wrapped around octamers of the evolutionarily conserved core histone proteins H3, H4, H2A, and H2B, forming nucleosomes—the fundamental unit of chromatin. Chromatin acts as a barrier to the transcriptional machinery, and therefore precise coordination of nucleosome organization is required for the passage of RNA polymerase II (RNAPII)1 (1–3). Nucleosome organization is regulated by chromatin remodelers, histone chaperones, and other complexes that enzymatically add, remove, or bind (“write,” “erase,” or “read,” respectively) post-translational modifications (PTMs) on histones (4–7). Together, these factors ensure that genes remain accessible to transcription factors, activators or coactivators, and RNAPII (5). Critically, these factors also function to restore chromatin structure following the passage of RNAPII during transcription elongation (3, 8).
Histone PTMs are established at gene loci in a context specific manner, typically defined by the position along a gene (i.e. promoters versus open reading frames (ORFs)), as well as the transcriptional status of that gene (4, 6, 9–14). The dual capacity of chromatin complexes to “read” and “write” histone PTMs confines certain PTM combinations to discrete regions within genomic loci (15–20). For example, histone H3 is often combinatorially modified by trimethylation on lysine 4 (H3K4me3) and acetylation, particularly on lysine 14 (H3K14ac), at the 5′-ends of actively transcribed genes (12, 13). NuA3, a conserved S. cerevisiae histone acetyltransferase (HAT) complex (21), specifically binds H3K4me3 that is generated by the Set1 histone methyltransferase (HMT), through the plant homeodomain (PHD) finger in the Yng1 subunit (an ortholog of human ING5) (19, 22–27). NuA3 then acetylates H3K14 on the same H3 molecule through the HAT domain of Sas3 (an ortholog of human MYST3) (19, 23–25). Subsequently, additional factors bind either NuA3-catalyzed acetylation or NuA3 subunits themselves, including the remodel the structure of chromatin (RSC) chromatin-remodeling complex (that contains an H3K14ac-binding bromodomain) and the facilitates chromatin transcription (FACT) histone chaperone complex (that binds to Sas3), to promote transcription initiation at a subset of genes (19, 25, 28–31).
Other histone PTMs positioned within gene bodies further facilitate transcription and maintain transcript fidelity (3, 8, 10, 32). In Saccharomyces cerevisiae, the Set2 HMT localizes to the bodies of actively transcribed genes (13, 33) by physically binding to the hyperphosphorylated C-terminal domain (CTD) of RNAPII (34–39). CTD binding by Set2 is required for the establishment of H3K36me3 (34). H3K36 methylation correlates with transcription elongation (13, 34, 35, 37, 40), and maintains chromatin integrity by recruiting complexes that collectively restore chromatin structure (3, 8, 32). For example, the histone deacetylase (HDAC) complex, Rpd3S, engages H3K36me2 via the chromodomain of Eaf3 and the PHD finger of Rco1 (18, 41–47). Rpd3S generates a hypoacetylated environment behind the elongating RNAPII, which compacts chromatin and represses intergenic or “cryptic” transcription (41, 44, 45, 48, 49). H3K36me3 also maintains chromatin integrity by blocking trans-histone exchange through at least two mechanisms: the steric reduction of histone chaperone affinity for histone targets (50) and the recruitment of chromatin remodelers that preserve H3K36me3/hypoacetylated histones (3, 51, 52). Specifically, H3K36me3 precludes Asf1 from depositing newly synthesized histones (50) and recruits the Isw1b ATP-dependent remodeler, via the proline-tryptophan-tryptophan-proline (PWWP) domain of the Ioc4 subunit, to position nucleosomes in a manner that stimulates Rpd3S activity (51–53). Recently, human PWWP domain proteins have been shown to bind H3K36me3 (20, 54) and perform a variety of functions, including the regulation of transcription (55, 56), DNA methylation guidance (57), and alternative splicing (58).
Using recent advances in mass spectrometry, we previously found that Ylr455w, an uncharacterized PWWP domain-containing protein (59–61), copurifies with stable members of the NuA3 HAT complex (19). We propose that Ylr455w be called Pdp3: PWWP domain protein in NuA3. Moreover, Pdp3 (Q09842) is also the name of a PWWP domain-containing protein in Schizosaccharomyces pombe that interacts with homologous subunits of the NuA3 complex (62).
Here, we find evidence that two functionally distinct forms of NuA3 exist. One form of NuA3 binds H3K4me3, through the PHD finger of Yng1, and acetylates H3K14 at the 5′-ends of actively transcribed genes to promote transcription initiation (19, 24, 26, 27). In contrast, an interaction between Pdp3 and NuA3 recruits the complex to chromatin, through the Pdp3 PWWP domain binding to H3K36me3. Deletion of the PDP3 gene decreases NuA3-directed transcription and results in growth defects when combined with transcription elongation mutants, suggesting Pdp3-associated NuA3 functions in the transcription elongation process. Although H3K36me2/3 can act as a repressive mark that protects chromatin integrity during transcription elongation (3, 8, 32), the work described in this study suggests that H3K36me3 can also act to positively regulate transcription elongation through the recruitment of the NuA3 complex via Pdp3.
EXPERIMENTAL PROCEDURES
S. cerevisiae Strains
All strains are described in supplemental Table S1.
Escherichia coli and S. cerevisiae Plasmids
All constructs are described in supplemental Table S2.
Peptide Sequences
All peptide sequences are listed in supplemental Table S3.
Primer Sequences
Primers were designed by Primer3 to target the 5′-end of our genes of interest. All primer sequences are listed in supplemental Table S4.
Mass Spectrometric Protein Identification
Pdp3-TAP protein complex purification was performed with S. cerevisiae grown to log phase in yeast extract peptone dextrose (YPD), essentially as described, to maintain complex integrity (61, 63). Proteins copurifying with Pdp3 were subjected to tandem MS analysis of peptides with a Thermo LTQ-XL mass spectrometer coupled to an Eksigent nanoLC 2D system as previously described (30). Spectral counts and proteins were identified with Mascot (Matrix Science, London, UK; version 2.3.01).
Database Searching
Tandem mass spectra were extracted by Thermo ExtractMSn version 1.0.0.8. Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.3.01). Mascot was set up to search the SwissProt_57.15 database (selected for S. cerevisiae, 57.15, 6973 entries), assuming the digestion enzyme was nonspecific. Mascot was searched with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 2.0 Da. The iodoacetamide derivative of cysteine was specified in Mascot as a fixed modification. S-carbamoylmethylcysteine cyclization at the N-terminus, oxidation of methionine, pyro-carbamidomethyl formation at the N-terminus, acetylation of asparagine, and the presence of proline at the N-terminus were specified in Mascot as variable modifications.
Criteria for Protein Identification
Scaffold (version Scaffold_4.0.1, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 20.0% probability by the Peptide Prophet algorithm (64). Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm (65). Proteins that contained similar peptides, and could not be differentiated based on MS/MS analysis alone, were grouped to satisfy the principles of parsimony. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (66) with the data set identifier PXD001156 (67–69).
Protein Expression
PDP3 constructs were made with an N-terminal HIS6-pfuMBP(60–434)-FLAG tag (pET28a derivative vector obtained from the G. Bowman Laboratory, Johns Hopkins University) and/or an N-terminal Thioredoxin-HIS6-S•tag tag (pET32a vector, Millipore). Proteins were exogenously expressed in chemically competent BL21 E. coli (Invitrogen, Carlsbad, CA) after overnight induction with 1 mm IPTG at 18–20 °C in LB medium. Point mutants were made with the QuikChange Site-Directed Mutagenesis Kit (Stratagene, LaJolla, CA) and expressed as described.
Protein Purification
For pull-down assays, BL21 cells (pET28a constructs) were resuspended in purification buffer (50 mm Tris pH 7.5, 500 mm NaCl, 40 mm imidazole, 10% glycerol, 2 mm β-ME, 1 mm phenylmethanesulfonyl fluoride (PMSF), and 2 mm benzamidine, pH 8.0) and lysed by sonication (Branson). Clarified lysate was nutated with Ni-NTA agarose resin (Invitrogen) for at least 1 h at 4 °C. Resin was washed with purification buffer and protein was eluted with purification buffer containing 300 mm imidazole. Protein was flash frozen in liquid nitrogen and stored at −80 °C. For fluorescence polarization assays, BL21 cells (pET32a constructs) were resuspended in purification buffer (50 mm Tris pH 7.7, 500 mm NaCl, 10% glycerol, 5 mm dithiothreitol (DTT), 1 mm PMSF, and 2 mm benzamidine) and lysed with a microfluidizer (Watts Fluidair). Clarified lysate was run through a BioScale Mini Profinity IMAC cartridge (BioRad, Hercules, CA) using an AKTA Purifier system (GE Healthcare). The cartridge was washed with purification buffer containing 12 mm imidazole and protein was eluted with purification buffer containing 125 mm imidazole. Protein was exchanged into gel filtration buffer (50 mm Tris pH 7.5, 500 mm NaCl, 10% glycerol, and 5 mm DTT) and separated by a Superdex 200 26/60 column (GE Healthcare) using an AKTA Purifier system (GE Healthcare). Monomeric protein was flash frozen in liquid nitrogen and stored at −80 °C.
