Abstract
Hypoxia is a critical microenvironmental factor that drives cancer progression through angiogenesis and metastasis. Glycoproteins, especially those on the plasma membrane, orchestrate this process; however, questions remain regarding hypoxia-perturbed protein glycosylation in cancer cells. We focused on the effects of hypoxia on the integrin family of glycoproteins, which are central to the cellular processes of attachment and migration and have been linked with cancer in humans. We employed electrostatic repulsion hydrophilic interaction chromatography coupled with iTRAQ labeling and LC-MS/MS to identify and quantify glycoproteins expressed in A431. The results revealed that independent of the protein-level change, N-glycosylation modifications of integrin α 3 (ITGA3) were inhibited by hypoxia, unlike in other integrin subunits. A combination of Western blot, flow cytometry, and cell staining assays showed that hypoxia-induced alterations to the glycosylation of ITGA3 prevented its efficient translocation to the plasma membrane. Mutagenesis studies demonstrated that simultaneous mutation of glycosites 6 and 7 of ITGA3 prevented its accumulation at the K562 cell surface, which blocked integrin α 3 and β 1 heterodimer formation and thus abolished ITGA3's interaction with extracellular ligands. By generating A431 cells stably expressing ITGA3 mutated at glycosites 6 and 7, we showed that lower levels of ITGA3 on the cell surface, as induced by hypoxia, conferred an increased invasive ability to cancer cells in vitro under hypoxic conditions. Taken together, these results revealed that ITGA3 translocation to the plasma membrane suppressed by hypoxia through inhibition of glycosylation facilitated cell invasion in A431.
As solid tumors grow, those areas distant from the existing blood vessels can become chronically or intermittently deprived of sufficient oxygen. These hypoxic conditions place tremendous pressure on tumor cells and drive the development of increasingly malignant and metastatic phenotypes (1, 2). As metastases are responsible for more than 90% of human cancer-related deaths, much research has aimed to define the underlying molecular mechanisms of the tumor cell response to a hypoxic microenvironment (3–5). However, despite the evident clinical relevance of metastasis, the full complexity of the process remains incompletely understood. An emerging area of interest is the importance of the carbohydrate structures in tumor cells, which have been linked to control of protein folding and stability, cell–cell recognition, adhesion, invasion, and metastatic potentials (6–11). The post-translational enzymatic addition of glycans (glycosylation) to proteins is a potent modulator of the functions of many receptors involved in cell growth, adhesion, and signal transduction (11–14) and is commonly seen in both in vitro and in vivo cancer models (15, 16). Furthermore, a growing body of studies has shown a clear correlation between aberrant glycosylation and human disease states, including cancers (11, 17, 18). Our previous proteomic and functional study revealed that glycosylation pathways in cell lines derived from human tumors are markedly altered by hypoxic conditions (5). Here we went on to investigate the significance of hypoxia-induced changes to glycosylation of specific proteins and to ask about the functional effects of those changes in tumor cells with regard to the metastatic phenotype.
The process of metastasis can be broadly broken down into several steps that include detachment from the primary tumor mass, exit from tissues into the blood vessel, transport through the blood, exit from the vessel at a distant site, and reestablishment in the new tissue (19). Central to the process is modulation of tumor cell adhesion to the extracellular matrix, other tumor or stromal cells, and the basement membrane, all of which involves members of the integrin family of adhesion molecules. Integrins are cell surface transmembrane glycoproteins that mediate cell–cell and cell–extracellular matrix attachment and communication, influencing cell cycle, motility, cytoskeletal organization, and morphology (20). Appropriate regulation of the glycosylation of integrins is crucial for their biological functions; for example, integrin α6β1 could not bind to its ligands or be normally transported to the cell surface after treatment of cells with tunicamycin, a glycosylation inhibitor (21), and cleavage of the N-glycans of purified integrin α5β1 blocked both heterodimer formation and ligand interaction (22). Aberrant changes in the N-glycosylation of integrins are often seen during carcinogenesis and have profound downstream effects on cell spreading and migration (23–25), both of which could feasibly impact the metastatic potential of tumor cells. Functional integrin heterodimers comprise large (α) and small (β) subunits. In mammals, 18 α and 8 β subunits have been characterized, which combine to generate 24 unique integrin heterodimers (26). Multiple integrin subunits are up-regulated under hypoxic conditions in a range of cell types (5, 27, 28). Among these, integrin α3β1 is an enigmatic member of the integrin family, and the regulation of its function also marks it as distinct from that of other integrins (29). It was originally identified as a promiscuous receptor for a range of ligands including collagen, laminin-1, fibronectin, and entactin (30, 31), but it has since been shown to favor laminin-5 and a5-containing laminins as ligands (32, 33), and perhaps thrombospondin-1 in certain cell types (34). There is much interest in the possible role of integrin α3β1 in the migration and metastasis of tumor cells (35). Early studies linked decreased expression of integrin α3β1 with the malignant phenotype in breast and lung cancer cells (36, 37), and more recently it was shown that expression of integrin α3β1 was down-regulated in transformed cells overexpressing the transcription factors c-myc or n-myc (38, 39). Other investigators suggested that integrin α3β1 might also stimulate invasiveness in certain tumors, particularly non-epithelial types that expressed laminin-5 (40, 41). Our own data showed that the overall protein expression level of ITGA31 was slightly induced by hypoxia in A431 (5); however, the changes of its glycosylation, and thus of its effect on cell migration, remain unstudied under hypoxia.
Considerable technical challenges exist for the accurate study of glycosylated proteins in complex biological systems, and to date several approaches have met with some success. The most commonly employed technique for the analysis of protein glycosylation is HPLC coupled to MS (42, 43). A number of HPLC protocols have been developed that enable the enrichment of glycopeptides from complex samples, including lectin affinity (44, 45), hydrazide reaction (46), ion exchange (47), hydrophilic-interaction chromatography (48), and TiO2 (49). For quantitative analysis of glycoproteins, Zhou et al. analyzed N-glycoprotein profiles in tear fluid using hydrazide-resin capture, iTRAQ labeling, and two-dimensional LC-MS/MS identification; a total of 43 unique N-glycoproteins were identified, and some were found to be modulated at several glycosites under disease conditions (50). In 2012, Palmisano et al. quantified formerly N-linked sialylated glycopeptides by means of TiO2 and salt-free fractionation with hydrophilic-interaction chromatography for enrichment and found that 300 unique sialylated glycopeptides were regulated during mouse brain development (51). Recently, the development of electrostatic repulsion hydrophilic interaction chromatography (ERLIC), a mixed-mode chromatography technique, has enabled glycopeptide and phosphopeptide enrichment based on both electrostatic and hydrophilic properties, which permits the identification of glycoproteins from complex mixtures via chromatographic enrichment and has been proved compatible with iTRAQ labeling (52–55).
We believed that application of these techniques to the quantitative assessment of protein glycosylation in cancer cells would lead to substantial advances in our understanding of the biology of human tumor cells. Therefore, in this study we employed ERLIC to enrich glycopeptides from tumor cell lysates and iTRAQ labeling and LC-MS/MS for the identification and quantification of N-glycosylated peptides. N-glycosylation modification of ITGA3 was detected down-regulated, and thus its secretion to plasma membrane was blocked under hypoxia, which was confirmed to enhance A431 cell invasion.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Reagents were purchased from Sigma Chemical Co. (St. Louis, MO) unless otherwise specified. Antibody against ITGA3 for WB and cell staining was from Proteintech (Chicago, IL) and BioLegend (San Diego, CA), respectively. The FITC-conjugated anti-ITGA3 and isotope control for flow cytometry was from AbD Serotec (Toronto, ON, Canada). Anti-actin (clone C4 MAB1501) and anti-ITGB1 were purchased from Millipore (Billerica, MA) and Santa Cruz (Santa Cruz, CA), respectively. Human laminin 5 was from Abcam (Cambridge, UK). Hygromycin B was bought from PAA Laboratories (Piscataway, NJ). EZ-Link Sulfo-NHS-LC-Biotin and avidin were purchased from Thermo Scientific (Rockford, IL). PNGase F was ordered from New England Biolabs (Ipswich, MA). Protease inhibitor mixture tablets were obtained from Roche (Basel, Switzerland).
Cell Culture and Hypoxic Conditions
The A431 human epithelial carcinoma cell line was purchased from ATCC and maintained in DMEM with 10% FBS. Normoxia, hypoxia, and reoxygenation treatments of A431 were performed as described previously (4, 5). Briefly, the cells cultured in serum-free media were exposed to normoxia (Nx) (21% O2, 5% CO2) in a normal incubator or hypoxia (Hx) (<0.1% O2, 5% CO2) in a modular incubator chamber (Billups-Rothenberg Inc., Del Mar, CA). For reoxygenation, the cells were exposed to Nx after a period of Hx culture and changed into fresh media. At least three independent biological replicates were pooled for the analyzed samples in each condition. The human chronic myelogenous leukemia cell line K562 was from ATCC and maintained in RPMI 1640 with 10% FBS.
