Abstract
In this study, the human cerebrospinal fluid (CSF) proteome was mapped using three different strategies prior to Orbitrap LC-MS/MS analysis: SDS-PAGE and mixed mode reversed phase-anion exchange for mapping the global CSF proteome, and hydrazide-based glycopeptide capture for mapping glycopeptides. A maximal protein set of 3081 proteins (28,811 peptide sequences) was identified, of which 520 were identified as glycoproteins from the glycopeptide enrichment strategy, including 1121 glycopeptides and their glycosylation sites. To our knowledge, this is the largest number of identified proteins and glycopeptides reported for CSF, including 417 glycosylation sites not previously reported. From parallel plasma samples, we identified 1050 proteins (9739 peptide sequences). An overlap of 877 proteins was found between the two body fluids, whereas 2204 proteins were identified only in CSF and 173 only in plasma. All mapping results are freely available via the new CSF Proteome Resource (http://probe.uib.no/csf-pr), which can be used to navigate the CSF proteome and help guide the selection of signature peptides in targeted quantitative proteomics.
Cerebrospinal fluid (CSF)1 surrounds and supports the central nervous system (CNS), including the ventricles and subarachnoid space (1). About 80% of the total protein amount in CSF derives from size-dependent filtration of blood across the blood-brain barrier (BBB), and the rest originate from drainage of interstitial fluid from the CNS (2–4). Because CSF is in direct contact with the CNS, it should be a promising source for finding biomarkers for diseases in the CNS (5).
Mapping studies characterizing the human CSF proteome and peptidome has previously been carried out using various experimental designs, including both healthy and disease-affected individuals (5–16). A total of 2630 proteins were detected in normal CSF by immunoaffinity depletion of high abundant proteins followed by strong cation exchange fractionation and LC-MS (5), whereas proteome and peptidome analyses of human CSF (collected for diagnostic purposes and turned out normal) by gel separation and trypsin digestion followed by LC-MS analysis have shown 798 proteins and 563 peptide products (derived from 91 precursor proteins) (6). In another publication, Pan et al. combined several proteomics studies in CSF from both normal subjects and subjects with neurological diseases and created a dataset of 2594 identified proteins (16). But in general, the availability and usefulness of published data from proteome mapping experiments is scarce, and the format of the data often makes searching and comparison across datasets difficult. Thus, organizing the data in online databases would greatly benefit the scientific community by making the data more accessible and easier to query. Current online databases containing MS data for CSF include the Sys-BodyFluid, with a total of 1286 CSF proteins from six studies (17). The proteome identifications database (PRIDE) (18) includes 19 studies on human CSF, but none reporting more than 103 identified proteins.
Glycosylation is one of the most common post-translational modifications (PTMs), and many known clinical biomarkers as well as therapeutic targets are glycoproteins (19–25). Furthermore, glycosylation plays important roles in cell communication, signaling, aging, and cell adhesion (26, 27). Nevertheless, there are few studies on glycoprotein identification in CSF. One study identified 216 glycoproteins in CSF using both lectin affinity and hydrazide chemistry (8), and another reported 36 N-linked and 44 O-linked glycosylation sites, from 23 and 22 glycoproteins respectively, by enriching for sialic-acid containing glycopeptides (28).
Considering the sparse information about the CSF proteome available in public repositories, we have combined several proteomics approaches to create a map of the global CSF proteome, the CSF glycoproteome, and the respective plasma proteome from a pool of 21 (20 for the plasma pool) neurologically healthy individuals. The large amount of data generated through these four datasets (with linked and complementary information) would not easily be accessible through existing repositories. We therefore developed the open access CSF Proteome Resource (CSF-PR, www.probe.uib.no/csf-pr), an online database including the detailed data from the four different proteomics experiments described in this study. CSF-PR will be particularly useful in guiding the selection of appropriate signature peptides for the development of targeted CSF protein assays.
EXPERIMENTAL PROCEDURES
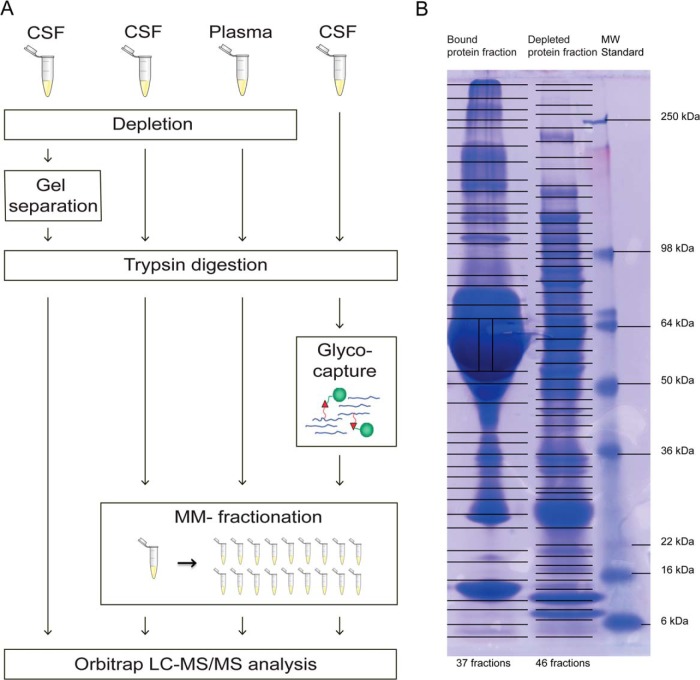
The workflow of the four different experiments performed in this study is displayed in Fig. 1A, and described in detail below.
Fig. 1.