Cellular Pull-down Assays
TAP-tagged S. cerevisiae strains were grown to mid-log phase in YPD, cryogenically lysed with a mixer mill (Retsch MM301), and stored at −80 °C. Cells (1 g per pull-down condition) were homogenized (ProScientific) in 650 mm extraction buffer (650 mm NaCl, 20 mm HEPES pH 7.9, 25% glycerol, 1.5 mm MgCl2, 0.2 mm EDTA, 1 mm PMSF, 0.2% Triton X-100, 1% bovine serum albumin (BSA), and 40 mm imidazole) at a ratio of 1 ml buffer per 1 g yeast and nutated for 1 h at 4 °C. Clarified extracts were diluted to 300 mm NaCl with “no-salt” extraction buffer, mixed with Ni-NTA agarose resin (Invitrogen) (100 μl per sample precoated with Pdp3 protein), and nutated for 30 min at 4 °C. Resin was washed five times with 300 mm wash buffer (300 mm KCl, 20 mm HEPES pH 7.9, 0.2% Triton X-100, 1% BSA, and 40 mm imidazole) and one time with buffer containing 10 mm NaCl and 4 mm HEPES pH 7.9. Resin was incubated in 2X SDS-PAGE loading buffer containing 300 mm imidazole for 10 min to elute Pdp3-bound proteins. Samples were boiled for 5 min, resolved on 8% SDS-polyacrylamide gels, transferred to polyvinylidene fluoride (PVDF) membrane, and probed with antibodies recognizing the PrA (DAKO: P0450, 1/1,500) and FLAG (SIGMA: F3165, 1/1,000) tags. Immunoblots were visualized using HRP-conjugated secondary antibodies and ECL solution (GE Healthcare). Inputs represent ∼0.02–0.05% of total yeast lysate.
Peptide Synthesis for Pull-down Assays
Peptides were synthesized as previously described by the C. D. Allis Laboratory (The Rockefeller University) or the UNC Peptide Synthesis and Arraying Core Facility (70).
Peptide Pull-down Assays
Steptavidin-coupled Dynabeads (Invitrogen) (25 μl per sample) were incubated with biotinylated histone peptides (1 μg per sample) in binding buffer (20 mm HEPES pH 7.9, 150 mm NaCl, 0.5 mm PMSF, 20% glycerol, 0.2% Triton X-100, and 1% BSA) for 1 h at room temperature. Unbound peptide was washed in binding buffer and beads were incubated with purified Pdp3 proteins (20 μg per sample) for 1 h at room temperature. Beads were washed three times for 5 min each with binding buffer, and one time with buffer containing 4 mm HEPES pH 7.9, 10 mm NaCl, 0.5 mm PMSF, 20% glycerol, and 0.2% Triton X-100. Peptide-bound proteins were eluted in boiling 2X SDS-PAGE loading buffer. Samples were resolved on 15% SDS-polyacrylamide gels, transferred to PVDF membrane, and probed with antibodies recognizing the FLAG (SIGMA: F3165, 1/1,000) and streptavidin (Molecular Probes: S-911, 1/10,000) tags. Immunoblots were visualized using HRP-conjugated secondary antibodies and ECL solution (GE Healthcare). Inputs represent 0.5 μg of Pdp3 proteins.
Peptide Synthesis for Fluorescence Polarization Assays
Fluorescent peptides were synthesized using standard Fmoc-solid phase peptide chemistry on a Prelude Peptide Synthesizer (Protein Technologies). Peptides were made on a 0.05 mmol scale with four equivalents of amino acids using Rink Amide AM resin (Novabiochem) to generate peptide amides. 5-Carboxyfluorescein (5-FAM) (Chempep) was coupled to the peptides using Lys(ivDde) (Chempep). The ivDde protecting group was orthogonally removed using standard deprotection procedures. The resulting peptides were typically cleaved using TFA/thioanisole/water/triisopropylsilane/phenol (87.5:2.5:2.5:2.5:5 v/v) and purified with a Varian Dynamax Microsorb C18 preparative column (Agilent). Purified peptide was lyophilized and its mass was confirmed with an Applied Biosystems Voyager DE-STR MALDI-TOF mass spectrometer (Invitrogen). Of note, to obtain the H3K79me3 5-FAM-linked peptide, it was necessary to install Fmoc-(FmocHmb)Phe-OH (Novabiochem) at F84.
Fluorescence Polarization Assays
Binding assays were performed as described in (71) with the following modifications. Full-length purified wild-type Pdp3, F18A, and W21A proteins were exchanged into FP buffer (50 mm Tris pH 7.5, 150 mm NaCl, and 5 mm DTT) and concentrated to ∼260–430 μm using Amicon Centrifugal Filter Units MWCO 30,000 (Millipore). Binding assays were performed in a 60 μl volume with 96 well half area black flat bottom nonbinding surface plates (Corning). Protein was serially diluted with FP Buffer in twofold increments and incubated with 120 nm of 5-FAM-labeled histone peptides. Following a 30 min equilibration period, fluorescence was detected at room temperature with an Infinite M1000 plate reader (Tecan, Durham, NC) using a 470 nm excitation filter and 527 ± 20 nm emission filter. Binding curves were analyzed by the total binding equation Y = Bmax* X/(Kd + X) + NS * X + Background, where Bmax = 1 and nonspecific (NS) and background variables are constrained to be equal between peptides, using Prism 5.0 (GraphPad Inc., San Diego, CA). Error bars represent the S.D. of a representative experiment (n = 2) performed in triplicate.
Yeast Strains and Cell Spotting Assays
S. cerevisiae strains were created using heterologous gene replacement (72). Strains were grown on YPD or synthetic complete (SC) media as indicated. BUR1 delete shuffle strains were grown on SC-Ura plates to maintain the wild-type BUR1 plasmid prior to plating on media containing the drug 5-Fluoroorotic acid (5-FOA) (73). For cell spotting assays, either 0.5 or 2 ODs of cells were fivefold serially diluted, spotted onto the appropriate plates, and incubated at 30 °C for 2–3 days as indicated.
Immunoblot Analyses for Chromatin Association Assay
5 ODs of cells were isolated and lysed by bead beating in SUTEB buffer (1% SDS, 8 m urea, 10 mm Tris pH 6.8, 10 mm EDTA, and 0.01% bromphenol blue) for 3 min. The lysates were then boiled for 10 min and the supernatant was clarified and isolated. 5 μl of extracts were resolved on 15% SDS-polyacrylamide gels and then transferred to PVDF membrane for 90 min at 45 mA. Blots were dried in methanol, washed in TBST (Tris buffered saline with 0.05% Tween 20), and then incubated overnight at 4 °C with the indicated antibodies: Protein A (Sigma Aldrich), G6PDH (Sigma Aldrich), H3 (in house), H4 (Abcam: ab10158), H3K14ac (Millipore: 07–353), H3K36me3 (Abcam: ab9050), and H3K4me3 (Active Motif: 39159). Immunoblots were visualized using HRP-conjugated secondary antibodies and ECL Prime solution (GE Healthcare).
Chromatin Association Assay
Strains were grown overnight in YPD to confluence. Each strain was diluted to 0.1 ODs in 50 mLs of YPD and grown to an OD of ∼0.8–1. 40–50 ODs of cells were then isolated, washed with water and SB buffer (1 m Sorbitol, 20 mm Tris.Cl pH 7.4), and frozen at −80 °C until ready for isolation. Cells were resuspended in 1 ml of PSB buffer (20 mm Tris.Cl pH 7.4, 2 mm EDTA, 100 mm NaCl, and 10 mm β-ME), followed by addition of 1 ml of SB buffer. Cells were then spheroplasted with Zymolyase (Seikagaku Biobusiness, Rockville, MD) for 30 min at room temperature. Spheroplasts were spun down at 2000 × g and washed twice with LB (0.4 m Sorbitol, 150 mm potassium acetate, 2 mm magnesium acetate, and 20 mm PIPES pH 6.8). TritonX-100 was added to LB (final concentration of 1%). Cells were lysed for 15 min on ice. Chromatin was isolated by spinning down lysates at 5000 × g for 15 min. The supernatant was collected and saved as the “soluble” fraction. The chromatin was washed once more with LB and then resuspended into an equal volume to that of the “soluble” fraction. Volume equivalents were resolved on 15% SDS-polyacrylamide gels and subjected to immunoblot analysis.
Relative Transcript Levels
Total RNA was prepared from TAP-tagged YNG1, PDP3, yng1Δ, and pdp3Δ S. cerevisiae strains via Trizol (Invitrogen) and digested with Turbo DNase (Invitrogen). cDNA was synthesized with the Superscript III First Strand Synthesis System (Invitrogen). Differences in transcript levels of control genes and NuA3 target genes were measured by qPCR using Power SYBR Green PCR Master Mix (Invitrogen) and a Real Time PCR system (Applied Biosystems, Foster City, CA, v2.1). The relative transcript levels from mutant strains as compared with wildtype were calculated using the relative standard curve method. Error bars represent the S.E. of a representative experiment (n = 4) done in triplicate. Statistical significance was determined by an unpaired two-tailed t test.
RESULTS
Pdp3 Interacts with Members of the NuA3 HAT Complex
Eaf6, Nto1, Sas3, Taf14, and Yng1 were previously identified as stable members of the S. cerevisiae NuA3 HAT complex (19, 21, 24, 25). Pdp3 (Ylr455w), an uncharacterized PWWP domain-containing protein (59–61), also showed modest association with the complex (19), as determined through isotopic differentiation of interactions as random or targeted (i-DIRT) technology (63). Interestingly, BRPF1, a component of human MOZ/MORF HAT complexes and homolog of yeast Nto1, contains a PWWP domain that is absent in Nto1 (23, 55, 56). We reasoned Pdp3 might function similarly to the PWWP domain of BRPF1 in the NuA3 complex. Genomically TAP-tagged Pdp3 was isolated from S. cerevisiae using a method that preserves complex integrity (61, 63). Proteins copurifying with Pdp3-TAP were resolved by SDS-PAGE and subjected to MS/MS analysis. Using a 95% protein confidence threshold, 96 proteins were identified by MS/MS (Fig. 1A, supplemental Tables S5 and S6). A functional classification of the proteins identified revealed 12 proteins with functions related to transcription (Fig. 1A, supplemental Table S5). The remaining 84 proteins were “nonspecific” copurifying proteins (e.g. ribosomal, metabolic, nucleolar, and heat shock) typically observed in these types of large-scale affinity enrichments (supplemental Table S6) (30, 63, 74, 75). Specific proteins copurifying with Pdp3-TAP include all stable members of NuA3 (Eaf6, Nto1, Sas3, Taf14, and Yng1) (19, 21, 24, 25), core histones (H3, H4, H2A, and H2B), and components of RNAPII (Rpb2, Rpb4). These copurifications suggest that Pdp3 is a component of the NuA3 complex and that Pdp3 is involved transcriptional regulation.
