Abstract
Glucagon-like peptide-1 (GLP-1) is an incretin hormone that regulates glucose homeostasis. Because of their direct stimulation of insulin secretion from pancreatic β cells, GLP-1 receptor (GLP-1R) agonists are now important therapeutic options for the treatment of type 2 diabetes. To better understand the mechanisms that control the insulinotropic actions of GLP-1, affinity purification and mass spectrometry (AP-MS) were employed to uncover potential proteins that functionally interact with the GLP-1R. AP-MS performed on Chinese hamster ovary cells or MIN6 β cells, both expressing the human GLP-1R, revealed 99 proteins potentially associated with the GLP-1R. Three novel GLP-1R interactors (PGRMC1, Rab5b, and Rab5c) were further validated through co-immunoprecipitation/immunoblotting, fluorescence resonance energy transfer, and immunofluorescence. Functional studies revealed that overexpression of PGRMC1, a novel cell surface receptor that associated with liganded GLP-1R, enhanced GLP-1-induced insulin secretion (GIIS) with the most robust effect. Knockdown of PGRMC1 in β cells decreased GIIS, indicative of positive interaction with GLP-1R. To gain insight mechanistically, we demonstrated that the cell surface PGRMC1 ligand P4-BSA increased GIIS, whereas its antagonist AG-205 decreased GIIS. It was then found that PGRMC1 increased GLP-1-induced cAMP accumulation. PGRMC1 activation and GIIS induced by P4-BSA could be blocked by inhibition of adenylyl cyclase/EPAC signaling or the EGF receptor–PI3K signal transduction pathway. These data reveal a dual mechanism for PGRMC1-increased GIIS mediated through cAMP and EGF receptor signaling. In conclusion, we identified several novel GLP-1R interacting proteins. PGRMC1 expressed on the cell surface of β cells was shown to interact with the activated GLP-1R to enhance the insulinotropic actions of GLP-1.
Glucagon-like peptide-1 (GLP-1)1 is a gastrointestinal hormone secreted by intestinal L cells upon food intake that is best known for its role in controlling glucose homeostasis. Acting through its cognate glucagon-like peptide-1 receptor (GLP-1R), GLP-1 has several important physiological and pharmacological functions. GLP-1 is best known for enhancing glucose-stimulated insulin secretion (GSIS) from the pancreatic β cells. Importantly, the insulinotropic properties of GLP-1 are maintained in patients with type 2 diabetes (1), which is characterized by insufficient insulin secretion from pancreatic β cells and an inability to maintain glucose homeostasis. Therefore, therapeutic strategies targeting GLP-1R have been developed to treat type 2 diabetes (2, 3). In addition to augmenting insulin secretion, GLP-1 has been known to improve glucose sensing, proinsulin biosynthesis, survival, and proliferation of β cells (3, 4) in a variety of experimental models. GLP-1 also has several extrapancreatic effects, including actions on the central nervous system to inhibit food intake (5), the stomach to decrease gastric emptying and gastric acid secretion (6), and the lungs to stimulate secretion of macromolecules from airways (7). Additionally, GLP-1 has an effect on the heart and possibly the kidney to modulate blood pressure and heart rate (8, 9).
The GLP-1R is a member of the B1 family of G protein–coupled receptors (secretin receptor family). In mammals, GLP-1R is expressed in multiple tissues, including pancreatic β cells and δ cells (10), hypothalamus, lung, stomach, heart, kidney (11), and thyroid (12), which in part explains its diverse actions. Upon ligand binding, the GLP-1R is capable of coupling to diverse cell signal transduction pathways, but it is best known for its actions on G protein Gs α and adenylate cyclase activity to increase intracellular cAMP. It is known that other proteins can affect GLP-1R activity in addition to G proteins, including β-arrestin and caveolin, which affect receptor internalization and trafficking. β-Arrestin 1 is also required for proper GLP-1-stimulated cAMP production (13–15). More recently, it was shown that another B1 family member, gastric inhibitory polypeptide receptor heterodimerizes with GLP-1R, decreasing GLP-1-induced β-arrestin recruitment and mobilization (16). Very recently, our group identified several novel potential GLP-1R interactors using a membrane-based split-ubiquitin yeast two-hybrid (MYTH) assay (17). Three β cell–expressing membrane-bound interactors, solute carrier family 15 member 4 (SLC15A4), amyloid β A4 precursor-like protein 1 (APLP1), and adaptor-related protein complex 2 subunit mu (AP2M1), were further selected for individual knockdown in mouse insulinoma (MIN6) β cells using small interfering RNAs (siRNAs). GLP-1-induced insulin secretion was significantly enhanced when these genes were silenced, suggesting that these interactor proteins attenuate GLP-1R activity. These findings demonstrated that GLP-1R protein interactions are complex and the interactors can have measurable effects on receptor trafficking and downstream signaling. Such interactions may in part explain the diverse tissue-specific effects of GLP-1 and offer avenues for controlling GLP-1 actions in a tissue-selective manner.
Although the MYTH system is well established (18) and has been applied to study G protein–coupled receptor interactomes (17), it is limited on two fronts. Firstly, it must be performed in yeast which is not an ideal representation of the mammalian system. Secondly, it is technically difficult to activate the receptor in MYTH, thus, effects of ligand stimulation on the receptor interactome cannot be assessed. Recently, affinity purification–mass spectrometry (AP-MS) has become a powerful tool for discovering and examining novel protein–protein interactions, including those between membrane-bound proteins in mammalian cells (19–21). In the current study, we applied AP-MS to discover novel GLP-1R interactors and employed a human GLP-1R harboring a FLAG® epitope. GLP-1R-Flag was expressed in either Chinese hamster ovary (CHO) cells or MIN6 β cells, and interactors were studied in the presence or absence of GLP-1.
EXPERIMENTAL PROCEDURES
Cell Culture and Animals
CHO and MIN6 cells were cultured with high-glucose DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. INS1 832/3 cells were kindly provided by Dr. Christopher B. Newgard. INS1 832/3 cells were cultured with RPMI 1640 supplemented with 10% FBS, 1% penicillin and streptomycin, and β-mercaptoethanol.
CD1 mice aged 8 to 12 weeks were purchased from Charles River Laboratories (Wilmington, MA). Unless otherwise indicated, male mice were used for primary islet isolation. All animal experiments were approved by the Animal Care Committee (University of Toronto), and animals were handled according to the Canadian Council of Animal Care guidelines.
Plasmids, Reagents, and Transfection
C-terminal Flag-tagged human GLP-1R plasmids and C-terminal HA-tagged interactor plasmids were provided by Eli Lilly. Transient transfection of plasmids into CHO cells, MIN6 cells, or INS1 832/3 cells was conducted using Lipofectamine 2000 (Invitrogen) according to the protocol supplied by the vendor. GLP-1 7–37 was purchased from Abcam (Cambridge, MA). EGFR inhibitor AG1478 was purchased from Cayman Chemical (Ann Arbor, Michigan). Protein kinase G inhibitor Rp-8-bromo-β-phenyl-1,N2-ethenoguanosine3,5-cyclic monophosphorothioate sodium salt hydrate (RP8), PGRMC1 inhibitor (AG-205), adenylyl cyclase inhibitor (MDL-12,330A hydrochloride), EPAC inhibitor (ESI-09), PI3K inhibitor (Wortmannin), and PGRMC1 ligand progesterone (P4) were purchased from Sigma. PGRMC1 surface ligand P4-BSA was purchased from Steraloids (Newport, RI). EGF was purchased from R&D Systems (Minneapolis, MN).
Affinity Purification and LC-MS/MS
To profile the GLP-1R interactome in CHO cells and MIN6 cells, we used non-transfected cells or GLP-1R-Flag transfected cells with or without GLP-1 7–37 treatment for affinity purification. CHO cells transfected with GLP-1R-Flag were incubated with DMEM with 0.5% BSA for 30 min. Next, CHO cells were stimulated with or without 10 nm GLP-1 for 5 min before protein extraction. MIN6 cells expressing GLP-1R-Flag were preincubated with Krebs-Ringer buffer (22) for 2 h, and this was followed by treatment of 10 nm GLP-1 7–37 for 5 min before protein extraction.
Twenty 100 × 20 mm dishes of CHO cells and ten 100 × 20 mm dishes of MIN6 cells from each condition were harvested with protein lysis buffer (10% glycerol, 50 mm Hepes, 150 mm NaCl, 2 mm EDTA, 0.25% n-dodecyl-b-d-maltoside, and complete protease inhibitor mixture (Roche)). The protein lysates (40 mg per condition for CHO cells and 20 mg per condition for MIN6 cells) were collected for anti-Flag co-immunoprecipitation according to the methods reported previously (19). Briefly, 50 μl of anti-Flag M2 beads (A2220, Sigma-Aldrich) were added to cell lysates for rotation at 4 °C overnight. Anti-Flag M2 beads were washed three times with 800 μl of wash buffer (10% glycerol, 50 mm Hepes, 150 mm NaCl, 2 mm EDTA, 0.25% n-dodecyl-b-d-maltoside, and complete protease inhibitor mixture (Roche)) and then washed with 800 μl of 50 mm ammonium bicarbonate (pH 8.0). Protein complexes were eluted using 100 μl of 500 mm ammonium hydroxide (pH 11) three times. The pooled eluents were dried using a SpeedVac (Thermo Scientific) to remove ammonium hydroxide and then washed with H2O. Co-IP eluents were suspended in 50 mm (NH4) HCO3 for in-solution trypsin digestion. In brief, we first added an end concentration of 25 mm DTT for 40 min at 55 °C and then added an end concentration of 80 mm iodoacetamide for 20 min at room temperature in darkness. 2 μl of 0.5 μg/μl trypsin (Promega, Madison, MI) was added to digest at 37 °C overnight. On day 2 the digests were dried with a SpeedVac and then resuspended in buffer A (95% H2O, 5% acetonitrile, 0.1% formic acid). Next, the tryptic mixture was injected for LC-MS/MS analysis (LTQ-XL linear ion trap mass spectrometer, Thermo Scientific) as previously reported (19). In brief, reverse phase analytical columns were made of 14 to 19 cm of reverse phase material (Jupiter 4 mm Proteo 90A, Phenomenex, Inc., Torrance, CA). Peptides were eluted via a 2-h gradient method in which aqueous buffer A was progressively mixed with higher proportions of organic buffer B (5% water, 95% acetonitrile, and 0.1% formic acid) in the reverse phase column by the high-performance liquid chromatograph with a flow rate of 20 to 50 nl/min. Peptide ions were dynamically selected for fragmentation using data-dependent acquisition by the operating software (the five most intense precursor ions of each MS scan were selected for subsequent MS/MS).