A, Overview of the workflow of the four different experiments in the study. One experiment included depletion, gel separation, and trypsin digestion on CSF (left), two experiments included depletion, trypsin digestion, and mixed-mode fractionation on CSF (middle left), and plasma (middle right), and one experiment included trypsin digestion, glyco-capture by solid-phase extraction of N-linked glycopeptides (SPEG), and mixed-mode fractionation (right). Orbitrap LC-MS/MS analysis was performed for all four experiments. Glyco-capture figure adapted from (33) B, Image of the gel after SDS-PAGE separation of the bound (left) and the depleted (middle) protein fraction. The gel MW standard is shown to the right with the masses marked. Lines across the gel indicate where the bands were excised and point toward the marked standard masses (right).
Biological Samples
CSF was collected by lumbar puncture of neurologically healthy (spinal anesthesia subjects (SAS) (29)) individuals who, under written informed consent, were to undergo spinal anesthesia for surgery at the Department of Anesthesia and Intensive Care Medicine, Haukeland University Hospital. Information about the subjects can be found in supplemental Table S1. Parallel CSF and plasma samples were collected according to the published consensus protocol for CSF collection and biobanking (30). None of the patients experienced traumatic CSF taps. CSF samples were centrifuged and cells removed prior to proteomics analysis. Plasma was collected in K2 EDTA tubes and centrifuged at room temperature at 2300 × g for 25 min. Aliquots of 500 μl CSF from each of the 21 individuals included in this study were combined into a pool. A similar pool was generated for the plasma samples from 20 of the patients, as plasma from one patient could not be obtained. The study was approved by The Regional Committee for Medical and Health Research Ethics of Western Norway.
Chemicals
If not otherwise stated, all chemicals and products were purchased from Sigma-Aldrich (St. Louis, MO). All protein concentrations were measured using a Qubit™ fluorometer kit (Invitrogen, Carlsbrad, CA) according to the vendor's instructions.
Sample Preparation of CSF for SDS-PAGE
3 ml (1400 μg protein) of the CSF pool was up-concentrated to 20 μl using 3 kDa ultracentrifugation filters (Amicon Ultra-4, Merck Millipore, Billerica, MA), prerinsed with deionized water (MilliQ). The concentrated sample was protein depleted of 14 high abundant proteins using the human Multiple Affinity Removal System (MARS HU-14) 4.6 mm x 50 mm LC column (Agilent Technologies, Santa Clara, CA) according to the vendor's recommendations. The two resulting protein fractions (depleted and bound) were then concentrated using ultracentrifugation filters as described above expect the filters were coated with 1 ml 0.1% N-octyl-Beta-d-glucopyranoside (NOG).
Each fraction was dissolved in LDS sample buffer (Invitrogen) containing 10 mm DTT (GE Healthcare, Amersham Biosciences, Buckinghamshire, UK) at 95¦°C for 5 min, alkylated by adding up to 20 mm iodoacetamine (IAA), followed by incubation in room temperature and in the dark for 20 min, prior to gel separation in two separate lanes using a lab-casted 20 cm 5–15% SDS-polyacrylamide gradient gel. Samples were separated at 60 V for 16 h in 1x electrode running buffer (25 mm Tris base, 192 mm Glysine, and 0.1% SDS diluted in milliQ water). After protein separation, the gel was stained with Coomassie Brilliant Blue (GE Healthcare). The lanes were cut into a total of 83 gel slices as described in Fig. 1B, 37 slices from the lane with the bound (high abundant) protein fraction and 46 slices from the lane with the depleted fraction. The gel slices were in-gel digested (supplemental File S1A) using between 120 and 240 ng of trypsin, depending on the size of the band, before MS/MS analysis.
Sample Preparation of CSF for Mixed Mode Fractionation
1.6 ml (750 μg protein) CSF was concentrated and immunoaffinity depleted as described above. The entire amount of the depleted fraction (∼40 μg) and an aliquot (100 μg) from the bound fraction was in-solution trypsin digested (Supplementary File 1B), using 0.8 and 2 μg trypsin, respectively. After digestion, the samples were desalted using C18 Oasis™ μElution plates (Waters, Milford, MA) as described elsewhere (31). The two samples were then fractionated using mixed mode reversed phase-anion exchange (MM RP-AX) HPLC as described by Phillips et al. (32). A Promix MP 250 mm x 2.1 mm id, pore size 300 Å column (SIELC Technologies, Prospect Heights, IL) connected to an Agilent Technology 1260 off-line LC-system was used. The desalted and dried samples were resuspended in 120 μl buffer A (20 mm Ammonium formate/3% ACN, pH 6.5) and loaded onto the column.
The setup for the LC was as follows. The flow was always 50 μl/min and the gradient length was 70 min. From 0–45 min buffer B (2 mm Ammonium formate/80% ACN) increased linearly from 15% to 60%, from 45–55 min 60% B, from 55–65 min 100% B, and from 65–70 min 15% B. The depleted CSF sample was separated into 80 fractions, where a fraction was collected every 0.87 min from 0–70 min (some fractions at each end of the gradient were later combined to give the total number of 66 fractions). Ten fractions were collected for the bound sample, one fraction the first 2 mins and then one fraction every 7 mins from 2–70 min.
Sample Preparation of Plasma for Mixed Mode Fractionation
For the characterization of the plasma proteome, the same experimental setup as for the CSF characterization using MM (RP-AX) fractionation was applied to 40 μl plasma, representing roughly 2400 μg protein.