Fig. 1.
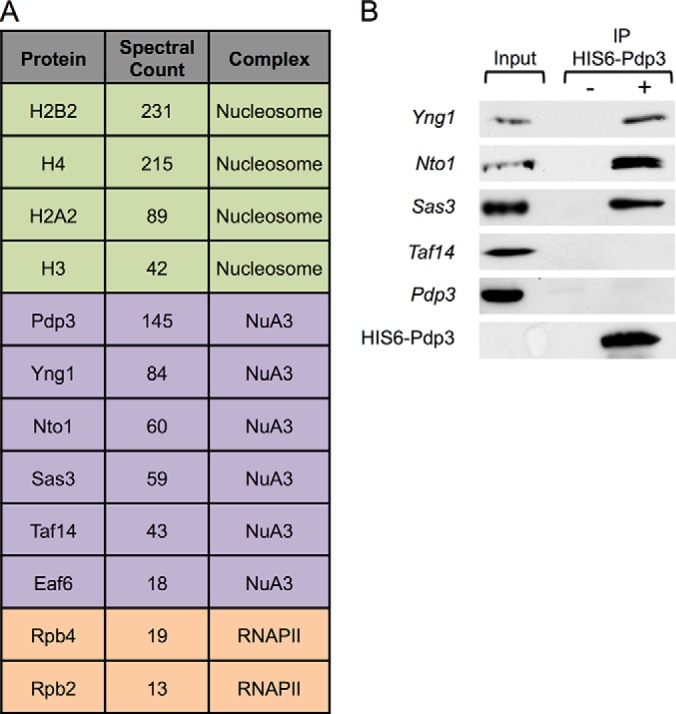
Pdp3/YLR455W is associated with the NuA3 complex, chromatin, and RNAPII. A, Proteins copurifying with Pdp3-TAP bait that are involved in transcriptional regulation. Bulk MS/MS data are reported for proteins affinity purified by Pdp3 after searching with Mascot. B, Whole cell extracts (WCEs) from the indicated TAP-tagged yeast strains were immunoprecipitated with HIS6-FLAG-Pdp3 treated (+) and untreated (-) resin. WCEs (inputs) and immunoprecipitated samples (IPs) were resolved by SDS-PAGE. The presence of NuA3 complex members was monitored by western blotting.
To confirm that Pdp3 is a member of the NuA3 complex, we tested the ability of full-length recombinant Pdp3 to pull down NuA3 proteins from cellular extracts (Fig. 1B). HIS6-FLAG-tagged Pdp3 was purified from E. coli and incubated with lysates from S. cerevisiae endogenously expressing TAP-tagged NuA3 subunits. Immunoprecipitated samples were resolved by SDS-PAGE and visualized with an antibody against the TAP tag. HIS6-FLAG-Pdp3 interacts with Yng1, Nto1, and Sas3 (Fig. 1B), supporting the MS/MS results (Fig. 1A, supplemental Table S5). Neither Taf14 nor Eaf6 were detected in recombinant Pdp3 pull-downs, which may indicate that these proteins do not interact with Pdp3 (Fig. 1B, data not shown); however, we cannot exclude the possibility that the TAP tag disrupts native interactions. Also, because Taf14 is a member of multiple chromatin-associated complexes (76), the quantity of soluble Taf14 available to be pulled down in this assay may be insufficient to detect.
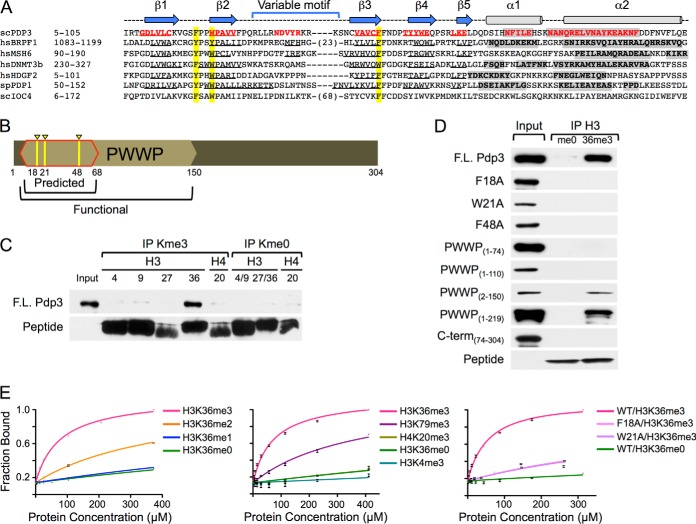
Pdp3 Specifically Engages H3K36me3 Through a Conserved PWWP Domain
H3K36me3 localizes to the body of actively transcribed genes, and is associated with transcription elongation in yeast and humans (13, 34, 35, 37, 40). BRPF1, a human homolog of yeast Nto1 (23), engages H3K36me3 through a PWWP domain (supplemental Fig. S1) (55, 56) that is absent in Nto1. Interestingly, structural modeling predicts that Pdp3 contains a PWWP domain with aromatic residues critical for methyl-lysine binding, as well as beta strands and alpha helices conserved in other PWWP-domain containing proteins that bind methyl-lysine (Figs. 2A, 2B, and supplemental Fig. S1). Because of the structural conservation with the BRPF1 PWWP domain, and the homology between the human and yeast HAT complexes, we reasoned that Pdp3 might bind H3K36me3. To test for this possibility, we interrogated the ability of full-length recombinant Pdp3 to bind to trimethylated histone peptides (Fig. 2C). Biotinylated peptides were immobilized on streptavidin resin and incubated with HIS6-FLAG-Pdp3. Immunoprecipitated samples were resolved by SDS-PAGE and visualized with antibodies against FLAG and streptavidin. Pdp3 preferentially engages H3K36me3 compared with all other trimethylated lysine residues tested (Fig. 2C).
Fig. 2.
NuA3 specifically interacts with H3K36me3 through the PWWP domain of Pdp3. A, Clustal W alignment of PWWP domain-containing proteins. Beta sheets (arrows/underlined sequence) and alpha helices (cylinders/gray shading) are annotated. Aromatic cage residues are highlighted in yellow. See also Supplemental Fig. S1. B, Schematic representation of the Pdp3 protein. The predicted (red outline) and functional (tan hexagon) PWWP domains are annotated. Aromatic cage residues F18, W21, and F48 are highlighted in yellow. C, Peptide pull-down assays were performed with full-length HIS6-FLAG-Pdp3 and biotinylated histone peptides. Purified protein (input) and immunoprecipitated samples (IPs) were resolved by SDS-PAGE. Binding was monitored by western blotting. D, Peptide pull-down assays were performed with full-length HIS6-FLAG-Pdp3, mutants F18A, W21A, and F48A, truncations PWWP(1–74), PWWP(1–110), PWWP(2–150), PWWP(1–219), and C-term(74–304), and biotinylated histone peptides. Purified proteins (inputs) and immunoprecipitated samples (me0 and 36me3) were resolved by SDS-PAGE. Binding was monitored by western blotting. E, Fluorescence polarization assays were used to measure binding affinities of full-length S·tag-Pdp3 and mutants F18A and W21A to the indicated 5-FAM-labeled histone peptides. Error bars represent the S.D. of a representative experiment (n = 2) performed in triplicate.
A Conserved Aromatic Cage within the Pdp3 PWWP Domain is Required for Binding H3K36me3
Like other Royal Family reader modules, PWWP domains employ an aromatic cage to interact with specific trimethylated histones (supplemental Fig. S1) (7, 20, 56, 77). For example, BRPF1 requires aromatic residues Y1096, Y1099, and F1147 to coordinate the trimethylammonium group of H3K36me3 (supplemental Fig. S1) (56). These aromatic residues are conserved in the PWWP domain of Pdp3 at positions F18, W21, and F48 (Figs. 2A, 2B, and supplemental Fig. 1). To determine whether Pdp3 uses an aromatic cage to bind H3K36me3, we tested the ability of H3K36me3 peptide to pull down full-length recombinant Pdp3 mutated at residues F18, W21, or F48 (Fig. 2D). All three mutations independently abolish the interaction between Pdp3 and H3K36me3 peptide (Fig. 2D), suggesting Pdp3 requires a conserved aromatic cage to bind chromatin.
The Pdp3 PWWP Domain is Necessary and Sufficient for Binding H3K36me3
We next wanted to determine whether the predicted PWWP domain of Pdp3 was sufficient for H3K36me3 binding. Uniprot defined the Pdp3 PWWP domain as the amino acid residues spanning 7–68 (Fig. 2B) (78). However, we found that truncated Pdp3 (PWWP1–74) is unable to bind H3K36me3 peptide (Fig. 2D), suggesting residues beyond the predicted PWWP domain are necessary for Pdp3 function. From the crystal structure of BRPF1 (56), we observed that two C-terminal alpha-helices support the aromatic cage and may be critical for engagement of H3K36me3 (supplemental Fig. S1). These alpha-helices are conserved in the structural model of Pdp3 (supplemental Fig. S1) (79), yet extend beyond the predicted PWWP domain (Figs. 2A, 2B). We created Pdp3 constructs to include increasing segments of the (modeled) C-terminal alpha-helices and tested the ability of H3K36me3 peptide to pull down PWWP1–110, PWWP2–150, and PWWP1–219. As shown in Fig. 2D, PWWP2–150 restores binding to H3K36me3 and thus represents the functional PWWP domain of Pdp3 (Fig. 2B). These data suggest Pdp3, and likely other PWWP domain proteins, require extended alpha-helical regions for aromatic cage stability and function.