Processing of Mass Spectrometric Data
Raw files with MS/MS spectra of each sample were used for creating peak lists with Xcalibur 2.0 SR2 (Thermo Scientific). Mascot (Matrix Science, London, UK) version 2.3.02 was used to perform database searching for each sample against the Mouse refseqV41 20100519 database (36,378 sequences, 16,698,579 residues) or the CHO RefV64cRapRev database (43,335 sequences, 23,505,858 residues). The mass tolerance was set as 3 Da for precursor ions and 0.6 Da for MS/MS fragment ions. One missed cleavage site was allowed for trypsin digestion. Fixed modifications were set as carbamidomethycysteine, and oxidation of methionine was set as a variable modification. The false discovery rate was calculated by using the “decoy” option in Mascot. The protein false discovery rate was controlled at less than 0.8% and the peptide false discovery rate was controlled at less than 1.5% by setting the significance threshold at 0.05, the ion score cutoff at 27, and the Mascot score cutoff at 50. We used the Prohits software to create the comparative tables for candidate GLP-1R interactors (23) while filtering out keratin protein. To facilitate the selection of GLP-1R interactors, the following criteria were established: (i) identified at least two times from three runs; (ii) not cytoskeleton protein or heat shock protein; (iii) not nucleus, mitochondrion, or endoplasmic reticulum proteins; and (iv) not identified in non-transfected control runs.
Network Analysis
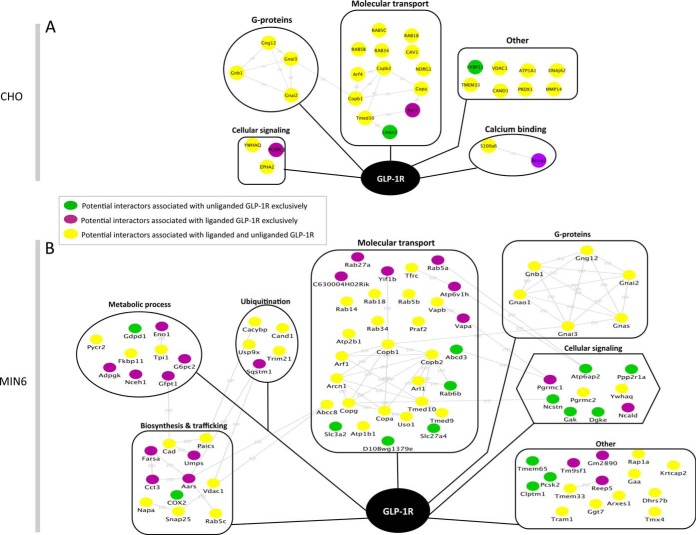
To understand the functional categories of these potential GLP-1R interactors, we created a comprehensive functional network of potential liganded and unliganded GLP-1R binding partners (24). In brief, potential GLP-1R interactors were input into the STRING functional protein association network database (version 9.1) (25). Protein functional interactions were visualized using Cytoscape (26). Biological functions associated with potential interactors were retrieved from the UniProt protein database and incorporated into Cytoscape for network analysis. Proteins identified exclusively under the liganded state were shown as purple nodes, those only identified under the unliganded state were displayed as green nodes, and proteins identified in both states were shown as yellow nodes (Fig. 2).
Fig. 2.
AP-MS was used to profile potential GLP-1R interactors and their functional clusters in CHO cells (A) and MIN6 cells (B). The potential GLP-1R binding proteins include those exclusively identified in the unliganded cells (green), in the liganded cells (purple), and in both (yellow). The gray lines represent the protein–protein (PP) functional interactions retrieved from the String database. The potential interactors are categorized into functional clusters according to the information retrieved from the UniProt protein database.
Co-immunoprecipitation and Western Blot
For co-IP, HA-tagged GLP-1R interactors and Flag-tagged GLP-1R plasmids were transfected into MIN6 cells using Lipofectamine 2000 (Invitrogen). After 24 h of transfection, cells were scraped and lysed using protein lysis buffer as described above. In another set, selected HA-tagged GLP-1R interactors were transfected into MIN6 cells, and proteins were extracted after 24 h. Anti-Flag-tag affinity gel (Sigma) was then used to pull down the bait protein and its interacting partners. Next, 2.5% input and 50% co-IP eluents from either co-transfection or transfection samples were loaded onto 10% SDS-PAGE gel for electrophoresis. Proteins were transferred to PVDF membranes, and a mouse anti-HA primary antibody (1:5000, Covance, Princeton, NJ) was used to immunoblot HA-tagged proteins overnight with an anti-mouse secondary antibody (1:10,000) subsequently applied for 1 h at room temperature. The membranes were imaged with the ECL Advance kit (GE Healthcare) on the Kodak Image Station 4000pro (Carestream Health, Inc., Rochester, NY). The intensity of protein bands on membranes was quantified using ImageJ software (National Institutes of Health) for both co-transfection of interactors and bait proteins and transfection of interactors alone. As the percentage of input or co-IP eluents loaded for western blot were known, the band intensities could be converted to the percentages of HA-tagged proteins interacting with Flag-tagged GLP-1R. The percentage of specific binding could be calculated by subtracting the unspecific binding intensities from the control group.
HTRF Measurement of Protein–Protein Interaction
HEK293 cells were transiently transfected in suspension with Fugene6 reagent (Promega) and expression constructs that encode Flag-tagged human GLP-1R and HA-tagged human interactor proteins. Transfectants were seeded at 2 × 104 per well to a tissue culture treated solid black microplate (Costar 3875). Cells were adhered for 2 days to account for phenotypic lag prior to HTRF detection of molecular interaction. Medium was removed, and cells were disrupted with ice-cold lysis buffer (5 mm HEPES, pH 7.4, 320 mm KF, 0.10% BSA fraction V, 0.025% SDS, 0.25% Nonidet P-40, 0.125% sodium deoxycholate, 0.5× EBSS, 1× Roche complete protease inhibitor). HTRF anti-tag toolbox reagents were purchased from Cisbio (Bedford, MA). The europium cryptate conjugated donor moiety, anti-Flag M2 monoclonal antibody, was diluted in lysis buffer and used at a final concentration of 0.25 nm. The d2 dye conjugated acceptor moiety, the anti-HA HAS01 monoclonal antibody, was diluted as above to 3.5 nm. Following 2 h of incubation at room temperature, the proximity of the tagged proteins was quantitated with a PerkinElmer Envision 2104 multilabel reader. Relative 665-nm fluorescence was normalized by 620 nm and compared with pcDNA3.1 parent vector controls.
Immunofluorescence
To study the colocalization of the GLP-1R-Flag and selected interactors, we used immunofluorescence microscopy with the Flag-tagged GLP-1R and HA-tagged interactors. In brief, MIN6 cells were fixed for 30 min in 4% paraformaldehyde at room temperature, washed three times with PBS, and permeabilized with 0.1% Triton. Blocking was performed with 3% BSA for 30 min at room temperature. Next, rabbit anti-Flag (1:250; Cell Signaling, Danvers, MA) and anti-HA (1:500; Roche, Laval, QC, Canada) were applied and incubated for 1 h at room temperature. Goat anti-rabbit FITC (1:500) and goat anti-mouse FITC (1:500) secondary antibody were added and incubated for 1 h at room temperature. The MIN6 cells were further stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (Molecular Probe®, Life technologies, Grand Island, NY). Slides were imaged using spinning disk confocal microscopy. Pearson's correlation coefficient (PCC) was calculated with Volocity 3D Image Analysis software (PerkinElmer Life Sciences) (17).
Glucose-stimulated Insulin Secretion, Total Insulin Content, and Quantitative RT-PCR
INS1 832/3 cells transfected with GLP-1R interactors or transfected with siRNAs (smart pool, Dharmacon, GE Healthcare, Ottawa, ON, Canada) were assessed for GSIS and GIIS. Briefly, INS1 832/3 cells were washed once with HEPES balanced salt solution (114 mmol/l NaCl, 4.7 mmol/l KCl, 1.2 mmol/l KH2PO4, 1.16 mmol/l MgSO4, 20 mmol/l HEPES, 2.5 mmol/l CaCl2, 25.5 mmol/l NaHCO3, and 0.2% bovine serum albumin (essentially fatty-acid free, pH 7.2)) with 3 mm glucose and preincubated for 2 h in the same buffer in 96-well plates. Insulin secretion was then measured using static incubation for 1 to 2 h in 100 μl of HEPES balanced salt solution containing 3 mm glucose (low-glucose), 15 mm glucose (high-glucose), or 15 mm glucose with 10 nm GLP-1 (high-glucose + GLP-1), or with compounds (5 μm AG-205, 1 μm P4, 1 μm P4-BSA, 100 nm AG1478, 30 μm RP8, 25 μm MDL-12, 30 μm ESI-09, 100 nm AG1478, 100 nm wortmannin). INS1 832/3 cells were lysed after GSIS for Bradford protein assay, western blot, and HTRF insulin assay for measuring total insulin content following the manufacturer's protocol (Cisbio) using a PheraStar Plus microplate reader (BMG LABTECH, Cary, NC).
MIN6 cells transfected with GLP-1R interactors were preincubated in KRB buffer (NaCl 115 mm, KCl 5 mm, NaHCO3 24 mm, CaCl2 2.5 mm, MgCl2 1 mm, HEPES 10 mm, BSA 0.1%, pH 7.4) for 2 h and then treated with the indicated concentrations of glucose (low-glucose, 0 mm glucose; high-glucose, 20 mm glucose), GLP-1, or compounds (5 μm AG-205, 1 μm P4, 1 μm P4-BSA) for 1 to 2 h. The supernatant was collected for insulin measurement using HTRF.
Mouse islets were isolated from pancreas for GSIS. Islets were preincubated in KRB buffer (NaCl 115 mm, KCl 5 mm, NaHCO3 24 mm, CaCl2 2.5 mm, MgCl2 1 mm, HEPES 10 mm, BSA 0.1%, pH 7.4) for 2 h and stimulated with the indicated concentrations of glucose (low-glucose, 2 mm glucose; high-glucose, 11.5 mm glucose), GLP-1, or compounds (1.5 μm AG-205, 1 μm P4, 1 μm P4-BSA) for 2 h. Supernatants were collected for insulin measurement using HTRF.