Preparation of CSF and Isolation of Glycopeptides by Solid-phase Extraction of N-linked Glycopeptides
Three milliliters (1200 μg protein) CSF was in-solution trypsin digested and further processed by solid-phase extraction of N-linked glycopeptides (SPEG) essentially as described in (33) and (34), but with some exceptions. The CSF was purified and concentrated to 15 μl using 3 kDa ultracentrifugation filters precoated with 1 ml 0.1% NOG. Dilution with 135 μl denaturation buffer (8 m Urea/0.4 M Ammonium Bicarbonate (ambic)/0.1% SDS (Bio-Rad Laboratories, Hercules, CA) followed, and then 120 mm Tris(2-carboxyethyl)phosphine (TCEP) was added to a final concentration of 10 mm and a one hour incubation at 37¦°C. IAA was added to a final concentration of 12 mm, and incubated for 30 min in the dark. The sample was then diluted with 0.1 m ambic until the urea concentration in the sample was below 1 m, and trypsin digested (1:50 ratio trypsin:protein, Trypsin porcine (Promega, Madison, WI)) overnight at 37¦°C. The next day the sample was desalted using Oasis C18, oxidized and desalted again essentially as described in (33), with some exceptions. 100% formic acid (FA) was used for acidification, Oasis plates were used for the desalting, the sample was dried after the first desalting to remove ACN and resuspended in 400 μl 0.1% TFA before oxidation. The sample was coupled to 4 mg (133 μl) magnetic hydrazide beads (BioClone Inc. San Diego, CA) and further processed as described in (34). Beads and supernatant were separated using a Dynal® magnetic bead separation rack (Invitrogen). The released and now deglycosylated peptides were collected the following day as previously described (33), except that the hydrazide resin was washed only once and with 200 μl ambic, and then acidified with 7 μl 5 m hydrochloric acid and 200 μl 0.1% FA before desalting by Oasis C18 as described in (34). The SPEG processing was done in three separate experiments (1 ml CSF for each), which were combined before further processing.
The digested and SPEG processed sample containing the deglycosylated peptides (former glycopeptides) were fractionated into 20 fractions using MM (RP-AX) as described above with the following set-up: one fraction was collected between 0–2 min, one between 2–7 min, from 7–55 min fractions were collected every 3 min, and from 55–70 min every 5 min. The entire peptide amount from each fraction was injected for LC-MS analysis.
MS Analysis
Dry samples were dissolved to a final concentration of 0.1–5% FA before analysis on an LTQ Orbitrap Velos Pro mass spectrometer (Thermo Fischer Scientific, Bremen, Germany) equipped with a nano spray Flex ion source (Thermo Fischer Scientific), coupled to a Dionex Ultimate NCS-3000 LC system (Thermo Fischer Scientific). Approximately 0.5 μg of digested protein was loaded and desalted on a precolumn (Acclaim PepMap 100, 2 cm × 75 μm i.d. nanoViper column, packed with 3 μm C18 beads) at a flow rate of 5 μl/min for 6 min using an isocratic flow of 0.1% FA (v/v) with 2% ACN (v/v). Peptides were separated during a biphasic ACN gradient from two nanoflow UPLC pumps with flow rate of 280 nl/min on the analytical column (Acclaim PepMap 100, 15 cm × 75 μm i.d. nanoViper column, packed with 2 μm C18 beads). Solvent A was 0.1% FA (v/v) with 2% ACN (v/v). Solvent B was 0.1% FA (v/v) with 90% ACN (v/v). The gradient was 0–61.5 min ramp from 8–38% B, 61.5–64.5 min ramp from 38–90% B, and 64.5–69.5 min 90% B, followed by column conditioning for 12 min with 5% B. Data dependent acquisition was utilized and collision-induced dissociation (CID) with normalized collision energy of 35% and wideband-activation enabled. Survey full scan MS spectra (from m/z 300–2000) were acquired with resolution r = 60,000 at m/z 400. The 15 ions with the highest intensity were selected for MS/MS fragmentation. MS data were acquired over 90 min.
Analysis of LC-MS/MS Data
The acquired raw files were searched against the human Swiss-Prot database (from December 2012 - 20,226 entries) using SearchGUI v1.10.4 (35) (search engines: OMSSA v2.1.9 (36) and X!Tandem CYCLONE (2010.12.01.1) (37)) and processed in PeptideShaker v0.19.0, http://peptide-shaker.googlecode.com (38). The search criteria were: carbamidomethylation of cystein as a fixed modification and oxidized methionine as a variable modification for all datasets. Deamidation of asparagine was set as a variable modification only for the glyco dataset. Precursor mass tolerance was 10 ppm, fragment mass tolerance 0.7 Da, and maximum number of missed cleavages by trypsin was 2. PeptideShaker converts search engine e-values into confidence values using the distribution of decoy matches as described by Nesvizhskii (39). For all four of our datasets a validation threshold of 1% false discovery rate was employed individually at the protein, peptide and peptide spectrum match (PSM) level. ProteoWizard v.3.0.3650 (40) with default settings was used to convert the raw files to peak lists. Gene ontology analyses were performed using PANTHER (41).
Analysis of Glyco-data
The resulting data from the mapping of glycoproteins and -peptides required some manual interpretation to decide which peptides that were true glycopeptides, and which were unspecifically bound to the hydrazide beads during the protocol. The detailed analysis is described in Supplemental File 1C. For a site to be considered as a true glycosite, the peptide had to have at least one deamidated asparagine and contain the N-glyco sequence motif [N][XP̂][ST] (where X can be any amino acid except proline). However, if the deamidation was confidently assigned only to an asparagine outside the motif, the peptide was not considered to be a glycopeptide. To investigate the chance that a random peptide has both a deamidated asparagine and the N-glyco sequence motif, we researched the data from the global mixed-mode fractionated and depleted CSF experiment, but this time with deamidation as variable modification. After filtering the results, we found that 0.4% of the peptides in this nonglyco experiment had both a deamidation and the N-glyco motif, indicating a 0.4% chance for a false positive glycopeptide by our approach. Considering that we in addition performed glycopeptide enrichment in the glyco experiment, this false positive rate would in reality be even lower. The data postprocessing, performed in PeptideShaker, provides the position of the deamidated asparagine in the peptide sequence and also a location confidence. The location confidence tells us how confident the software is in assigning the modification to the specific amino acid (very confident, confident, doubtful or random, see Supplemental File 1C for details).
