Abstract
Human milk contains a rich set of soluble, reducing glycans whose functions and bioactivities are not well understood. Because human milk glycans (HMGs) have been implicated as receptors for various pathogens, we explored the functional glycome of human milk using shotgun glycomics. The free glycans from pooled milk samples of donors with mixed Lewis and Secretor phenotypes were labeled with a fluorescent tag and separated via multidimensional HPLC to generate a tagged glycan library containing 247 HMG targets that were printed to generate the HMG shotgun glycan microarray (SGM). To investigate the potential role of HMGs as decoy receptors for rotavirus (RV), a leading cause of severe gastroenteritis in children, we interrogated the HMG SGM with recombinant forms of VP8* domains of the RV outer capsid spike protein VP4 from human neonatal strains N155(G10P[11]) and RV3(G3P[6]) and a bovine strain, B223(G10P[11]). Glycans that were bound by RV attachment proteins were selected for detailed structural analyses using metadata-assisted glycan sequencing, which compiles data on each glycan based on its binding by antibodies and lectins before and after exo- and endo-glycosidase digestion of the SGM, coupled with independent MSn analyses. These complementary structural approaches resulted in the identification of 32 glycans based on RV VP8* binding, many of which are novel HMGs, whose detailed structural assignments by MSn are described in a companion report. Although sialic acid has been thought to be important as a surface receptor for RVs, our studies indicated that sialic acid is not required for binding of glycans to individual VP8* domains. Remarkably, each VP8* recognized specific glycan determinants within a unique subset of related glycan structures where specificity differences arise from subtle differences in glycan structures.
Human milk offers nutrition, innate immune protection, and other developmental benefits to infants (1, 2). In addition to essential nutrients and bioactive antibodies, human milk uniquely possesses a rich pool of free-reducing glycans (oligosaccharides), most of which are unique to human milk (3, 4). Depending on the blood group status and the lactation stage of an individual, the concentration of human milk glycans (HMGs)1 larger than lactose varies between 5 and 15 g/l, making them the third largest component of human milk after lactose and lipids (5). Over the past decades, more than 100 structurally distinct HMGs have been identified (6–9). All of these glycans originate from a lactose that is extended by type 1 (Galβ1–3GlcNAc) or type 2 (Galβ1–4GlcNAc) N-acetyllactosamine in either linear or branch forms and further modified with α-linked fucose and/or N-acetylneuraminic acid. It has been shown that HMGs are only minimally digested in the upper gastrointestinal tract and are transported intact into the lower parts of intestine (10, 11). Additionally, ∼1% to 2% of HMGs are excreted via an infant's urine and seem to appear in the circulation (12, 13).
Accumulated evidence has indicated that HMGs play multiple biological roles. In addition to having well-known prebiotic effects that promote the growth of beneficial microflora in the intestine (14, 15), HMGs are suggested to competitively interfere with pathogen attachment to the host cell surface by acting as soluble decoy receptors (16–18), and such anti-adhesive effects are often glycan specific (19). For example, α1–2 fucosylated HMGs, which arise mainly from individuals that are Secretor(+), were observed to prevent the adherence of Campylobacter jejuni to epithelial cells (20) and were associated with protection against diarrhea caused by Campylobacter, caliciviruses, and Escherichia coli toxin in breastfed infants (21–23). Sialylated HMGs were exclusive receptors for influenza viruses (24–26) and showed a capacity to inhibit cholera toxin B (27), Vibrio cholera (28), enterotoxigenic E. coli, and uropathogenic E. coli strains (29, 30). It was also proposed that HMGs might serve as anti-inflammatory components and thus contribute to the lower incidence of necrotizing enterocolitis in breastfed infants. This idea is supported by the observations that the acidic fraction of HMG inhibits leukocyte rolling, adhesion, and activation (31) and disialyllacto-N-tetraose prevents necrotizing enterocolitis in neonatal rats (32). Furthermore, a variety of cytoprotective activities of HMGs have been reported against Clostridium difficile toxins (33), Helicobacter pylori (34, 35), Streptococcus pneumonia (36), Entamoeba histolytica (37), and HIV-1-gp120 (38). Although the numerous in vitro and in vivo data provide important information about the function of HMGs, these studies have typically used HMG fraction mixtures or a small panel of defined HMGs, and therefore the bioactive HMGs were not or poorly identified.
In order to better understand the interactions of HMGs with various microorganisms, it is necessary to examine the entire milk metaglycome and identify the specific bioactive components, which is not possible via traditional methods that mainly focus on compositional analysis of HMGs (39). To find an efficient route for establishing the function–structure relationship of HMGs, we applied a “shotgun glycomics” approach to generate a shotgun glycan microarray (SGM) from isolated human milk glycans of a Lewis-positive, non-secretor individual (25, 40). The functional recognition studies, along with metadata-assisted glycan sequencing (MAGS), revealed novel epitopes/receptors for anti-TRA-1 antibodies, influenza viruses, and minute viruses of mice. Our work represented the first natural glycan microarray of HMGs containing over 100 glycans. Notably, the antibody binding data showed a lack of α1,2-fucosylated HMGs on this SGM, confirming that the donor was a non-secretor (41, 42).
Here we describe our studies in which we prepared a SGM containing over 200 isolated HMG targets from pooled human milk of mixed Lewis and Secretor phenotypes and investigated the binding of rotavirus (RV) cell attachment protein to them. Human RVs are the leading cause of severe gastroenteritis in children, responsible for an estimated 453,000 deaths each year worldwide (43). As with many other pathogens, RV infection is initiated by the interaction with specific cellular glycans. The VP8* domain of the RV outer capsid spike protein VP4 mediates this process (44), but the identity of VP8* receptors is quite controversial. It was believed that VP8* recognized either terminal sialic acid or internal sialic acid, mainly based on crystallographic and NMR studies (45–48). However, recently a human strain (HAL1166) with a P[14] VP8* was found to bind to A-type histo-blood group antigen (49), a neonatal strain with a P[11] VP8* bound to type 2 precursor glycans (50), and several other P types recognized secretor-related antigens Lewis b and H type 1 (51). These studies indicate that sialic acid might not be required by all RVs and that the glycan receptors are genotype-dependent. The infectivity of a porcine RV was inhibited by sialyl HMGs in vitro (52); however, there are limited data on human RVs. Here, we demonstrate that the VP8* of two different human neonatal RVs and an additional bovine strain bound to HMGs independent of sialic acid and that each VP8* demonstrated a unique glycan-binding specificity.
EXPERIMENTAL PROCEDURES
Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and used without further purification unless otherwise indicated. HPLC solvents were purchased from Thermo Fisher Scientific (Waltham, MA). Free reducing glycans used as standards were obtained from Sigma-Aldrich and V-LABS, Inc. (Covington, LA). 2-Amino-N-(2-aminoethyl)benzamide (AEAB) was synthesized in the lab. Human milk samples were obtained from the Mothers Milk Bank (Austin, TX). Biotinylated lectins were from Vector Labs (Burlingame, CA). Cyanine 5- and Alexa488-labeled Streptavidin, as well as Alexa488- and Alexa633-labeled secondary antibodies, were from Invitrogen (Grand Island, NY). Neuraminidase (Arthrobacter ureafaciens), α1–2 fucosidase, β1–3 galactosidase, and Jack bean β-galactosidase were from Roche Applied Science (Indianapolis, IN), Megazyme (Wicklow, Ireland), New England BioLabs (Ipswich, MA), and ProZyme (Hayward, CA), respectively. Endo-β-galactosidase from Bacteroides fragilis was purchased from Roche Applied Science GmbH (Germany). Mouse monoclonal antibodies against blood group H1 antigen [17–206] (ab3355), Lewis a (Lea) blood group [7LE] (ab3967), and Lewis b (Leb) blood group [2–25LE] (ab3968) were purchased from Abcam (Cambridge, MA). Anti-CD15 clone W6D3 (Lewis x, Lex) antibody and mouse anti-CD15s (CSLEX1) antibody against Sialyl-Lewis x (SLex) were obtained from BD Biosciences (San Jose, CA). Anti-type-1 chain antibody (clone 6A7) and anti-Lewis y (Ley) antibody (clone UoN30) were kind gifts from Dr. Irving Weissman (Stanford University, Stanford, CA) and Dr. Ian Spendlove (The University of Nottingham, Nottingham, England), respectively. An anti-Lex antibody (IgM -clone 5F1) was developed as a monoclonal antibody through production of a hybridoma from spleens of mice that had been infected with Schistosoma mansoni, using methods described previously (53). Carbograph SPE column was purchased from Grace Davison Discovery Sciences (Deerfield, IL). Sephadex G-25 resin was from GE Healthcare (Cleveland, OH). Whatman DEAE-cellulose (DEAE-52) resin and Biogel P-10 fine gel were from Sigma-Aldrich and Bio-Rad (Hercules, CA), respectively. N-Hydroxysuccinimide-activated NEXTERION® Slide H slides were purchased from Schott North America (Louisville, KY).
Isolation of Human Milk Glycans for Lewis Blood Group Analysis
To evaluate the Lewis Blood Group antigens in samples from 10 individual donors, free glycan fractions were prepared using an adaptation of a previously reported protocol (54). An aliquot (5 ml) of each milk sample was centrifuged at 4,000 rpm for 30 min to remove the fat. The solution phase was extracted with 4 volumes of chloroform-methanol (2:1, v/v) and centrifuged at 4,000 rpm for 30 min. The upper layer was collected and concentrated to 2 ml by a SpeedVac concentrator (Thermo Scientific), and this was followed by ethanol precipitation (2 volumes) at 4 °C overnight. The bulk of proteins and the lactose were removed after centrifugation, and the clear supernatant was freeze-dried. The crude HMGs were then fractionated into neutral and acidic glycans using a Carbograph SPE column (150 mg) as described previously (54). The neutral glycans that presumably contained the Lewis Blood Group determinants were conjugated with AEAB as described (25), and the excess AEAB was removed by a BioBasic SEC-60 column (7.8 × 150 mm, Thermo Scientific). The final products were quantified by fluorescence and printed in replicates of n = 4 at a concentration of 200 μm.
Isolation and Fractionation of Human Milk Glycans
The isolation and fractionation of HMGs followed our published procedures, with some modifications (25). Human milk (1 l comprising 100-ml samples from 10 different donors) was defatted via centrifugation at 6,000 × g for 30 min (4 °C). The skimmed milk was filtered through glass wool, mixed with 2 volumes of ethanol, and allowed to stand at 4 °C overnight to precipitate the bulk of the lactose and proteins. After centrifugation, the supernatant was concentrated via rotary evaporation (BUCHI, Flawil, Switzerland) and left at 4 °C for 2 days to allow the lactose to precipitate further, and the lactose was then removed via filtration. The clarified crude glycans were fractionated on a column (6 × 100 cm) of Sephadex G-25 using water as the mobile phase. Eluents were monitored for hexose using phenol-sulfuric acid assay, and the fractions containing glycans larger than lactose were then pooled and applied to a column (2 × 15 cm) of DEAE-cellulose equilibrated with 2 mm pyridine acetate, yielding neutral, monosialyl, and disialyl fractions through sequential elution with 2 mm, 20 mm, and 200 mm pyridine acetate, respectively (55). The resulting fractions were lyophilized and conjugated with AEAB. Next, the three HMG fractions were further fractionated on a Biogel P-10 fine column (2 × 170 cm) into 14 fractions: 4 neutral fractions, 3 monosialyl fractions, 3 disialyl fractions, and 4 low-molecular-weight sialyl fractions. All of the sialyl materials and part of the neutral materials were subjected to HPLC separation.