Sample Preparation, Glycopeptide Enrichment, iTRAQ Labeling, and LC-MS/MS Analysis
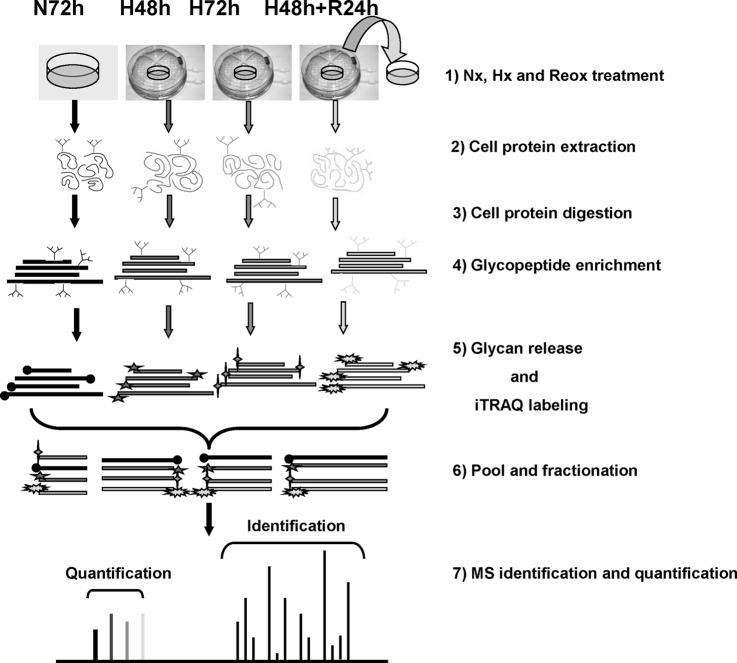
We employed an integrated strategy to allow the identification and quantification of glycoproteins in human cancer cell lines exposed to a range of oxygenation conditions (Fig. 1). Firstly we used ERLIC to enrich glycopeptides from the total tryptic peptides from A431 that had been subjected to Nx (72 h), Hx (48 h or 72 h), or Hx (48 h)/reoxygenation (24 h). The enriched glycopeptides were then treated with PNGase F to remove their N-glycans and labeled with iTRAQ reagents. These labeled deglyco-peptides were fractionated by ERLIC again and then identified and quantified using a Q-Star Elite mass spectrometer, because ERLIC has been proved as a more convenient and more effective alternative to strong cation exchange for the fractionation of iTRAQ-labeled peptides (55).
Fig. 1.
Experimental workflow for iTRAQ analysis of N-linked glycoprotein from A431 cells.
Nonenzymatic deamidation occurs in peptides under basic conditions commonly used in treatment with trypsin and PNGase F. Our previously published results showed that digesting peptides in pH 6.0 and releasing glycans in pH 5.0 significantly reduced the artifacts of glycosite identification from deamidated peptides introduced in sample preparation (56). Solutions with pH 6.0 and with pH 5.0 were used to extract A431 proteins and remove glycans of peptides after enrichment, respectively. Cellular proteins were extracted by 8 m urea in 20 mm phosphate buffer (pH 6) solution with protease inhibitor mixture (1:50). 4 mg of proteins from cells exposed to each oxygenation condition were reduced, alkylated, and digested by trypsin as described previously (5). For glycopeptide enrichment, the peptides were suspended in 80% ACN with 0.1% FA and passed through an ERLIC column (PolyLC, Columbia, MD; 4.6 × 200 mm, 5-μm particle size, 300-Å pore size) with a 78-min gradient at a flow rate of 0.5 ml/min, as per our previous research (0% buffer B for 10 min, 0% to 8% buffer B for 20 min, 8% to 12% buffer B for 6 min, and 100% buffer B for 42 min; A, 80% ACN/0.1% FA; B, 10% ACN/2% FA) (52). The peptides from the 36–78-min gradient were pooled and dried for deglycosylation. Phosphate buffer (pH 5) was used to dissolve the glycopeptides and for releasing N-linked glycans by PNGase F according to its instruction manual. The peptides without glycans were dissolved in 1 m TEAB (pH 8.5) and labeled with the isobaric tags (Applied Biosystems, Foster City, CA) according to the manufacturer's protocol as follows: the Nx-treated sample was labeled with 114; the samples under Hx for 48 h or 72 h were labeled with 115 or 116, respectively; and the reoxygenated sample was labeled with 117.
The labeled sample was fractionated on an ERLIC column (PolyLC; 4.6 × 200 mm, 5-μm particle size, 300-Å pore size) again using a Shimadzu Prominence UFLC system (Kyoto, Japan). Fractionation was performed with a 60-min gradient at a flow rate of 0.9 ml/min (0% buffer B for 5 min, 0% to 28% buffer B for 40 min, 28% to 100% buffer B for 5 min, and 100% buffer B for 10 min; A, 10 mm CH3COONH4 in 85% ACN/1% FA; B, 30% ACN/0.1% FA). A total of 30 fractions were collected and desalted using Sep-Pak C18 cartridges. Each fraction was dried and reconstituted in 40 μl of 3% ACN and 0.1% formic acid for Q-Star Elite LC-MS/MS analysis as previously described (4, 5). Briefly, peptides were separated on a home-packed nanobored C18 column with a picofrit nanospray tip (75-μm inner diameter × 15 cm, 5-μm particles) (New Objectives, Woburn, MA) at a flow rate of 300 nL/min with a 90-min gradient (13% to 15% B for 1 min, 15% to 23% B for 14 min, 23% to 40% B for 55 min, 40% to 65% B for 5 min, 65% to 90% B for 3 min, 90% B for 2 min, 90% to 13% B for 1 min, and 13% B for 9 min; A, 0.1% FA; B, 80% ACN in 0.1% FA). MS data were acquired in positive ion mode with a mass range of 300–1600 m/z using Analyst QS 2.0 software (Applied Biosystems). Peptides with +2 to +4 charge states were selected for MS/MS. The three most abundant peptides were selected for MS/MS and dynamically excluded for 30 s with a mass tolerance of 0.03 Da. The peak areas of the iTRAQ reporter ions were used to quantify proteins in the samples.
Database Searching and Criteria
Protein identification and quantification were performed using ProteinPilotTM software 3.0 with revision number 114732 (Applied Biosystems) by searching the combined raw data from the two runs of labeled sample against the concatenated “target” (UniProt human database, downloaded on June 14, 2012, including 87,187 sequences) databases. The Paragon and Pro Group algorithms in the ProteinPilot software were used for peptide identification and isoform-specific quantification. User-defined parameters were as follows: (i) sample type, iTRAQ 4-plex (peptide-labeled); (ii) cysteine alkylation, methyl methanethiosulfonate; (iii) digestion, trypsin; (iv) instrument, QStar Elite ESI; (v) special factors, emphasis on N deamidation; (vi) species, none; (vii) specify processing, quantitative; (viii) I.D. focus, biological modifications; and (ix) search effort, thorough I.D. The resulting data were automatically bias-corrected to account for potential reporter variations due to the possible unequal quantities of proteins in the different labeled samples. During bias correction, the software identifies the median average protein ratio and corrects it to unity, and then it applies this factor to all quantitation results. For iTRAQ quantitation, the peptide for quantification was automatically selected by the Pro Group algorithm to calculate the reporter peak area, error factor, and p value.
Glycopeptide Identification and Quantitation
Only the peptides with identification confidence ≥ 95% were adopted as identification. The glycopeptides were picked up according to the motif of NXS/T/C (where X is any amino acid except P) with deamidation of N to D. The identified glycosites were matched with the glycosites annotated in the UniProt database by our in-house Perl program for evaluation. The results contained information about glycosite position in the amino acid sequence and the type annotation of the glycosite in the UniProt database (“with reference support,” “potential,” or “unknown”). The ratio of unknown glycosites or those with reference support partially indicated the quality of the glycosite identification. To further confirm the quality of the identification, a manual check for the spectrum of each glycopeptide was performed. Sometimes database searches return some false-positive identifications of deamidated peptides because the 13C peaks of amidated peptides can be wrongly assigned as the monoisotopic peaks of the corresponding deamidated peptides (56). The close N or Q to the motif of glycosylation can also cause false-positive identification if the evidence is not enough to confirm which one is really deamidated.