RT-PCR and quantitative PCR were performed as previously described (27). In brief, total RNA from cells was extracted using the RNeasy Plus Kit (Qiagen, Vehlo, Netherland) and converted to cDNA using a reverse transcription kit (Sigma). Power SYBR green PCR master mix was used for qPCR according to the manufacturer's protocol (Applied Biosystems, Grand Island, NY). We used ViiA7 Real-Time PCR system software (Invitrogen) to conduct the data analysis for quantitative PCR data. Primer sequences can be found in supplemental Table S3.
cAMP and Calcium Imaging
To measure the effects of selected GLP-1R interactors on GLP-1 stimulated cAMP response, we used an HTRF assay kit (Cisbio). In brief, His-V5 tagged GLP-1R (GLP-1R-His-V5) stably expressed CHO cells (RC2 cells) were transfected with empty vector (pcDNA3.1) or PGRMC1 for 48 h. RC2 cells were then preincubated with DMEM containing 0.5% BSA and treated with different concentrations of GLP-1 in DMEM containing 0.5% BSA and 1 μm phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine for 30 min. At the end of the test period, treatment buffers were incubated with cAMP-D2 and cryptate conjugate tracers for 1 h and then quantified using the PHERAstar Plus microplate reader (BMG).
INS1 832/3 cells transfected with plasmids for 2 days were washed twice in imaging buffer (10 mm NaCl, 5 mm KCl, 2 mm CaCl2, 1 mm MgCl2, 5 mm NaHCO3, 10 mm HEPES, pH 7.4) and then incubated with dye buffer containing 2 μm Fluo4AM (Molecular Probe, Invitrogen) and 2 μm Hoechst 33342 (Molecular Probe, Invitrogen) for 30 min. Cells were then treated sequentially with imaging buffers containing 0 mm glucose, 15 mm glucose, and 15 mm glucose with 1 nm GLP-1. Calcium images were acquired and analyzed on a Thermo Fisher Cellomics ArrayScan VTI HCS Reader using iDEV software.
Statistics
All data are expressed as the mean ± S.E. and were analyzed via analysis of variance followed by Tukey's post hoc multiple comparison tests. Student's t test was used where appropriate. The significance was accepted at a p value less than 0.05. The statistical analysis was conducted using IBM SPSS statistics software.
RESULTS
AP-MS Identified Potential GLP-1R Interacting Proteins in CHO Cells and MIN6 Cells
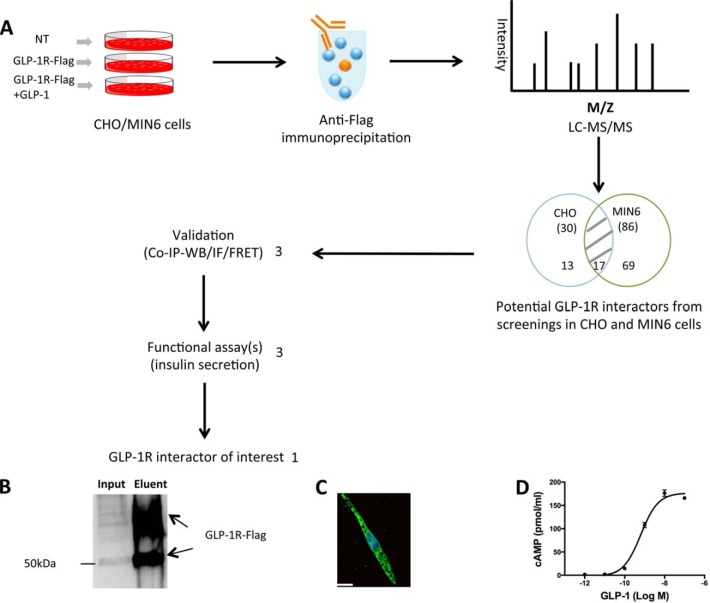
To discover novel GLP-1R interactors, we used AP-MS to profile the GLP-1R interactome in both CHO cells and MIN6 β cells. The flow chart in Fig. 1A demonstrates the strategies used to screen, select, and characterize putative GLP-1R interactors. For the screening components, we used non-transfected cells, GLP-1R-Flag transfected cells, and GLP-1-stimulated GLP-1R-Flag transfected cells. CHO cells were transfected with GLP-1R-Flag plasmid, and we confirmed that GLP-1R-Flag was pulled down from protein lysates before MS analysis by immunoprecipitation and western blot (Fig. 1B). We also showed that GLP-1R-Flag was expressed in CHO cells using immunofluorescence (Fig. 1C). In addition, GLP-1R-Flag-expressing CHO cells showed a typical dose response to GLP-1-stimulated cAMP accumulation (Fig. 1D). The EC50 for GLP-1R-Flag is 0.64 nm, which is close to the EC50 for normal GLP-1R (0.5 nm) (28). After anti-Flag affinity purification and LC-MS/MS analysis followed by database searching with filtering criteria, we identified both our bait GLP-1R and its potential interacting proteins in CHO and MIN6 cells. For CHO cells, we identified 27 potential proteins associated with the unliganded GLP-1R and 28 associated with the liganded GLP-1R (Fig. 2A, supplemental Table S1). Among these, 25 potential interactors were identified in both the liganded and unliganded states. Interestingly, only two potential interactors (FKBP11 and LMAN) were found exclusively in the unliganded state, and three (RER1, ANXA2, and PGRMC1) in the liganded state. In MIN6 cells, AP-MS revealed 65 potential interactors associated with unliganded GLP-1R and 71 potential interactors associated with liganded GLP-1R (Fig. 2B, supplemental Table S2). Among these, 50 potential interactors were identified in both the liganded and unliganded states, 15 potential interactors were found only in the unliganded status, and 21 were identified only in the liganded state. Thus more interactors were identified in a β cell line, with a significantly greater proportion of those interactors associated with the active GLP-1R than with unliganded GLP-1R. All protein and peptide identification information can be found in supplemental Table S5. The MS/MS spectra are shown for proteins identified with one unique peptide (supplemental Figs. S8 and S9).
Fig. 1.
Discovery of novel GLP-1R interactors. A, flow chart showing the AP-MS strategy used to identify novel GLP-1R interactors. Non-transfected (NT) CHO/MIN6 cells were used as the negative control group. CHO/MIN6 cells expressing GLP-1R-Flag were used with or without 10 nm GLP-1 stimulation for 5 min. Protein lysates from each condition were used for anti-Flag co-IP, and co-IP eluents were then used for in-solution trypsin digestion and LC-MS/MS (LTQ-XL) to profile potential GLP-1R interactors. The process was repeated three times for each condition. Among 99 potential GLP-1R interactors, 3 were selected for further validation by co-IP and western blot, immunofluorescence, and FRET. Three validated GLP-1R interactors were then selected for assessment of their effects on GIIS, leading to the discovery of one novel GLP-1R interactor of interest. B, anti-Flag immunoprecipitation and anti-Flag western blot showing that CHO cells expressed GLP-1R-Flag. C, representative immunofluorescent image showing GLP-1R-Flag expressed in CHO cells. Bar = 14 μm. D, cAMP accumulation in CHO cells expressing GLP-1R-Flag in response to GLP-1.
When the GLP-1R interactors discovered in CHO and MIN6 cells were combined (99 in total), 17 were identified in the two cell lines (PGRMC1, Rab5b, Rab5c, Rab18, 14–3-3 protein τ, TMED10, VDAC1, Rab34, COPA, COPB1, COPB2, TMEM33, CAND1, GNB1, GNG12, GNAi3, GNAi2), 13 were found only in CHO cells, and 69 were found only in MIN6 cells. Interestingly, among the 17 potential GLP-1R interactors identified from both cell lines, 3 novel interactors (PGRMC1, Rab5b, and Rab5c) were cell membrane proteins and known to be involved in G protein–coupled receptor functions (supplemental Fig. S1). PGRMC1 was reported to regulate membrane progestin receptor α cell surface expression and functions (29), and Rab5b and Rab5c associate with GPCR trafficking and early endosomal localization (30). In addition, these three interactors showed significant association with liganded GLP-1R (p < 0.01 for PGRMC1, p < 0.05 for Rab5b and Rab5c, n = 8–11; supplemental Fig. S1), indicating that they may affect liganded GLP-1R actions. For these reasons, a focus was put on these three interactors.
Co-IP and Western Blot Validated Selected GLP-1R Interacting Proteins in MIN6 Cells
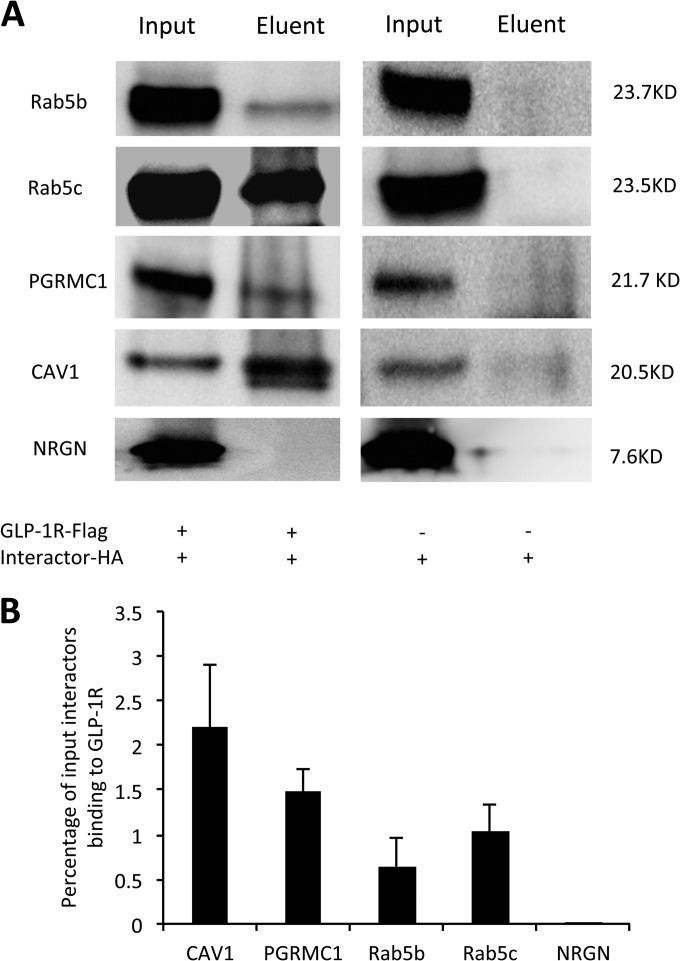
For further assessment, in addition to PGRMC1, Rab5b, and Rab5c, we included CAV1 as a positive control for further assessment because it is a known GLP-1R interactor and we found it in our screen. We also included neurogranin (NRGN) as a negative control, as it was found in our previous study not to bind to GLP-1R (17). We conducted both co-transfection of HA-tagged interactors and GLP-1R-Flag and transfection of HA-tagged interactors alone into MIN6 cells. Anti-Flag co-IP and anti-HA western blot showed that human GLP-1R-Flag interacted with four HA-tagged interactors (Rab5b, Rab5c, CAV1, and PGRMC1) in MIN6 β cells (Fig. 3A) that were not likely to be explained by unspecific binding between HA-tagged interactors and anti-Flag affinity gels. Band intensity analysis normalized to input showed that 2.2% CAV1, 1.48% PGRMC1, 0.64% Rab5b, and 1.04% Rab5c bound to the GLP-1R-Flag (Fig. 3B). Thus, co-IP and western blot methods validated both the known GLP-1R interactor (CAV1) and three novel GLP-1R interactors.