RESULTS AND DISCUSSION
In this study, we performed three LC-MS experiments using different analytical fractionation strategies to create a library of proteins and peptides identified in human CSF. For all three experiments, aliquots of a CSF pool from 21 neurologically healthy volunteers were used (Fig. 1A). Glycopeptide enrichment in combination with MM (RP-AX) peptide fractionation was applied in one experiment to map the glycopeptides in CSF. Immunoaffinity depletion combined with either SDS-PAGE (Fig. 1B) or MM (RP-AX) fractionation was used for the two other strategies to map the global CSF proteome. In addition, immunoaffinity depletion combined with MM (RP-AX) peptide fractionation was applied to a plasma pool from 20 of the same individuals (Fig. 1A). The number of proteins, peptides and spectra identified in all the experiments is summarized in Table I.
Table I. The number of peptides and proteins identified in CSF and plasma.
The number of proteins, peptides and spectra identified in the four CSF datasets and in plasma are indicated. “SPEG” means all proteins and peptides identified in the glyco enrichment experiment (including unspecifically bound), while “Glyco” means all proteins and peptides verified as truly glycosylated by having a deamidation of an asparagine and a glycopattern. “Peptides” means unique peptide sequences.
| Data set | Maximal Protein Set | Protein groups | Peptides | Spectra |
|---|---|---|---|---|
| Mixed-Mode | 2779 | 2632 | 21,003 | 170,400 |
| SDS-PAGE | 1883 | 1718 | 18,955 | 266,826 |
| SPEG | 679 | 598 | 2594 | 14,213 |
| Glyco | 520 | 508 | 1121 | 4631 |
| CSF Total | 3081 | 2875 | 28,811 | 451,439 |
| Plasma | 1050 | 946 | 9739 | 126,164 |
CSF MM Glyco Mapping
A total of 2594 peptide sequences and 598 protein groups were identified in the N-glycopeptide enrichment experiment. The maximal protein set (MPS), which is the sum of all the proteins in all of the protein groups in the dataset, was 679 whereof 520 were true glycoproteins, i.e. having one or more true glycopeptides. The 520 true glycoproteins were represented by 1121 former glycopeptide sequences, and 846 specific glycosylation sites (because of miscleavages some sites were identified in more than one peptide), and this is to our knowledge the most comprehensive mapping of glycosylation in CSF.
PeptideShaker and manual inspection of the peptides, as described in detail in Supplemental File S1C, was used for assignment of the specific site of deamidation. This assignment is straightforward when the number of deamidations matches the number of asparagines in the peptide sequence. Most of the glycopeptides that we identified were in this category, having only one asparagine in the sequence (665 peptides). Several other glycopeptides however, had more than one asparagine in the peptide sequence (459 peptides). With multiple possibilities for a deamidation, Peptideshaker uses PTM scores (Ascore (42) and d-score (43)) to assign the modification to the correct asparagine, and thereby determining the position of the glyco-unit. When the scoring does not give certainty about the position of the deamidation (more than one asparagine in the peptide and not sufficient information from the MS/MS spectra), it is doubtfully or randomly assigned. 202 of our glycopeptides were in this category. Considering the low probability of a peptide having both a deamidation and a glycomotif and not being a former glycopeptide (calculated to be 0.4% by our approach), and the fact that we performed glycopeptide enrichment, we find it very likely that also these peptides are true glycopeptides and that the position of the deamidation is on the asparagine in the glycomotif.
Previously, the most comprehensive CSF glycoprotein study identified 216 glycoproteins in CSF by both lectin affinity and hydrazide chemistry as enrichment strategies prior to MS analysis (8). After crosschecking our identified glycopeptides and glycosylation sites against the Swiss-Prot database (as described in Supplemental File 1C), we found that 417 of the identified glycosylation sites were not verified in the database. Most of these were as of April 2013 listed as potential (349 sites), probable (three sites), or by similarity (four sites). The remaining new sites had no annotation in Swiss-Prot. All peptides where novel glycosylation sites have been identified are listed in supplemental Table S2. This is an important contribution to the field of glycoproteomics.
For some of the proteins several new glycosylation sites were identified, as for example for neuronal cell adhesion molecule (nine new sites), protocadherin Fat 2 (nine new sites), and protocadherin-9 (four new sites). Protocadherins are known to be involved in brain development (reviewed in (44)), and knowing the glycosylation characteristics for such proteins is relevant in order to determine their biochemistry and function.
CSF SDS-PAGE Global Mapping
The SDS-PAGE fractionation approach (Fig. 1) resulted in the identification of 18,955 peptide sequences and a maximal protein set of 1883 when combining the results from both the depleted and bound protein fractions f (Table I). As expected, the depleted fraction accounted for almost all the proteins identified, however, the analysis of the bound fraction gave an additional 912 peptides and 77 proteins (MPS) of which the majority was immunoglobulins. The total number of identified proteins (MPS) from the bound fraction was 441.