Biophysical Characterization of the Interaction between Pdp3 and H3K36me3
To biophysically quantitate the specificity of the interaction between Pdp3 and H3K36me3, we performed fluorescence polarization assays using full-length recombinant S·tag-Pdp3 and 5-FAM-labeled histone peptides (Fig. 2E, Table I). As expected from our pull-down assays, Pdp3 favors binding to the H3K36me3 peptide over other known targets of PWWP domain proteins, such as H3K79me3 and H4K20me3, with a Kd of 69.51 ± 3.7 μm (Fig. 2E, Table I) (54, 80). Notably, this value is one of the lowest reported dissociation constants for PWWP domain proteins that bind histones (supplemental Table S7) (54, 56, 57, 80, 81). We also generated Pdp3 mutants predicted to disrupt the aromatic cage and hence interaction with H3K36me3 (F18A and W21A). These mutants reduced H3K36me3 peptide binding ∼10-fold compared with wild-type Pdp3 (Fig. 2E, Table I). Pdp3 also showed weak binding to H3K36me2 peptide with a Kd of ∼414 μm (Fig. 2E, Table I). Importantly, the ∼sixfold increase in specificity of Pdp3 for H3K36me3, over H3K36me2, suggests Pdp3 has a distinct function from the Rpd3S HDAC complex, which preferentially engages H3K36me2 (18, 41, 42, 45, 82).
Table I. Dissociation constants obtained from fluorescence polarization assays. The total binding equation was used to calculate dissociation constants with Graphpad Prism software. Kd values were tabulated from two independent experiments (performed in triplicate) and averaged. Error represents the S.D. of two independent experiments.
| Pdp3 | Peptide | Kd(μm) |
|---|---|---|
| WT | H3K36me3 | 69.5+/−3.7 |
| WT | H3K36me2 | 414+/−23 |
| WT | H3K36me1 | ≫1000 |
| WT | H3K36me0 | ≫1000 |
| WT | H3K4me3 | ≫1000 |
| WT | H3K79me3 | 434+/−49 |
| WT | H4K20me3 | ≫1000 |
| F18A | H3K36me3 | 667+/−68 |
| W21A | H3K36me3 | 703+/−73 |
Pdp3 Requires H3K36me3 to Bind Chromatin
To further understand the biological role of Pdp3, we next determined whether Pdp3 binds H3K36me3 in vivo. We deleted the H3K36me3 methyltransferase, SET2, in the PDP3-TAP background and used a chromatin association assay to separate soluble proteins from those that bind to chromatin. For added controls, we also deleted the H3K4me3 methyltransferase, SET1, and the NuA3 H3K4me3 binding protein, YNG1. Strikingly, in the absence of Set2, and thus H3K36me3, we observe an almost complete ablation of Pdp3 binding to chromatin (Fig. 3A), indicating H3K36me3 is critical for Pdp3 localization in vivo. In contrast, Pdp3 remains bound to chromatin in the absence of Set1 (H3K4me3) and Yng1 (Fig. 3A). The observation that Pdp3 engages chromatin independently of both H3K4me3 and Yng1 suggests that Pdp3 targets NuA3 to active coding regions via an interaction with H3K36me3 (13, 14, 50). Interestingly, in the absence of Yng1, we note a slight decrease in the amount of chromatin-bound Pdp3 (Fig. 3A), as well as lower levels of cellular Pdp3 (supplemental Fig. S2A). Together, these data suggest Pdp3 is moderately unstable in yng1Δ cells, perhaps because of an inability to be incorporated into NuA3.
Fig. 3.
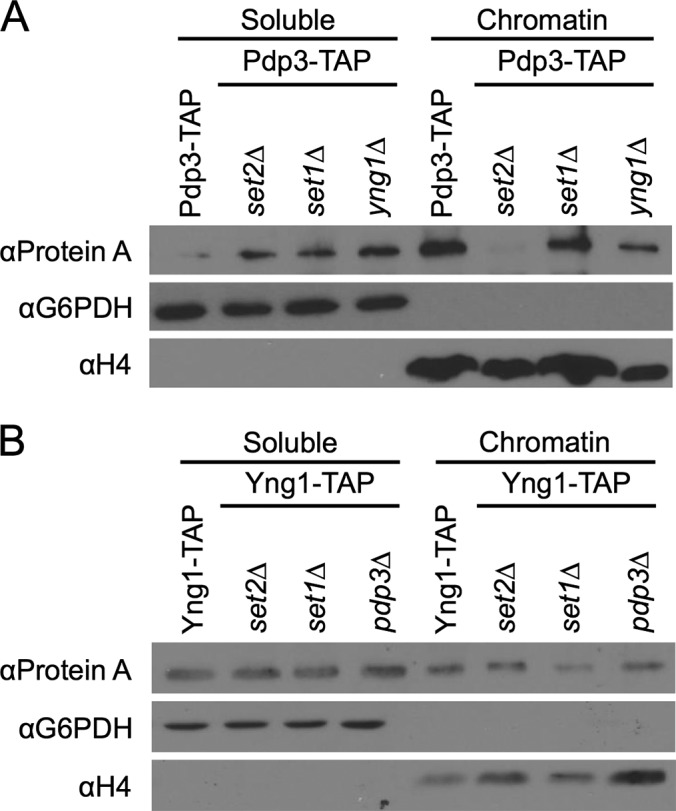
Pdp3, but not Yng1, requires H3K36me3 for chromatin association in vivo. A, TAP-tagged PDP3 yeast strains and B, TAP-tagged YNG1 yeast strains were biochemically fractionated into chromatin-associated proteins and soluble proteins. Fractions were probed for the presence of Pdp3 and Yng1, respectively. G6PDH and H4 serve as both loading and fractionation controls.
We next deleted SET1, SET2, and PDP3 in the YNG1-TAP background. In contrast to Pdp3, Yng1 remains bound to chromatin in the absence of either Set1 or Set2 (Fig. 3B). Furthermore, Yng1 levels are not significantly altered in pdp3Δ cells (supplemental Fig. S2B). These results are consistent with past studies (26, 27, 83), and further suggest that while Yng1 can engage chromatin through both methyl-dependent and -independent means, the interaction between Pdp3 and chromatin requires H3K36me3.
Pdp3 is Required for NuA3-regulated Transcription
Mutation of the Yng1 PHD finger results in genome-wide mislocalization of NuA3 and decreased transcription of NuA3-regulated loci (19). To further assess the function of Pdp3 in vivo, we performed RT-qPCR with wild-type, pdp3Δ, and yng1Δ cells, and calculated the relative transcript levels of NuA3-regulated loci (Fig. 4A). As expected, yng1Δ cells showed a slight but significant decrease in the relative transcript levels of NuA3 target genes, indicating a positive role for Yng1 in NuA3 activity at these genes (Fig. 4A) (19, 84, 85). Interestingly, pdp3Δ cells also showed a reduced level of NuA3 target gene transcript (Fig. 4A). These data indicate that NuA3 binding to both H3K4me3, via Yng1, and H3K36me3, via Pdp3, is required for proper NuA3-directed transcription.
Fig. 4.
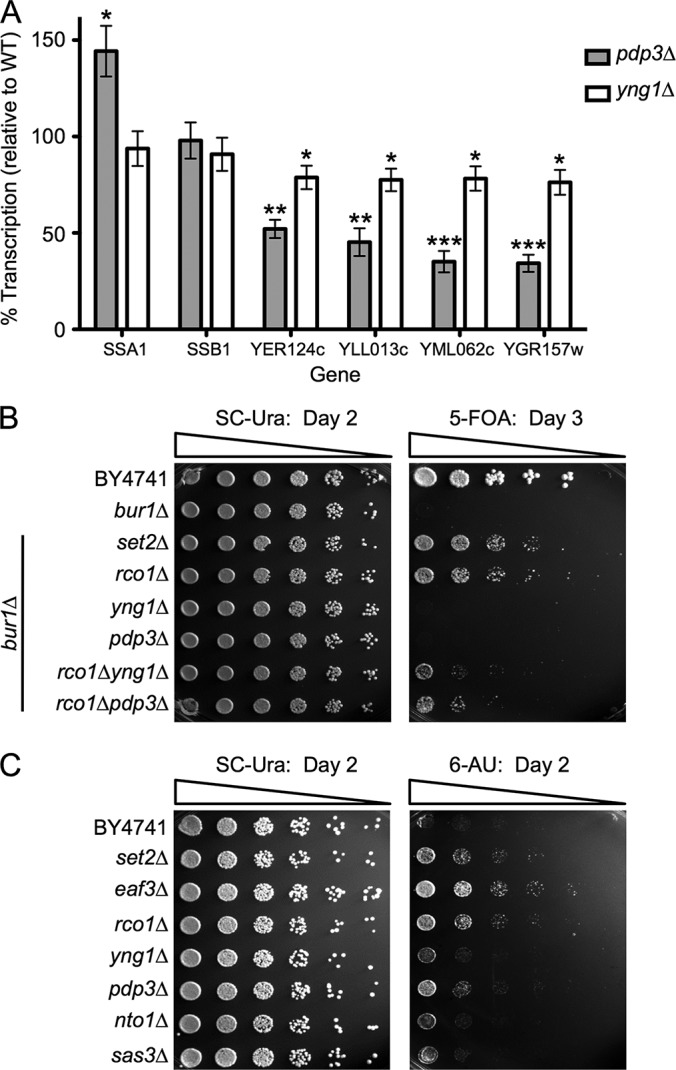
NuA3 function in transcription requires both Yng1 and Pdp3. A, RT-qPCR analysis of WT, pdp3Δ, and yng1Δ cells to determine the relative transcript levels of NuA3-target genes and nontarget genes (SSA1 and SSB1). Transcript levels were normalized to Actin expression. Error bars represent the S.E. of a representative experiment (n = 4) performed in triplicate. Asterisks indicate statistical significance as determined by an unpaired two-tailed t test. * = p < .05, ** = p < .01, *** = p < .001. B, 2 ODs of the indicated yeast strains were 5-fold serially diluted on SC-Ura (left) or SC-Ura + 5-FOA (right) plates and grown at 30 °C for 2 or 3 days, respectively. C, 0.5 ODs of the indicated yeast strains were 5-fold serially diluted onto SC-Ura (left) or SC-Ura + 150 μg/ml 6-AU (right) plates and grown at 30 °C for 2 days.