Purification of HMGs via High-performance Liquid Chromatography
For multidimensional HPLC purification of HMGs, we used a Shimadzu CBM-20A system coupled with an SPD-20A UV detector and an RF-10AXL fluorescence detector. For the first-dimensional purification, the 14 HMG fractions were separated via normal phase HPLC on a Zorbax NH2 column (250 × 9.4 mm, Agilent, Santa Clara, CA). The mobile phase contained acetonitrile and ammonium acetate, pH 4.5, in water, and linear gradients were optimized for neutral and acidic fractions. The effluents were monitored by UV absorption at 330 nm and/or fluorescence at 420 nm with excitation 330 nm, and fractions were collected every minute. For the second-dimensional purification, the normal phase fractions were pooled based on the elution profile and further purified via reversed phase HPLC on a porous graphitic carbon (Hypercarb) column (150 × 4.6 mm, Thermo Scientific) using gradients of acetonitrile/water containing 0.1% trifluoroacetic acid as the mobile phase. Individual peaks were collected and characterized by an Ultraflex-II MALDI-TOF/TOF mass spectrometry system (Bruker Daltonics, Billerica, MA). Fractions containing more than one composition based on the molecular ion were further re-purified on the porous graphitic carbon column in an optimized gradient. The resulting collection of glycans comprised the HMG-tagged glycan library (TGL) of 247 individual glycan targets having common compositions. Lactose-AEAB was used as the standard for the quantification of the AEAB derivatives.
Printing of the Human Milk SGM and Binding Assays
Printing of the HM-SGM, binding assays, and data analysis have been described previously (25, 56, 57). The glycans were printed on N-hydroxysuccinimide-activated slides as four arrays of 247 glycans plus 13 controls in replicates of four with a piezorray printer (PerkinElmer Life Sciences, Waltham, MA). This unique SGM was designated HM-SGM-v2. Glycan-binding proteins at various concentrations were applied to separate subarrays in 250 μl of binding buffer (20 mm Tris-HCl, 150 mm NaCl, 0.2 mm CaCl2, and 0.2 mm MgCl2, pH 7.4, containing 1% BSA and 0.05% Tween-20) in the wells formed on the slide with a silicone grid (four wells per slide). After incubation for 1 h at room temperature, the wells were washed with wash buffer (20 mm Tris-HCl, 150 mm NaCl, 0.2 mm CaCl2, and 0.2 mm MgCl2, pH 7.4, containing 0.05% Tween-20), and lectins were detected with cyanine5- (0.5 μg/ml) or Alexa488-labeled streptavidin (5 μg/ml) in the same binding buffer. Anti-carbohydrate monoclonal antibodies were detected using Alexa633- or Alexa488-labeled goat anti-mouse IgG (5 μg/ml). After washing to remove excess reagent, the fluorescence of bound protein was determined using a ProScanArray Scanner (PerkinElmer Life Sciences) equipped with four lasers covering an excitation range from 488 to 633 nm. The data were analyzed with the ScanArray Express software (PerkinElmer Life Sciences) and reported as a histogram of relative fluorescence units versus print identification number that identified the glycan target.
Preparation of GST-tagged RV Viral Protein VP8*
The gene sequences of VP8* (amino acids 64–225) of neonatal strains N155(G10P[11]) and RV3(G3P[6]) and bovine strain B223(G10P[11]) of RVs were synthesized (Epoch Life Science, Sugarland, TX) and cloned into expression vector pGEX-2T (GE Healthcare). The recombinant N-terminal GST-tagged VP8*s were expressed in E. coli BL21 (DE3) (Novagen, Madison, WI) and purified using Thermo Scientific Pierce glutathione agarose. The GST-VP8*s were concentrated using Spin-X UF 20 centrifugal concentrators (Corning, Corning, NY) and further purified via size exclusion chromatography on a High Load 16/60 Superdex 200 (GE Healthcare) column with 10 mm Tris, pH 7.4, 100 mm NaCl at 4 °C. The concentration of purified GST-VP8* was determined by measuring absorbance at 280 nm and using absorption coefficients of 76,270 M−1 cm−1 (N155), 73,000 M−1 cm−1 (RV3), and 73,830 M−1 cm−1 (B223), calculated using Vector NTI 11 software (Invitrogen).
Identification of HMGs That Bind RV Attachment Proteins
The GST-tagged RV attachment proteins were applied to the HM-SGM-v2 at different concentrations in 250 μl of binding buffer, incubated, and processed as described above in the wells formed on the slide with the silicon grid (four wells per slide). After washing, the bound protein was detected using Alexa488-labeled anti-GST antibody.
Structural Analysis of Selected HMGs via MAGS
Based on the binding of the GST-tagged RV attachment proteins to HMGs on the HM-SGM-v2, 32 fractions were selected and printed in replicates of n = 4 on a slide comprising 14 individual subarrays that were separated into individual wells for analysis using a silicon grid. This RV-MAGS microarray comprised the 32 selected glycans, 12 defined milk glycans, 2 related structures obtained from the Consortium for Functional Glycomics (CFG), a biotin control, and a buffer control. The subarrays were interrogated with lectins and antibodies with defined specificities as described. Further, exoglycosidase digestions were performed on the array, and selected lectins/antibodies were assayed to detect the loss of terminal residues. The on-array exoglycosidase digestions were carried out at 37 °C in buffers recommended by the suppliers of the enzymes. After rehydration in the digestion buffer for 5 min, 70 μl of reaction mixture was added to each well, and the slide was incubated at 37 °C in a humidified chamber. Reaction conditions were optimized for each exoglycosidase and are listed in supplemental Table S1. The logic used to predict structures based on lectin/antibody binding before and after glycosidase digestions is summarized in the supplemental material for each of the 32 glycans printed on the RV-MAGS microarray. In order to confirm the structures of HMGs determined by antibody/lectin binding, or in some cases to complete the analyses because of the unavailability of a defined glycan binding protein with appropriate specificity, we carried out MSn analyses on 28 of the 32 glycans, and a companion report2 presents the details of these structural analyses.
RESULTS
Binding of RV Cell Attachment Proteins to the CFG Defined Glycan Microarray
Glycan microarrays are a unique tool for rapidly identifying possible receptors for pathogen attachment (58, 59). A variety of viral proteins or whole viruses, including several RV VP8*s, have been analyzed on glycan arrays, which provided important information to direct biological and structural studies. The VP8*s of N155 and B223 have been analyzed on the CFG defined glycan microarray (50, 60), which contains a wide assortment of glycans, including N-glycans, but is not useful for identifying HMGs that bind to viruses, because the CFG microarray contains only a few of the simple HMGs. Nevertheless, we analyzed the VP8* RV3 on the CFG version 5.0 array at several concentrations (supplemental Fig. S1) and compared the differences in specificity of these proteins for the polylactosamine structures presented on the CFG array in supplemental Table S2. On the CFG array, all three VP8* domains bind predominantly to bi- and tri-antennary N-glycans with extended polylactosamines (LacNAc2 to LacNAc6). Each VP8* domain bound to unique polylactosamine structures. The VP8* from bovine strain B233 bound only polylactosamines that terminated in a type 2 LacNAc, whereas the related human-bovine reassortant strain N155 bound the same type 2–terminating polylactosamines as strain B233 but also bound polylactosamines terminating in GlcNAcβ1–3Gal, suggesting that this protein may bind internal sequences of polylactosamine. The VP8* from the human strain RV3 bound not only polylactosamines having a terminal GlcNAcβ1–3Gal, but also a variety of shorter glycans with terminal β-linked GlcNAc and type 1 LacNAc (Galβ1–3GlcNAc). Although it is interesting that the different VP8* domains could distinguish different synthetic polylactosamine structures, the significance of these observations is not known, because little is known about the quantitative expression, occurrence, or distribution of such multiantennary N-glycans with extended polylactosamine. Because the CFG array possesses only a few glycans related to the HMGs, we developed a human milk shotgun glycan microarray to explore in more detail the recognition of glycans by RV VP8* domains.
Preparation and Characterization of Human Milk Shotgun Glycan Microarray (HM-SGM-v2)
Because information on the Lewis blood types of the donors was not available, the AEAB derivatives of neutral glycans from 5 ml of each of 10 milk samples were printed as a microarray and assayed with antibodies specific for the H-type 1, Lea, and Leb epitopes (Fig. 1). The large variations in the relative amounts of these determinants observed among the 10 milk samples are consistent with previous observations (61) that concentrations of individual free glycans in breast milk vary with respect to donors and lactation stages. The fact that all of the determinants related to the Lewis blood groups were represented among the 10 individual samples indicated that the pooled sample (1 l) was representative of the human milk free glycan glycome. The human milk shotgun glycan microarray reported previously was from a single non-secretor individual and lacked α1–2-linked fucose (25).
Fig. 1.
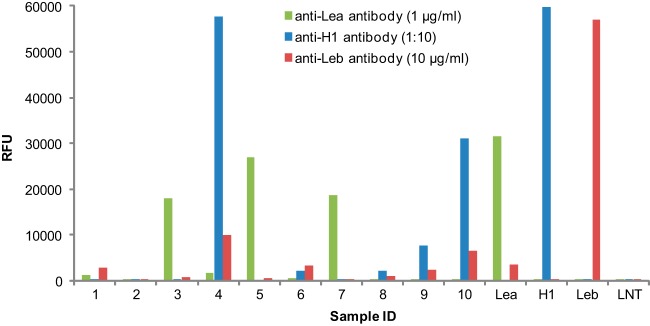
Lewis blood groups detected in 10 human milk samples by glycan microarray analysis. The neutral HMG mixtures from 10 human milk samples were printed on NHS-activated slides at 200 μm as quantified by total fluorescence using LNnT-AEAB as a standard. Four defined AEAB-labeled glycans were printed at 100 μm to serve as controls: Lea, Galβ1–3(Fucα1–4)GlcNAcβ1–3Galβ1–4Glc; H1, Fucα1–2Galβ1–3GlcNAcβ1–3Galβ1–4Glc; Leb, Fucα1–2Galβ1–3(Fucα1–4)GlcNAcβ1–3Galβ1–4Glc; and LNT, Galβ1–3GlcNAcβ1–3Galβ1–4Glc. The array was assayed with anti-Lea antibody (1 μg/ml), anti-H type 1 antibody (1:10 dilution), and anti-Leb antibody (10 μg/ml).
One liter of pooled milk was processed through multidimensional HPLC as summarized in supplemental Fig. S2. The resulting TGL is summarized in supplemental Table S3 and lists the 247 isolated HMG targets, the amounts obtained, intact masses as determined via MALDI-TOF analysis, and the predicted compositions expressed as molar ratios of H = hexose (Glc or Gal), n = HexNAc (GlcNAc), F = 2-deoxy-hexose (Fuc), and S = sialic acid (N-acetylneuraminic acid). It should be noted that the amount of each fraction represents the amount obtained and not the amount existing in the human milk. The TGL (247 glycan targets) was printed as a shotgun glycan microarray with known standards identified as HMG-1 through HMG-260 (print I.D. numbers 1 through 260). The array comprised 96 uncharged components (39% of the total 247 target glycans) corresponding to print I.D. numbers 1–96, where the targets with print I.D. numbers 89 to 96 were high-molecular-weight (>2,200 Da), mixed, neutral glycans. The TGL generated 111 monosialylated glycan targets (45% of the total 247 target glycans) corresponding to print I.D. numbers 97 to 207; the bulk of the targets possessed a single mass, with only 8 targets having two or three different masses. The disialylated glycan targets corresponded to print I.D. numbers 208 to 234. These 27 targets (11% of the total 247 target glycans) were confirmed as disialylated glycans based on their compositions as determined by MALDI analysis, and the majority of this class presented a single mass. Finally, we included 13 unresolved fractions of sialylated glycans that were primarily monosialyl glycans corresponding to print I.D. numbers 235 to 247. The size of purified glycans ranged from 2 residues (2 Hex, 506.23[M+H]) up to 14 residues (H6N4F4, 2573.85[M+Na]) with higher molecular masses up to ∼4,000 that were observed in the unresolved mixtures. The unresolved fractions of neutral and sialylated glycans were not present in sufficient quantity for further purification.