Protein Distribution on the SDS-PAGE Gel and Observed and Theoretical MW
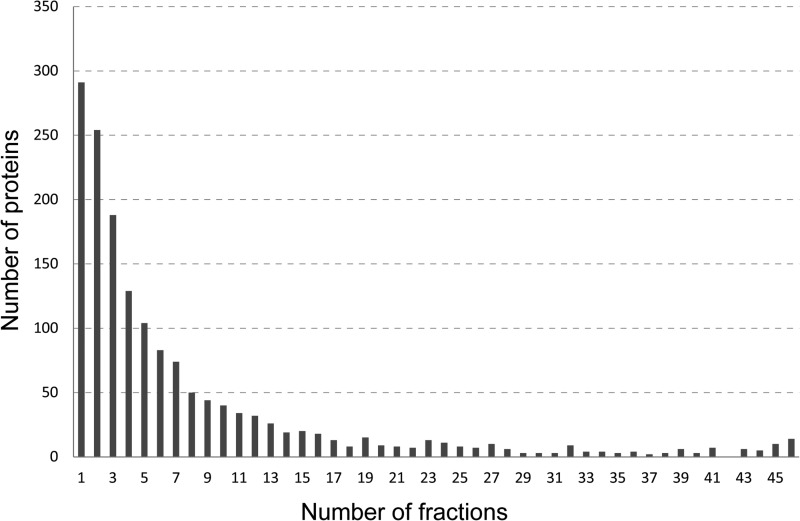
By using the average precursor intensity it was possible to track the intensity distribution of the peptides (representing corresponding proteins) across the gel. Fig. 2 shows the number of proteins with peptides identified in only one fraction (vertical gel slice), two fractions, three fractions etc., up to all 46 fractions in the depleted gel lane. As illustrated in the figure, most proteins were identified in only one, two, or three fractions, and there is a decreasing trend in the number of proteins (y-axis) as the number of fractions (x-axis) increases. However, there is a slight increase toward the end of the x-axis for proteins found in all or almost all 46 fractions, that is proteins identified across the whole molecular weight (MW) area of the gel.
Fig. 2.
Number of proteins (y axis) with peptides observed in from 1 to 46 fractions (x axis) from the immunoaffinity depleted and gel separated sample.
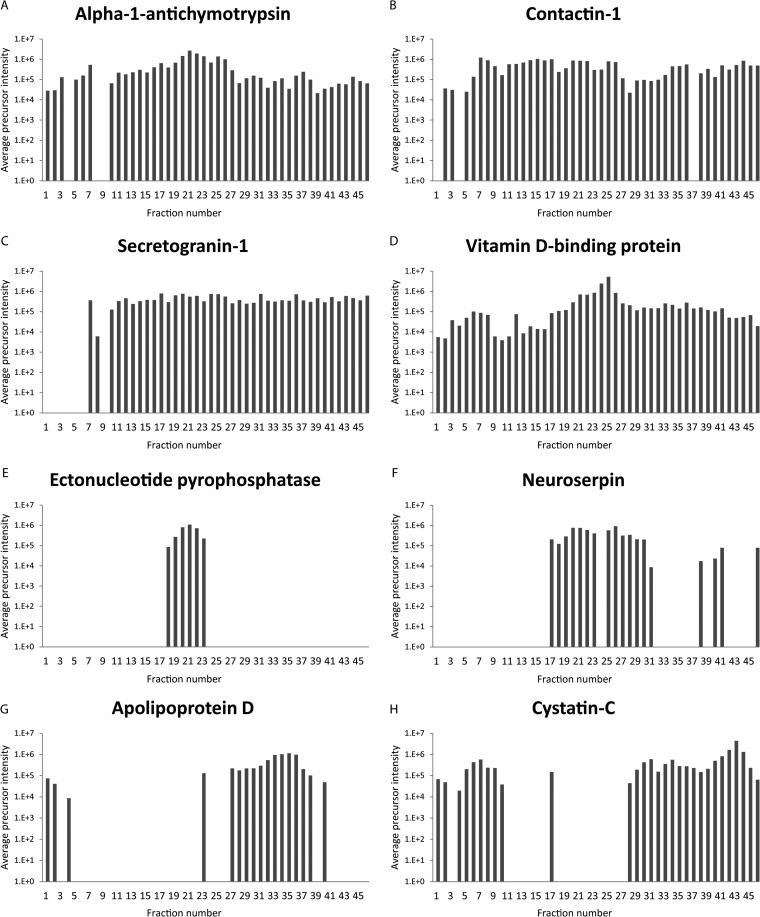
The 24 most widespread proteins (present in 45 or 46 fractions, listed in supplemental Table S3) have theoretical molecular masses ranging from 15.9 kDa (Transthyretin) to 263.5 kDa (Fibronectin). Most of them are present in high amounts in CSF and high-abundant proteins tend to create smears in the gel, which could explain why these proteins are widespread in the gel. Alpha-1-antichymotrypsin, contactin-1, secretogranin-1, and vitamin d-binding protein (VDBP) are some of the proteins found in all or almost all fractions of the gel (Fig. 3A–3D). All these proteins have been suggested as biomarker candidates for multiple sclerosis and/or other neurological disorders (45–52). Secretogranin-1 is a proprotein (protein precursor) known to be proteolytically processed in vivo giving rise to biologically active peptide fragments (53, 54). VDBP is a liver synthesized carrier glycoprotein that transports vitamin D (55), and it has over 80 variants listed in UniProt.
Fig. 3.
Examples of various average precursor intensity distributions (logarithmic scale) across the 46 fractions from the depleted and gel separated sample. Fig. shows data for the proteins alpha-1-antichymotrypsin A, contactin-1 B, secretogranin-1 C, vitamin d-binding protein D, ectonucleotid pyrophosphatase E, neuroserpin F, apolipoprotein D G, and cystatin-C H.
The many fragments and isoforms previously described for these proteins can also explain their wide distribution in the gel. Investigating such proteins in a proteomics experiment, for example, in biomarker discovery and verification studies, clearly presents challenges. Hence, considerations should be taken when selecting sample processing methods and peptides to use for targeted quantification experiments in CSF. During size dependent fractionation, different proteoforms can be present in different MW areas, and experimental results could vary depending on what MW area is included in the sample processing. Thus, our results contribute to guiding the targeted quantification strategies for proteins observed as different size variants in CSF.
For other proteins found in multiple fractions the distribution pattern in the gel varied (Fig. 3E–3H), often with distinct MW areas with high intensity observations separated by stretches without any evidence of the protein. This could point toward the presence of different size isoforms of the protein or indicate that the protein exists both with and without PTMs increasing the MW.