Fig. 3.
Validation of selected GLP-1R interactors by co-IP and western blot. A, anti-Flag co-IP and anti-HA western blot of selected interactors from MIN6 cells co-transfected with GLP-1R-Flag and interactor-HA (left panel) or transfected with interactor-HA alone (right panel). B, the binding between selected interactors and GLP-1R was evaluated by calculating the percentage of input interactors binding to the GLP-1R.
HTRF Measurement Demonstrates Interactions between Selected Interactors and GLP-1R
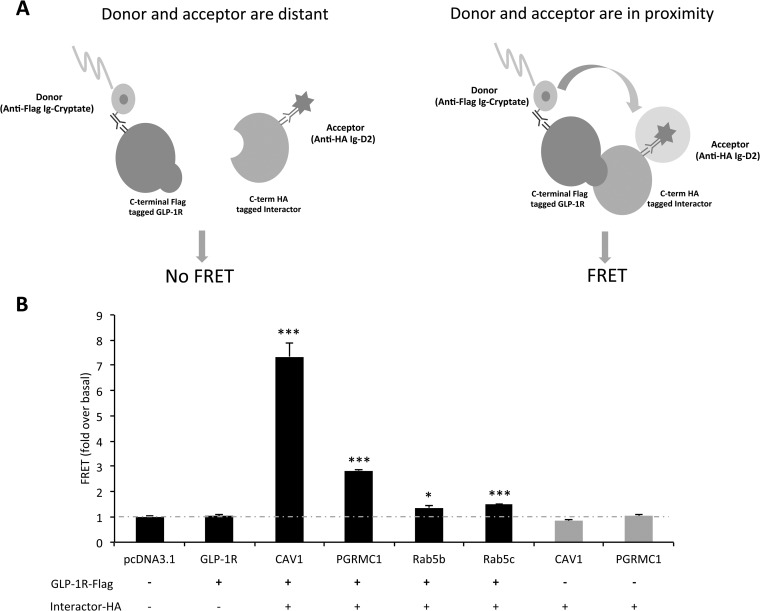
HTRF technology, combining fluorescence resonance energy transfer (FRET) with time-resolved measurement, was also employed to measure protein–protein interactions (Fig. 4A). The interactions between GLP-1R-Flag and HA-tagged CAV1, PGRMC1, Rab5b, and Rab5c were further confirmed with the fold change ranging from 1.37 to 7.35 when compared with the pcDNA3.1 control group (***p < 0.001, *p < 0.05, n = 3 per group, Fig. 4B). Additional control experiments revealed that the FRET was GLP-1R dependent and that both epitopes were required to be co-expressed within cells in order to produce a signal of protein–protein interaction (supplemental Figs. S6A and S6B).
Fig. 4.
Validation of selected GLP-1R interactor by FRET. A, the principle of FRET for measuring protein–protein interaction. Biotinylated anti-Flag antibody linked to donor molecule (cryptate) and anti-HA antibody linked to acceptor molecule (d2) were incubated with cells expressing GLP-1R-Flag and interactor-HA. If the interactor of interest does not bind to GLP-1R-Flag, the donor and acceptor are not in close enough proximity to generate a FRET signal. If the interactor of interest binds to GLP-1R-Flag, two fluorophores (donor and acceptor) are in close enough proximity for fluorescent energy to resonate between them following excitation, and a FRET signal will be observed at 665 nm. Thus, HTRF technology combines the advantages of FRET and time-resolved measurement (low background) to measure protein–protein interactions. B, HTRF measurement of protein–protein interactions further confirmed the interaction between GLP-1R and CAV1, PGRMC1, Rab5b, and Rab5c (***p < 0.001, *p < 0.05, n = 3 per group).
Immunofluorescence Supports Colocalization of Selected Interactors and GLP-1R in MIN6 Cells
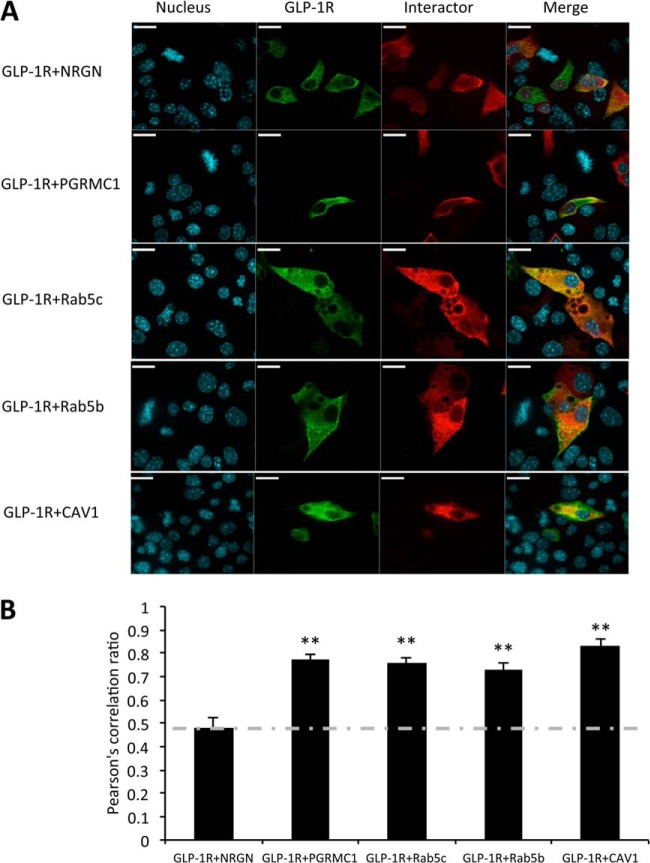
Immunofluorescence microscopy was used to assess the subcellular distribution of selected HA-tagged interactors and the GLP-1R-Flag in MIN6 cells. The stained nuclei are indicated in blue, and the immunostained proteins are shown in green (GLP-1R-Flag) and red (HA-tagged interactors) (Fig. 5A). Among the four interactors, PGRMC1 was observed to be mainly expressed on the plasma membrane, whereas Rab5b, Rab5c, and CAV1 were localized to both plasma membrane and cytoplasm. Pearson's correlation coefficient was calculated to quantify the degree of colocalization between each interactor and the GLP-1R. HA-tagged neurogranin was used as a negative control and was confirmed to not interact with GLP-1R as reported in our previous study (17). We found that PGRMC1, Rab5b, Rab5c, and CAV1 had significantly increased colocalization with GLP-1R (average PCC = 0.77, 0.75, 0.73, and 0.83 respectively) relative to neurogranin (average PCC = 0.47) (p < 0.01, n = 20 to 30 cells from three independent studies) (Fig. 5B). These results suggest that PGRMC1, Rab5b, Rab5c, and CAV1 colocalize with GLP-1R in MIN6 β cells, providing additional support for the interaction between these proteins and GLP-1R.
Fig. 5.
The interaction between GLP-1R and selected interactors as assessed by immunofluorescence. A, immunofluorescence images showing colocalization of selected interactors (red) and GLP-1R (green) in MIN6 cells. NRGN (neurogranin), a known non-interactive protein of GLP-1R, was used as the negative control. Bar = 26 μm. B, Pearson's correlation coefficient (PCC) analysis showed the colocalization between selected interactors and GLP-1R (**p < 0.01). PCC = 1, perfect colocalization; PCC = 0, absence of colocalization. n = 20 to 30 cells from three independent studies.
Overexpression of GLP-1R Interactors Affected Insulin Secretion in INS1 832/3 Cells
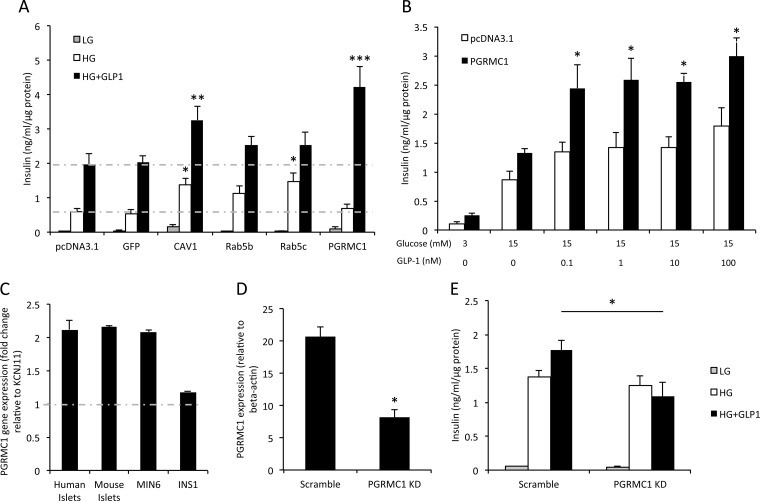
To assess how novel GLP-1R interacting proteins may affect GLP-1-mediated functions, we overexpressed four HA-tagged GLP-1R interactors (Rab5B, Rab5C, CAV1, and PGRMC1) in INS1 832/3 β cells and assessed their effects on GIIS. It was found that overexpression of CAV1 and PGRMC1 significantly increased the 10 nm GIIS relative to the negative control (pcDNA3.1 alone) (p = 0.007 for CAV1, ***p < 0.001 for PGRMC1, n = 3 per group, Fig. 6A). Overexpression of CAV1 and Rab5c increased GSIS in INS1 832/3 cells (p = 0.041 for CAV1, p = 0.038 for Rab5c), but overexpression of Rab5b did not significantly increase GSIS (p > 0.05). These results suggest that one novel GLP-1R interactor (PGRMC1) potentiates GIIS, and it was therefore selected for further mechanistic studies.