For quantification, only the glycopeptides with less than 20% area error were adopted. The mean ± S.D. of the quantification ratio for all identified glycopeptides from the same protein was calculated as its fold change. For individual glycopeptides, the fold change was calculated from all quantifications from different scans. A paired t test was used for statistical analysis between two groups. The significance level was set at p < 0.05.
Cell Membrane Fractionation by Ultracentrifugation
Cell membrane purification was performed as described previously (53), with minor modifications: cells were collected via centrifugation at 4 °C, suspended in HES buffer (20 mm HEPES, pH 7.4, 1 mm EDTA, 250 mm sucrose) supplemented with protease inhibitor (Roche Diagnostics), and homogenized by repeated passage through a 27-gauge needle until most of the cells were lysed with intact nuclei released. The nuclei, remaining debris, and unbroken cells were removed by centrifugation at 1000 × g for 10 min at 4 °C. Resulting supernatants were transferred to an ultracentrifuge tube and centrifuged at 100,000 × g for 1 h at 4 °C. The pellets containing membrane fractions were resuspended in Na2CO3 (0.1 m, pH 11), sonicated for 30 s on ice, and centrifuged at 100,000 × g for 1 h at 4 °C. After two washes with Milli-Q water, membranes were collected via centrifugation at 100,000 × g for 30 min at 4 °C. The membrane pellet was then dissolved in 8 m urea solution, and the protein content was determined with a 2-D Quant kit (GE Healthcare) according to the manufacturer's instructions.
Cell Surface Biotinylation
K562 cells were washed three times with ice-cold PBS (pH 8.0) to remove amine-containing medium and proteins from the collected cells. The cells were then suspended at a concentration of ∼25 × 106 cells/ml in PBS (pH 8.0). Freshly prepared 10 mm Sulfo NHS-LC-Biotin reagent was added (200 μl/ml cell suspension) and incubated with the cells at room temperature for 30 min. Finally the cells were washed three times with ice-cold PBS with 25 mm Tris-HCl to remove and quench excess biotin-labeling reagent and its by-products. The biotinylated cells were then exposed to lysis buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, CompleteTM EDTA-free protease inhibitor mixture) before incubation with avidin at room temperature for 1 h. After the beads had been washed with lysis buffer three times, reducing SDS-PAGE loading buffer was used to elute the biotinylated proteins for WB as described below.
Cell Labeling for Microscopy and Flow Cytometry
For microscopy, A431 cells were stained with anti-ITGA3 (1:50, BioLegend) as described before (5). For flow cytometry, collected cells were washed three times with ice-cold PBS and then incubated with 5% human serum in PBS for 30 min on ice for blocking. Isotype control antibody (negative control) or FITC-conjugated anti-ITGA3 antibody was added and incubated for 1 h on ice. After three washes with 1% BSA in PBS, flow cytometry analyses were performed using a FACSCalibur instrument (BD Biosciences) operated with CELLQuestPro software.
Release of N-glycans from Glycoproteins in Solution
N48 and H48 A431 cell proteins were extracted in lysis buffer (1% SDS, 50 mm Tris-HCl, pH 8). The proteins with 0.5% SDS were denatured under 100 °C for 10 min in 40 mm DTT. After cooling, PNGase F (1:200 v/w) was added into the denatured proteins in 1×G7 buffer with 1% Nonidet P-40 (supplied with PNGase F kit). The reaction was stopped with heating at 100 °C for 10 min after incubation at 37 °C for 6 h. 20 μg of total proteins were used for WB.
Immunoprecipitation and Western Blotting
Cells were washed three times with ice-cold PBS before the addition of lysis buffer (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, CompleteTM EDTA-free protease inhibitor mixture). 31-gauge needles were used to facilitate cell lysis. The cell lysates were centrifuged at 12,000 × g for 15 min at 4 °C. The supernatants were collected and incubated with 3 μl of ITGA3 antibody overnight, and then 15 μl of protein G-Sepharose was added for an additional hour at 4 °C. After three washes with lysis buffer, SDS-PAGE loading buffer was used to elute proteins from the Sepharose beads for WB. The complexes were resolved by 7% SDS-PAGE and then transferred to nitrocellulose membrane and immunoblotted using antibodies as indicated in the figures. Proteins of interest were detected using the Invitrogen ECL system according to the manufacturer's instructions. For general Western blotting, the protein concentration of cell lysates dissolved in 1% SDS in 40 mm Tris-HCl (pH 8.0) was quantified using BCA assay to enable the use of equal total protein amounts from each sample for comparison.
Construction and Mutagenesis of ITGA3 Expression Vectors
Full-length ITGA3 cDNA was obtained from the A431 cDNA library via PCR as mentioned before (5). There are two isoforms of ITGA3 in mammalian cells: isoform 1 is widely expressed, whereas isoform 2 is expressed only in brain and heart. Thus we designed primers for the common isoform 1 of ITGA3 (supplemental Table S4). The ITGA3 gene was inserted into the expression vector pCDNA3.1+/Hygromycin. For mutagenesis, the primers were designed by the software QuickChange Primer Design from Agilent Technologies (supplemental Table S5). The mutation of glycosites within ITGA3 was performed using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) according to the manufacturer's protocol. All sequences of WT-ITGA3 and its mutants were confirmed by DNA sequencing. For stable expression of WT or mutated ITGA3, K562 and A431 cells were transfected using lipofectamine 2000 (Invitrogen) at a ratio of 1:5 (w/w) according to the manufacturer's instructions. Successfully transfected cells were then selected by culture in standard medium with 0.4 mg/ml hygromycin B. After 2 weeks, to get monoclones with WT or mutated ITGA3 expression in K562, the cells were diluted into at most one cell per well in 96-well plates. For A431 monoclones, cells were picked up from the plates by means of trypsin digestion. The positive clones were confirmed by WB.
Knockdown of ITGA3 Expression in A431
The primer sequences for shRNA knockdown of ITGA3 or luciferase (control) were generated by the Invitrogen BLOCK-iT™ RNAi Designer (supplemental Table S5). These fragments were then inserted into shRNA vector pSuperior-retro-puro after annealing according to the manufacturer's instructions. The shRNA plasmids with positive insertions were transfected into A431 with lipofectamin 2000 (Invitrogen) according to the manufacturer's protocol. Cells were then selected with standard medium with 0.5 μg/ml puromycin. After 1 week, the monoclones were picked up from the plates via trypsin digestion. The positive clones were confirmed by WB.
Cell Spreading Assay
96-well plates were coated with 2 μg/ml laminin 5, or 0.5% BSA in DMEM as a negative control. K562 cells were seeded at 5 × 104 live cells per well and incubated for indicated times at 37 °C before imaging at ×100 magnification via light microscopy. To assess the spreading of K562 cells on an in vitro extracellular matrix, A431 cells were cultured on glass coverslips for 3 to 5 days before removal by sequential washing with 1% Triton X-100 in PBS, 2 m urea in 1 m NaCl, and 8 m urea (all containing protease inhibitor mixture). The resulting coverslips were washed with PBS, blocked with BSA, and used in cell spreading assays (32).
Transwell Invasion Assay
24-well Transwell plates containing 8-μm-pore polycarbonate filters (Corning Costar, Cambridge, MA) were used for invasion assay. Transwell insert membranes were coated with 100 μl of laminin 5 and incubated overnight at 4 °C. Triplicate assays were performed for each cell line. 4 × 104 cells in 100 μl of serum-free medium were seeded in each top Transwell insert, and 600 μl of media with serum were added in the lower chamber. After 24 h of incubation, the cells without migration were swabbed from the upper wells using cotton buds. Cells that had migrated through the membranes were fixed with 4% formaldehyde for 20 min and then stained with crystal violet for image capture and counting under a light microscope. For quantification, the membranes were cut carefully from the inserts. The cells on the membranes were solubilized by 200 μl of 0.5% Triton X-100 overnight at room temperature. The absorbance was measured at A595 using a microplate reader (Tecan Magellan, Männedorf, Switzerland).
Statistical Analysis
Values are expressed as mean ± S.D. Paired t testing was used for statistical analysis between two groups. The significance level was set at p < 0.05.