In many cases, we discovered differences between theoretical and observed MW of the identified proteins. The protein neuropilin and tolloid-like protein 2, which is an accessory subunit of neuronal glutamate receptors, were only found in the lower mass regions of the gel, although its theoretical mass is 59 kDa according to UniProt, suggesting the middle mass region. Also other proteins, such as fibulin-2, TGF beta-1, and multimerin-2 were found at much lower MW areas than the theoretical, suggesting they have naturally occurring fragments in CSF. On the contrary, biglycan, serglycin, and apolipoprotein A-I were observed with MW higher than the theoretical mass. Biglycan and serglycin are proteoglycans with glycosaminoglycan chains attached, likely to slow down the proteins migration in the gel, which could explain this behavior. Another explanation for this unexpected migration could be that these proteins form stable complexes not denatured in our SDS-PAGE conditions and therefore appear with higher MW.
Nontryptic Peptides
Information about semi- or nontryptic peptides, i.e. peptide sequences where trypsin digestion only appears likely for one of the peptide's end points, or none of them, respectively, is also available from this study. The existence of such nontryptic peptides can indicate that the protein appears in a truncated form or has peptide fragments present in CSF. As we identified 57 proteins with protease or peptidase in their UniProt-assigned protein name, it is likely that the presence of the nontryptic peptides could be caused by the activity of these and potentially other proteases present in CSF. Nontryptic peptides could also be the result of in-source fragmentation of tryptic peptides (56) and thus not have a biological explanation. Nevertheless, it is important to know which peptides that are observed as truncated when selecting signature peptides for targeted proteomics experiments. The proteins present in many fractions generally have a higher number of nontryptic peptides associated to them, indicating the existence of truncation products for these proteins, a possible explanation for the spread observed in the SDS-PAGE gel.
As an example, for the already mentioned protein secretogranin-1, 27 of the 99 validated peptides from our study were nontryptic. Secretogranin-1 has a theoretical mass of 78 kDa, but is observed in the gel in close to all fractions below the 148 kDa standard (phosphorylase), as can be observed in Fig. 3C. A CSF peptidome (and proteome) study performed by Zougman et al. (6) identified 52 endogenous peptides from secretogranin-1, the highest number for all the 91 proteins which they identified. This further suggests that a high number of molecular weight proteoforms can exist for certain CSF proteins.
CSF Mixed-mode Global Mapping
As a complement to the SDS-PAGE approach to identify CSF proteins, we used a MM (RP-AX) HPLC separation strategy in combination with immunoaffinity depletion. A total of 21,003 peptide sequences and 2779 proteins were identified from the depleted and bound protein fraction. Analysis of the depleted fraction resulted in 18,947 unique peptides and 2661 proteins and the bound fraction in 845 unique peptides and two unique proteins (Ig alpha-2 chain C region and Ig mu heavy chain disease protein). The total number of proteins that were observed from the bound fraction in this experiment was 118, compared with 441 in the SDS-PAGE experiment. Extra caution should be taken when interpreting the quantitative results for these proteins in samples depleted using the MARS hu-14 approach.
Three Proteome Mapping Strategies
From our combined study using three different approaches we found that all separation strategies provided unique information. The MM (RP-AX) fractionation strategy clearly resulted in the most identified proteins (2779 proteins), compared with the gel approach (1883 proteins), however, in the latter we identified 284 proteins not found in the MM (RP-AX) approach (compared with 1180 unique for MM). In addition, the gel approach provided important information about the size distribution of the proteins identified.
When comparing the identified glycopeptides with the identifications from the two global CSF mapping experiments, we found that 18 proteins were uniquely identified from the glyco experiment (supplemental Table S4). Most of these were proteins with several potential PTMs, such as glycosylations, phosphorylations, acetylations, and/or disulfide bonds, which could make them difficult to identify by the other two approaches. We also found 148 peptide sequences that overlapped between the glyco and global experiments for CSF (supplemental Table S5). This suggests that these N-glyco sites are partially occupied, and exist in CSF both as glycosylated and nonglycosylated. Some of the proteins with several partially occupied sites were prostaglandin, major prion protein, complement factor H, and IgGFc-binding protein. This information is important when designing targeted quantitative assays as two different forms of the peptide/protein can be quantified. Signature peptides representing both nonglycosylated asparagine and formerly glycosylated (deamidated asparagine after PNGase F treatment) forms should in these cases be included in the assay.
The Normal Cerebrospinal Fluid Proteome
The most extensive mapping of proteins in normal CSF published to date was performed by Schutzer et al. This study provided 2630 identified proteins with IPI identifiers. As our proteins are identified by UniProt accession numbers, we used the Protein Identifier Cross-Reference (PICR) conversion tool (57) to convert the IPI identifiers to UniProt accession numbers to be able to compare the two studies. We were able to confidently (status: identical) convert 2134 of the 2630 IPI identifications to UniProt accession numbers. By comparing these to our maximal protein set of 3081, we found that 1489 proteins overlapped between the two studies, 645 proteins were unique for the Schutzer study and 1592 were unique for our study (supplemental Table S6). Reasons why a relatively high number of proteins did not overlap between the two studies could be because of the loss of proteins during the conversion from IPI identifiers to UniProt accession numbers, differences in experimental approaches between the two studies, and differences in instrumentation used.
The patient samples in this study were collected from neurologically healthy patients, who agreed to donate CSF before undergoing spinal anesthesia for orthopedic surgery typically in the hips and lower limbs. An alternative to this patient group would be to investigate CSF from completely healthy volunteers, but such samples are hard to obtain because of ethical aspects as discussed in (29). The proteins present in CSF is expected to be qualitatively similar between these two groups, as this is even the case when comparing control patients to patients with various neurological diseases (31, 47, 58, 59). The concentration of certain proteins would however expect to vary, but this would mainly influence quantitative measurements, and not a qualitative study like the one described here. So in this regard, the choice of neurologically healthy patients appears to be appropriate for the purpose of mapping the CSF proteome, and we anticipate that the protein content in the database also is highly representable for other patient groups.