Fig. 6.
PGRMC1 effect on insulin secretion in INS1 832/3 cells. A, effects of selected GLP-1R interactors on insulin secretion in INS1 cells (*p < 0.05, **p < 0.01, ***p < 0.001, n = 3). B, PGRMC1 effect on GIIS (*p < 0.05, n = 3). C, endogenous PGRMC1 gene expression in human islets, mouse islets, MIN6 and INS1 cells. The expression of PGRMC1 was evaluated by the fold change relative to KCNJ11 based on previous microarray results (53–55). D, quantitative PCR showing efficient knockdown of PGRMC1 in INS1 832/3 cells using rat PGRMC1 siRNA transfection (*p < 0.05, n = 3). E, rat PGRMC1 knockdown effect on insulin secretion in INS1 832/3 cells (*p < 0.05, n = 3).
PGRMC1 Modulates GLP-1-induced Glucose-stimulated Insulin Secretion
PGRMC1, a member of the membrane-associated progesterone receptor family, is a single transmembrane protein with a cytochrome b5-like heme/steroid binding domain (31). It is known that progesterone (P4) binds to PGRMC1 with high affinity in mammalian cells and that PGRMC1 mediates P4 function (32–34). We observed that the overexpression of PGRMC1 increased GIIS significantly with multiple GLP-1 doses in INS1 832/3 cells (Fig. 6B). In brief, PGRMC1 overexpression increased GIIS 70% to 80% under 0.1 nm to 100 nm GLP-1 treatment (p = 0.033 for 0.1 nm, p = 0.004 for 1 nm, p = 0.027 for 10 nm, and p = 0.021 for 100 nm GLP-1; n = 3 to 4 independent experiments per group). We also showed that the PGRMC1 gene was significantly expressed in human islets, mouse islets, MIN6, and INS1 832/3 β cells (comparable to the expression level of the major subunit of the KATP-sensitive potassium channel KCNJ11) (Fig. 6C). To better assess the effect of reducing PGRMC1 expression in β cells, siRNA studies were performed on INS1 832/3 cells knocking down PGRMC1 gene expression by ∼60% (p < 0.05, Fig. 6D). It was found that PGRMC1 knockdown significantly reduced GIIS in INS1 832/3 cells (p = 0.019, n = 3 per group, Fig. 6E). These results indicated that PGRMC1 might play an important role in modulating GIIS and might be crucial in maintaining the response of β cells to GLP-1-stimulated insulin secretion.
Effects of PGRMC1 Activator and Inhibitor on GIIS
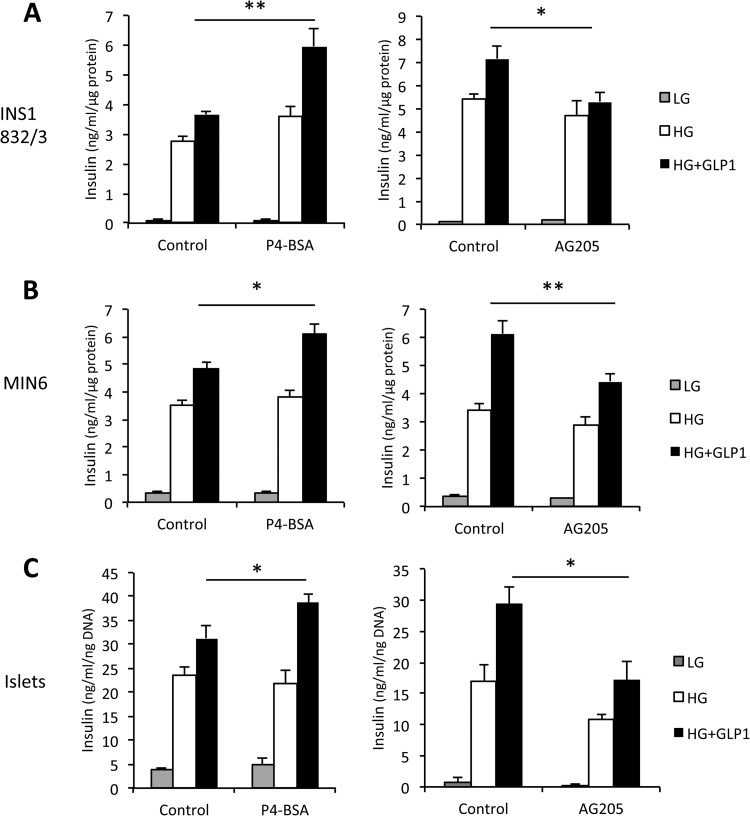
Based on the premise that PGRMC1 is a membrane-bound progesterone receptor that interacts with GLP-1R, we wanted to determine whether progesterone ligands affected GIIS. To assess this, we tested two PGRMC1 activators (P4, which is membrane permeable, and P4-BSA, which cannot penetrate into cells) on GIIS in INS1 832/3 cells, MIN6 cells, and primary mouse islets. We found that a 2-h treatment of 1 μm P4-BSA significantly increased GIIS in INS1 832/3 cells, MIN6 cells, and primary mouse islets relative to its vehicle control (p = 0.005 for INS1 832/3 cells, p = 0.011 for MIN6 cells, p = 0.038 for islets, n = 3 to 4 per group, Figs. 7A–7C). Interestingly, 2-h treatment of 1 μm P4 (the permeable ligand) had no significant effects on GSIS in INS1 832/3 cells, MIN6 cells, and mouse islets relative to its vehicle control (p > 0.05, n = 3 to 4 per group, supplemental Fig. S3). To further assess the relationship between PGRMC1 and GLP-1R, we examined the effect of a PGRMC1 inhibitor (AG-205) on GIIS. A 2-h treatment of 5 μm AG-205 significantly decreased GIIS in INS1 832/3 cells, MIN6 cells, and mouse islets (p = 0.021 for INS1 832/3 cells, p = 0.005 for MIN6 cells, p = 0.02 for islets, n = 3 to 4 per group, Figs. 7A–7C). These results suggest that PGRMC1 activity affects GIIS in INS1 β cells and primary mouse β cells, and the activation of membrane-bound progesterone receptor, including PGRMC1, may potentiate the insulinotropic actions of GLP-1 in pancreatic β cells.
Fig. 7.
Modulation of PGRMC1 activity affects insulin secretion in INS1 832/3 cells (A), MIN6 cells (B), and isolated primary mouse islets (C). The PGRMC1 surface ligand P4-BSA (1 μm) and AG205 (5 μm) were used to activate and inhibit PGRMC1, respectively (*p < 0.05, **p < 0.01, n = 3 to 4). Control = vehicle control group (1 μm fatty-acid-free BSA or dimethyl sulfoxide).
PGRMC1 May Potentiate GIIS through cAMP, EGFR, and PI3K Signaling
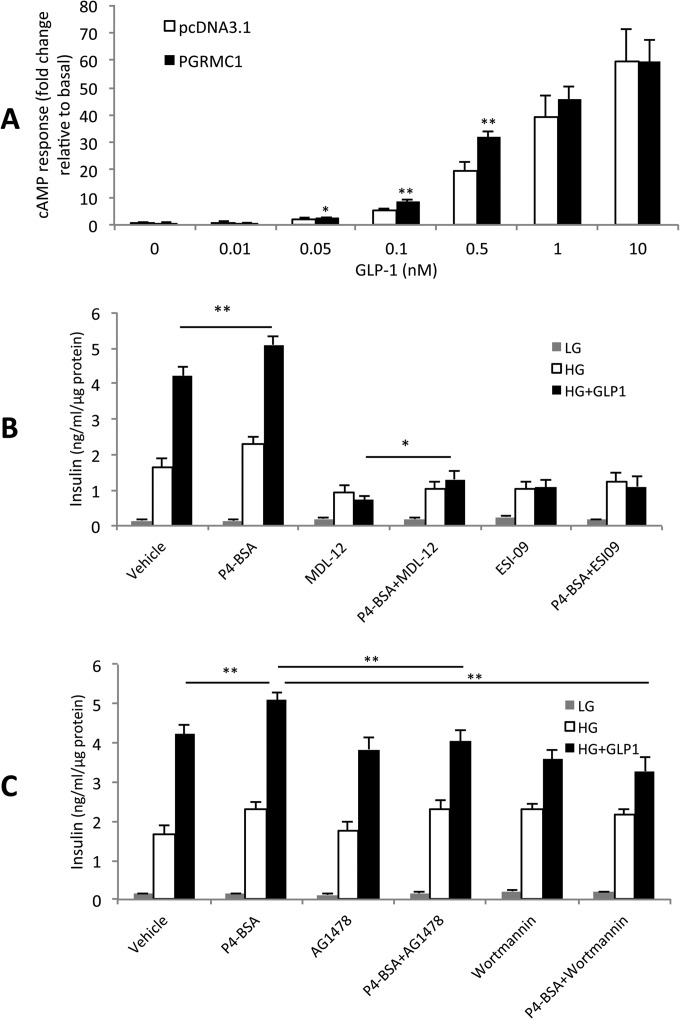
To explore how PGRMC1 increased GIIS, we first studied the effects of PGRMC1 overexpression on GLP-1-induced cAMP accumulation, because cAMP signaling is a well-known mechanism underlying GLP-1-induced insulin secretion. In RC2 cells, we found significant effects of PGRMC1 overexpression on 0.5 nm, 0.1 nm, and 0.05 nm GLP-1-induced cAMP accumulation after 30 min relative to pcDNA3.1 control (p = 0.004 for 0.5 nm, p = 0.001 for 0.1 nm, and p = 0.046 for 0.05 nm GLP-1 treatment; n = 3 to 6 per group; Fig. 8A). However, overexpression of PGRMC1 for 2 days did not alter the total insulin content in INS1 832/3 cells (n = 3 per group, supplemental Fig. S2B). To explore other potential mechanisms of enhanced GIIS, we also measured the effects of adenylyl cyclase inhibitor (MDL-12), EPAC inhibitor (ESI-09), EGFR inhibitor (AG1478), and PI3K inhibitor (wortmannin) on PGRMC1 surface ligand (P4-BSA)-increased GIIS in INS1 832/3 cells (Figs. 8B and 8C). Although it was found that EGFR-PI3K signaling was involved in GLP-1R activation-induced antagonism of Kv channels (35), there is no direct evidence that EGFR-PI3K signaling affects GLP-1-induced insulin secretion. Interestingly, 1-h treatment of adenylyl cyclase inhibitor (MDL-12) blocked the effect of GLP-1 on GSIS (p < 0.001), but 1 μm P4-BSA treatment still significantly increased GIIS in the presence of MDL-12 (p = 0.034, n = 3 per group, Fig. 8B), indicating that another mechanism independent of cAMP might be involved in the insulinotropic role of PGRMC1. However, EPAC inhibitor (ESI-09) treatment fully blocked the effect of P4-BSA on insulin secretion (p > 0.05, n = 3 per group, Fig. 8B). The EGFR inhibitor (AG1478) treatment significantly blocked the effect of P4-BSA-increased GIIS (p = 0.002, n = 3 per group) but retained the effect of GLP-1 on GSIS (Fig. 8C). PI3 kinase is the downstream target of EGFR and EPAC, and the inhibition of PI3K by wortmannin significantly blocked the effect of P4-BSA on GIIS (p < 0.001, n = 3 per group) but retained the effect of GLP-1 on GSIS (Fig. 8C). In addition, we also assessed the effect of EGFR inhibitor (AG1478) and PKG inhibitor RP8 (PKG is downstream of PGRMC1) on PGRMC1 overexpression-increased GIIS in INS1 832/3 cells and found that treatment of 100 nm AG1478 significantly reduced PGRMC1 overexpression-increased GIIS (p = 0.02, n = 3 per group), whereas no significant effects of RP8 were observed (supplemental Fig. S4A). These results collectively suggest that PGRMC1 potentiates GIIS by modulating both cAMP and EGFR-PI3K signaling.