PDP3 and NuA3 have Genetic Links to SET2 and Promote Transcription Elongation
Because Pdp3 binds H3K36me3 and is required for proper transcription in vivo, we predicted that Pdp3 would show genetic interactions with genes in the Set2/H3K36me pathway, and might influence transcription elongation. One genetic assay that has been used to measure the influence of Set2/H3K36 methylation on transcription elongation is the BUR1 bypass assay. Bur1, a cyclin-dependent kinase, acts as a positive regulator of transcription through the phosphorylation of several components of elongating RNAPII, including the CTD of RNAPII and the Spt5 C-terminal repeat domain (86–88). Under normal conditions, deletion of BUR1 is lethal; however, deletion of SET2 or RCO1, a unique member of Rpd3S, bypasses this lethality (34, 44). To explore whether Pdp3 and/or Yng1 contribute to transcription elongation, we examined deletions of SET2, RCO1, PDP3, and YNG1 in the bur1Δ background. Although the bur1Δset2Δ and bur1Δrco1Δ strains bypass the lethality of BUR1 deletion as expected, neither bur1Δpdp3Δ nor bur1Δyng1Δ cells are viable (Fig. 4B). This suggests that either the bypass phenotype is too weak to observe, or Pdp3 and Yng1 function as positive elongation factors, and thus would not display a positive growth phenotype in this assay. To distinguish between these two possibilities, we created triple mutant strains with PDP3 or YNG1 deleted in the bur1Δrco1Δ background. Because this background shows a bypass phenotype, we could now observe positive or negative growth changes resulting from the absence of Pdp3 or Yng1. Significantly, the triple mutant strains both show a decrease in growth as compared with the bur1Δrco1Δ background (Fig. 4B). These data suggest that Pdp3 and Yng1 positively regulate transcription elongation, and are consistent with the idea that NuA3 functions both at promoters and at gene bodies.
To further connect NuA3 activity to the transcription elongation pathway, we asked whether the absence of NuA3 complex members resulted in sensitivity or resistance to the transcription elongation inhibitor 6-azauracil (6-AU). Strains containing a deletion of SET2, or downstream effectors, show a 6-AU resistance phenotype (34, 44). As expected, strains lacking Set2, Rco1, and Eaf3, a member of the Rpd3S HDAC and NuA4 HAT complexes, display resistance to 6-AU as compared with wild-type (Fig. 4C). Of all tested NuA3 members, only pdp3Δ cells show resistance to 6-AU, a result consistent with the idea that Pdp3 functions in the Set2/H3K36 methylation pathway (Fig. 4C).
PDP3 is not Synthetically Lethal with GCN5
It has previously been determined that NuA3 complex members, Sas3 and Yng1, display synthetic lethality with the HAT, Gcn5 (89). These data indicate Gcn5 and Sas3 likely collaborate to promote gene activation and/or transcription elongation, as Gcn5 is found in both the promoters and transcribed regions of genes (90, 91). Given the physical interaction and functional overlap between NuA3 and Pdp3, we wanted to determine if Pdp3 is also synthetically lethal with Gcn5. We created GCN5 shuffle strains that allowed us to delete YNG1 or PDP3 in a gcn5Δ background. Upon shuffling out the wild-type GCN5 plasmid, only gcn5Δyng1Δ cells display synthetic lethality (supplemental Fig. S3). Surprisingly, gcn5Δpdp3Δ cells show no deleterious phenotype (supplemental Fig. S3), indicating PDP3 does not genetically interact with GCN5. These results suggest that Pdp3-containg NuA3 participates in an alternate transcriptional pathway that is distinct from the role of Gcn5 at promoters and in gene bodies.
DISCUSSION
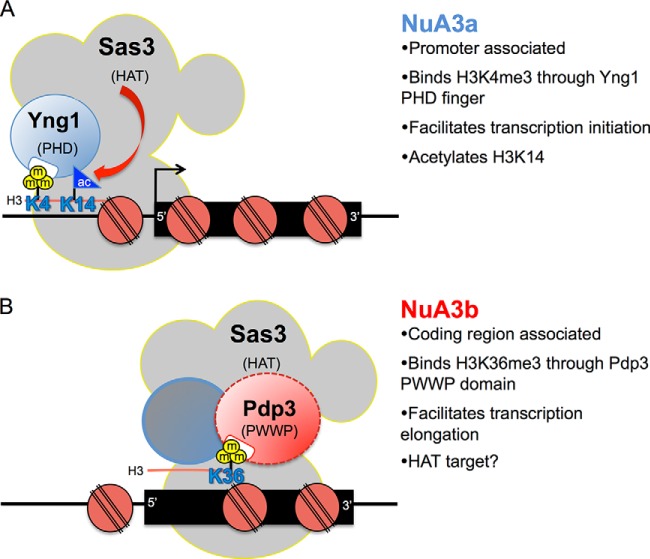
In this study, we characterize a unique form of the NuA3 HAT complex that contains the PWWP domain protein, Pdp3. Using mass spectrometric, biochemical, and genetic approaches, our collective findings suggest that NuA3 exists in two functionally distinct forms: NuA3a and NuA3b (Fig. 5). This nomenclature has previously been used to distinguish different variations of related protein complexes. For example, although the Isw1a and Isw1b ATP-dependent chromatin remodeling complexes both have the same catalytic protein, Isw1, Isw1b contains a unique PWWP domain protein, Ioc4, required for targeting remodeling activity to H3K36me3 enriched nucleosomes (51, 52). For NuA3 complexes, we hypothesize that promoter-associated NuA3a contains the proteins Eaf6, Nto1, Sas3, Taf14, and Yng1, and specifically associates with H3K4me3 using the PHD finger of Yng1 (Fig. 5A) (19, 24–27). NuA3a subsequently acetylates H3K14 through the HAT domain of Sas3, initiating transcription at a subset of genes (Fig. 5A) (19, 24, 25). In contrast, NuA3b, via the H3K36me3-binding PWWP domain protein Pdp3, links to H3K36 methylation-associated transcription elongation in gene bodies (Fig. 5B). Accordingly, our previous mass spectrometry (i-DIRT) data support the model that NuA3b is compositionally distinct from NuA3a (19). However, we cannot exclude the alternate possibility that Pdp3 is a member of both NuA3a and NuA3b. To this end, human H3K36me3-binding BRPF proteins resemble a fusion of yeast Nto1 and Pdp3, as if these separate NuA3 proteins are physically linked within the human MOZ/MORF complexes (92). This brings up the intriguing possibility that the PWWP domain of Pdp3 (and potentially BRPFs) has a regulated capacity to bind H3K36me3, becoming active or inactive in a context dependent manner.
Fig. 5.
The NuA3 HAT complex has two functionally distinct forms that participate in transcription. A, Model of the NuA3a HAT complex. Yng1 binds to H3K4me3 through its PHD finger, thereby recruiting NuA3a to the promoter regions of actively transcribed genes. Sas3 then acetylates H3K14, leading to transcription initiation at a subset of genes (19, 24–27). B, Model of the NuA3b HAT complex. NuA3b contains a unique member, Pdp3. Pdp3 binds to H3K36me3 through its PWWP domain, thereby recruiting NuA3b to the coding regions of actively transcribed genes. Although the function of NuA3b is not fully defined, we speculate that Sas3 may acetylate histones or nonhistone proteins to facilitate transcription elongation.
Our data clearly place Pdp3 and NuA3b within the Set2-dependent transcriptional elongation pathway. Pdp3 binds to H3K36me3, a Set2-catalyzed histone PTM found almost exclusively within transcriptionally active gene bodies (13, 33–35, 37, 40); and deletion of the PDP3 gene results in growth defects when combined with transcription elongation mutants. The interaction between Pdp3 and Rpb4, as determined from our mass spectrometry studies, further supports a role for NuA3b in transcriptional elongation, given the links between Rpb4 and actively transcribing RNAPII in S. pombe (93). Although our data suggest that NuA3b positively regulates transcription elongation, the exact function of NuA3b remains unclear. Because NuA3-directed transcription is decreased in pdp3Δ cells, NuA3b could participate in acetylation-dependent nucleosome eviction within the ORF, similar to the proposed function of Gcn5 at coding regions (90, 94, 95). Although both Gcn5 and Sas3 favor acetylation of H3K14 (19, 24, 25, 96–99), unlike NuA3a, NuA3b is not synthetically lethal with deletion of GCN5, suggesting Sas3, within the context of NuA3b, has a distinct function from Gcn5 (perhaps acetylating other factors in the transcription elongation complex or an alternate lysine(s) in histones) (Fig. 5B). This result, and the finding that HBO1 (a related human MYST family HAT) can switch between H4 and H3 acetylation depending on its association with JADE1/2/3 or BRPF1, supports the idea that NuA3b may target non-H3K14 substrates (100). It is also possible that Gcn5-mediated H3 acetylation has a distinct function from NuA3b acetylation at H3K14 in ORFs. Future work is needed to determine the precise mechanism of NuA3b function in the transcription elongation process.