RESULTS AND DISCUSSION
Analysis of Identified Glycoproteins Enriched via ERLIC
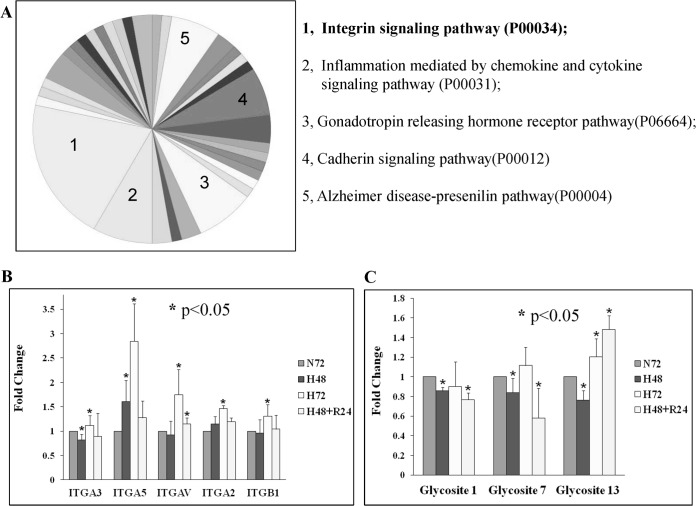
Protein and peptide identification summary results are listed in supplemental Tables S1 and S2. A total of 1209 proteins were identified with at least two unique peptides (confidence ≥ 95%). For glycopeptides, only those with confidence ≥ 95% were adopted as identifications and used for further analysis. Glycopeptides were detected by the motif NXS/T/C (where X is any amino acid except P and N is deamidated). In total, 241 glycopeptides with 215 glycosites were identified, belonging to 146 proteins. Matching these glycosites with those provided in the UniProt database, we found that 93 glycosites already had “reference support,” 50 sites were classified as “potential,” and only 72 (33.5%) were unknown (Table I and supplemental Table S3_1–3). Putative false-positive N-glycosites were highlighted by matching with the potential false N-glycosites obtained from A431 whole proteome analysis, in which the proteins were digested at pH 8.0 without PNGase F treatment. In all, 26 glycosites from this study were also identified in the whole proteome analysis, with 24 of these in the “unknown” group and 2 in “reference support.” The deamidation of asparagine is accelerated when the following amino acid is glycine, which can easily cause false-positive glycosylation identification during sample preparation (57), and accordingly 12 of these deamidated N peptides (46.2%) were in N-G-S/T sequences (supplemental Table S3_1–3). To further confirm the identification, the MS/MS spectra of peptides with putative N-glycosite identification were checked manually one by one. In total, 180 spectra with fragmentation matching evidence were provided with higher confidence, including all 93 glycosites with UniProt reference support, 48 of 50 UniProt potential sites, and 39 of 72 UniProt unknown sites (supplemental Table S3_1–3). The Functional Annotation Clustering tool from DAVID Bioinformatics Resources 6.7 was used to group the 146 proteins (58). It indicated that 101 of the 146 proteins we identified with N-glycosites were known N-linked glycoproteins, with 84 being membrane-associated and 70 of those belonging to the plasma membrane. The dominant functional category was cell adhesion (supplemental Fig. S1). As a major family of adhesion proteins, integrins play an important role in cell adhesion and migration. Accordingly, pathway analysis of the 146 proteins using the Panther classification system revealed that the integrin signaling pathway was also the dominant category (Fig. 2A). Within the integrin category we identified seven integrin subunits, and we quantified the glycopeptides of these subunits that were modulated by hypoxia and reoxygenation (supplemental Table S4 and Fig. 2B). Only five items of integrin subunit quantification information are presented in Fig. 2B because ITGA6 and ITGB6 had no statistical quantification information to represent their changes due to hypoxia (supplemental Table S4). With 72-h hypoxia, all five integrin subunits had an increase in glycosylation with p < 0.05; ITGA3 had the slightest increase with a ratio of 1.11 ± 0.2, and the others had ratios greater than 1.3. The quantification results were consistent with our findings in the A431 whole proteome analysis for 72-h hypoxia (ITGA3: 1.14 ± 0.21) (5). Interestingly, with 48-h hypoxia, only ITGA3 showed a significant decrease in glycosylation modification with p < 0.05 (0.82 ± 0.12). The integrin quantification results were confirmed by a new iTRAQ analysis with 2 MS running in another biological repeat: ITGA3 was down-regulated at hour 48 (0.9 ± 0.22) and slightly up-regulated at hour 72 (1.17 ± 0.26) (supplemental Table S4). Our previous study showed that the total protein expression level of integrin subunits (ITGA5/3/6/V, ITGB1/4) under hypoxia was up-regulated (5), and therefore the increased level of glycosylated integrin subunits found here (ITGA5/2/V and ITGB1) might be attributable to whole expression level up-regulation; however, the level of glycosylated ITGA3 decreased at hour 48, whereas its total protein level increased (ITGA3: 1.14 ± 0.12 and 1.15 ± 0.09 for iTRAQ and LC–multiple reaction monitoring–MS/MS, respectively). Three glycosites of ITGA3 (glycosite 1/7/13) were identified in our iTRAQ data; Fig. 2C indicates that all three sites were significantly less glycosylated (p < 0.05) under 48-h hypoxia. In 2011, Parker et al. enriched and quantified iTRAQ-labeled N-glycopeptides during myocardial ischemia and reperfusion injury via sequential enrichment with TiO2 followed by IP-ZIC-HILIC (Zwitterionic hydrophilic interaction chromatography with ion-pairing) for the unbound fraction. More than 400 N-linked glycopeptides were statistically quantified, and 80 glycosylation sites were found altered, in which the glycosylation of several integrin subunits (ITGA6/7/V and ITGB1) was also found up-regulated following ischemia and reperfusion injury (59). These results were consistent with our findings.
Table I. Analysis of glycoproteins isolated via ERLIC enrichment and Q-Star Elite identification.
| Glycoprotein number | Glycopeptide number | Glycosite number | Glycosite characterization |
||
|---|---|---|---|---|---|
| With reference support | Potential | Unknown | |||
| 146 | 241a | 215 | 93 | 50 | 72 |
a All glycopeptides had ≥95% identification confidence, and one site might be repeatedly identified in different peptides.
Fig. 2.
The glycosylation of integrin subunits was modulated by hypoxia. A, Panther pathway analysis results for all identified putative glycoproteins. B, iTRAQ analysis results for identified glycopeptide changes of five integrin subunits modulated by hypoxia. C, iTRAQ analysis results for the changes in three identified glycopeptides in ITGA3 responsible for hypoxia.
Translocation of ITGA3 to the Plasma Membrane Was Suppressed under Hypoxia
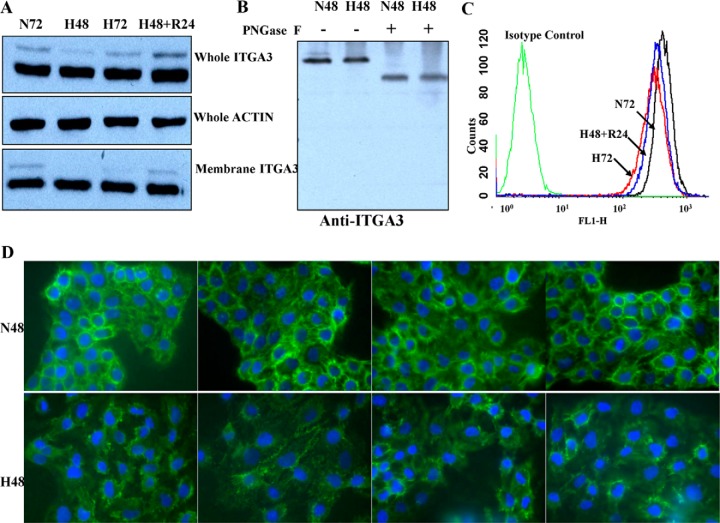
WB was performed to confirm the levels of ITGA3 protein in lysates from A431 cells under normoxia, hypoxia, and reoxygenation conditions. As shown in Fig. 3A, two different MW isoforms of ITGA3 were detected; the abundance of the lower MW isoform did not show any significant changes induced by hypoxia, whereas the higher MW isoform seemed to decrease obviously after 48 h of hypoxia and slightly after 72 h, with clear recovery of expression level upon reoxygenation. These results clearly showed that the real changes in abundance of the higher MW ITGA3 isoform were identified by glycopeptide quantification, whereas they had been obscured by the relatively more abundant low-MW isoform in our previous whole proteome quantification (5). Because ITGA3 undergoes extensive glycosylation in the endoplasmic reticulum and Golgi during translocation to the plasma membrane, we hypothesized that the higher MW ITGA3 isoform would be destined for the plasma membrane and therefore more relevant for tumor cell attachment and migration. To test our hypothesis, we adopted PNGase F to treat the cell proteins from Nx and Hx. Fig. 3B shows that both ITGA3 isoforms disappeared and combined as one band with a lower MW after PNGase F treatment to remove N-glycans. This result clearly indicates that both isoforms had glycosylation modifications and that the higher MW isoform had more glycosylation than the lower MW one. Membrane fraction WB results confirmed that the decrease in the higher MW ITGA3 isoform in response to hypoxia was more pronounced in plasma membrane fractions than in whole cell lysates (Fig. 3A, lowest panel). Whole A431 cells exposed to the different oxygenation conditions were then surface labeled with ITGA3-specific FITC-conjugated antibodies, and the intensity of ITGA3 expression at the cell surface was quantified via flow cytometry. The results further confirmed that less membrane ITGA3 was present following 72 h of hypoxia than after normoxia for 72 h. In agreement with the WB results, membrane ITGA3 levels recovered in the cells undergoing 24 h of reoxygenation after 48-h Hx treatment (Fig. 3B). A decrease of membrane ITGA3 in cell staining was also observed via fluorescence microscopy (Fig. 3C). Thus, though hypoxia did not markedly affect the overall expression level of ITGA3 in A431 cells, it decreased ITGA3 glycosylation, which is associated with reduced abundance of ITGA3 at the plasma membrane.