Fig. 8.
Potential pathways involved in PGRMC1's effect on insulin secretion. A, PGRMC1 overexpression increased GLP-1-induced cAMP accumulation in a stable CHO cell line expressing hGLP-1R (RC2 cells) (*p < 0.05, **p < 0.01, n = 3 to 6). B, effect of cAMP pathway inhibition on P4-BSA increased GIIS (n = 3). Adenylyl cyclase (AC) inhibitor MDL-12 (25 μm) and EPAC inhibitor ESI-09 (30 μm) were used to inhibit the cAMP pathway. EPAC inhibition fully blocked the effect of P4-BSA (p > 0.05, P4-BSA+ESI-09 versus ESI-09), whereas AC inhibition only blocked the GLP-1 effect (***p < 0.001, MDL12 versus Veh) but retained the effect of P4-BSA on insulin secretion (*p < 0.05, P4-BSA + MDL-12 versus MDL-12). C, inhibition of EGFR signaling blocked P4-BSA increased GIIS (**p < 0.01, P4-BSA + AG1478 versus P4-BSA; **p < 0.01, P4-BSA + wortmannin versus P4-BSA, n = 3 per group). EGFR inhibitor AG1478 (100 nm) and EGFR downstream PI3K inhibitor wortmannin (100 nm) were used to block EGFR signaling. Vehicle = vehicle control group (1 μm fatty-acid-free BSA). The MDL-12, ESI-09, AG1478, and wortmannin groups all contained the vehicle.
DISCUSSION
Affinity Purification and Mass Spectrometry Revealed Potential GLP-1R Interactors
AP-MS has been employed by others to study receptor–protein interactions. Husi et al. studied the NMDA receptor complex (36) and revealed three proteins (NF1, Rsk-2, and L1) associated with learning impairment in mouse. Becamel et al. studied the 5HT receptor interactome (37) and discovered PSD95 and Veli3 as novel 5HT(2C) receptor interactors. Furthermore, Raju et al. studied the GABAA receptor complex (38) and discovered GABARAP as a novel autoantigen in a rare neurological autoimmune disease called Stiff-Person Syndrome. Because GLP-1R is an emerging target to treat type 2 diabetes, as well as cardiovascular and neurodegenerative disorders, we employed AP-MS to better understand the complex nature of GLP-1R signaling. In the current study, we revealed a total of 99 potential GLP-1R interactors from MIN6 and CHO cells (69 exclusively from MIN6, 13 from CHO, and 17 in common). The observation of more potential GLP-1R interactors in MIN6 than CHO cells (86 versus 30) could be due to the fact that MIN6 cells have an endogenous GLP-1R complex (17). In MIN6 cells, according to their biological function, liganded GLP-1R interactors were associated with metabolic processes, biosynthesis, and trafficking and ubiquitination, whereas unliganded GLP-1R interactors were associated with cellular signaling (Fig. 2B), suggesting that the GLP-1R interactome profile adapts to the receptor activity. Compared with the yeast two-hybrid system, AP-MS has the advantages of allowing study of protein–protein interactions in mammalian cells or tissues and the capacity to test the effects of ligands/compounds on the receptor interactome profile. When we compared the interactomes revealed by AP-MS and MYTH, we found that they shared similar functional categories. The majority of interactors from both AP-MS and MYTH are associated with transport and trafficking (44.4% versus 23.7% for AP-MS versus MYTH). This was followed by interactors associated with cellular signaling/G-protein signaling (22.2% versus 18.4% for AP-MS versus MYTH) and metabolic processes (9.1% versus 15.7% for AP-MS versus MYTH). Furthermore, we discovered interactors from the same functional/structural families including ATPases (ATP6V0B and ATP13A1 from MYTH screening; ATP1b1, ATP6v1h, and ATP2b1 from AP-MS screens) and progesterone receptor (PAQR6 from MYTH screening and PGRMC1 from AP-MS screens). Interestingly, we did not find specific common interactors in MYTH and AP-MS, which could be a result of the natures of the different methodologies. AP-MS targets the entire receptor complex, within which both direct and indirect interactions are revealed, whereas MYTH only identifies proteins with direct binary interaction with the receptor. Tissue hosts could also contribute to the difference: mammalian β cell settings were used in AP-MS, whereas in MYTH we used yeast as host and human fetal cDNA library as prey. Importantly, some proteins revealed in the current study were known GLP-1R interactors, such as caveolin-1 and G protein α and β subunits, indicating that the AP-MS method is effective in identifying novel GLP-1R interactors.
The Role of PGRMC1 in Modulating Insulin Secretion in Pancreatic β Cells
As mentioned, PGRMC1 belongs to the MAPR family (39) and binds progesterone with high affinity in mammalian cells. It also has the ability to bind other molecules, including testosterone, glucocorticoids, heme, and cholesterol metabolites (32, 33). Recently it was discovered that PGRMC1 binds σ-2 receptor ligands, indicating that the PGRMC1 complex is a putative σ-2 receptor-binding site (40). Although PGRMC1 is clearly involved in membrane-mediated progesterone signaling, it is unclear whether PGRMC1 is a specific progesterone receptor (32) or a component of the membrane progesterone receptor protein complex that acts as an adaptor protein (29). Some studies have shown that progesterone does not bind to bacterially expressed PGRMC1 proteins (41), but it is possible that the bacterially expressed PGRMC1 lacks proper folding to bind progesterone (32). Also, partially purified PGRMC1-fusion protein isolated from spontaneously immortalized granulosa cells or human granulosa/luteal cells specifically binds progesterone with high affinity (34, 42). One recent study revealed that PGRMC1 overexpression enhances cell surface expression and functions of membrane progesterone receptor α, suggesting that PGRMC1 also acts as an adaptor protein for seven transmembrane receptors to modulate receptor functions (29). It is interesting that another membrane progesterone receptor, PAQR6, was revealed to bind GLP-1R in our previous MYTH screening study (17) and that overexpression of PAQR6 increased GIIS in β cells (supplemental Fig. S5). Although PGRMC1 is highly expressed in pancreatic β cells, no previous studies have revealed its role in regulating β cell insulin secretion. Nonetheless, the PGRMC1 ligand (progesterone) was found to enhance islet insulin secretion and plasma insulin responses to glucose administration in rat (43, 44), mouse (45), and monkey (46). A recent study revealed that progesterone may increase insulin secretion by promoting incretin (GLP-1 and GIP) secretion that is mediated by progesterone receptors on the cell membrane of GLUTag cells (45), an immortalized cell model of intestinal l-cells.
PGRMC1 Increased GIIS by Regulating cAMP, EGFR and PI3K Pathways
Based on the positive actions of PGRMC1/P4-BSA on GIIS, it is important to better understand the mechanisms behind these effects. To address this we first focused on cAMP, the major known downstream signaling molecule of GLP-1R in most cell types (47). Our study revealed that recombinant PGRMC1 expression had a modest effect on augmenting GLP-1-induced cAMP accumulation, whereas PGRMC1 knockdown blocked the cAMP response to GLP-1 stimulation (supplemental Fig. S2A). In addition, inhibition of EPAC (downstream of cAMP; stimulates calcium-dependent insulin exocytosis (48)) blocked the effects of both GLP-1 and PGRMC1 ligand (P4-BSA) on insulin secretion. These results collectively indicate that endogenous PGRMC1 is a component of the liganded GLP-1R complex and potentiates cAMP signaling to increase GIIS in β cells. PGRMC1 might increase GLP-1-induced cAMP response by enhancing membrane GLP-1R expression; PGRMC1 can regulate endocytosis and trafficking of membrane proteins in yeast (41), stabilizes receptors on cell membranes (49), and acts as an adaptor protein to enhance seven-transmembrane protein expression on the cell surface (29). Interestingly, P4-BSA retains the ability to increase insulin secretion after blocking cAMP accumulation, suggesting that other mechanisms independent of the cAMP pathway contribute to PGRMC1-increased GIIS. As PGRMC1 is known to interact with EGFR (49), which is trans-activated by GLP-1R signaling through c-Src (50), and as increased EGFR activity can increase insulin secretion through increased calcium influx (51), we further explored the role of EGFR signaling in PGRMC1-modulated GIIS. We demonstrated the novel findings that EGF increased GIIS in INS1 β cells (supplemental Fig. S4B) and that inhibition of EGFR or PI3K (downstream of EGFR) blocked the effect of the PGRMC1 activator (P4-BSA) to impair GIIS. Our group has shown previously that GLP-1R agonist (exendin-4) blocks voltage-dependent K+ currents through both cAMP and EGFR-PI3K pathways (35, 52) and that antagonism of Kv channels increases GSIS (52). However, no direct evidence showed that the EGFR-PI3K pathway mediated GLP-1-induced insulin secretion. Therefore, our results indicate a novel role of PGRMC1 in enhancing GIIS by modulating both cAMP and EGFR/PI3K signaling to potentiate calcium flux and insulin exocytosis (supplemental Fig. S7, Fig. 9).