A recurring question in chromatin biology is how the addition or removal of methylation groups on specific lysine residues can alter functional properties of the associated chromatin. In yeast, Set2 catalyzes all three forms of H3K36 methylation (33). To date, H3K36me1 is suggested to function in DNA replication (101), and although H3K36me2 and me3 are both linked to transcriptional elongation, H3K36me2 is essential for Rpd3S recruitment (18, 41–47), whereas H3K36me3 is implicated in nucleosomal positioning via Isw1b and repression of trans-histone exchange (50–52). Interestingly, unlike H3K36me2, the establishment of H3K36me3 specifically requires Set2 association with the CTD of RNAPII, and H3K36me3 is positively correlated with the rate of transcription (13). Consistent with H3K36me3 links to highly transcribed genes, we speculate that H3K36me3, via association with NuA3b, may have an alternate function on NuA3-regulated genes that is distinct from former pathways determined for H3K36 methylation. The finding that the HDAC Rpd3S is linked to H3K36me2, whereas the HAT complex NuA3b is linked to CTD-dependent H3K36me3, suggests that differential methylation of H3K36 may act as a “chromatin switch” to regulate overall levels of transcription. For example, on lowly transcribed genes, the H3K36me2 state may predominate and recruit Rpd3S to maintain a more repressed chromatin environment suitable for low-level transcription. Conversely, on highly transcribed genes, H3K36me3 may predominate and recruit NuA3b to facilitate nucleosome disruption for RNAPII elongation (on genes regulated by NuA3). Alternatively, both H3K36me2 and me3 may be coordinating HDAC and HAT activities together, within the same coding region, to properly modulate the progression of RNAPII during transcriptional elongation. Although speculative, these intriguing models await testing.
Finally, NuA3 joins a growing list of HAT complexes that can localize to both promoters and gene bodies by modulating complex components (90, 94, 102–104). Whether NuA3b engages a dual H3K4me/H3K36me signature, through the combinatorial action of the PHD finger of Yng1 and the PWWP domain of Pdp3, to fine-tune transcription initiation and elongation pathways, remains an important question for future studies. It is worth noting that mutations of NuA3 human homologs (23, 56) are associated with oral squamous cell carcinoma (105) and acute myeloid leukemia (106). Given that HATs and many other chromatin-bound protein complexes are gaining favor as pharmaceutical targets (107–110), future studies into the PWWP domain of Pdp3 and NuA3b may elucidate how chromatin effectors contribute to human disease, thereby opening up new targets for drug discovery.
Supplementary Material
Acknowledgments
We thank the UAMS Proteomics Facility and the Johns Hopkins Proteomics Facility for mass spectrometric support, the PRIDE Team for assistance in dissemination of the mass spectrometry data, and the following laboratories for their assistance: J. Sohn, fluorescence polarization assays; K.L. Reddy, real time PCR; C. Wolberger, protein expression; D.J. Leahy, protein purification; P.A. Cole and D. Meyers, peptide purification; M.H. Kuo, GCN5 plasmids; M. Keogh and S. Buratowski, BUR1 shuffle strains; C.D. Allis and UNC Peptide Synthesis and Arraying Core Facility, peptides; members of the Taverna laboratory, including A. Raman for reagent preparation, R. Papazyan and K. Stephens for technical assistance, and A.M. Cieniewicz for critical reading of the manuscript, as well as members of the Strahl laboratory, including S. Rothbart for reagent preparation.
Footnotes
Author contributions: T.M.G., S.L.M., A.J.T., B.D.S., and S.D.T. designed research; T.M.G., S.L.M., and S.D.B. performed research; T.M.G., S.L.M., J.A.C., B.C.D., and H.W. contributed new reagents or analytic tools; T.M.G., S.L.M., S.D.B., H.W., A.J.T., B.D.S., and S.D.T. analyzed data; T.M.G., S.L.M., A.J.T., B.D.S., and S.D.T. wrote the paper.
* This work was supported by National Institutes of Health grants R01DA025755 (SDT & AJT), R01GM106024 (SDT and AJT), R01GM68088 (BDS), U01GM094588, U54 RR020839, P30GM103450, P20GM103429, and UL1TR000039, and National Science Foundation grants 90046096 (TMG), 0953430 (HW), 1330320 (BDS).
 This article contains Supplemental Figs. S1 to S3 and Tables S1 to S7.
This article contains Supplemental Figs. S1 to S3 and Tables S1 to S7.
Contributions: T.M.G. and S.L.M. designed and performed research, analyzed data, and wrote the manuscript; S.D.B. performed research and analyzed data; J.A.C. and B.C.D. designed and contributed materials; H.W. contributed materials and analyzed data; A.J.T., B.D.S. and S.D.T. designed research, analyzed data, and wrote the manuscript.
1 The abbreviations used are:
- RNAPII
- RNA polymerase II
- PTM
- Post-translational modification
- HAT
- Histone acetyltransferase
- HDAC
- Histone deacetylase
- HMT
- Histone methyltransferase
- ac
- Acetylation
- me
- Methylation
- K
- Lysine
- CTD
- C-terminal domain
- PHD finger
- Plant homeodomain finger
- PWWP domain
- Proline-tryptophan-tryptophan-proline domain
- ORF
- Open reading frame
- MS/MS
- Tandem mass spectrometry
- LTQ
- Linear trap quadrupole
- MALDI-TOF
- Matrix-assisted laser desorption/ionization- time-of-flight
- i-DIRT
- Isotopic differentiation of interactions as random or targeted
- RT-qPCR
- Reverse transcription-quantitative polymerase chain reaction
- TAP
- Tandem affinity purification
- 5-FAM
- 5-Carboxyfluorescein
- 6-AU
- 6-Azauracil
- 5-FOA
- 5-Fluoroorotic acid.
REFERENCES
- 1. Kulaeva O. I., Gaykalova D. A., Pestov N. A., Golovastov V. V., Vassylyev D. G., Artsimovitch I., Studitsky V. M. (2009) Mechanism of chromatin remodeling and recovery during passage of RNA polymerase II. Nat. Struct. Mol. Biol. 16, 1272–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kulaeva O. I., Hsieh F. K., Studitsky V. M. (2010) RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc. Natl. Acad. Sci. U. S. A. 107, 11325–11330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Smolle M., Workman J. L., Venkatesh S. (2013) reSETting chromatin during transcription elongation. Epigenetics 8, 10–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gardner K. E., Allis C. D., Strahl B. D. (2011) Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Petesch S. J., Lis J. T. (2012) Overcoming the nucleosome barrier during transcript elongation. Trends Genet. 28, 285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Strahl B. D., Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 [DOI] [PubMed] [Google Scholar]
- 7. Taverna S. D., Li H., Ruthenburg A. J., Allis C. D., Patel D. J. (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J. S., Shilatifard A. (2007) A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat. Res. 618, 130–134 [DOI] [PubMed] [Google Scholar]
- 9. Barrera L. O., Ren B. (2006) The transcriptional regulatory code of eukaryotic cells–insights from genome-wide analysis of chromatin organization and transcription factor binding. Curr. Opin. Cell Biol. 18, 291–298 [DOI] [PubMed] [Google Scholar]
- 10. Fuchs S. M., Laribee R. N., Strahl B. D. (2009) Protein modifications in transcription elongation. Biochim. Biophys. Acta 1789, 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Heyse K. S., Weber S. E., Lipps H. J. (2009) Histone modifications are specifically relocated during gene activation and nuclear differentiation. BMC Genomics 10, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang G., Lin J. C., Wei V., Yoo C., Cheng J. C., Nguyen C. T., Weisenberger D. J., Egger G., Takai D., Gonzales F. A., Jones P. A. (2004) Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc. Natl. Acad. Sci. U. S. A. 101, 7357–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., Young R. A. (2005) Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 14. Rando O. J. (2007) Global patterns of histone modifications. Curr. Opin. Genet. Dev. 17, 94–99 [DOI] [PubMed] [Google Scholar]
- 15. Bian C., Xu C., Ruan J., Lee K. K., Burke T. L., Tempel W., Barsyte D., Li J., Wu M., Zhou B. O., Fleharty B. E., Paulson A., Allali-Hassani A., Zhou J. Q., Mer G., Grant P. A., Workman J. L., Zang J., Min J. (2011) Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30, 2829–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Choy J. S., Tobe B. T., Huh J. H., Kron S. J. (2001) Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 276, 43653–43662 [DOI] [PubMed] [Google Scholar]
- 17. Klein B. J., Lalonde M. E., Cote J., Yang X. J., Kutateladze T. G. (2013) Crosstalk between epigenetic readers regulates the MOZ/MORF HAT complexes. Epigenetics 9, 186–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li B., Gogol M., Carey M., Lee D., Seidel C., Workman J. L. (2007) Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316, 1050–1054 [DOI] [PubMed] [Google Scholar]
- 19. Taverna S. D., Ilin S., Rogers R. S., Tanny J. C., Lavender H., Li H., Baker L., Boyle J., Blair L. P., Chait B. T., Patel D. J., Aitchison J. D., Tackett A. J., Allis C. D. (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vermeulen M., Eberl H. C., Matarese F., Marks H., Denissov S., Butter F., Lee K. K., Olsen J. V., Hyman A. A., Stunnenberg H. G., Mann M. (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 [DOI] [PubMed] [Google Scholar]
- 21. Eberharter A., John S., Grant P. A., Utley R. T., Workman J. L. (1998) Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods 15, 315–321 [DOI] [PubMed] [Google Scholar]
- 22. Briggs S. D., Bryk M., Strahl B. D., Cheung W. L., Davie J. K., Dent S. Y., Winston F., Allis C. D. (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 15, 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doyon Y., Cayrou C., Ullah M., Landry A. J., Cote V., Selleck W., Lane W. S., Tan S., Yang X. J., Cote J. (2006) ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol. Cell 21, 51–64 [DOI] [PubMed] [Google Scholar]