Fig. 3.
Less ITGA3 was detected on the plasma membrane of A431 cells under hypoxia. WB results showed that only the higher MW isoform of ITGA3 was down-regulated by hypoxia (A), and it disappeared and combined with the smaller isoform as one band with a lower MW after N-glycan removal (B). This decrease was amplified in the membrane fractions (lowest panel in A). Lower levels of membrane ITGA3 on A431 cells under hypoxia were confirmed by flow cytometry (C) and immunofluorescence cell staining (D).
N-linked Glycosylation at Sites 6 and 7 Is Essential for the Translocation of ITGA3 to the Plasma Membrane
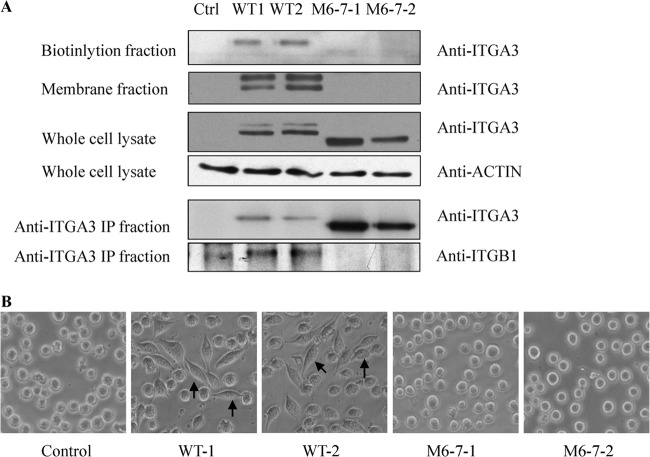
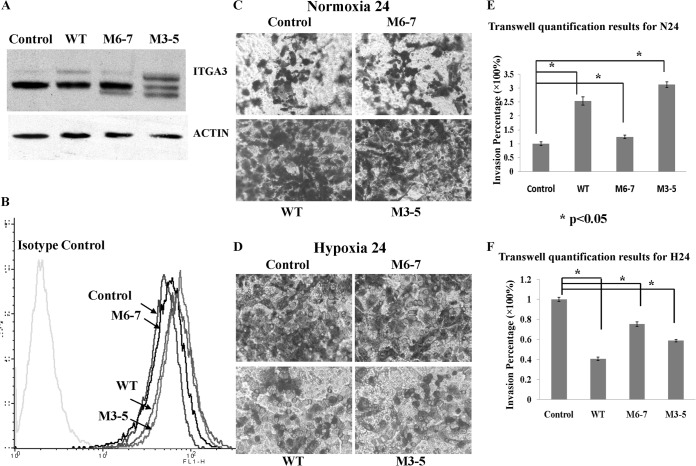
To study the functional effects of the changes to ITGA3 glycosylation induced by hypoxia, we first constructed a stable cell line expressing either WT ITGA3 or ITGA3 mutated at its potential glycosites. K562 cells do not express ITGA3 but do express its binding partner ITGB1 (60, 61), which renders them a good model for measuring changes in the biological function of ITGA3 following heterodimerization to form the active α3β1 integrin receptor. The potential N-glycosylation sites of human ITGA3 are (Asn-Xaa-Ser/Thr), Asn-86, Asn-107, Asn-265, Asn-500, Asn-511, Asn-573, Asn-605, Asn-656, Asn-697, Asn-841, Asn-857, Asn-926, Asn-935, and Asn-969, which are all located in the extracellular segment. We constructed nine potential N-glycosite mutants as shown in supplemental Fig. S2; those sites within the β-propeller domain were chosen as mutation targets following the report that glycosites in this domain are required for membrane translocation of another integrin family member, ITGA5 (9). All primers for ITGA3 expression and mutation are listed in supplemental Table S5. The pcDNA3.1+/Hygromycin plasmids containing WT or mutated ITGA3 sequences were transfected into K562 cells. Surface ITGA3 expression was measured on transfected cells via flow cytometry. Supplemental Fig. S3 shows that some cell subpopulations with membrane ITGA3 were detected in the pool of cells transfected with WT and M6-, M7-, and M3-5-containing plasmids, but cells expressing M6-7 had no membrane ITGA3. Membrane ITGA3 was also detected on M3/4/5-transfected cells (data not shown). This suggested that the secretion of ITGA3 might be blocked by the simultaneous mutation of glycosites 6 and 7. To confirm this conclusion, two WT ITGA3-positive clones and two M6-7-ITGA3-expressing clones were selected for more detailed analysis. As shown in Fig. 4A, WB revealed that the WT ITGA3 clones expressed both higher and lower MW isoforms of ITGA3, as in A431 cells; however, the M6-7 clones expressed only one lower MW form of ITGA3, consistent with less glycosylation. From this we hypothesized that M6-7 ITGA3 would be absent on the plasma membrane because it might not complete intact glycosylation modification through Golgi to form the isoform with a higher MW, and this was confirmed by Western blotting of membrane fractions of the transfected cells. No membrane ITGA3 bands were detected in the two clones of M6-7, but two bands were developed in the membrane fractions of WT clones with more enrichment for the bigger isoforms. The WB results from biotin-labeled fractions of membrane surface proteins further confirmed the lack of membrane ITGA3 in the two M6-7-expressing cell lines. In this result, mainly the bands with lower MWs in WT cell lines were detected after enrichment, and the higher bands were developed with overexposure after more samples were loaded (data not shown). It is possible that the biotin reagent (EZ-Link Sulfo-NHS-LC-Biotin) only reacts efficiently with primary amines exposed on surface, but the primary amines in isofoms with higher glycosylation are blocked by glycans, which causes inefficient labeling. The immunoprecipitation results similarly showed an absence of membrane ITGA3 in the cell lines of clones expressing M6-7-mutated ITGA3: using an immunoprecipitation antibody recognizing the C terminus of ITGA3, we found that only ITGB1 was detected in WT ITGA3 immunoprecipitation products, and no ITGB1 was bound with M6-7 ITGA3 in its immunoprecipitation products, even with more M6-7 ITGA3 precipitation. The impact of the absence of integrin α3β1 heterodimers on the cell membrane was then functionally assessed in a cell spreading assay. As expected, transfection of WT ITGA3 enabled rapid K562 cell adhesion and spreading onto plates coated with the α3β1 ligand laminin 5 within 20 min (Fig. 4B), which was abolished by the addition of an anti-ITGA3 antibody, but not by normal mouse IgG (data not shown). In contrast, expression of the ITGA3 glycosylation mutant M6-7 did not rescue cell spreading even when cells were cultured for more than 2 h on plates coated with laminin 5 (Fig. 4B). The transfection of M3-5 did induce cell attachment and spreading on laminin 5–coated plates, as did overexpression of the WT ITGA3 in K562 cells (data not shown). The same pattern of results was observed on plates coated with A431-derived extracellular matrix (data not shown). Together these results suggest that N-glycosylation at sites 6 and 7 in the ITGA3 subunit is essential for its translocation to the cell surface and its biological functions. In endoplasmic reticulum, newly synthesized ITGA3 precursors undergo N-glycosylation and folding facilitated by the chaperone calnexin and are assembled with ITGB1 and other proteins as a complex. The properly folded and assembled complex is then transported to the Golgi, where its high-mannose glycans are processed to complex types and mature integrin α3β1 is formed finally. A lack of glycosylation at sites 6 and 7 might cause a failure to reach the final conformation for the complex formation and thus prevent the traffic of ITGA3 to plasma membrane. Nicolaou et al. reported that a gain of glycosylation at amino acid 349 of ITGA3 via mutation impeded its heterodimerization with ITGB1 and thus impaired its expression at the cell surface, which provided strong support to explain the effects of ITGA3 conformation on its traffic and functions (11). The molecular mechanism that controls specific site glycosylation still remains unclear. Our attempts to prove that the glycosylation of sites 6 and 7 is reduced simultaneously in hypoxia have failed so far. The partners that favor integrin α3β1 complex assembly might be good targets for hypoxia regulation to control ITGA3 transport processes.