To demonstrate the utility of the HM-SGM-v2 and collect metadata on the individual structures, we interrogated the array with eight biotinylated lectins. No binding was observed with Concanavalin A, Vicia villosa lectin, and Maackia amurensis lectin I, which recognize mannose (62), terminal GalNAc (63), and terminal Neu5Acα2–3Galβ1–4GlcNAc (64), respectively. These observations are consistent with the literature and our previous observations (6–9, 25) on the lectin specificities and the absence or extremely low abundance of mannose, GalNAc, and the Neu5Acα2–3Galβ1–4GlcNAc determinant in human milk free glycans. The other five biotinylated lectins, Aleuria aurantia lectin (AAL), Ricinus communis agglutinin-I (RCA-I), Erythrina cristagalli lectin (ECL), Sambucus nigra agglutinin (SNA), and Ulex europaeus agglutinin-I (UEA-I), and nine anti-carbohydrate antibodies against various Lewis- and Secretor-related antigens were tested on the HM-SGM-v2. The results of these analyses are shown in Fig. 2, Fig. 3, and supplemental Table S4. Together, the lectin and antibody results demonstrate the efficiency of printing the glycans and provide significant structural information on the glycans of the human milk free glycome.
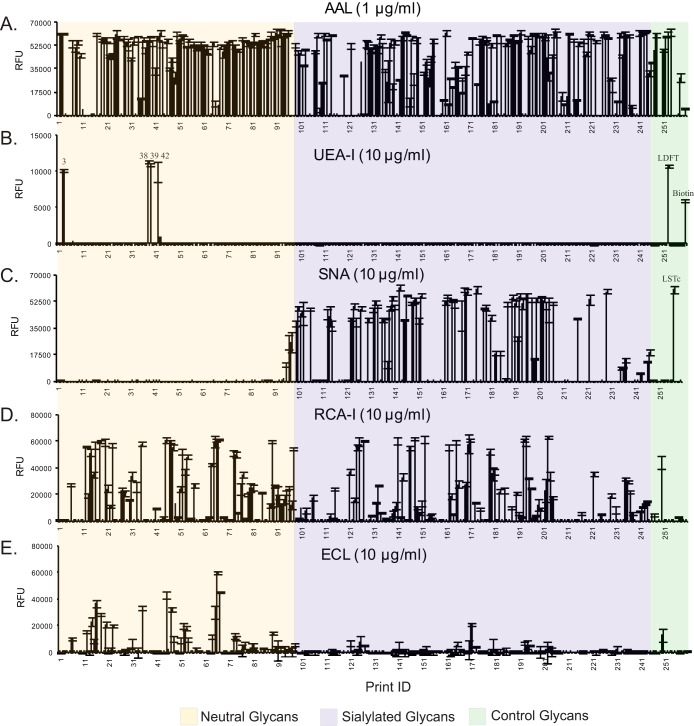
Fig. 2.
Plant lectin binding to HM-SGM-v2. The HM-SGM-v2 was characterized with (A) AAL (1 μg/ml), (B) UEA-I (10 μg/ml), (C) SNA (10 μg/ml), (D) RCA-I (10 μg/ml), and (E) ECL (10 μg/ml). A total of 260 samples were printed on the microarray. Glycans 1–88 are neutral glycans, 89–96 are unresolved fractions of neutral glycans, 97–207 are monosialyl glycans, 208–234 are disialyl glycans, 235–247 are unresolved fractions of sialylated glycans, 248–259 are controls of structurally defined glycans, and 260 is biotin control. All glycans were printed at the same concentration; however, unresolved fractions will not represent single glycans.
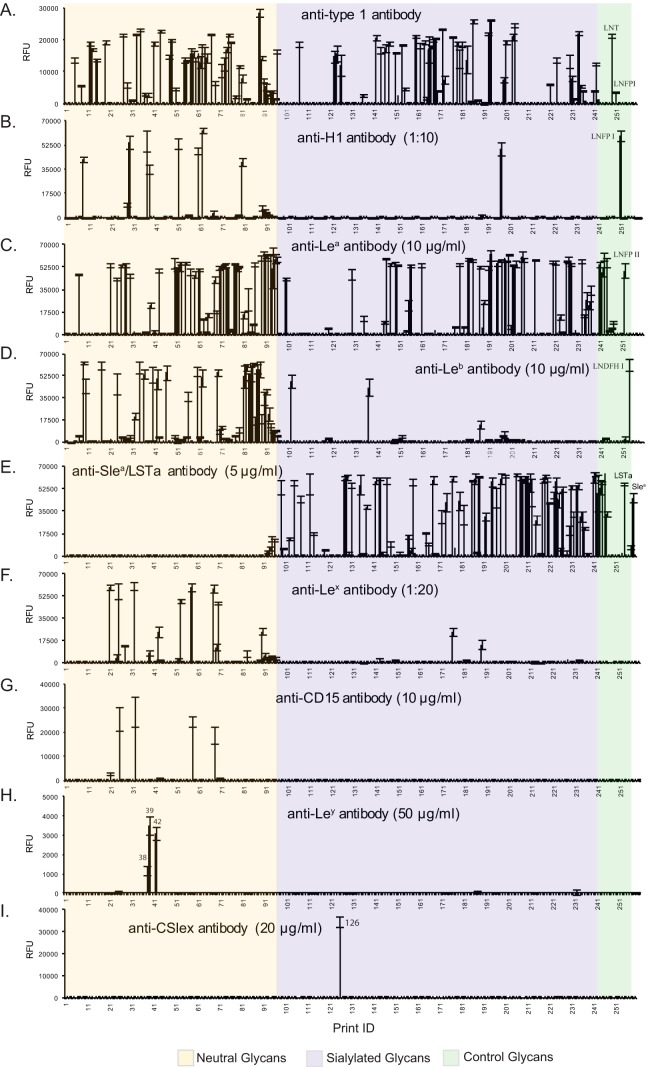
Fig. 3.
Antibody binding to HM-SGM-v2. The HM-SGM-v2 was characterized with (A) anti-type 1 antibody (concentration unknown), (B) anti-H type 1 antibody (1:10 dilution), (C) anti-Lea antibody (10 μg/ml), (D) anti-Leb antibody (10 μg/ml), (E) anti-SLea/LSTa (NeuAcα2–3Galβ1–3GlcNAcβ1–3Galβ1–4Glc antibody, 5 μg/ml), (F) anti-Lex antibody (1:20 dilution), (G) anti-CD15 antibody (10 μg/ml), (H) anti-Ley antibody (50 μg/ml), and (I) anti-CSlex antibody (20 μg/ml). A total of 260 samples were printed on the microarray. Glycans 1–88 are neutral glycans, 89–96 are unresolved fractions of neutral glycans, 97–207 are monosialyl glycans, 208–234 are disialyl glycans, 235–247 are unresolved fractions of sialylated glycans, 248–259 are controls of structurally defined glycans, and 260 is biotin control. All glycans were printed at the same concentration; however, unresolved fractions will not represent single glycans.
Specific Lectins and Antibodies Identify Determinants of the Human Milk Free Glycan Glycome
Approximately 193 glycan targets (78%), including a majority of the unresolved mixtures, exhibited strong binding with AAL (Fig. 2A), which is specific for α-linked fucose (65). Only five HMGs (3, 38, 39, 42, and LDFT, a control Ley determinant) on the HM-SGM-v2 were recognized by UEA-I (Fig. 2B), which binds Fucα1–2Galβ1–4GlcNAc (H type 2) and Fucα1–2Galβ1–4(Fucα1–3)GlcNAc (Ley) determinants. Examination of supplemental Tables S3 and S4 reveals that HMG-3 was fucosylated lactose (H2F1), and the UEA-I binding indicated that it must have been the 2-fucosyllactose (2-FL, Fucα1–2Galβ1–4Glc). Interestingly, the other three UEA-I-binding glycans all carry multiple fucose residues (H4N2F3 for 38 and 39 and H4N2F4 for 42). The antibody binding data indicate that HMG-38 possessed H1, HMG-39 possessed Lea, and HMG-42 possessed Leb epitopes (Figs. 3B–3D and supplemental Table S4). As each of these three glycans was also bound by the anti-Ley antibody (Fig. 3H), all of these minor glycans must have a branched hexaose core (H4N2) with a common difucosylated Ley determinant. Thus, we did not observe any H type 2 determinants (Fucα1–2Galβ1–4GlcNAc) in the glycans other than 2-FL (Fucα1–2Galβ1–4Glc) in the pooled human milk samples used in this study.
Three lectins that are specific for type 2 structures (Galβ1–4GlcNAc) showed that the HM-SGM-v2 was rich in type 2 glycans. Consistent with their known specificities, SNA, which binds Neu5Acα2–6Galβ1–4GlcNAc, bound to sialylated glycans (66) (Fig. 2C); RCA-I, which binds Galβ1–4GlcNAc and Neu5Acα2–6Galβ1–4GlcNAc (67), bound to both neutral and sialyl HMGs; and ECL, which is specific for Galβ1–4GlcNAc over Galβ1–3GlcNAc (68), primarily recognized only neutral glycans (Figs. 2D and 2E). Conversely, the type 1 structures (Galβ1–3GlcNAc) were detected with an anti–type 1 monoclonal antibody that bound 33 neutral glycan targets and 36 sialylated glycan targets with signals of over 5,000 relative fluorescence units (Fig. 3A). Although this anti–type 1 antibody also recognizes α1–2-fucosylated type 1 motif (H type 1, Fucα1–2Galβ1–3GlcNAc) with lower affinity, the majority of this binding was actually to the type 1 glycans, as the anti-H type 1 antibody only bound to 10 glycans (see below). These observations are consistent with the reported predominance of type 1 glycans as a feature specific to human milk (69).
Fucosylation is very common for the HMGs, and this property is mainly the result of two fucosyltransferases that prefer type 1 chain acceptors. FUT2 encodes the enzyme that synthesizes the H type 1, and FUT3 encodes the enzyme that synthesizes the Lea (13, 70); in combination these two enzymes are responsible for synthesis of the Leb epitope. It should be noted that FUT1, which encodes the α1–2 fucosyltransferase responsible for H-blood group generation, does not contribute significantly to α1–2 fucosylation of HMGs. To reveal the Lewis- and Secretor-related structures on the HM-SGM-v2, we interrogated the microarray with several blood-group-related monoclonal antibodies. The monoclonal antibody to H type 1 bound to nine neutral glycans and one sialylated glycan, as well as the control Fucα1–2Galβ1–3GlcNAcβ1–6Galβ1–4Glc (Fig. 3B). In contrast to the low population of terminal H1 structures, the anti-Lea antibody was bound by 31 neutral glycans and 35 sialylated glycans, accounting for 30% of purified HMGs (Fig. 3C). In addition, 54 out of 138 purified sialylated HMGs bound the anti-SLea/NeuAcα2–3Galβ1–3GlcNAcβ1–3Galβ1–4Glc antibody and may contain the SLea moiety, because the majority of the binders are fucosylated (Fig. 3E). Nevertheless, it is certain that Lea is a highly abundant structure in the human milk free glycome. The anti-Leb antibody showed strong binding to 24 neutral glycans and 2 sialylated glycans (Fig. 3D). The rare occurrence of α1,2-fucose in sialylated HMGs was presumably due to the fact that both α1,2-fucose and sialic acid are chain-terminating residues.