- 24. Howe L., Kusch T., Muster N., Chaterji R., Yates J. R., 3rd, Workman J. L. (2002) Yng1p modulates the activity of Sas3p as a component of the yeast NuA3 Hhistone acetyltransferase complex. Mol. Cell. Biol. 22, 5047–5053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. John S., Howe L., Tafrov S. T., Grant P. A., Sternglanz R., Workman J. L. (2000) The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14, 1196–1208 [PMC free article] [PubMed] [Google Scholar]
- 26. Martin D. G., Baetz K., Shi X., Walter K. L., MacDonald V. E., Wlodarski M. J., Gozani O., Hieter P., Howe L. (2006) The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 26, 7871–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin D. G., Grimes D. E., Baetz K., Howe L. (2006) Methylation of histone H3 mediates the association of the NuA3 histone acetyltransferase with chromatin. Mol. Cell. Biol. 26, 3018–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishimi Y., Kikuchi A. (1991) Identification and molecular cloning of yeast homolog of nucleosome assembly protein I, which facilitates nucleosome assembly in vitro. J. Biol. Chem. 266, 7025–7029 [PubMed] [Google Scholar]
- 29. Park Y. J., Chodaparambil J. V., Bao Y., McBryant S. J., Luger K. (2005) Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 280, 1817–1825 [DOI] [PubMed] [Google Scholar]
- 30. Smart S. K., Mackintosh S. G., Edmondson R. D., Taverna S. D., Tackett A. J. (2009) Mapping the local protein interactome of the NuA3 histone acetyltransferase. Protein Sci. 18, 1987–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. VanDemark A. P., Xin H., McCullough L., Rawlins R., Bentley S., Heroux A., Stillman D. J., Hill C. P., Formosa T. (2008) Structural and functional analysis of the Spt16p N-terminal domain reveals overlapping roles of yFACT subunits. J. Biol. Chem. 283, 5058–5068 [DOI] [PubMed] [Google Scholar]
- 32. Wagner E. J., Carpenter P. B. (2012) Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 13, 115–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Strahl B. D., Grant P. A., Briggs S. D., Sun Z. W., Bone J. R., Caldwell J. A., Mollah S., Cook R. G., Shabanowitz J., Hunt D. F., Allis C. D. (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 22, 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kizer K. O., Phatnani H. P., Shibata Y., Hall H., Greenleaf A. L., Strahl B. D. (2005) A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell. Biol. 25, 3305–3316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krogan N. J., Kim M., Tong A., Golshani A., Cagney G., Canadien V., Richards D. P., Beattie B. K., Emili A., Boone C., Shilatifard A., Buratowski S., Greenblatt J. (2003) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23, 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li J., Moazed D., Gygi S. P. (2002) Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 277, 49383–49388 [DOI] [PubMed] [Google Scholar]
- 37. Schaft D., Roguev A., Kotovic K. M., Shevchenko A., Sarov M., Shevchenko A., Neugebauer K. M., Stewart A. F. (2003) The histone 3 lysine 36 methyltransferase, SET2, is involved in transcriptional elongation. Nucleic Acids Res. 31, 2475–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xiao T., Hall H., Kizer K. O., Shibata Y., Hall M. C., Borchers C. H., Strahl B. D. (2003) Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 17, 654–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li B., Howe L., Anderson S., Yates J. R., 3rd, Workman J. L. (2003) The Set2 histone methyltransferase functions through the phosphorylated carboxyl-terminal domain of RNA polymerase II. J. Biol. Chem. 278, 8897–8903 [DOI] [PubMed] [Google Scholar]
- 40. Morris S. A., Shibata Y., Noma K., Tsukamoto Y., Warren E., Temple B., Grewal S. I., Strahl B. D. (2005) Histone H3 K36 methylation is associated with transcription elongation in Schizosaccharomyces pombe. Eukaryotic Cell 4, 1446–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Carrozza M. J., Li B., Florens L., Suganuma T., Swanson S. K., Lee K. K., Shia W. J., Anderson S., Yates J., Washburn M. P., Workman J. L. (2005) Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 [DOI] [PubMed] [Google Scholar]
- 42. Huh J. W., Wu J., Lee C. H., Yun M., Gilada D., Brautigam C. A., Li B. (2012) Multivalent di-nucleosome recognition enables the Rpd3S histone deacetylase complex to tolerate decreased H3K36 methylation levels. EMBO J. 31, 3564–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Joshi A. A., Struhl K. (2005) Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol. Cell 20, 971–978 [DOI] [PubMed] [Google Scholar]
- 44. Keogh M. C., Kurdistani S. K., Morris S. A., Ahn S. H., Podolny V., Collins S. R., Schuldiner M., Chin K., Punna T., Thompson N. J., Boone C., Emili A., Weissman J. S., Hughes T. R., Strahl B. D., Grunstein M., Greenblatt J. F., Buratowski S., Krogan N. J. (2005) Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123, 593–605 [DOI] [PubMed] [Google Scholar]
- 45. Li B., Jackson J., Simon M. D., Fleharty B., Gogol M., Seidel C., Workman J. L., Shilatifard A. (2009) Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J. Biol. Chem. 284, 7970–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sun B., Hong J., Zhang P., Dong X., Shen X., Lin D., Ding J. (2008) Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J. Biol. Chem. 283, 36504–36512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu C., Cui G., Botuyan M. V., Mer G. (2008) Structural basis for the recognition of methylated histone H3K36 by the Eaf3 subunit of histone deacetylase complex Rpd3S. Structure 16, 1740–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li B., Gogol M., Carey M., Pattenden S. G., Seidel C., Workman J. L. (2007) Infrequently transcribed long genes depend on the Set2/Rpd3S pathway for accurate transcription. Genes Dev. 21, 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lickwar C. R., Rao B., Shabalin A. A., Nobel A. B., Strahl B. D., Lieb J. D. (2009) The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PloS One 4, e4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Venkatesh S., Smolle M., Li H., Gogol M. M., Saint M., Kumar S., Natarajan K., Workman J. L. (2012) Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 [DOI] [PubMed] [Google Scholar]
- 51. Maltby V. E., Martin B. J., Schulze J. M., Johnson I., Hentrich T., Sharma A., Kobor M. S., Howe L. (2012) Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol. Cell. Biol. 32, 3479–3485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smolle M., Venkatesh S., Gogol M. M., Li H., Zhang Y., Florens L., Washburn M. P., Workman J. L. (2012) Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 19, 884–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee C. H., Wu J., Li B. (2013) Chromatin remodelers fine-tune H3K36me-directed deacetylation of neighbor nucleosomes by Rpd3S. Mol. Cell 52, 255–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wu H., Zeng H., Lam R., Tempel W., Amaya M. F., Xu C., Dombrovski L., Qiu W., Wang Y., Min J. (2011) Structural and histone binding ability characterizations of human PWWP domains. PloS One 6, e18919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Laue K., Daujat S., Crump J. G., Plaster N., Roehl H. H., Tubingen Screen C., Kimmel C. B., Schneider R., Hammerschmidt M. (2008) The multidomain protein Brpf1 binds histones and is required for Hox gene expression and segmental identity. Development 135, 1935–1946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vezzoli A., Bonadies N., Allen M. D., Freund S. M., Santiveri C. M., Kvinlaug B. T., Huntly B. J., Gottgens B., Bycroft M. (2010) Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat. Struct. Mol. Biol. 17, 617–619 [DOI] [PubMed] [Google Scholar]
- 57. Dhayalan A., Rajavelu A., Rathert P., Tamas R., Jurkowska R. Z., Ragozin S., Jeltsch A. (2010) The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 285, 26114–26120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pradeepa M. M., Sutherland H. G., Ule J., Grimes G. R., Bickmore W. A. (2012) Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 8, e1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrin-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 60. Maurer-Stroh S., Dickens N. J., Hughes-Davies L., Kouzarides T., Eisenhaber F., Ponting C. P. (2003) The Tudor domain ‘Royal Family’: Tudor, plant Agenet, Chromo, PWWP, and MBT domains. Trends Biochem.l Sci. 28, 69–74 [DOI] [PubMed] [Google Scholar]
- 61. Tackett A. J., Dilworth D. J., Davey M. J., O'Donnell M., Aitchison J. D., Rout M. P., Chait B. T. (2005) Proteomic and genomic characterization of chromatin complexes at a boundary. J. Cell Biol. 169, 35–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wang Y., Kallgren S. P., Reddy B. D., Kuntz K., Lopez-Maury L., Thompson J., Watt S., Ma C., Hou H., Shi Y., Yates J. R., 3rd, Bahler J., O'Connell M. J., Jia S. (2012) Histone H3 lysine 14 acetylation is required for activation of a DNA damage checkpoint in fission yeast. J. Biol. Chem. 287, 4386–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tackett A. J., DeGrasse J. A., Sekedat M. D., Oeffinger M., Rout M. P., Chait B. T. (2005) I-DIRT, a general method for distinguishing between specific and nonspecific protein interactions. J. Proteome Res. 4, 1752–1756 [DOI] [PubMed] [Google Scholar]