Possible Sample Contaminants
A total of 26 keratin proteins were found in our CSF dataset (nine in the glycodataset) and four in the plasma dataset. By comparing our identified proteins against the list of common Repository of Adventitious Proteins (cRAP, http://www.thegpm.org/cRAP/index.html) provided by the Global Proteome Machine, representing common laboratory dust/skin contaminants, we found that at least ten human keratin proteins and one human salivary protein are likely to be present in our CSF dataset because of contamination by skin, hair, or saliva during sample collection or processing. For the plasma dataset, no proteins matched this list. The remaining keratins in our dataset are likely to either be naturally present in CSF and plasma in small amounts, or be the result of contamination during CSF collection or in the further laboratory processing. When fractionating samples to such an extensive degree as in this study, even small amounts of contaminants could be identified. The CSF samples included in this study were chosen based on not showing signs of blood contamination when visually inspecting the samples or when examined by SRM against hemoglobin β, indicating a very low degree of blood contamination. Despite this, hemoglobin α, β, and δ were identified from the pool made from these samples. Likely, this is because of the extensive fractionation applied in the three experiments, making even marginal protein traces detectable.
Plasma Mixed Mode Mapping
In order to compare the CSF proteome to the plasma proteome, 40 μl of plasma (2400 μg protein) from the same individuals were processed and analyzed in the same way as for the MM (RP-AX) CSF strategy. A total of 9739 peptide sequences and 1050 proteins (MPS) were identified.
When comparing the proteins identified in CSF with the proteins identified in plasma, we found that 2204 were only observed in CSF, 173 were unique for plasma and 877 were observed in both body fluids. The proteins found uniquely in plasma were mainly intracellular and involved in metabolic and cellular processes (gene ontology data not shown), which could be explained by association to cells or platelets, not found in CSF. It is likely that the proteins found in both fluids are mainly plasma derived, because CSF is believed to be, at least in part, an ultrafiltrate of plasma (reviewed in (60)), and also because CNS-derived proteins are expected to have a low abundance in plasma because of dilution. Proteins detected in CSF only could potentially also be present in plasma, but be below the detection limit of the instrumentation used. However, most of the proteins uniquely found in CSF are probably not plasma derived.
To further investigate the origin of the identified CSF proteins, we utilized a recent study from our group (Aasebø et al.) where the effect of blood contamination in CSF samples was analyzed (61). Proteins that were affected in concentration by blood spike-in were considered likely to be blood derived, whereas proteins unaffected were likely CNS derived. The over 800 CSF proteins identified in this experiment were compared with our study, and we found that 267 of our total number of 3081 CSF proteins were affected by blood contamination, 147 were uncertain and 393 were unaffected. This indicates that most of the proteins identified in our study are CNS derived, although a large number of proteins could not be classified as they were not identified in the paper by Aasebø et al. For the glycodataset specifically, 125 proteins were affected, 67 uncertain, and 203 were unaffected.
CSF Proteome Resource
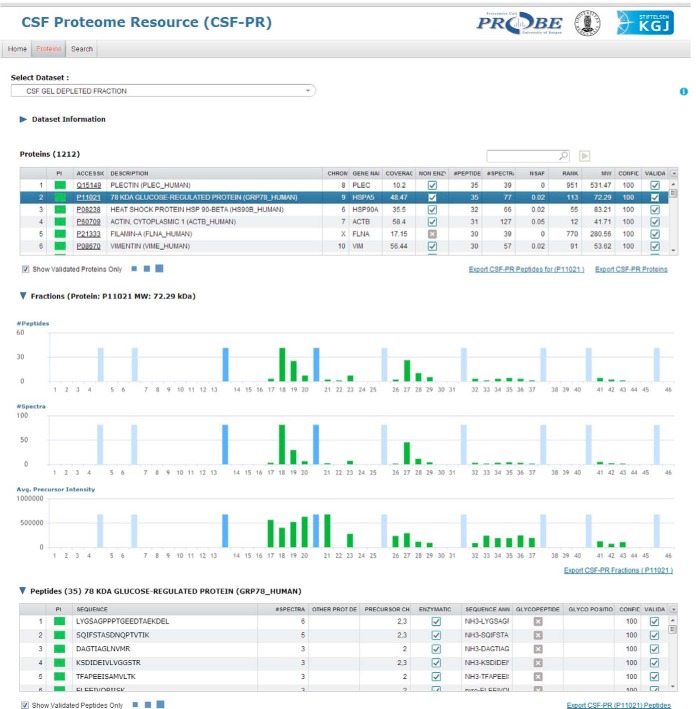
One of the goals of this comprehensive CSF proteome mapping study was to create a publicly available database that could be used to easily browse the CSF proteome as characterized by our extensive LC-MS analyses. This database, the CSF Proteome Resource (CSF-PR), is available at http://probe.uib.no/csf-pr, where all the datasets (glyco, SDS-PAGE, mixed mode, and plasma) are accessible (Fig. 4). Information retrieved from our database should be useful for proteomic studies in CSF and the CNS, and particularly for guiding the selection of suitable signature peptides for targeted quantification.
Fig. 4.
Overview of the CSF Proteome Resource (CSF-PR), showing a selected experiment and the proteins identified in this experiment, and the distribution of the selected protein across the gel fractions is shown in green bars at the bottom, with gel standards marked by light blue bars. The slightly darker blue bars represent the range where the protein's theoretical molecular mass would suggest that the protein should occur.
CSF-PR serves as a freely available reference library where information about proteins and peptides identified in CSF can be retrieved. Protein name, accession number or peptide sequence can be searched for in a single experiment, or across the whole resource. Contained within the resource is also information about protein inference, MW, confidence (a transposition of the search engine e-values), sequence coverage, number of peptides and spectra, glycosylation patterns, and whether nontryptic peptides have been identified for the protein.