Lex- and Ley-containing HMGs were detected by interrogating the array with monoclonal antibodies including the commercial anti-CD15 antibody, an anti-Lex monoclonal antibody (clone 5F1) developed in our lab, and an anti-Ley monoclonal antibody whose specificities were defined using the CFG glycan microarray as shown in supplemental Fig. S5. It is known that a different fucosyltransferase that is not FUT3 adds the α1–3 linked fucose on Lex and Ley structures. This enzyme prefers the type 2 precursor and is constitutively expressed in all milk donors (70, 71); the Lex determinant is found to be more common in the milk of non-secretors (72). Previously, we noticed that the widely used anti-CD15 antibody does not recognize all Lex-containing HMGs (25). Indeed, it only bound to 4 glycans on the HM-SGM-v2 (Fig. 3G), whereas the clone 5F1 anti-Lex monoclonal antibody recognized more than 15 glycans (Fig. 3F), including the 4 glycans bound by the anti-CD15 antibody. Interestingly, neither of the anti-Lex antibodies recognized an internal Lex or Lex on a branched type 2 determinant (Galβ1–4(Fucα1–3)GlcNAcβ1–6Gal-R) as shown in supplemental Fig. S5.
In contrast to the common occurrence of the Lex structure, the Ley determinant, detected with the anti-Ley antibody, was only found on three minor glycans that were also bound by UEA-I. The extremely low abundance of Ley-containing HMGs is consistent with previous observations (6–9), but there is still no direct evidence to identify which α1,2-fucosyltransferase is responsible for adding the α1,2-fucose on 2-FL and Ley determinant; the FUT2 enzyme is believed to be responsible for the H type 1 and Leb determinants in HMGs (73). Finally, we tested the commercial anti-SLex antibody CSLEX1 (anti-CD15s), because HMGs were reported to affect leukocyte rolling mediated by SLex on PSGL-1 (31). As shown in Fig. 3I, it is remarkable that one glycan (HMG-126) was strongly bound by CSLEX1, suggesting that this glycan contains the SLex determinant. Consistent with this possibility, MS analysis indicated that HMG-126 has a hexasaccharide core with one fucose and one sialic acid. However, the abundance of this glycan was low—we recovered only 6 nmol in our TGL—and to our knowledge this determinant has not been conclusively identified previously in human milk (6–9). Taken together, the lectin and antibody results demonstrated the structural diversity of HM-SGM-v2 and also revealed the occurrence of typical determinants expected in the human milk free glycome.
Binding of RV Cell Attachment Proteins to Human Milk SGM-v2
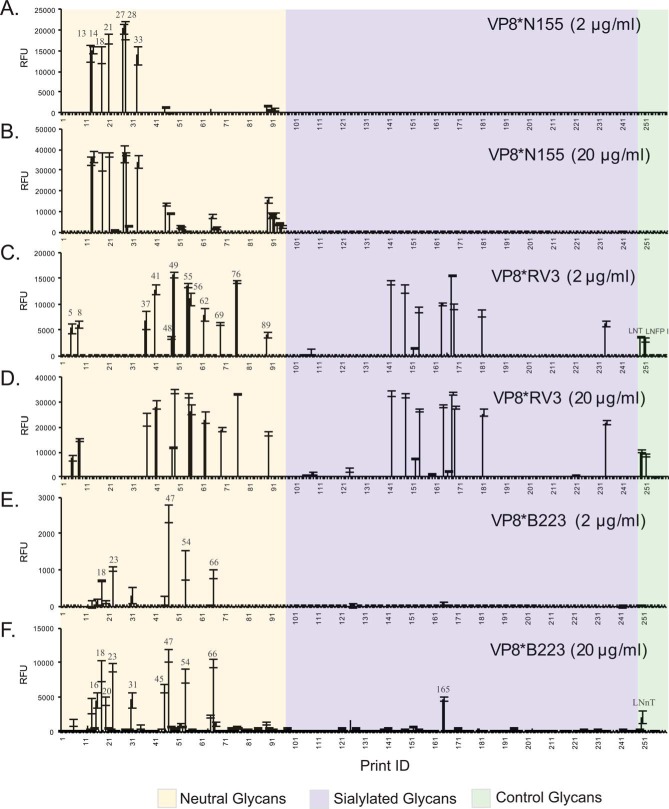
We analyzed the recombinant, GST fusion-tagged VP8* domains from the same RV strains at various concentrations on the HM-SGM-v2 where VP8*N155 strongly bound to seven neutral glycans (HMG-13, -14, -18, -21, -27, -28, and -33) at 2 μg/ml (Fig. 4A). Additional binding was observed at higher protein concentrations (20 μg/ml (Fig. 4B) and 200 μg/ml (Fig. 5A)), including several purified neutral glycans and all of the unresolved high-molecular-weight neutral glycan targets (print I.D.s 89 to 96).
Fig. 4.
Recombinant RV VP8* binding to HM-SGM-v2. The HM-SGM-v2 was screened with GST-tagged RV VP8. A, VP8*N155 (2 μg/ml); B, VP8*N155 (20 μg/ml); C, VP8*RV3 (2 μg/ml); D, VP8*RV3 (20 μg/ml); E, VP8*B223 (2 μg/ml); F, VP8*B223 (20 μg/ml). A total of 260 glycans was printed on the microarray. Glycans 1–88 are neutral glycans, 89–96 unresolved fractions of neutral glycans, 97–207 are monosialyl glycans, 208–234 are disialyl glycans, 235–247 are unresolved fractions of sialylated glycans, 248–259 are controls of structurally defined glycans, and 260 is biotin control. All glycans were printed at the same concentration; however, unresolved fractions will not represent single glycans.
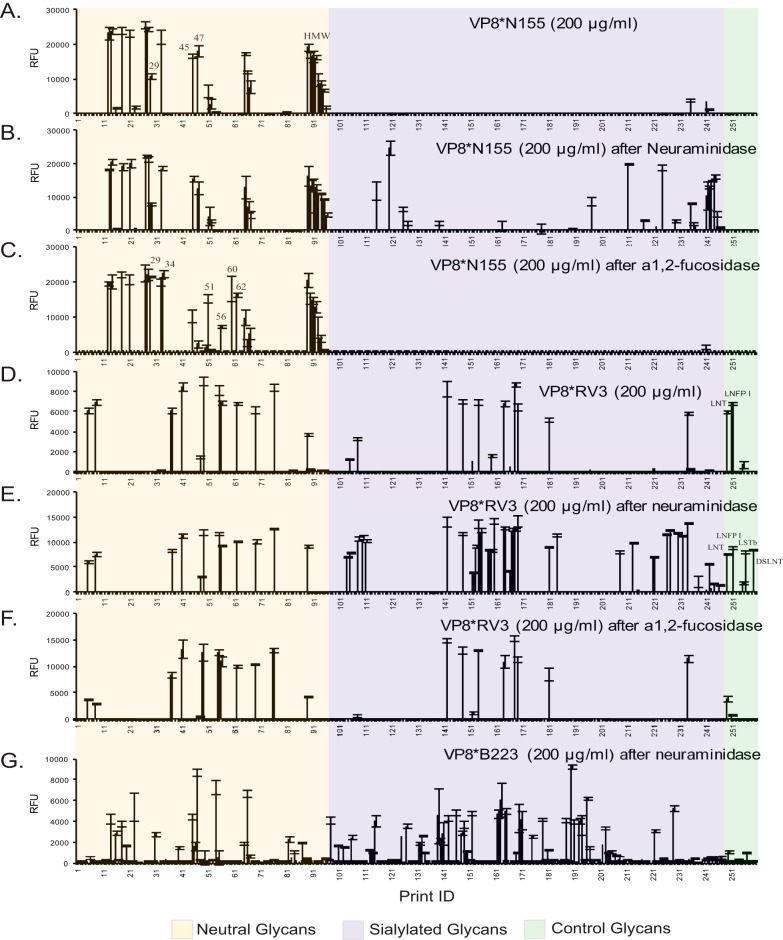
Fig. 5.
Recombinant RV VP8* binding to enzyme-treated HM-SGM-v2. The HM-SGM-v2 was treated with either neuraminidase or α1–2-fucosidase and assayed with GST-tagged RV VP8. A, VP8*N155 (200 μg/ml); B, VP8*N155 (200 μg/ml) after the array was treated by neuraminidase; C, VP8*N155 (200 μg/ml) after the array was treated by α1–2-fucosidase; D, VP8*RV3 (200 μg/ml); E, VP8*RV3 (200 μg/ml) after the array was treated by neuraminidase; F, VP8*RV3 (200 μg/ml) after the array was treated by α1,2-fucosidase; G, VP8*B223 (200 μg/ml) after the array was treated by neuraminidase. A total of 260 glycans was printed on the microarray. Glycans 1–88 are neutral glycans, 89–96 are unresolved fractions of neutral glycans, 97–207 are monosialyl glycans, 208–234 are disialyl glycans, 235–247 are unresolved fractions of sialylated glycans, 248–259 are controls of structurally defined glycans, and 260 is biotin control. All glycans were printed at the same concentration; however, unresolved fractions will not represent single glycans.
In contrast to VP8*N155, the binding of G3P[6] strain VP8*RV3 scattered over the entire array, which included 12 neutral glycans, 8 sialylated glycans, and 2 control glycans, Galβ1–3GlcNAcβ1-6Galβ1–4Glc and Fucα1–2Galβ1–3GlcNAcβ1–6Galβ1–4Glc (Fig. 4C). For RV3, increasing the protein concentration changed the signal intensity but not the binding profile (Figs. 4D and 5D). Interestingly, no glycans were recognized by both VP8* proteins, which raises the possibility that N155 and RV3 use completely different cell surface glycans as receptors. To determine whether the binders for the individual VP8*s share any common structural motif, we examined the lectin and antibody binding data and found that most of the N155 binders were recognized by RCA-I (type 2 glycans), whereas all of the RV3 binders were recognized by the anti–type 1 antibody (supplemental Table S5), which is consistent with the absence of overlap among glycan determinants recognized by N155 and RV3.
Because many human RVs were reported to recognize internal sialic acids but not terminal sialic acid, we inspected our metadata from defined lectin and antibody binding to the eight sialylated RV3 binders (HMG-142, -148, -152, -154, -164, -168, -169, and -181) seen in Figs. 4C and 4D. The MALDI data showed that they all had an octasaccharide or larger core structure (see supplemental Table S3 for compositions) and were all bound by the anti–type 1 antibody (supplemental Table S4), suggesting that these sialylated binders were multi-branched glycans containing at least one neutral branch and one sialylated branch. We treated the HMG-SGM-v2 with Arthrobacter neuraminidase, and sialic acid removal was confirmed by the complete elimination of SNA binding. Interrogation of the neuraminidase-digested array with both VP8*N155 and VP8*RV3 indicated that about 20 new binders for each VP8* were found among the previously sialylated glycans (compare Fig. 5A with Fig. 5B and Fig. 5D with Fig. 5E; data compiled in supplemental Table S5). The binding of the original eight sialylated RV3 binders was not affected, despite the loss of sialic acid, providing direct evidence that RV3 did not bind to the sialylated branch on the HMGs. The gain of binding to the standards NeuAcα2–3Galβ1–3GlcNAcβ1–3Galβ1–4Glc and Galβ1–3(NeuAcα2–6)GlcNAcβ1–3Galβ1–4Glc further indicated that the terminal type 1 chain is one of the recognition motifs for VP8*RV3.
HMGs terminating with α1–2 fucose have been suggested to be the receptors of various viruses. To test whether the removal of α1–2 fucose would affect the binding of N155 and RV3, we treated the HM-SGM-v2 array with α1–2 fucosidase. The two VP8*s were then analyzed on the fucosidase-digested array. For VP8*N155, six neutral glycans (HMG-29, -34, -51, -56, -60, and -62) that were not binders or weak binders (HMG-29) were strongly bound after the enzyme treatment (compare Fig. 5A with Fig. 5C; data compiled in supplemental Table S5), whereas the original binding glycans were not affected. A close inspection of the new binders revealed that all of them contained either a terminal H type 1 or a terminal Leb determinant, which could be converted to terminal type 1 and Lea determinants, respectively, by α1–2 fucosidase digestion. For VP8*RV3, no additional binders were revealed after α1–2 fucosidase digestion (compare Fig. 5D with Fig. 5F; data compiled in supplemental Table S5). Overall, these results indicated that the binding of N155 and RV3 was not Secretor-dependent.