Fig. 4.
N-linked glycosylation sites 6 and 7 were essential for the translocation of ITGA3 to the plasma membrane in K562 cells. A, WB revealed a single ITGA3 band in K562-M6-7 mutants, with an absence of ITGA3 in membrane or biotinylation fractions. Accordingly, no binding of ITGA3 and ITGB1 was detected in immunoprecipitation experiments. B, transfection of WT ITGA3 into K562 cells rescued cell spreading on laminin 5, whereas no cell spreading was detected in cells expressing M6-7. Arrows indicate spread cells on laminin 5–coated plates.
Decreased Surface ITGA3 Enables Increased A431 Cell Migration under Hypoxic Conditions
Integrin α3β1 is important for cell migration, but both positive and negative effects have been reported (62–64), with one study even finding that knockdown of ITGA3 in keratinocytes compromises collective migration but enhances single-cell migration (65). To mimic the changes in ITGA3 induced by hypoxia, the cDNAs of WT and mutant cells were transfected into A431 to determine the effect of less membrane ITGA3 on cell invasion. As shown in Fig. 5A, WBs of lysates from A431-WT cells had the same two bands as the control-plasmid transfected cells, but they showed a marked increase in expression of the higher MW isoform. A431-M3-5 cell lysates had four bands: two for the original WT ITGA3 isoforms basally expressed by A431, and two lower isoforms following the original ones arising from partially glycosylated M3-5 ITGA3 mutants. A431-M6-7 produced only three bands: two for basal WT ITGA3 expression, and the third with a lower MW representing expression of the less glycosylated M6-7 ITGA3. This phenomenon was consistent with the expression of WT and mutant ITGA3s in K562. Moreover, it seems that the overexpression of WT and M3-5 causes a greater increase in ITGA3 isoforms with higher MWs than of lower isoforms. The membrane ITGA3 in each stable cell line was detected via flow cytometry. In Fig. 5B, it is clear that the WT and M3-5 belong to the same population with similar membrane ITGA3 expression that is higher than in the population of control and M6-7. The functional effects of differing ITGA3 expression in the cells were then evaluated by means of transwell invasion assay with human laminin 5 coating under normoxic conditions for 24 h. Markedly, much more transmembrane was observed in cells overexpressing WT (2.5-fold ± 0.2-fold) or M3-5 (3.1-fold ± 0.1-fold) ITGA3 than in control cells transfected with pcDNA3.1+ (Fig. 5C). Under normoxic conditions, A431-M6-7 cells exhibited invasive ability similar to that of the control cells, in accordance with their similar membrane levels of ITGA3 (Figs. 5B and 5C). It has been reported that integrin α3β1 promotes radiation-induced migration of meningioma cells and that suppression of integrin α3β1 inhibits tumorigenesis and invasion in breast cancer cells (66). Our transwell assay on the ITGA3 knockdown cells also supported the same conclusion. Less total and membrane ITGA3 expression in A431 suppressed cell invasion through laminin 5–coated membranes under normoxia (supplemental Fig. S4). In contrast, the transwell invasion assay performed under hypoxia revealed that the invasion of cells with WT and M3-5 overexpression was substantially less than that of control cells (0.41 ± 0.02 and 0.59 ± 0.01), whereas cells transfected with the M6-7 mutant had a higher relative invasion rate (0.75 ± 0.02 of control cells) than WT or M3-5 overexpression cells (Fig. 5D). In these assays, M3-5 had an invasive ability similar to that of WT because it had normal membrane secretion of ITGA3 even with three glycosite mutations. M6-7 had behavior similar to that of control cells because of the lack of membrane ITGA3. These transwell invasion assay results clearly showed that under normoxia the extent of cell invasion positively correlated with the level of plasma membrane ITGA3, whether in ITGA3 overexpression or knockdown A431 cells. Under hypoxia, however, the situation was different; A431-WT transfected cells, with higher levels of plasma membrane ITGA3, had less invasion than A431-M6-7 with less plasma membrane ITGA3. Thus we conclude that less ITGA3 translocation to the cell membrane, as induced by hypoxia, increases the invasion of A431 cells in suboptimal oxygenation conditions.
Fig. 5.
A431-M6-7 cells with less membrane ITGA3 had more invasive ability than A431-WT under hypoxia. A, B, whole or membrane ITGA3 in stable cell lines transfected with blank plasmids (control), WT, M6-7, or M3-5 ITGA3 was detected via Western blotting or flow cytometry, respectively. C, D, transwell images for each stable cell line under normoxia for 24 h and hypoxia for 24 h, respectively. E, F, transwell quantification results for each stable cell line under Nx and Hx measured with crystal violet A595. For normoxia, 0.5 μg/ml laminin 5 was coated with transwell membranes, and the medium with 5% FBS was added into the lower chambers. For hypoxia, to improve invasion, 1 μg/ml laminin 5 was coated with transwell membranes. 2.5% FBS and 10% FBS in the medium were adopted for cell suspension in the upper inserts and cell migratory stimulus in the lower chambers, respectively. The plates were subjected to normoxia for 2 h for cell attachment after cell seeding and then put into hypoxia chambers.
CONCLUSIONS
In this study, we quantified N-glycoproteins from normoxic, hypoxic, and reoxygenated A431 cells using iTRAQ-based proteomic approaches. Taking into account the clinical significance of metastasis, we focused on the integrin family of cell adhesion molecules and in particular ITGA3. We detected substantially decreased levels of N-glycosylated ITGA3 under hypoxia, independent of changes in protein level. We showed that specific glycosylation sites within ITGA3 were required for its normal trafficking to the cell surface, and thus for its dimerization with ITGB1 and functional effects. Thus we have revealed a new pathway through which hypoxia can potentially modulate tumor cell behavior in order to favor metastasis. More broadly, this study highlights the power of glycoproteomics to enrich the data generated by more conventional proteomic studies and to elucidate the true functional significance of modulations to glycoproteins in both health and disease states.
In addition, why simultaneous mutations of glycosites 6 and 7 cause ITGA3 deficiency in plasma membrane is an intriguing question. The conformation changes due to a lack of glycans on sites 6 and 7 should be the keys to open the mysterious doors to reveal the interaction of integrin α3β1 complex. It will be our future work to find ITGA3 partners that help with its traffic to plasma membrane and to confirm whether these partners are down-regulated by hypoxia in A431. This could give clues regarding the underlying mechanism in metastatic cascades and provide new insights in studies on cancer cell hypoxia.
Supplementary Material
Acknowledgments
We thank Lucy Robinson for critically reviewed the manuscript.
Footnotes
Author contributions: S.S., Y.R. and S.L. designed research; Y.R. and P.H. performed research; Y.R. and P.H. analyzed data; Y.R. and S.S. wrote the paper.
* This work is supported by the Singapore Ministry of Health's National Medical Research Council (NMRC/CBRG/0004/2012).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ITGA
- integrin α
- Nx
- normoxia
- Hx
- hypoxia
- iTRAQ
- isobaric tag for relative and absolute quantification
- ERLIC
- electrostatic repulsion hydrophilic interaction chromatography
- WB
- Western blot
- PBS
- phosphate-buffered saline
- FBS
- fetal bovine serum
- FA
- formic acid
- ACN
- acetonitrile
- ITGB
- integrin β
- WT
- wild type
- MW
- molecular weight.