- 64. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 65. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 66. Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cote R. G., Griss J., Dianes J. A., Wang R., Wright J. C., van den Toorn H. W., van Breukelen B., Heck A. J., Hulstaert N., Martens L., Reisinger F., Csordas A., Ovelleiro D., Perez-Rivevol Y., Barsnes H., Hermjakob H., Vizcaino J. A. (2012) The PRoteomics IDEntification (PRIDE) Converter 2 framework: an improved suite of tools to facilitate data submission to the PRIDE database and the ProteomeXchange consortium. Mol. Cell. Proteomics 11, 1682–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang R., Fabregat A., Rios D., Ovelleiro D., Foster J. M., Cote R. G., Griss J., Csordas A., Perez-Riverol Y., Reisinger F., Hermjakob H., Martens L., Vizcaino J. A. (2012) PRIDE Inspector: a tool to visualize and validate MS proteomics data. Nat. Biotechnol. 30, 135–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rothbart S. B., Krajewski K., Strahl B. D., Fuchs S. M. (2012) Peptide microarrays to interrogate the “histone code.” Methods Enzymol. 512, 107–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Rothbart S. B., Krajewski K., Nady N., Tempel W., Xue S., Badeaux A. I., Barsyte-Lovejoy D., Martinez J. Y., Bedford M. T., Fuchs S. M., Arrowsmith C. H., Strahl B. D. (2012) Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat. Struct. Mol. Biol. 19, 1155–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Janke C., Magiera M. M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., Knop M. (2004) A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 [DOI] [PubMed] [Google Scholar]
- 73. Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. (1987) 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 154, 164–175 [DOI] [PubMed] [Google Scholar]
- 74. Byrum S. D., Raman A., Taverna S. D., Tackett A. J. (2012) ChAP-MS: a method for identification of proteins and histone posttranslational modifications at a single genomic locus. Cell Reports 2, 198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Byrum S. D., Taverna S. D., Tackett A. J. (2013) Purification of a specific native genomic locus for proteomic analysis. Nucleic Acids Res. 41, e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kabani M., Michot K., Boschiero C., Werner M. (2005) Anc1 interacts with the catalytic subunits of the general transcription factors TFIID and TFIIF, the chromatin remodeling complexes RSC and INO80, and the histone acetyltransferase complex NuA3. Biochem. Biophys. Res. Commun. 332, 398–403 [DOI] [PubMed] [Google Scholar]
- 77. Yap K. L., Zhou M. M. (2010) Keeping it in the family: diverse histone recognition by conserved structural folds. Crit. Rev. Biochem. Mol. Biol. 45, 488–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Dimmer E. C., Huntley R. P., Alam-Faruque Y., Sawford T., O'Donovan C., Martin M. J., Bely B., Browne P., Mun Chan W., Eberhardt R., Gardner M., Laiho K., Legge D., Magrane M., Pichler K., Poggioli D., Sehra H., Auchincloss A., Axelsen K., Blatter M. C., Boutet E., Braconi-Quintaje S., Breuza L., Bridge A., Coudert E., Estreicher A., Famiglietti L., Ferro-Rojas S., Feuermann M., Gos A., Gruaz-Gumowski N., Hinz U., Hulo C., James J., Jimenez S., Jungo F., Keller G., Lemercier P., Lieberherr D., Masson P., Moinat M., Pedruzzi I., Poux S., Rivoire C., Roechert B., Schneider M., Stutz A., Sundaram S., Tognolli M., Bougueleret L., Argoud-Puy G., Cusin I., Duek-Roggli P., Xenarios I., Apweiler R. (2012) The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 40, D565–D570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 80. Qiu Y., Zhang W., Zhao C., Wang Y., Wang W., Zhang J., Zhang Z., Li G., Shi Y., Tu X., Wu J. (2012) Solution structure of the Pdp1 PWWP domain reveals its unique binding sites for methylated H4K20 and DNA. Biochem. J. 442, 527–538 [DOI] [PubMed] [Google Scholar]
- 81. Wen H., Li Y., Xi Y., Jiang S., Stratton S., Peng D., Tanaka K., Ren Y., Xia Z., Wu J., Li B., Barton M. C., Li W., Li H., Shi X. (2014) ZMYND11 links histone H3.3K36me3 to transcription elongation and tumor suppression. Nature 508, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Youdell M. L., Kizer K. O., Kisseleva-Romanova E., Fuchs S. M., Duro E., Strahl B. D., Mellor J. (2008) Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 28, 4915–4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chruscicki A., Macdonald V. E., Young B. P., Loewen C. J., Howe L. J. (2010) Critical determinants for chromatin binding by Saccharomyces cerevisiae Yng1 exist outside of the plant homeodomain finger. Genetics 185, 469–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Doolin M. T., Johnson A. L., Johnston L. H., Butler G. (2001) Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40, 422–432 [DOI] [PubMed] [Google Scholar]
- 85. Tadauchi T., Matsumoto K., Herskowitz I., Irie K. (2001) Post-transcriptional regulation through the HO 3′-UTR by Mpt5, a yeast homolog of Pumilio and FBF. EMBO J. 20, 552–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Keogh M. C., Podolny V., Buratowski S. (2003) Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. Biol. 23, 7005–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Liu Y., Warfield L., Zhang C., Luo J., Allen J., Lang W. H., Ranish J., Shokat K. M., Hahn S. (2009) Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 29, 4852–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chu Y., Sutton A., Sternglanz R., Prelich G. (2006) The BUR1 cyclin-dependent protein kinase is required for the normal pattern of histone methylation by SET2. Mol. Cell. Biol. 26, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Howe L., Auston D., Grant P., John S., Cook R. G., Workman J. L., Pillus L. (2001) Histone H3 specific acetyltransferases are essential for cell cycle progression. Genes Dev. 15, 3144–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Govind C. K., Zhang F., Qiu H., Hofmeyer K., Hinnebusch A. G. (2007) Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol. Cell 25, 31–42 [DOI] [PubMed] [Google Scholar]
- 91. Sterner D. E., Belotserkovskaya R., Berger S. L. (2002) SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. U. S. A. 99, 11622–11627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ullah M., Pelletier N., Xiao L., Zhao S. P., Wang K., Degerny C., Tahmasebi S., Cayrou C., Doyon Y., Goh S. L., Champagne N., Cote J., Yang X. J. (2008) Molecular architecture of quartet MOZ/MORF histone acetyltransferase complexes. Mol. Cell. Biol. 28, 6828–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kimura M., Suzuki H., Ishihama A. (2002) Formation of a carboxy-terminal domain phosphatase (Fcp1)/TFIIF/RNA polymerase II (pol II) complex in Schizosaccharomyces pombe involves direct interaction between Fcp1 and the Rpb4 subunit of pol II. Mol. Cell. Biol. 22, 1577–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ginsburg D. S., Govind C. K., Hinnebusch A. G. (2009) NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol. Cell. Biol. 29, 6473–6487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Sanso M., Vargas-Perez I., Quintales L., Antequera F., Ayte J., Hidalgo E. (2011) Gcn5 facilitates Pol II progression, rather than recruitment to nucleosome-depleted stress promoters, in Schizosaccharomyces pombe. Nucleic Acids Res. 39, 6369–6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Balasubramanian R., Pray-Grant M. G., Selleck W., Grant P. A., Tan S. (2002) Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 277, 7989–7995 [DOI] [PubMed] [Google Scholar]
- 97. Grant P. A., Eberharter A., John S., Cook R. G., Turner B. M., Workman J. L. (1999) Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274, 5895–5900 [DOI] [PubMed] [Google Scholar]
- 98. Kuo Y. M., Andrews A. J. (2013) Quantitating the specificity and selectivity of Gcn5-mediated acetylation of histone H3. PloS One 8, e54896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhang W., Bone J. R., Edmondson D. G., Turner B. M., Roth S. Y. (1998) Essential and redundant functions of histone acetylation revealed by mutation of target lysines and loss of the Gcn5p acetyltransferase. EMBO J. 17, 3155–3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Lalonde M. E., Avvakumov N., Glass K. C., Joncas F. H., Saksouk N., Holliday M., Paquet E., Yan K., Tong Q., Klein B. J., Tan S., Yang X. J., Kutateladze T. G., Cote J. (2013) Exchange of associated factors directs a switch in HBO1 acetyltransferase histone tail specificity. Genes Dev. 27, 2009–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Pryde F., Jain D., Kerr A., Curley R., Mariotti F. R., Vogelauer M. (2009) H3 k36 methylation helps determine the timing of cdc45 association with replication origins. PloS One 4, e5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Kremer S. B., Gross D. S. (2009) SAGA and Rpd3 chromatin modification complexes dynamically regulate heat shock gene structure and expression. J. Biol. Chem. 284, 32914–32931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Kristjuhan A., Svejstrup J. Q. (2004) Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23, 4243–4252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Wyce A., Xiao T., Whelan K. A., Kosman C., Walter W., Eick D., Hughes T. R., Krogan N. J., Strahl B. D., Berger S. L. (2007) H2B ubiquitylation acts as a barrier to Ctk1 nucleosomal recruitment prior to removal by Ubp8 within a SAGA-related complex. Mol. Cell 27, 275–288 [DOI] [PubMed] [Google Scholar]
- 105. Cengiz B., Gunduz E., Gunduz M., Beder L. B., Tamamura R., Bagci C., Yamanaka N., Shimizu K., Nagatsuka H. (2010) Tumor-specific mutation and downregulation of ING5 detected in oral squamous cell carcinoma. Int.J. Cancer 127, 2088–2094 [DOI] [PubMed] [Google Scholar]
- 106. Avvakumov N., Cote J. (2007) The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 26, 5395–5407 [DOI] [PubMed] [Google Scholar]
- 107. Gunduz M., Gunduz E., Rivera R. S., Nagatsuka H. (2008) The inhibitor of growth (ING) gene family: potential role in cancer therapy. Curr. Cancer Drug Targets 8, 275–284 [DOI] [PubMed] [Google Scholar]
- 108. Helin K., Dhanak D. (2013) Chromatin proteins and modifications as drug targets. Nature 502, 480–488 [DOI] [PubMed] [Google Scholar]
- 109. Natoli G. (2009) Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harbor Perspectives In Biology 1, a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Taverna S. D., Cole P. A. (2010) Drug discovery: reader's block. Nature 468, 1050–1051 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.