In addition to the two human neonatal RV strains, we analyzed a bovine isolate, B223, which also belonged to the G10[P11] genotype on the HMG-SGM-v2. At 20 μg/ml, VP8*B223 bound to 10 neutral HMGs, one sialylated glycan (HMG-165), and the control LNnT (Fig. 4F). Examination of the lectin binding data (supplemental Table S4) revealed that all of the VP8*B223 binders were RCA-I and ECL positive, which indicated that a type 2 chain was involved in the recognition of VP8*B223. When the protein concentration was reduced to 2 μg/ml, the binding to LNnT was no longer observed (Fig. 4E), suggesting that Galβ1–4GlcNAc (type 2 glycan) alone is not a strong receptor for B223. Whereas VP8*RV3 did not bind to any glycans in common with N155 and B223, the two G10[P11] strains had some common binders at high protein concentrations (supplemental Table S5), which is consistent with the gene origins of two strains (supplemental Table S6) and the overlap in specificity for polylactosamines on the CFG glycan microarray. The removal of sialic acid by neuraminidase revealed more binders for B223 (Fig. 5G), but the binding to HMG-165 was not affected (compare Fig. 4F and Fig. 5G; data compiled in supplemental Table S5). Together with the data from the virus binding to the CFG glycan microarray, our results show that the bovine G10[P11] strain B223 does not recognize any sialylated glycans in agreement with previous observations (74).
Structural Analysis of Selected RV VP8* Binders
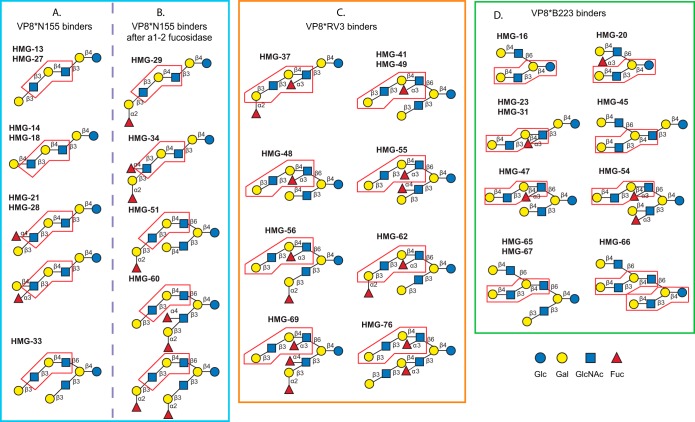
Interrogation of the HMG-SGM-v2 with lectins and antibodies provided significant structural information regarding the isolated free milk glycome and the VP8*-bound glycans. However, in order to identify a binding motif for these viral attachment proteins and understand the potential role of human milk glycans as decoy receptors for the corresponding RVs, definitive structural analysis was required. We retrieved 32 HMGs or glycan targets from the TGL in order to determine their structures via MAGS, as reported previously (25, 75). The 32 HMGs included 7 glycans bound by VP8*N155 (HMG-13, -14, -18, -21, -27, -28, and -33), 4 glycans bound by VP8*N155 after they were treated by α1,2-fucosidase digestion (HMG-29, -34, -51, and -60), 11 glycans bound by VP8*RV3 (HMG-5, -8, -37, -41, -48, -49, -55, -56, -62, -69, and -76), and 10 glycans bound by VP8*B223 (HMG-16, -20, -23, -31, -45, -47, -54, -65, -66, and -67). Only neutral RV binders were selected, because our neuraminidase digestion showed that sialic acid was not involved in the binding. It should be noted that five of the B223 binders, HMG-45, -47, -65, -66, and -67, are also bound by VP8*N155 at high protein concentrations. The HPLC profile and MALDI spectra of the 32 glycans are shown in supplemental Fig. S3. These selected glycan targets were printed on a separate RV-MAGS array along with 14 structurally defined control glycans. The 14 controls, whose structures are shown in supplemental Fig. S4, are defined neutral glycans of human milk. Binding to these glycans was used to monitor the behaviors of reagents and to direct the structural predictions.
Because some of the 14 controls were not included in the HM-SGM-v2 array, the RV-MAGS array was first interrogated with the five lectins (AAL, RCA-I, ECL, UEA-I, and Griffonia simplicifolia II) and six monoclonal antibodies (anti–type 1, -H1, -Lea, -Leb, -Lex, and -CD15). These binding data were used to assign non-reducing terminal determinants (Table I, supplemental Table S7). Next, the RV-MAGS array was treated with three linkage-specific β-galactosidases and one α-1,2-fucosidase. The binding of G. simplicifolia II after treatment with specific β1,3- and β1,4-galactosidase provided additional evidence of type 1 and type 2 termini in addition to ECL and anti–type 1 antibody binding. The endo-β1,4-galactosidase cleaved internal β1,4-linkage, exposing terminal GlcNAc that was detected by G. simplicifolia II and was particularly helpful in defining linear structures. The α1–2-fucosidase-treated array was tested with anti-Lea antibody to verify that the Leb determinant was indeed converted to the Lea determinant by this enzyme, because a group of unknowns were VP8* N155 binders only after α1–2 fucosidase digestion. Finally, all of the collected binding data were combined with MS data to predict the structures of the 32 unknown glycan targets. It was not difficult to resolve the structure of glycans with a tetrasaccharide (H3N1, 1 LacNAc + 1 Lac) or hexasaccharide (H4N2, 2 LacNAc + 1 Lac) core. However, for the octasaccharides (H5N3, 3 LacNAc + 1 Lac) or larger glycans (H6N4, 4 LacNAc + 1 Lac), it was not possible to define all of the branches and the linkages based on the binding data before and after glycosidase digestions. Thus, to confirm or complete the structural analyses of the RV binding glycans, 28 glycan targets were retrieved from the TGL and analyzed via MSn. Based on all of the metadata, including binding data and MSn analyses of aliquots of material from the TGL, we predicted the structures of all 32 unknown HMGs that bound RV VP8* domains, and a detailed description of the logic used to predict the structure of each glycan is shown in the supplemental material. The detailed MSn analyses of 32 AEAB derivatives of selected HMGs are discussed in a separate companion manuscript.2 Six glycan pairs were found to be identical (HMG-13 and -27, HMG-14 and -18, and HMG-41 and -49) or to contain the same structures (HMG-21 and -28, HMG-23 and -31, and HMG-65 and -67). This was due to overlap of fractions during the two-dimensional HPLC. In addition, several target glycans comprised contaminating isomeric structures, which are also described in the supplemental material and in detail in the companion manuscript.2 The major components that were relevant to the interactions and possessed a common motif corresponding to the three different VP8* proteins are shown in Fig. 6. Interestingly, among the 32 glycan targets analyzed (see the supplemental material), we identified 40 structures, and 21 novel glycans that had not been previously described as free glycans in human milk based on the structures reported in recent reviews on milk glycan structures (6–9, 76). The results of structural analyses demonstrate that glycans bound by specific RV VP8* domains possess distinct determinants, highlighted in Fig. 6, that are likely to be important in these protein–glycan interactions.
Table I. Heat map of lectin and antibody binding data for structural analysis.
A heatmap summary of the binding data from analyses of the RV-MAGS subarray with different lectins and anti-carbohydrate antibodies at indicated concentration. A total of 46 glycans were printed on this array, including 14 control glycans and 32 selected RV VP8* binders. The color-coded numbers highlight the binding intensity (the darker the color, the higher the binding intensity). The untreated RV-MAGS slides were tested with 5 lectins and 6 antibodies. The slides treated with β1–3 galactosidase (β1–3 galse) and β1–4 galactosidase (β1–4 galse) were tested with GSL-II. The slide treated with α1–2-fucosidase (α1,2-fucase) was assayed with anti-Lea antibody.
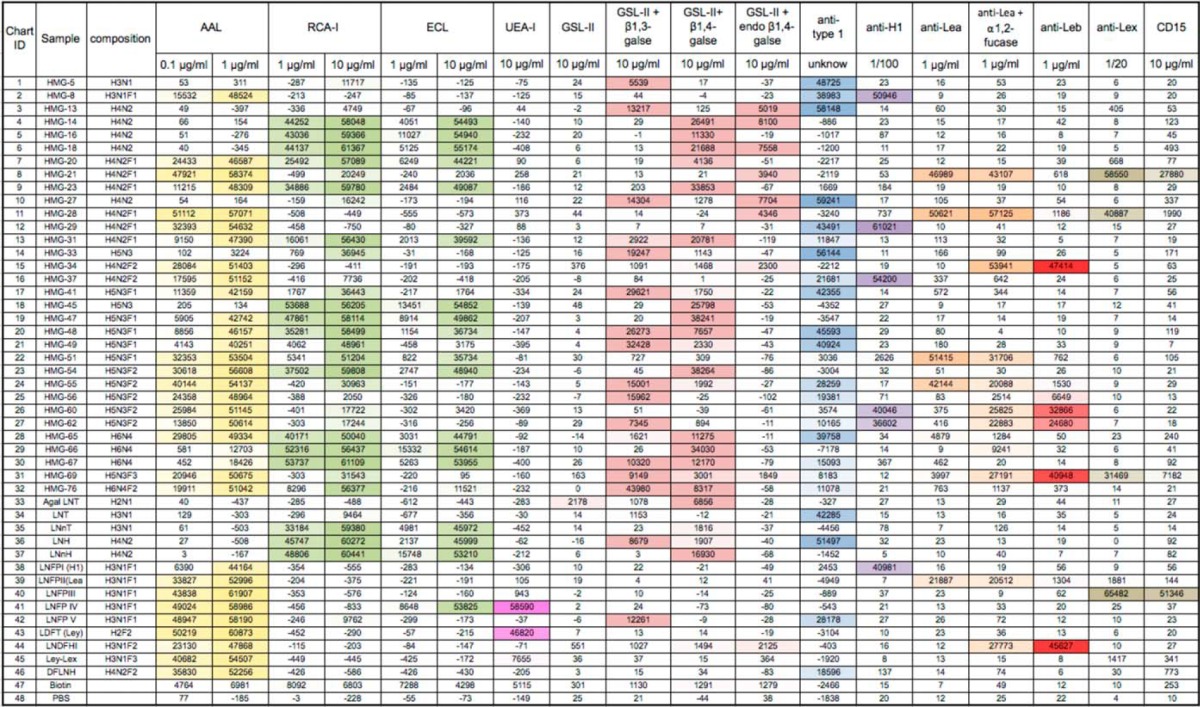
Fig. 6.
The predicted structures of 32 selected human milk glycans. The 32 selected HMGs include (A) seven glycan targets identified as VP8*N155 binders, (B) four glycan targets identified as VP8*N155 binders after treated by α1,2-fucosidase, (C) nine glycan targets identified as VP8*RV3 binders, and (D) 10 glycan targets identified as VP8*B223 binders. The structures shown are based on the predominant structure(s) in each glycan target determined using the metadata (lectin and antibody binding and tandem MS analysis) available on each.
DISCUSSION
We analyzed VP8* domains from three strains of RV with different clinical characteristics (supplemental Table S6) for binding on HM-SGM-v2. Human N155 belongs to the G10P[11] genotype and has been associated with a high incidence of gastrointestinal symptoms and asymptomatic infections in neonates in India (77–80). RV3 belongs to the G3P[6] genotype, associated with asymptomatic infections in neonates and found to be protective against subsequent severe gastroenteritis in these infants (81). Bovine RV B223 also belongs to the G10P[11] genotype (82) and has been described as a common cause of diarrhea in cattle (83–86). The human G10P[11] isolates are naturally occurring bovine-human reassortants and share a high degree of identity with the bovine G10[P11] isolates in their VP4 gene (50, 87). Studies on P[11] RVs and RV3 are important in the context of their public health impact and vaccine development (88).