REFERENCES
- 1. Subarsky P., Hill R. P. (2003) The hypoxic tumour microenvironment and metastatic progression. Clin. Exp. Metastasis 20, 237–250 [DOI] [PubMed] [Google Scholar]
- 2. Majmundar A. J., Wong W. J., Simon M. C. (2010) Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsai Y. P., Wu K. J. (2012) Hypoxia-regulated target genes implicated in tumor metastasis. J. Biomed. Sci. 19, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Park J. E., Tan H. S., Datta A., Lai R. C., Zhang H., Meng W., Lim S. K., Sze S. K. (2010) Hypoxic tumor cell modulates its microenvironment to enhance angiogenic and metastatic potential by secretion of proteins and exosomes. Mol. Cell. Proteomics 9, 1085–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ren Y., Hao P., Dutta B., Cheow E. S., Sim K. H., Gan C. S., Lim S. K., Sze S. K. (2013) Hypoxia modulates A431 cellular pathways association to tumor radioresistance and enhanced migration revealed by comprehensive proteomic and functional studies. Mol. Cell. Proteomics 12, 485–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Varki A. (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3, 97–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dwek R. A. (1995) Glycobiology: “towards understanding the function of sugars”. Biochem. Soc. Trans. 23, 1–25 [DOI] [PubMed] [Google Scholar]
- 8. Saxon E., Bertozzi C. R. (2001) Chemical and biological strategies for engineering cell surface glycosylation. Annu. Rev. Cell Dev. Biol. 17, 1–23 [DOI] [PubMed] [Google Scholar]
- 9. Isaji T., Sato Y., Zhao Y., Miyoshi E., Wada Y., Taniguchi N., Gu J. (2006) N-glycosylation of the beta-propeller domain of the integrin alpha5 subunit is essential for alpha5beta1 heterodimerization, expression on the cell surface, and its biological function. J. Biol. Chem. 281, 33258–33267 [DOI] [PubMed] [Google Scholar]
- 10. Isaji T., Sato Y., Fukuda T., Gu J. (2009) N-glycosylation of the I-like domain of beta1 integrin is essential for beta1 integrin expression and biological function: identification of the minimal N-glycosylation requirement for alpha5beta1. J. Biol. Chem. 284, 12207–12216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nicolaou N., Margadant C., Kevelam S. H., Lilien M. R., Oosterveld M. J., Kreft M., van Eerde A. M., Pfundt R., Terhal P. A., van der Zwaag B., Nikkels P. G., Sachs N., Goldschmeding R., Knoers N. V., Renkema K. Y., Sonnenberg A. (2012) Gain of glycosylation in integrin alpha3 causes lung disease and nephrotic syndrome. J. Clin. Invest. 122, 4375–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshimura M., Ihara Y., Matsuzawa Y., Taniguchi N. (1996) Aberrant glycosylation of E-cadherin enhances cell-cell binding to suppress metastasis. J. Biol. Chem. 271, 13811–13815 [DOI] [PubMed] [Google Scholar]
- 13. Gu J., Taniguchi N. (2008) Potential of N-glycan in cell adhesion and migration as either a positive or negative regulator. Cell Adhes. Migr. 2, 243–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo H. B., Johnson H., Randolph M., Pierce M. (2009) Regulation of homotypic cell-cell adhesion by branched N-glycosylation of N-cadherin extracellular EC2 and EC3 domains. J. Biol. Chem. 284, 34986–34997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kannagi R., Izawa M., Koike T., Miyazaki K., Kimura N. (2004) Carbohydrate-mediated cell adhesion in cancer metastasis and angiogenesis. Cancer Sci. 95, 377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Izawa M., Kumamoto K., Mitsuoka C., Kanamori C., Kanamori A., Ohmori K., Ishida H., Nakamura S., Kurata-Miura K., Sasaki K., Nishi T., Kannagi R. (2000) Expression of sialyl 6-sulfo Lewis X is inversely correlated with conventional sialyl Lewis X expression in human colorectal cancer. Cancer Res. 60, 1410–1416 [PubMed] [Google Scholar]
- 17. Kim Y. J., Varki A. (1997) Perspectives on the significance of altered glycosylation of glycoproteins in cancer. Glycoconj. J. 14, 569–576 [DOI] [PubMed] [Google Scholar]
- 18. Vogt G., Chapgier A., Yang K., Chuzhanova N., Feinberg J., Fieschi C., Boisson-Dupuis S., Alcais A., Filipe-Santos O., Bustamante J., de Beaucoudrey L., Al-Mohsen I., Al-Hajjar S., Al-Ghonaium A., Adimi P., Mirsaeidi M., Khalilzadeh S., Rosenzweig S., de la Calle Martin O., Bauer T. R., Puck J. M., Ochs H. D., Furthner D., Engelhorn C., Belohradsky B., Mansouri D., Holland S. M., Schreiber R. D., Abel L., Cooper D. N., Soudais C., Casanova J. L. (2005) Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat. Genet. 37, 692–700 [DOI] [PubMed] [Google Scholar]
- 19. Woodhouse E. C., Chuaqui R. F., Liotta L. A. (1997) General mechanisms of metastasis. Cancer 80, 1529–1537 [DOI] [PubMed] [Google Scholar]
- 20. Huttenlocher A., Horwitz A. R. (2011) Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 3, a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chammas R., Veiga S. S., Line S., Potocnjak P., Brentani R. R. (1991) Asn-linked oligosaccharide-dependent interaction between laminin and gp120/140. An alpha 6/beta 1 integrin. J. Biol. Chem. 266, 3349–3355 [PubMed] [Google Scholar]
- 22. Zheng M., Fang H., Hakomori S. (1994) Functional role of N-glycosylation in alpha 5 beta 1 integrin receptor. De-N-glycosylation induces dissociation or altered association of alpha 5 and beta 1 subunits and concomitant loss of fibronectin binding activity. J. Biol. Chem. 269, 12325–12331 [PubMed] [Google Scholar]
- 23. Gu J., Taniguchi N. (2004) Regulation of integrin functions by N-glycans. Glycoconj. J. 21, 9–15 [DOI] [PubMed] [Google Scholar]
- 24. Bellis S. L. (2004) Variant glycosylation: an underappreciated regulatory mechanism for beta1 integrins. Biochim. Biophys. Acta 1663, 52–60 [DOI] [PubMed] [Google Scholar]
- 25. Miyoshi E., Noda K., Ko J. H., Ekuni A., Kitada T., Uozumi N., Ikeda Y., Matsuura N., Sasaki Y., Hayashi N., Hori M., Taniguchi N. (1999) Overexpression of alpha1–6 fucosyltransferase in hepatoma cells suppresses intrahepatic metastasis after splenic injection in athymic mice. Cancer Res. 59, 2237–2243 [PubMed] [Google Scholar]
- 26. Leitinger B., Hohenester E. (2007) Mammalian collagen receptors. Matrix Biol. 26, 146–155 [DOI] [PubMed] [Google Scholar]
- 27. Arimoto-Ishida E., Sakata M., Sawada K., Nakayama M., Nishimoto F., Mabuchi S., Takeda T., Yamamoto T., Isobe A., Okamoto Y., Lengyel E., Suehara N., Morishige K., Kimura T. (2009) Up-regulation of alpha5-integrin by E-cadherin loss in hypoxia and its key role in the migration of extravillous trophoblast cells during early implantation. Endocrinology 150, 4306–4315 [DOI] [PubMed] [Google Scholar]
- 28. Ryu M. H., Park H. M., Chung J., Lee C. H., Park H. R. (2010) Hypoxia-inducible factor-1alpha mediates oral squamous cell carcinoma invasion via upregulation of alpha5 integrin and fibronectin. Biochem. Biophys. Res. Commun. 393, 11–15 [DOI] [PubMed] [Google Scholar]
- 29. Weitzman J. B., Pasqualini R., Takada Y., Hemler M. E. (1993) The function and distinctive regulation of the integrin VLA-3 in cell adhesion, spreading, and homotypic cell aggregation. J. Biol. Chem. 268, 8651–8657 [PubMed] [Google Scholar]
- 30. Elices M. J., Urry L. A., Hemler M. E. (1991) Receptor functions for the integrin VLA-3: fibronectin, collagen, and laminin binding are differentially influenced by Arg-Gly-Asp peptide and by divalent cations. J. Cell Biol. 112, 169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dedhar S., Jewell K., Rojiani M., Gray V. (1992) The receptor for the basement membrane glycoprotein entactin is the integrin alpha 3/beta 1. J. Biol. Chem. 267, 18908–18914 [PubMed] [Google Scholar]
- 32. Carter W. G., Ryan M. C., Gahr P. J. (1991) Epiligrin, a new cell adhesion ligand for integrin alpha 3 beta 1 in epithelial basement membranes. Cell 65, 599–610 [DOI] [PubMed] [Google Scholar]