Fig. 9.
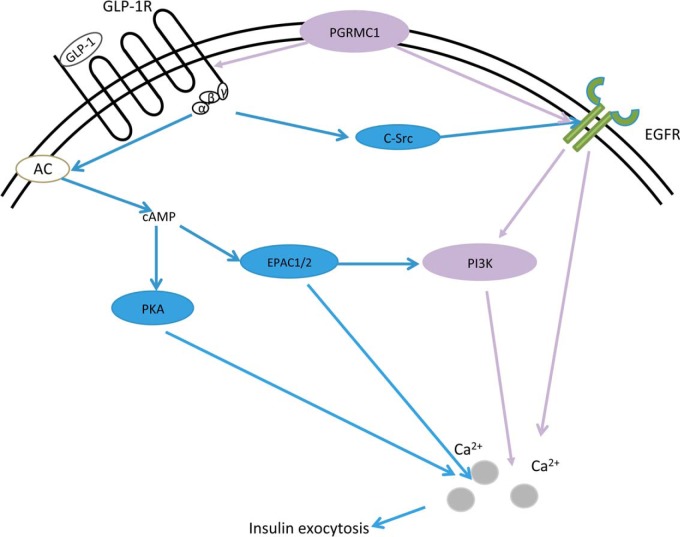
A schematic graph displaying the putative mechanism underlying PGRMC1's effect on GIIS. The diagram shows the classical role of GLP-1 in regulating insulin secretion in pancreatic β cells (blue color), mainly through cAMP, protein kinase A, and EPAC signaling. It is also known that GLP-1R may transactivate EGFR via c-Src to modulate insulin expression and β cell proliferation. Our study discovered the novel GLP-1R complex component PGRMC1 and its role in increasing GIIS through potentiating cAMP/EPAC signaling and activating EGFR and its downstream PI3K activity (purple color). Both cAMP and EGFR/PI3K signaling may collectively modulate downstream calcium flux and insulin exocytosis in pancreatic β cells.
It is possible that the phosphorylation status of the interactors could modify GPCR signaling. In the future, it would be of interest to assess phosphorylation changes of the interactors.
CONCLUSION
Our proteomics-based study uncovered a sizable network of potential GLP-1R interactors. Among three novel GLP-1R interactors associated with the liganded receptor, PGRMC1 displayed the most robust effect in modulating GIIS in pancreatic β cells. The mechanisms underlying PGRMC1 activation/overexpression-increased GIIS included cAMP and EGFR-PI3K signaling. This study revealed PGRMC1 as a novel component of the liganded GLP-1R complex, and its roles in β cells might allow for the design of novel drugs targeting PGRMC1 to maintain glucose homeostasis and possibly treat type 2 diabetes.
Supplementary Material
Acknowledgments
We thank Yuewei Qian and members of his laboratory for molecular biology support.
Footnotes
Author contributions: M.Z., K.W.S., and M.B.W. designed research; M.Z., M.R., A.D.S., X.H., Y.L., A.B., F.S.W., and J.H. performed research; M.Z., M.R., A.D.S., X.H., H.Y.G., S.A., K.W.S., and M.B.W. contributed new reagents or analytic tools; M.Z., M.R., A.D.S., X.H., F.F.D., and M.B.W. analyzed data; M.Z., F.F.D., and M.B.W. wrote the paper; M.R., A.D.S., F.S.W., S.F., L.W., K.W.S., F.F.D., and M.B.W. revised the paper; S.A. helped to design the experiment.
* This work was supported by grants from the Canadian Institutes of Health Research (MOP 102588) to M.B.W. and from Eli Lilly and Company through the Lilly Research Award Program to M.B.W. and K.S. M.Z. was supported by a Banting and Best Diabetes Centre (BBDC) Postdoctoral Fellowship. X.H. was supported by the BBDC and the Ontario Graduate Studentship program. J.H. was supported by a Shanghai Jiao Tong University Wuxi Research Institute Operating Grant. S.F. was supported by the BBDC-Novo Nordisk graduate student award.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- GLP-1
- glucagon-like peptide-1
- GLP-1R
- glucagon-like peptide-1 receptor
- AP-MS
- affinity purification–mass spectrometry
- cAMP
- cyclic adenosine monophosphate
- CAV1
- caveolin-1
- CHO
- Chinese hamster ovary
- co-IP
- co-immunoprecipitation
- EGFR
- epidermal growth factor receptor
- EPAC
- exchange protein directly activated by cAMP
- FRET
- fluorescence resonance energy transfer
- GIIS
- GLP-1-induced insulin secretion
- GSIS
- glucose-stimulated insulin secretion
- HTRF
- homogeneous time-resolved fluorescence
- INS1
- rat pancreatic β cell
- MIN6
- mouse pancreatic β cell
- MYTH
- membrane-based split-ubiquitin yeast two-hybrid
- P4
- progesterone
- PGRMC1
- progesterone receptor membrane component 1
- PCC
- Pearson's correlation coefficient
- PI3K
- phosphoinositide 3-kinase
- Rab5b
- Ras-related protein Rab-5B
- Rab5c
- Ras-related protein Rab-5
- RC2
- GLP-1R-His-V5 stable expressed Chinese hamster ovary
- RP8
- Rp-8-bromo-β-phenyl-1,N2-ethenoguanosine3,5-cyclic monophosphorothioate sodium salt hydrate.
REFERENCES
- 1. Nauck M. A., Heimesaat M. M., Orskov C., Holst J. J., Ebert R., Creutzfeldt W. (1993) Preserved incretin activity of glucagon-like peptide 1 [7–36 amide] but not of synthetic human gastric inhibitory polypeptide in patients with type-2 diabetes mellitus. J. Clin. Invest. 91, 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pratley R. E., Gilbert M. (2008) Targeting incretins in type 2 diabetes: role of GLP-1 receptor agonists and DPP-4 inhibitors. Rev. Diabetic Studies 5, 73–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drucker D. J. (2013) Incretin action in the pancreas: potential promise, possible perils, and pathological pitfalls. Diabetes 62, 3316–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fehmann H. C., Habener J. F. (1992) Insulinotropic hormone glucagon-like peptide-I(7–37) stimulation of proinsulin gene expression and proinsulin biosynthesis in insulinoma beta TC-1 cells. Endocrinology 130, 159–166 [DOI] [PubMed] [Google Scholar]
- 5. Kinzig K. P., D'Alessio D. A., Seeley R. J. (2002) The diverse roles of specific GLP-1 receptors in the control of food intake and the response to visceral illness. J. Neurosci. 22, 10470–10476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nauck M. A., Niedereichholz U., Ettler R., Holst J. J., Orskov C., Ritzel R., Schmiegel W. H. (1997) Glucagon-like peptide 1 inhibition of gastric emptying outweighs its insulinotropic effects in healthy humans. Am. J. Physiol. 273, E981–E988 [DOI] [PubMed] [Google Scholar]
- 7. Richter G., Feddersen O., Wagner U., Barth P., Goke R., Goke B. (1993) GLP-1 stimulates secretion of macromolecules from airways and relaxes pulmonary artery. Am. J. Physiol. 265, L374–L381 [DOI] [PubMed] [Google Scholar]
- 8. Moreno C., Mistry M., Roman R. J. (2002) Renal effects of glucagon-like peptide in rats. Eur. J. Pharmacol. 434, 163–167 [DOI] [PubMed] [Google Scholar]
- 9. Yamamoto H., Lee C. E., Marcus J. N., Williams T. D., Overton J. M., Lopez M. E., Hollenberg A. N., Baggio L., Saper C. B., Drucker D. J., Elmquist J. K. (2002) Glucagon-like peptide-1 receptor stimulation increases blood pressure and heart rate and activates autonomic regulatory neurons. J. Clin. Invest. 110, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Richards P., Parker H. E., Adriaenssens A. E., Hodgson J. M., Cork S. C., Trapp S., Gribble F. M., Reimann F. (2014) Identification and characterisation of glucagon-like peptide-1 receptor expressing cells using a new transgenic mouse model. Diabetes 63, 1224–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bullock B. P., Heller R. S., Habener J. F. (1996) Tissue distribution of messenger ribonucleic acid encoding the rat glucagon-like peptide-1 receptor. Endocrinology 137, 2968–2978 [DOI] [PubMed] [Google Scholar]
- 12. Gier B., Butler P. C., Lai C. K., Kirakossian D., DeNicola M. M., Yeh M. W. (2012) Glucagon like peptide-1 receptor expression in the human thyroid gland. J. Clin. Endocrinol. Metab. 97, 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jorgensen R., Martini L., Schwartz T. W., Elling C. E. (2005) Characterization of glucagon-like peptide-1 receptor beta-arrestin 2 interaction: a high-affinity receptor phenotype. Mol. Endocrinol. 19, 812–823 [DOI] [PubMed] [Google Scholar]
- 14. Sonoda N., Imamura T., Yoshizaki T., Babendure J. L., Lu J. C., Olefsky J. M. (2008) Beta-arrestin-1 mediates glucagon-like peptide-1 signaling to insulin secretion in cultured pancreatic beta cells. Proc. Natl. Acad. Sci. U.S.A. 105, 6614–6619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Syme C. A., Zhang L., Bisello A. (2006) Caveolin-1 regulates cellular trafficking and function of the glucagon-like Peptide 1 receptor. Mol. Endocrinol. 20, 3400–3411 [DOI] [PubMed] [Google Scholar]
- 16. Schelshorn D., Joly F., Mutel S., Hampe C., Breton B., Mutel V., Lutjens R. (2012) Lateral allosterism in the glucagon receptor family: glucagon-like peptide 1 induces G-protein-coupled receptor heteromer formation. Mol. Pharmacol. 81, 309–318 [DOI] [PubMed] [Google Scholar]
- 17. Huang X., Dai F. F., Gaisano G., Giglou K., Han J., Zhang M., Kittanakom S., Wong V., Wei L., Showalter A. D., Sloop K. W., Stagljar I., Wheeler M. B. (2013) The identification of novel proteins that interact with the GLP-1 receptor and restrain its activity. Mol. Endocrinol. 27, 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kittanakom S., Chuk M., Wong V., Snyder J., Edmonds D., Lydakis A., Zhang Z., Auerbach D., Stagljar I. (2009) Analysis of membrane protein complexes using the split-ubiquitin membrane yeast two-hybrid (MYTH) system. Methods Mol. Biol. 548, 247–271 [DOI] [PubMed] [Google Scholar]
- 19. Ahmed S. M., Daulat A. M., Angers S. (2011) Tandem affinity purification and identification of heterotrimeric g protein-associated proteins. Methods Mol. Biol. 756, 357–370 [DOI] [PubMed] [Google Scholar]
- 20. Chen G. I., Gingras A. C. (2007) Affinity-purification mass spectrometry (AP-MS) of serine/threonine phosphatases. Methods 42, 298–305 [DOI] [PubMed] [Google Scholar]