We show that the three RV VP8* domains interacted with specific glycans in human milk and that the recognition was restricted to distinct structural sets of HMGs. Our detailed structural analyses on selected VP8*-binding glycans revealed common features they each share for recognition by a specific VP8*. Analysis of the seven glycan targets that bound VP8*N155 with high affinity (2 μg/ml) resulted in the identification of five glycan structures (Fig. 6A). HMG-21 and -28 were mixtures of the same two isomers. HMG-13 and -27 were both para-Lacto-N-hexaose, and HMG-14 and -18 were both para-Lacto-N-neohexaose, whereas HMG-33 was a branched octaose. The non-reducing terminal LacNAc in all of the linear VP8*N155-binding glycans can be type 1 (HMG-13/27), type 2 (HMG-14/18), Lea, or Lex (HMG-21/28). The other VP8*N155 binder, HMG-33, has a branched structure, isolacto-N-octaose, that contains two type 1 branches. To better understand the glycan motif recognized by VP8*N155, we also characterized the structure of four HMGs that were bound by VP8*N155 after α1–2 fucosidase treatment (Fig. 6B). HMG-29 and -34 were fucosylated linear hexaoses that were converted to HMG-13/27 and HMG-21/28, respectively, after loss of the α1–2 fucose. HMG-51 was converted by α1–2 fucosidase to a structure closely related to HMG-33, whereas a component of HMG-60 was converted to HMG-33, and all became VP8*N155 binders. It is apparent that VP8*N155 can accommodate either a type 1 or a type 2 terminal LacNAc and α1–4 fucose (Lea fucose) or α1–3 fucose (Lex fucose) modification, respectively, at the terminal LacNAcs, but the presence of a terminal α1–2 fucose blocks the binding. These data suggest that an internal trisaccharide sequence (highlighted in Figs. 6A and 6B) might represent a common determinant in all of the glycans bound by this VP8* domain.
VP8*RV3 bound to a completely different, but related, group of HMGs. Of the 11 glycan targets analyzed, 8 glycans were identified that shared a common determinant, and the obvious common feature in all 8 VP8*RV3 binders was the presence of a type 1 or an H-type 1 terminal determinant and an internal Lex determinant (Fig. 6C). This is consistent with the observation that VP8*RV3 does not bind to terminal type 2 glycan determinants on the defined CFG array (supplemental Fig. S1 and supplemental Table S2). All of the VP8*RV3-binding glycans contain a common motif, a pentasaccharide containing an internal Lex-trisaccharide (highlighted in Fig. 6C). Comparison of the VP8*N155 and VP8*RV3 domains revealed the exquisite specificity of these glycan binding proteins. For example, if HMG-29, which was bound by VP8*N155 after α1–2 fucosidase treatment, is modified with an internal Lex fucose (HMG-37), it becomes a binder for VP8*RV3. Similarly, if the α1–3 fucose residue (Lex fucose) is absent from HMG-41/49, the VP8*RV3-binder becomes HMG-33, which binds VP8*N155, and if the α1–3 fucose residue (Lex fucose) is lacking from HMG-48, the VP8*RV3-binder becomes α1–2-fucosidase-treated HMG-51 that binds VP8*N155. Thus, it appears from these data that VP8*RV3 requires the internal Lex because it does not bind to the same structures lacking the α1–3 fucose that are binders of VP8*N155. This also indicates that the internal α1–3 fucose blocks the recognition of VP8*N155. Furthermore, all of the branched VP8*RV3 binders contain two type 1 branches except for HMG-48, which has a type 2 chain on the 3-arm, but is actually a weak binder (Figs. 4C and 4D). We conclude that VP8*RV3 is specific for the type 1 terminal sequence with an internal Lex on the 6-arm of branched HMGs and that it prefers a type 1 chain on the 3-branch. The binding of VP8*RV3 to HMG-5 (Galβ1–3GlcNAcβ1–6Galβ1–4Glc) and HMG-8 (Fucα1–2Galβ1–3GlcNAcβ1–6Galβ1–4Glc) shown in Figs. 4C and 4D seems inconsistent with this conclusion, but their interaction with VP8*RV3 was atypical, as described in the supplemental material.
VP8*B223 did not bind to any HMG smaller than a hexaose. The smallest binder and minimal recognition unit, HMG-16, is LNnH, which is a component of all of the branched binders (Fig. 6D). HMG-45 is a free isoLacto-N-neooctaose, which is a core structure that has not been previously described in human milk based on recent reviews of the literature (6–9). HMG-47 and -54 share another new core structure that also has never been described before as a human milk glycan. This core structure is an extension of LNnH and contains exclusively type-2 LacNAc units. The fucose modifications on HMG-47 and HMG-54 did not interfere with B223 binding, but interestingly the internal Lex determinant did not cause recognition by VP8*RV3, which is consistent with the required type 1 terminal structure preferred by VP8*RV3 (compare HMG-47 with HMG-48). The only linear glycan in this group was HMG-23/31, which is a binder for VP8*B223; it differs from HMG-14/18 only by a Fucα1–3GlcNAc (internal Lex determinant), but this modification eliminates binding by VP8*N155. As mentioned above, HMG-45, -47, -65/67, and -66 are also bound by VP8*N155 at high protein concentrations. Such an observation is not very surprising because VP8*N155 also binds to LNnT at 200 μg/ml, and these four B223 binders contain an LNnT-like arm. VP8*B223 does not bind to any of the VP8*N155 binders. A tetrasaccharide component (highlighted in Fig. 6D) that is common to all of the VP8*B223 binding glycans might be an important determinant in this specific recognition.
These analyses of the three VP8* domains, together with previous data from the defined CFG glycan microarray (50, 60), present a relatively complex picture of their binding specificities, which have an interesting similarity to those of another class of glycan binding proteins, the galectins. These VP8* domains, like many galectins, bind polylactosamines but have distinct underlying specificities for smaller determinants (89–91). The relationship of the structural similarities of the VP8* domains to galectins (92) and their similarities in binding properties is unknown.
The shotgun glycan microarray is a functional glycomics approach to understanding the functional recognition of human milk glycans. From the large array of 247 unknown glycans and glycan fractions, we selected and printed 32 for further analysis on the HM-SGM-v2. Identification of these glycans is not trivial, and we approached this problem using a variety of classical methods to accumulate metadata on each individual glycan. We have termed this approach metadata-assisted glycan sequencing (25, 75) and have again demonstrated its utility in determining the structures of VP8*-binding glycans (Fig. 6). Of the 40 glycans whose structures are reported, 21 had not been previously described as free glycans in human milk (6–9). As the majority of the glycans displayed in Fig. 6 are among the 21 previously unidentified HMGs, they might not have been identified if we had not observed them as isolated glycans binding to these VP8* domains.
It was surprising to discover that the VP8* of these three different rotavirus strains bound such closely related glycan structures while retaining unique specificities that were determined by comparing the detailed structures of the corresponding binding glycans. The VP8*RV3 requires the internal Lex determinant, whereas the N155 protein cannot tolerate it, but its presence does not affect binding of the related B223. It will be interesting to fully characterize these protein–glycan interactions by determining crystal structures of these proteins bound to these complex HMGs. Future studies on the role(s) of these HMGs as decoy receptors or the role(s) of the glycan determinants as virus receptors on cell surface glycoconjugates will require larger quantities of the glycans to be evaluated as inhibitors of virus infection in vitro or the glycomic analysis of cellular glycomes where cells or tissues susceptible to rotavirus infection are shown to possess the corresponding determinants.
None of the VP8* rotavirus attachment proteins required sialic acid for binding; in fact, the presence of terminal sialic acid prevented binding in some cases. Recent observations show that not all rotaviruses require a sialic acid determinant for binding (49, 74, 93). Based on the studies reported here, it is important that the so-called neuraminidase-insensitive or sialic-acid-independent rotaviruses be analyzed for their glycan binding specificities on similar shotgun glycan microarrays comprising milk glycans or glycans released from cells or tissues susceptible to rotavirus infection.
Supplementary Material
Acknowledgments
We acknowledge The Consortium for Functional Glycomics, funded by NIGMS GM62116 and GM98791, for services provided by the Glycan Array Synthesis Core (the Scripps Research Institute, La Jolla, CA) that produced the mammalian glycan microarray and the Protein-Glycan Interaction Core (Emory University School of Medicine, Atlanta, GA) that assisted with analysis of samples on the array. The MSn structural studies carried out were supported by a partnership arrangement between The UNH-Glycomics Center and Glycan Connections, LLC, Lee, NH. The authors dedicate this manuscript to Professor Akira Kobata in commemoration of his lifelong contributions to the purification, identification, and structural analysis of the human milk glycome.
Footnotes
Author contributions: R.D.C. and D.F.S. designed research; Y.Y., Y.L., X.S., and D.J.A. performed research; L.H., S.R., M.L.M., D.J.A., B.P., M.K.E., and V.N.R. contributed new reagents or analytic tools; Y.Y., Y.L., X.S., D.J.A., V.N.R., R.D.C., and D.F.S. analyzed data; Y.Y., L.H., S.R., B.P., M.K.E., R.D.C., and D.F.S. wrote the paper.
* This work was supported by National Institutes of Health Grant Nos. R01GM085448 (D.F.S.), P41GM103694 (R.D.C.), R01AI080656 (M.K.E.), R01AI105101 (M.K.E.), and R37AI36040 (B.V.V.P.) and by Robert A. Welch Foundation Grant No. Q1279 (B.V.V.P.).
 This article contains supplemental material.
This article contains supplemental material.
2 David J. Ashline, Ying Yu, Yi Lasanajak, Xuezheng Song, Liya Hu, Sasirekha Ramani, Venkataram Prasad, Mary K. Estes, Richard D. Cummings, David F. Smith, and Vernon N. Reinhold.
1 The abbreviations used are:
- HMG
- human milk glycan
- SGM
- shotgun glycan microarray
- RV
- rotavirus
- MSn
- multi-stage mass spectrometry
- MAGS
- metadata-assisted glycan sequencing
- Lea
- Lewis a
- Leb
- Lewis b
- Lex
- Lewis x
- SLex
- sialyl-Lewis x
- Ley
- Lewis y
- AEAB
- 2-amino-N-(2-amino-ethyl)-benzamide
- TGL
- tagged glycan library
- HM
- human milk
- CFG
- Consortium for Functional Glycomics
- HM-SGM-v2
- human milk shotgun glycan microarray, version 2
- AAL
- Aleuria aurantia lectin
- RCA-I
- Ricinus communis agglutinin-I
- ECL
- Erythrina cristagalli lectin
- UEA-I
- Ulex europaeus agglutinin-I
- GST
- glutathione S-transferase
- LNnT
- Galβ1–4GlcNAcβ1–6Galβ1–4Glc
- SNA
- Sambucus nigra agglutinin.