- 33. Delwel G. O., de Melker A. A., Hogervorst F., Jaspars L. H., Fles D. L., Kuikman I., Lindblom A., Paulsson M., Timpl R., Sonnenberg A. (1994) Distinct and overlapping ligand specificities of the alpha 3A beta 1 and alpha 6A beta 1 integrins: recognition of laminin isoforms. Mol. Biol. Cell 5, 203–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guo N., Templeton N. S., Al-Barazi H., Cashel J. A., Sipes J. M., Krutzsch H. C., Roberts D. D. (2000) Thrombospondin-1 promotes alpha3beta1 integrin-mediated adhesion and neurite-like outgrowth and inhibits proliferation of small cell lung carcinoma cells. Cancer Res. 60, 457–466 [PubMed] [Google Scholar]
- 35. Kreidberg J. A. (2000) Functions of alpha3beta1 integrin. Curr. Opin. Cell Biol. 12, 548–553 [DOI] [PubMed] [Google Scholar]
- 36. Pignatelli M., Hanby A. M., Stamp G. W. (1991) Low expression of beta 1, alpha 2 and alpha 3 subunits of VLA integrins in malignant mammary tumours. J. Pathol. 165, 25–32 [DOI] [PubMed] [Google Scholar]
- 37. Adachi M., Taki T., Huang C., Higashiyama M., Doi O., Tsuji T., Miyake M. (1998) Reduced integrin alpha3 expression as a factor of poor prognosis of patients with adenocarcinoma of the lung. J. Clin. Oncol. 16, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 38. Barr L. F., Campbell S. E., Bochner B. S., Dang C. V. (1998) Association of the decreased expression of alpha3beta1 integrin with the altered cell: environmental interactions and enhanced soft agar cloning ability of c-myc-overexpressing small cell lung cancer cells. Cancer Res. 58, 5537–5545 [PubMed] [Google Scholar]
- 39. Judware R., Culp L. A. (1997) Concomitant down-regulation of expression of integrin subunits by N-myc in human neuroblastoma cells: differential regulation of alpha2, alpha3 and beta1. Oncogene 14, 1341–1350 [DOI] [PubMed] [Google Scholar]
- 40. Plopper G. E., Domanico S. Z., Cirulli V., Kiosses W. B., Quaranta V. (1998) Migration of breast epithelial cells on laminin-5: differential role of integrins in normal and transformed cell types. Breast Cancer Res. Treat. 51, 57–69 [DOI] [PubMed] [Google Scholar]
- 41. Ura H., Denno R., Hirata K., Yamaguchi K., Yasoshima T. (1998) Separate functions of alpha2beta1 and alpha3beta1 integrins in the metastatic process of human gastric carcinoma. Surg. Today 28, 1001–1006 [DOI] [PubMed] [Google Scholar]
- 42. Morelle W., Canis K., Chirat F., Faid V., Michalski J. C. (2006) The use of mass spectrometry for the proteomic analysis of glycosylation. Proteomics 6, 3993–4015 [DOI] [PubMed] [Google Scholar]
- 43. Pan S., Chen R., Aebersold R., Brentnall T. A. (2011) Mass spectrometry based glycoproteomics—from a proteomics perspective. Mol. Cell. Proteomics 10, R110.003251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Madera M., Mechref Y., Novotny M. V. (2005) Combining lectin microcolumns with high-resolution separation techniques for enrichment of glycoproteins and glycopeptides. Anal. Chem. 77, 4081–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ongay S., Boichenko A., Govorukhina N., Bischoff R. (2012) Glycopeptide enrichment and separation for protein glycosylation analysis. J. Sep. Sci. 35, 2341–2372 [DOI] [PubMed] [Google Scholar]
- 46. Zhang H., Li X. J., Martin D. B., Aebersold R. (2003) Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21, 660–666 [DOI] [PubMed] [Google Scholar]
- 47. Lewandrowski U., Zahedi R. P., Moebius J., Walter U., Sickmann A. (2007) Enhanced N-glycosylation site analysis of sialoglycopeptides by strong cation exchange prefractionation applied to platelet plasma membranes. Mol. Cell. Proteomics 6, 1933–1941 [DOI] [PubMed] [Google Scholar]
- 48. Alpert A. J. (1990) Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. J. Chromatogr. 499, 177–196 [DOI] [PubMed] [Google Scholar]
- 49. Larsen M. R., Jensen S. S., Jakobsen L. A., Heegaard N. H. (2007) Exploring the sialiome using titanium dioxide chromatography and mass spectrometry. Mol. Cell. Proteomics 6, 1778–1787 [DOI] [PubMed] [Google Scholar]
- 50. Lei Z., Beuerman R. W., Chew A. P., Koh S. K., Cafaro T. A., Urrets-Zavalia E. A., Urrets-Zavalia J. A., Li S. F., Serra H. M. (2009) Quantitative analysis of N-linked glycoproteins in tear fluid of climatic droplet keratopathy by glycopeptide capture and iTRAQ. J. Proteome Res. 8, 1992–2003 [DOI] [PubMed] [Google Scholar]
- 51. Palmisano G., Parker B. L., Engholm-Keller K., Lendal S. E., Kulej K., Schulz M., Schwammle V., Graham M. E., Saxtorph H., Cordwell S. J., Larsen M. R. (2012) A novel method for the simultaneous enrichment, identification, and quantification of phosphopeptides and sialylated glycopeptides applied to a temporal profile of mouse brain development. Mol. Cell. Proteomics 11, 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hao P., Guo T., Sze S. K. (2011) Simultaneous analysis of proteome, phospho- and glycoproteome of rat kidney tissue with electrostatic repulsion hydrophilic interaction chromatography. PLoS One 6, e16884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang H., Guo T., Li X., Datta A., Park J. E., Yang J., Lim S. K., Tam J. P., Sze S. K. (2010) Simultaneous characterization of glyco- and phosphoproteomes of mouse brain membrane proteome with electrostatic repulsion hydrophilic interaction chromatography. Mol. Cell. Proteomics 9, 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Alpert A. J. (2008) Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal. Chem. 80, 62–76 [DOI] [PubMed] [Google Scholar]
- 55. Hao P., Qian J., Ren Y., Sze S. K. (2011) Electrostatic repulsion-hydrophilic interaction chromatography (ERLIC) versus strong cation exchange (SCX) for fractionation of iTRAQ-labeled peptides. J. Proteome Res. 10, 5568–5574 [DOI] [PubMed] [Google Scholar]
- 56. Hao P., Ren Y., Alpert A. J., Sze S. K. (2011) Detection, evaluation and minimization of nonenzymatic deamidation in proteomic sample preparation. Mol. Cell. Proteomics 10, O111.009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wright H. T. (1991) Sequence and structure determinants of the nonenzymatic deamidation of asparagine and glutamine residues in proteins. Protein Eng. 4, 283–294 [DOI] [PubMed] [Google Scholar]
- 58. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 59. Parker B. L., Palmisano G., Edwards A. V., White M. Y., Engholm-Keller K., Lee A., Scott N. E., Kolarich D., Hambly B. D., Packer N. H., Larsen M. R., Cordwell S. J. (2011) Quantitative N-linked glycoproteomics of myocardial ischemia and reperfusion injury reveals early remodeling in the extracellular environment. Mol. Cell. Proteomics 10, M110.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hemler M. E., Huang C., Schwarz L. (1987) The VLA protein family. Characterization of five distinct cell surface heterodimers each with a common 130,000 molecular weight beta subunit. J. Biol. Chem. 262, 3300–3309 [PubMed] [Google Scholar]
- 61. Takada Y., Huang C., Hemler M. E. (1987) Fibronectin receptor structures in the VLA family of heterodimers. Nature 326, 607–609 [DOI] [PubMed] [Google Scholar]
- 62. Nguyen B. P., Ryan M. C., Gil S. G., Carter W. G. (2000) Deposition of laminin 5 in epidermal wounds regulates integrin signaling and adhesion. Curr. Opin. Cell Biol. 12, 554–562 [DOI] [PubMed] [Google Scholar]
- 63. Zhou H., Kramer R. H. (2005) Integrin engagement differentially modulates epithelial cell motility by RhoA/ROCK and PAK1. J. Biol. Chem. 280, 10624–10635 [DOI] [PubMed] [Google Scholar]
- 64. Goldfinger L. E., Hopkinson S. B., deHart G. W., Collawn S., Couchman J. R., Jones J. C. (1999) The alpha3 laminin subunit, alpha6beta4 and alpha3beta1 integrin coordinately regulate wound healing in cultured epithelial cells and in the skin. J. Cell Sci. 112 (Pt 16), 2615–2629 [DOI] [PubMed] [Google Scholar]
- 65. Wen T., Zhang Z., Yu Y., Qu H., Koch M., Aumailley M. (2010) Integrin alpha3 subunit regulates events linked to epithelial repair, including keratinocyte migration and protein expression. Wound Repair Regen. 18, 325–334 [DOI] [PubMed] [Google Scholar]
- 66. Mitchell K., Svenson K. B., Longmate W. M., Gkirtzimanaki K., Sadej R., Wang X., Zhao J., Eliopoulos A. G., Berditchevski F., Dipersio C. M. (2010) Suppression of integrin alpha3beta1 in breast cancer cells reduces cyclooxygenase-2 gene expression and inhibits tumorigenesis, invasion, and cross-talk to endothelial cells. Cancer Res. 70, 6359–6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.