- 21. Anastas J. N., Biechele T. L., Robitaille M., Muster J., Allison K. H., Angers S., Moon R. T. (2012) A protein complex of SCRIB, NOS1AP and VANGL1 regulates cell polarity and migration, and is associated with breast cancer progression. Oncogene 31, 3696–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allister E. M., Robson-Doucette C. A., Prentice K. J., Hardy A. B., Sultan S., Gaisano H. Y., Kong D., Gilon P., Herrera P. L., Lowell B. B., Wheeler M. B. (2013) UCP2 regulates the glucagon response to fasting and starvation. Diabetes 62, 1623–1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu G., Zhang J., Larsen B., Stark C., Breitkreutz A., Lin Z. Y., Breitkreutz B. J., Ding Y., Colwill K., Pasculescu A., Pawson T., Wrana J. L., Nesvizhskii A. I., Raught B., Tyers M., Gingras A. C. (2010) ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 28, 1015–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Miteva Y. V., Cristea I. M. (2014) A proteomic perspective of SIRT6 phosphorylation and interactions, and their dependence on its catalytic activity. Mol. Cell. Proteomics 13, 168–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. von Mering C., Huynen M., Jaeggi D., Schmidt S., Bork P., Snel B. (2003) STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saito R., Smoot M. E., Ono K., Ruscheinski J., Wang P. L., Lotia S., Pico A. R., Bader G. D., Ideker T. (2012) A travel guide to Cytoscape plugins. Nat. Methods 9, 1069–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hardy A. B., Fox J. E., Giglou P. R., Wijesekara N., Bhattacharjee A., Sultan S., Gyulkhandanyan A. V., Gaisano H. Y., MacDonald P. E., Wheeler M. B. (2009) Characterization of Erg K+ channels in alpha- and beta-cells of mouse and human islets. J. Biol. Chem. 284, 30441–30452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dai F. F., Zhang Y., Kang Y., Wang Q., Gaisano H. Y., Braunewell K. H., Chan C. B., Wheeler M. B. (2006) The neuronal Ca2+ sensor protein visinin-like protein-1 is expressed in pancreatic islets and regulates insulin secretion. J. Biol. Chem. 281, 21942–21953 [DOI] [PubMed] [Google Scholar]
- 29. Thomas P., Pang Y., Dong J. (2014) Enhancement of cell surface expression and receptor functions of membrane progestin receptor alpha (mPRalpha) by progesterone receptor membrane component 1 (PGRMC1): evidence for a role of PGRMC1 as an adaptor protein for steroid receptors. Endocrinology 155, 1107–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drake M. T., Shenoy S. K., Lefkowitz R. J. (2006) Trafficking of G protein-coupled receptors. Circulat. Res. 99, 570–582 [DOI] [PubMed] [Google Scholar]
- 31. Mifsud W., Bateman A. (2002) Membrane-bound progesterone receptors contain a cytochrome b5-like ligand-binding domain. Genome Biol. 3, RESEARCH0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peluso J. J. (2013) Progesterone receptor membrane component 1 and its role in ovarian follicle growth. Front. Neurosci. 7, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thomas P. (2008) Characteristics of membrane progestin receptor alpha (mPRalpha) and progesterone membrane receptor component 1 (PGMRC1) and their roles in mediating rapid progestin actions. Front. Neuroendocrinol. 29, 292–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Peluso J. J., Romak J., Liu X. (2008) Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone's antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations. Endocrinology 149, 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. MacDonald P. E., Wang X., Xia F., El-kholy W., Targonsky E. D., Tsushima R. G., Wheeler M. B. (2003) Antagonism of rat beta-cell voltage-dependent K+ currents by exendin 4 requires dual activation of the cAMP/protein kinase A and phosphatidylinositol 3-kinase signaling pathways. J. Biol. Chem. 278, 52446–52453 [DOI] [PubMed] [Google Scholar]
- 36. Husi H., Ward M. A., Choudhary J. S., Blackstock W. P., Grant S. G. (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 3, 661–669 [DOI] [PubMed] [Google Scholar]
- 37. Becamel C., Alonso G., Galeotti N., Demey E., Jouin P., Ullmer C., Dumuis A., Bockaert J., Marin P. (2002) Synaptic multiprotein complexes associated with 5-HT(2C) receptors: a proteomic approach. EMBO J. 21, 2332–2342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raju R., Rakocevic G., Chen Z., Hoehn G., Semino-Mora C., Shi W., Olsen R., Dalakas M. C. (2006) Autoimmunity to GABAA-receptor-associated protein in stiff-person syndrome. Brain 129, 3270–3276 [DOI] [PubMed] [Google Scholar]
- 39. Petersen S. L., Intlekofer K. A., Moura-Conlon P. J., Brewer D. N., Del Pino Sans J., Lopez J. A. (2013) Novel progesterone receptors: neural localization and possible functions. Front. Neurosci. 7, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu J., Zeng C., Chu W., Pan F., Rothfuss J. M., Zhang F., Tu Z., Zhou D., Zeng D., Vangveravong S., Johnston F., Spitzer D., Chang K. C., Hotchkiss R. S., Hawkins W. G., Wheeler K. T., Mach R. H. (2011) Identification of the PGRMC1 protein complex as the putative sigma-2 receptor binding site. Nat. Commun. 2, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cahill M. A. (2007) Progesterone receptor membrane component 1: an integrative review. J. Steroid Biochem. Mol. Biol. 105, 16–36 [DOI] [PubMed] [Google Scholar]
- 42. Peluso J. J., Liu X., Gawkowska A., Johnston-MacAnanny E. (2009) Progesterone activates a progesterone receptor membrane component 1-dependent mechanism that promotes human granulosa/luteal cell survival but not progesterone secretion. J. Clin. Endocrinol. Metab. 94, 2644–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ashby J. P., Shirling D., Baird J. D. (1978) Effect of progesterone on insulin secretion in the rat. J. Endocrinol. 76, 479–486 [DOI] [PubMed] [Google Scholar]
- 44. Costrini N. V., Kalkhoff R. K. (1971) Relative effects of pregnancy, estradiol, and progesterone on plasma insulin and pancreatic islet insulin secretion. J. Clin. Invest. 50, 992–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Flock G. B., Cao X., Maziarz M., Drucker D. J. (2013) Activation of enteroendocrine membrane progesterone receptors promotes incretin secretion and improves glucose tolerance in mice. Diabetes 62, 283–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beck P. (1970) Reversal of progesterone-enhanced insulin production by human chorionic somatommammotropin. Endocrinology 87, 311–315 [DOI] [PubMed] [Google Scholar]
- 47. Willard F. S., Sloop K. W. (2012) Physiology and emerging biochemistry of the glucagon-like peptide-1 receptor. Exp. Diabetes Res. 2012, 470851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Holz G. G. (2004) Epac: a new cAMP-binding protein in support of glucagon-like peptide-1 receptor-mediated signal transduction in the pancreatic beta-cell. Diabetes 53, 5–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ahmed I. S., Rohe H. J., Twist K. E., Craven R. J. (2010) Pgrmc1 (progesterone receptor membrane component 1) associates with epidermal growth factor receptor and regulates erlotinib sensitivity. J. Biol. Chem. 285, 24775–24782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Buteau J., Foisy S., Joly E., Prentki M. (2003) Glucagon-like peptide 1 induces pancreatic beta-cell proliferation via transactivation of the epidermal growth factor receptor. Diabetes 52, 124–132 [DOI] [PubMed] [Google Scholar]
- 51. Lee H. Y., Yea K., Kim J., Lee B. D., Chae Y. C., Kim H. S., Lee D. W., Kim S. H., Cho J. H., Jin C. J., Koh D. S., Park K. S., Suh P. G., Ryu S. H. (2008) Epidermal growth factor increases insulin secretion and lowers blood glucose in diabetic mice. J. Cell. Mol. Med. 12, 1593–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. MacDonald P. E., Sewing S., Wang J., Joseph J. W., Smukler S. R., Sakellaropoulos G., Wang J., Saleh M. C., Chan C. B., Tsushima R. G., Salapatek A. M., Wheeler M. B. (2002) Inhibition of Kv2.1 voltage-dependent K+ channels in pancreatic beta-cells enhances glucose-dependent insulin secretion. J. Biol. Chem. 277, 44938–44945 [DOI] [PubMed] [Google Scholar]
- 53. Basford C. L., Prentice K. J., Hardy A. B., Sarangi F., Micallef S. J., Li X., Guo Q., Elefanty A. G., Stanley E. G., Keller G., Allister E. M., Nostro M. C., Wheeler M. B. (2012) The functional and molecular characterisation of human embryonic stem cell-derived insulin-positive cells compared with adult pancreatic beta cells. Diabetologia 55, 358–371 [DOI] [PubMed] [Google Scholar]
- 54. Van de Velde S., Hogan M. F., Montminy M. (2011) mTOR links incretin signaling to HIF induction in pancreatic beta cells. Proc. Natl. Acad. Sci. U.S.A. 108, 16876–16882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Prentice K. J., Luu L., Allister E. M., Liu Y., Jun L. S., Sloop K. W., Hardy A. B., Wei L., Jia W., Fantus I. G., Sweet D. H., Sweeney G., Retnakaran R., Dai F. F., Wheeler M. B. (2014) The furan fatty acid metabolite CMPF is elevated in diabetes and induces beta cell dysfunction. Cell Metab. 19, 653–666 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.