REFERENCES
- 1. Walker A. (2010) Breast milk as the gold standard for protective nutrients. J. Pediatr. 156, S3–S7 [DOI] [PubMed] [Google Scholar]
- 2. German J. B., Dillard C. J., Ward R. E. (2002) Bioactive components in milk. Curr. Opin. Clin. Nutr. Metab. Care 5, 653–658 [DOI] [PubMed] [Google Scholar]
- 3. Urashima T., Saito T., Nakamura T., Messer M. (2001) Oligosaccharides of milk and colostrum in non-human mammals. Glycoconj. J. 18, 357–371 [DOI] [PubMed] [Google Scholar]
- 4. Tao N., Wu S., Kim J., An H. J., Hinde K., Power M. L., Gagneux P., German J. B., Lebrilla C. B. (2011) Evolutionary glycomics: characterization of milk oligosaccharides in primates. J. Proteome Res. 10, 1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bode L. (2012) Human milk oligosaccharides: Every baby needs a sugar mama. Glycobiology 22, 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kobata A. (2010) Structures and application of oligosaccharides in human milk. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 731–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tadasu Urashima M. K., Terabayashi T., Fukuda K., Ohnishi M., Kobata A. (2011) Milk oligosaccharides. In Oligosaccharides: Sources, Properties, and Applications (Gordon N. S. ed.), pp. 1–58, Nova Science Publishers, Inc., New York [Google Scholar]
- 8. Wu S., Tao N., German J. B., Grimm R., Lebrilla C. B. (2010) Development of an annotated library of neutral human milk oligosaccharides. J. Proteome Res. 9, 4138–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu S., Grimm R., German J. B., Lebrilla C. B. (2011) Annotation and structural analysis of sialylated human milk oligosaccharides. J. Proteome Res. 10, 856–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engfer M. B., Stahl B., Finke B., Sawatzki G., Daniel H. (2000) Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am. J. Clin. Nutr. 71, 1589–1596 [DOI] [PubMed] [Google Scholar]
- 11. Gnoth M. J., Kunz C., Kinne-Saffran E., Rudloff S. (2000) Human milk oligosaccharides are minimally digested in vitro. J. Nutr. 130, 3014–3020 [DOI] [PubMed] [Google Scholar]
- 12. Gnoth M. J., Rudloff S., Kunz C., Kinne R. K. (2001) Investigations of the in vitro transport of human milk oligosaccharides by a Caco-2 monolayer using a novel high performance liquid chromatography-mass spectrometry technique. J. Biol. Chem. 276, 34363–34370 [DOI] [PubMed] [Google Scholar]
- 13. Rudloff S., Kunz C. (2012) Milk oligosaccharides and metabolism in infants. Adv. Nutr. 3, 398S–405S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sela D. A., Mills D. A. (2010) Nursing our microbiota: molecular linkages between bifidobacteria and milk oligosaccharides. Trends Microbiol. 18, 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garrido D., Barile D., Mills D. A. (2012) A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 3, 415S–421S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bode L. (2009) Human milk oligosaccharides: prebiotics and beyond. Nutr. Rev. 67 Suppl 2, S183–S191 [DOI] [PubMed] [Google Scholar]
- 17. Hickey R. M. (2012) The role of oligosaccharides from human milk and other sources in prevention of pathogen adhesion. Int. Dairy J. 22, 141–146 [Google Scholar]
- 18. Newburg D. S., Ruiz-Palacios G. M., Morrow A. L. (2005) Human milk glycans protect infants against enteric pathogens. Annu. Rev. Nutr. 25, 37–58 [DOI] [PubMed] [Google Scholar]
- 19. Bode L., Jantscher-Krenn E. (2012) Structure-function relationships of human milk oligosaccharides. Adv. Nutr. 3, 383S–391S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ruiz-Palacios G. M., Cervantes L. E., Ramos P., Chavez-Munguia B., Newburg D. S. (2003) Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J. Biol. Chem. 278, 14112–14120 [DOI] [PubMed] [Google Scholar]
- 21. Newburg D. S., Ruiz-Palacios G. M., Altaye M., Chaturvedi P., Meinzen-Derr J., Guerrero Mde L., Morrow A. L. (2004) Innate protection conferred by fucosylated oligosaccharides of human milk against diarrhea in breastfed infants. Glycobiology 14, 253–263 [DOI] [PubMed] [Google Scholar]
- 22. Morrow A. L., Ruiz-Palacios G. M., Altaye M., Jiang X., Guerrero M. L., Meinzen-Derr J. K., Farkas T., Chaturvedi P., Pickering L. K., Newburg D. S. (2004) Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J. Pediatr. 145, 297–303 [DOI] [PubMed] [Google Scholar]
- 23. Newburg D. S., Ruiz-Palacios G. M., Altaye M., Chaturvedi P., Guerrero M. L., Meinzen-Derr J. K., Morrow A. L. (2004) Human milk alpha1,2-linked fucosylated oligosaccharides decrease risk of diarrhea due to stable toxin of E. coli in breastfed infants. Adv. Exp. Med. Biol. 554, 457–461 [DOI] [PubMed] [Google Scholar]
- 24. Bradley K. C., Jones C. A., Tompkins S. M., Tripp R. A., Russell R. J., Gramer M. R., Heimburg-Molinaro J., Smith D. F., Cummings R. D., Steinhauer D. A. (2011) Comparison of the receptor binding properties of contemporary swine isolates and early human pandemic H1N1 isolates (Novel 2009 H1N1). Virology 413, 169–182 [DOI] [PubMed] [Google Scholar]
- 25. Yu Y., Mishra S., Song X., Lasanajak Y., Bradley K. C., Tappert M. M., Air G. M., Steinhauer D. A., Halder S., Cotmore S., Tattersall P., Agbandje-McKenna M., Cummings R. D., Smith D. F. (2012) Functional glycomic analysis of human milk glycans reveals the presence of virus receptors and embryonic stem cell biomarkers. J. Biol. Chem. 287, 44784–44799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gambaryan A. S., Tuzikov A. B., Piskarev V. E., Yamnikova S. S., Lvov D. K., Robertson J. S., Bovin N. V., Matrosovich M. N. (1997) Specification of receptor-binding phenotypes of influenza virus isolates from different hosts using synthetic sialylglycopolymers: non-egg-adapted human H1 and H3 influenza A and influenza B viruses share a common high binding affinity for 6′-sialyl(N-acetyllactosamine). Virology 232, 345–350 [DOI] [PubMed] [Google Scholar]
- 27. Idota T., Kawakami H., Murakami Y., Sugawara M. (1995) Inhibition of cholera toxin by human milk fractions and sialyllactose. Biosci. Biotechnol. Biochem. 59, 417–419 [DOI] [PubMed] [Google Scholar]
- 28. Coppa G. V., Zampini L., Galeazzi T., Facinelli B., Ferrante L., Capretti R., Orazio G. (2006) Human milk oligosaccharides inhibit the adhesion to Caco-2 cells of diarrheal pathogens: Escherichia coli, Vibrio cholerae, and Salmonella fyris. Pediatr. Res. 59, 377–382 [DOI] [PubMed] [Google Scholar]
- 29. Martin-Sosa S., Martin M. J., Hueso P. (2002) The sialylated fraction of milk oligosaccharides is partially responsible for binding to enterotoxigenic and uropathogenic Escherichia coli human strains. J. Nutr. 132, 3067–3072 [DOI] [PubMed] [Google Scholar]
- 30. Lin A. E., Autran C. A., Espanola S. D., Bode L., Nizet V. (2013) Human milk oligosaccharides protect bladder epithelial cells against uropathogenic Escherichia coli invasion and cytotoxicity. J. Infect. Dis. 209, 389–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bode L., Rudloff S., Kunz C., Strobel S., Klein N. (2004) Human milk oligosaccharides reduce platelet-neutrophil complex formation leading to a decrease in neutrophil beta 2 integrin expression. J. Leukoc. Biol. 76, 820–826 [DOI] [PubMed] [Google Scholar]
- 32. Jantscher-Krenn E., Zherebtsov M., Nissan C., Goth K., Guner Y. S., Naidu N., Choudhury B., Grishin A. V., Ford H. R., Bode L. (2012) The human milk oligosaccharide disialyllacto-N-tetraose prevents necrotising enterocolitis in neonatal rats. Gut 61, 1417–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. El-Hawiet A., Kitova E. N., Kitov P. I., Eugenio L., Ng K. K., Mulvey G. L., Dingle T. C., Szpacenko A., Armstrong G. D., Klassen J. S. (2011) Binding of Clostridium difficile toxins to human milk oligosaccharides. Glycobiology 21, 1217–1227 [DOI] [PubMed] [Google Scholar]
- 34. Boren T., Falk P., Roth K. A., Larson G., Normark S. (1993) Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262, 1892–1895 [DOI] [PubMed] [Google Scholar]
- 35. Simon P. M., Goode P. L., Mobasseri A., Zopf D. (1997) Inhibition of Helicobacter pylori binding to gastrointestinal epithelial cells by sialic acid-containing oligosaccharides. Infect. Immun. 65, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Andersson B., Porras O., Hanson L. A., Lagergard T., Svanborg-Eden C. (1986) Inhibition of attachment of Streptococcus pneumoniae and Haemophilus influenzae by human milk and receptor oligosaccharides. J. Infect. Dis. 153, 232–237 [DOI] [PubMed] [Google Scholar]
- 37. Jantscher-Krenn E., Lauwaet T., Bliss L. A., Reed S. L., Gillin F. D., Bode L. (2012) Human milk oligosaccharides reduce Entamoeba histolytica attachment and cytotoxicity in vitro. Br. J. Nutr. 108, 1839–1846 [DOI] [PubMed] [Google Scholar]
- 38. Hong P., Ninonuevo M. R., Lee B., Lebrilla C., Bode L. (2009) Human milk oligosaccharides reduce HIV-1-gp120 binding to dendritic cell-specific ICAM3-grabbing non-integrin (DC-SIGN). Br. J. Nutr. 101, 482–486 [DOI] [PubMed] [Google Scholar]
- 39. Ruhaak L. R., Lebrilla C. B. (2012) Advances in analysis of human milk oligosaccharides. Adv. Nutr. 3, 406S–414S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Song X., Lasanajak Y., Xia B., Heimburg-Molinaro J., Rhea J. M., Ju H., Zhao C., Molinaro R. J., Cummings R. D., Smith D. F. (2011) Shotgun glycomics: a microarray strategy for functional glycomics. Nat. Methods 8, 85–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Henry S., Oriol R., Samuelsson B. (1995) Lewis histo-blood group system and associated secretory phenotypes. Vox Sang. 69, 166–182 [DOI] [PubMed] [Google Scholar]
- 42. Thurl S., Munzert M., Henker J., Boehm G., Muller-Werner B., Jelinek J., Stahl B. (2010) Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br. J. Nutr. 104, 1261–1271 [DOI] [PubMed] [Google Scholar]
- 43. Tate J. E., Burton A. H., Boschi-Pinto C., Steele A. D., Duque J., Parashar U. D. (2012) 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect. Dis. 12, 136–141 [DOI] [PubMed] [Google Scholar]
- 44. Lopez S., Arias C. F. (2006) Early steps in rotavirus cell entry. Curr. Top Microbiol. Immunol. 309, 39–66 [DOI] [PubMed] [Google Scholar]
- 45. Blanchard H., Yu X., Coulson B. S., von Itzstein M. (2007) Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*). J. Mol. Biol. 367, 1215–1226 [DOI] [PubMed] [Google Scholar]
- 46. Haselhorst T., Fleming F. E., Dyason J. C., Hartnell R. D., Yu X., Holloway G., Santegoets K., Kiefel M. J., Blanchard H., Coulson B. S., von Itzstein M. (2009) Sialic acid dependence in rotavirus host cell invasion. Nat. Chem. Biol. 5, 91–93 [DOI] [PubMed] [Google Scholar]
- 47. Yu X., Coulson B. S., Fleming F. E., Dyason J. C., von Itzstein M., Blanchard H. (2011) Novel structural insights into rotavirus recognition of ganglioside glycan receptors. J. Mol. Biol. 413, 929–939 [DOI] [PubMed] [Google Scholar]
- 48. Yu X., Dang V. T., Fleming F. E., von Itzstein M., Coulson B. S., Blanchard H. (2012) Structural basis of rotavirus strain preference toward N-acetyl- or N-glycolylneuraminic acid-containing receptors. J. Virol. 86, 13456–13466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hu L., Crawford S. E., Czako R., Cortes-Penfield N. W., Smith D. F., Le Pendu J., Estes M. K., Prasad B. V. (2012) Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485, 256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ramani S., Cortes-Penfield N. W., Hu L., Crawford S. E., Czako R., Smith D. F., Kang G., Ramig R. F., Le Pendu J., Prasad B. V., Estes M. K. (2013) The VP8* domain of neonatal rotavirus strain G10P[11] binds to type II precursor glycans. J. Virol. 87, 7255–7264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang P., Xia M., Tan M., Zhong W., Wei C., Wang L., Morrow A., Jiang X. (2012) Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol. 86, 4833–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hester S. N., Chen X., Li M., Monaco M. H., Comstock S. S., Kuhlenschmidt T. B., Kuhlenschmidt M. S., Donovan S. M. (2013) Human milk oligosaccharides inhibit rotavirus infectivity in vitro and in acutely infected piglets. Br. J. Nutr. 110, 1233–1242 [DOI] [PubMed] [Google Scholar]
- 53. Mandalasi M., Dorabawila N., Smith D. F., Heimburg-Molinaro J., Cummings R. D., Nyame A. K. (2013) Development and characterization of a specific IgG monoclonal antibody toward the Lewis x antigen using splenocytes of Schistosoma mansoni-infected mice. Glycobiology 23, 877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ninonuevo M. R., Park Y., Yin H., Zhang J., Ward R. E., Clowers B. H., German J. B., Freeman S. L., Killeen K., Grimm R., Lebrilla C. B. (2006) A strategy for annotating the human milk glycome. J. Agric. Food Chem. 54, 7471–7480 [DOI] [PubMed] [Google Scholar]
- 55. Smith D. F., Zopf D. A., Ginsburg V. (1978) Fractionation of sialyl oligosaccharides of human milk by ion-exchange chromatography. Anal. Biochem. 85, 602–608 [DOI] [PubMed] [Google Scholar]
- 56. Song X., Lasanajak Y., Xia B., Smith D. F., Cummings R. D. (2009) Fluorescent glycosylamides produced by microscale derivatization of free glycans for natural glycan microarrays. ACS Chem. Biol. 4, 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Heimburg-Molinaro J., Song X., Smith D. F., Cummings R. D. (2011) Preparation and analysis of glycan microarrays. Curr. Protoc. Protein Sci. Chapter 12, Unit 12 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rillahan C. D., Paulson J. C. (2011) Glycan microarrays for decoding the glycome. Annu. Rev. Biochem. 80, 797–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Heimburg-Molinaro J., Tappert M., Song X., Lasanajak Y., Air G., Smith D. F., Cummings R. D. (2012) Probing virus-glycan interactions using glycan microarrays. Methods Mol. Biol. 808, 251–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu Y., Huang P. W., Jiang B. M., Tan M., Morrow A. L., Jiang X. (2013) Poly-LacNAc as an age-specific ligand for rotavirus P[11] in neonates and infants. PLoS One 8, e78113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chaturvedi P., Warren C. D., Altaye M., Morrow A. L., Ruiz-Palacios G., Pickering L. K., Newburg D. S. (2001) Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology 11, 365–372 [DOI] [PubMed] [Google Scholar]
- 62. So L. L., Goldstein I. J. (1968) Protein-carbohydrate interaction. 13. The interaction of concanavalin A with alpha-mannans from a variety of microorganisms. J. Biol. Chem. 243, 2003–2007 [PubMed] [Google Scholar]
- 63. Tollefsen S. E., Kornfeld R. (1984) The B4 lectin from Vicia villosa seeds interacts with N-acetylgalactosamine residues on erythrocytes with blood group Cad specificity. Biochem. Biophys. Res. Commun. 123, 1099–1106 [DOI] [PubMed] [Google Scholar]
- 64. Wang W. C., Cummings R. D. (1988) The immobilized leukoagglutinin from the seeds of Maackia amurensis binds with high affinity to complex-type Asn-linked oligosaccharides containing terminal sialic acid-linked alpha-2,3 to penultimate galactose residues. J. Biol. Chem. 263, 4576–4585 [PubMed] [Google Scholar]
- 65. Fukumori F., Takeuchi N., Hagiwara T., Ohbayashi H., Endo T., Kochibe N., Nagata Y., Kobata A. (1990) Primary structure of a fucose-specific lectin obtained from a mushroom, Aleuria aurantia. J. Biochem. 107, 190–196 [DOI] [PubMed] [Google Scholar]
- 66. Shibuya N., Goldstein I. J., Broekaert W. F., Nsimba-Lubaki M., Peeters B., Peumans W. J. (1987) The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha 2–6)Gal/GalNAc sequence. J. Biol. Chem. 262, 1596–1601 [PubMed] [Google Scholar]
- 67. Baenziger J. U., Fiete D. (1979) Structural determinants of Ricinus communis agglutinin and toxin specificity for oligosaccharides. J. Biol. Chem. 254, 9795–9799 [PubMed] [Google Scholar]
- 68. Kaladas P. M., Kabat E. A., Iglesias J. L., Lis H., Sharon N. (1982) Immunochemical studies on the combining site of the D-galactose/N-acetyl-D-galactosamine specific lectin from Erythrina cristagalli seeds. Arch. Biochem. Biophys. 217, 624–637 [DOI] [PubMed] [Google Scholar]
- 69. Urashima T., Asakuma S., Leo F., Fukuda K., Messer M., Oftedal O. T. (2012) The predominance of type I oligosaccharides is a feature specific to human breast milk. Adv. Nutr. 3, 473S–482S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mollicone R., Candelier J. J., Reguigne I., Couillin P., Fletcher A., Oriol R. (1994) Molecular genetics of alpha-L-fucosyltransferase genes (H, Se, Le, FUT4, FUT5 and FUT6). Transfus. Clin. Biol. 1, 91–97 [DOI] [PubMed] [Google Scholar]
- 71. Koda Y., Soejima M., Kimura H. (2001) The polymorphisms of fucosyltransferases. Leg. Med. 3, 2–14 [DOI] [PubMed] [Google Scholar]
- 72. Kobata A. (2000) A journey to the world of glycobiology. Glycoconj. J. 17, 443–464 [DOI] [PubMed] [Google Scholar]
- 73. Blank D., Dotz V., Geyer R., Kunz C. (2012) Human milk oligosaccharides and Lewis blood group: individual high-throughput sample profiling to enhance conclusions from functional studies. Adv. Nutr. 3, 440S–449S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ciarlet M., Estes M. K. (1999) Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80 (Pt 4), 943–948 [DOI] [PubMed] [Google Scholar]
- 75. Smith D. F., Cummings R. D. (2013) Application of microarrays for deciphering the structure and function of the human glycome. Mol. Cell. Proteomics 12, 902–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kobata A. (2010) Structures and application of oligosaccharides in human milk. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 731–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ramani S., Sowmyanarayanan T. V., Gladstone B. P., Bhowmick K., Asirvatham J. R., Jana A. K., Kuruvilla K. A., Kumar M., Gibikote S., Kang G. (2008) Rotavirus infection in the neonatal nurseries of a tertiary care hospital in India. Pediatr. Infect. Dis. J. 27, 719–723 [DOI] [PubMed] [Google Scholar]
- 78. Banerjee I., Gladstone B. P., Le Fevre A. M., Ramani S., Iturriza-Gomara M., Gray J. J., Brown D. W., Estes M. K., Muliyil J. P., Jaffar S., Kang G. (2007) Neonatal infection with G10P[11] rotavirus did not confer protection against subsequent rotavirus infection in a community cohort in Vellore, South India. J. Infect. Dis. 195, 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gladstone B. P., Ramani S., Mukhopadhya I., Muliyil J., Sarkar R., Rehman A. M., Jaffar S., Gomara M. I., Gray J. J., Brown D. W., Desselberger U., Crawford S. E., John J., Babji S., Estes M. K., Kang G. (2011) Protective effect of natural rotavirus infection in an Indian birth cohort. N. Engl. J. Med. 365, 337–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Iturriza Gomara M., Kang G., Mammen A., Jana A. K., Abraham M., Desselberger U., Brown D., Gray J. (2004) Characterization of G10P[11] rotaviruses causing acute gastroenteritis in neonates and infants in Vellore, India. J. Clin. Microbiol. 42, 2541–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Bishop R. F., Barnes G. L., Cipriani E., Lund J. S. (1983) Clinical immunity after neonatal rotavirus infection. A prospective longitudinal study in young children. N. Engl. J. Med. 309, 72–76 [DOI] [PubMed] [Google Scholar]
- 82. Woode G. N., Kelso N. E., Simpson T. F., Gaul S. K., Evans L. E., Babiuk L. (1983) Antigenic relationships among some bovine rotaviruses: serum neutralization and cross-protection in gnotobiotic calves. J. Clin. Microbiol. 18, 358–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Gulati B. R., Nakagomi O., Koshimura Y., Nakagomi T., Pandey R. (1999) Relative frequencies of G and P types among rotaviruses from Indian diarrheic cow and buffalo calves. J. Clin. Microbiol. 37, 2074–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Fukai K., Maeda Y., Fujimoto K., Itou T., Sakai T. (2002) Changes in the prevalence of rotavirus G and P types in diarrheic calves from the Kagoshima prefecture in Japan. Vet. Microbiol. 86, 343–349 [DOI] [PubMed] [Google Scholar]
- 85. Varshney B., Jagannath M. R., Vethanayagam R. R., Kodhandharaman S., Jagannath H. V., Gowda K., Singh D. K., Rao C. D. (2002) Prevalence of, and antigenic variation in, serotype G10 rotaviruses and detection of serotype G3 strains in diarrheic calves: implications for the origin of G10P11 or P11 type reassortant asymptomatic strains in newborn children in India. Arch. Virol. 147, 143–165 [DOI] [PubMed] [Google Scholar]
- 86. Reidy N., Lennon G., Fanning S., Power E., O'Shea H. (2006) Molecular characterisation and analysis of bovine rotavirus strains circulating in Ireland 2002–2004. Vet. Microbiol. 117, 242–247 [DOI] [PubMed] [Google Scholar]
- 87. Ramani S., Iturriza-Gomara M., Jana A. K., Kuruvilla K. A., Gray J. J., Brown D. W., Kang G. (2009) Whole genome characterization of reassortant G10P[11] strain (N155) from a neonate with symptomatic rotavirus infection: identification of genes of human and animal rotavirus origin. J. Clin. Virol. 45, 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Danchin M., Kirkwood C. D., Lee K. J., Bishop R. F., Watts E., Justice F. A., Clifford V., Cowley D., Buttery J. P., Bines J. E. (2013) Phase I trial of RV3-BB rotavirus vaccine: a human neonatal rotavirus vaccine. Vaccine 31, 2610–2616 [DOI] [PubMed] [Google Scholar]
- 89. Stowell S. R., Arthur C. M., Slanina K. A., Horton J. R., Smith D. F., Cummings R. D. (2008) Dimeric galectin-8 induces phosphatidylserine exposure in leukocytes through polylactosamine recognition by the C-terminal domain. J. Biol. Chem. 283, 20547–20559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stowell S. R., Arthur C. M., Mehta P., Slanina K. A., Blixt O., Leffler H., Smith D. F., Cummings R. D. (2008) Galectin-1, -2, and -3 exhibit differential recognition of sialylated glycans and blood group antigens. J. Biol. Chem. 283, 10109–10123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Stowell S. R., Arthur C. M., Dias-Baruffi M., Rodrigues L. C., Gourdine J. P., Heimburg-Molinaro J., Ju T., Molinaro R. J., Rivera-Marrero C., Xia B., Smith D. F., Cummings R. D. (2010) Innate immune lectins kill bacteria expressing blood group antigen. Nat. Med. 16, 295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Dormitzer P. R., Sun Z. Y., Wagner G., Harrison S. C. (2002) The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21, 885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Fleming F. E., Bohm R., Dang V. T., Holloway G., Haselhorst T., Madge P. D., Deveryshetty J., Yu X., Blanchard H., von Itzstein M., Coulson B. S. (2014) Relative roles of GM1 ganglioside, N-acylneuraminic acids, and alpha2beta1 integrin in mediating rotavirus infection. J. Virol. 88, 4558–4571 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.