The Protein Inference (PI) column, found in both the protein and peptide Tables in CSF-PR, indicates the degree of protein inference complexity for the given protein (group) or peptide, that is, if a peptide is unique for a given protein or shared between multiple proteins (62). The color scheme goes from green (single peptide to protein mapping), via yellow (peptide maps to more than one protein, but these proteins are assumed closely related), to red (peptide maps to more than one protein, and the proteins are assumed unrelated). Note that the distinction between related and unrelated proteins should not be taken as fact, but rather used as an indication for further inspection of the protein groups.
Information about the distribution of proteins in the SDS-PAGE gel is also displayed, and peptides identified as formerly glycosylated are marked with the position of the glycosylation. The presence of nontryptic peptides is also annotated, pointing toward truncated versions of the peptide and again the protein. This might indicate that the peptides are not suitable as surrogates for proteins in proteomic analyses.
Furthermore, mapping the corresponding plasma proteome was important in order to characterize whether the CSF proteins are detected in plasma. The ultimate goal with many discovery studies using CSF is to verify and validate the candidate proteins in plasma, a more easily accessible body fluid with respect to diagnostics and prognostics.
Taken together, CSF-PR is a useful tool for providing an overview of the CSF proteome, and for guiding the selection of appropriate signature peptides for proteins to be quantified in targeted quantification experiments (Fig. 5).
Fig. 5.
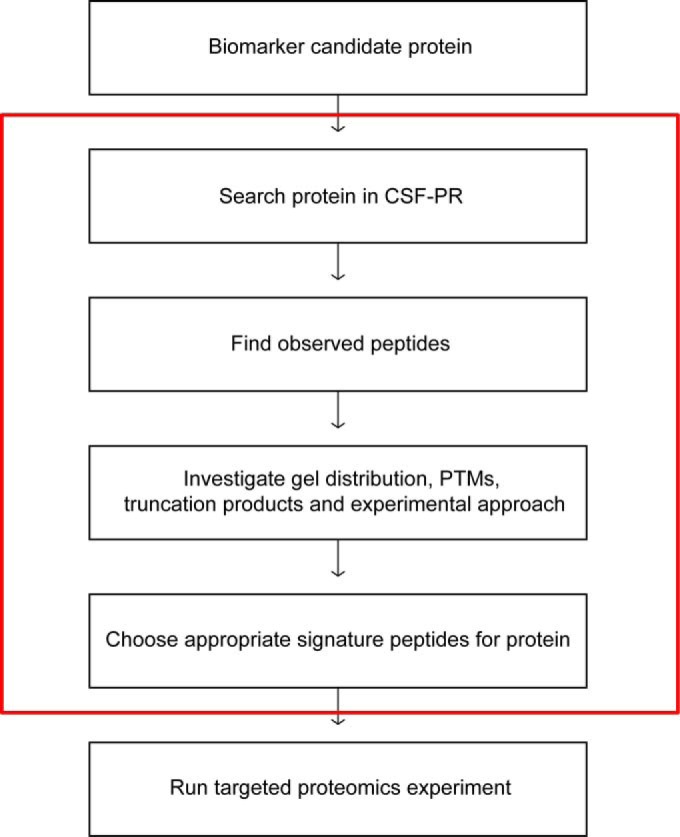
A proposed workflow for how CSF-PR can be used to guide the peptide selection for a targeted quantitative experiment, such as SRM. Everything inside the red box can be done in CSF-PR.
CONCLUSION
We consider the results from our proteomic characterization of CSF and the creation of the CSF-PR database as an important contribution to the field of CSF proteomics research. Especially as an aid in the challenging selection of signature peptides for targeted protein quantification assays, and to examine if proteins found in human blood samples (also provided in the dataset) or proteins from animal brain/CSF experiments also are present in human CSF, and approximately at what level. Our study contains the highest number of proteins (3081) and glycopeptides (1121) ever reported for CSF, including 417 glycosylation sites not previously referenced in Swiss-Prot. The migration and distribution pattern of CSF proteins in SDS-PAGE is also given, indicating the presence of many isoforms and PTMs affecting the MW. CSF-PR differs from other online databases and resources in that it is optimized for CSF, and significantly extends upon the limited information known about the critical CSF proteome. This is important information as this proteome is likely to reflect the status of the CNS. Accordingly, our objective is to continuously add proteomic experiments to CSF-PR, and to further develop its functions and use in the future.
Supplementary Material
Footnotes
Author contributions: A.G., H.V., E.O., K.M., J.A.O., H.B., and F.S.B. designed research; A.G., H.V., and H.G. performed research; Y.F., M.B., H.B., and F.S.B. contributed new reagents or analytic tools; A.G., H.V., Y.F., E.O., J.A.O., H.B., and F.S.B. analyzed data; A.G., H.V., E.O., M.B., K.M., J.A.O., H.B., and F.S.B. wrote the paper; Y.F. main resource developer; M.B. provided the samples.
* The study was supported by Western Norway Regional Health Authority, the Meltzer Foundation, Kjell Alme's Legacy for Research in Multiple Sclerosis, the Frank Mohn Foundation, the Research Council of Norway and the Kristian Gerhard Jebsen Foundation. The data deposition to the ProteomeXchange Consortium was supported by PRIDE Team, EBI.
 This article contains supplemental File S1A–C and Tables S1 to S6.
This article contains supplemental File S1A–C and Tables S1 to S6.
SUPPORTING INFORMATION: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (63), in seven separate datasets with the dataset identifiers PXD000651- PXD000657. Part 1 PXD000651 – CSF, gel separated, bound fraction Part 2 PXD000652 – CSF, gel separated, depleted fraction Part 3 PXD000653 – CSF, glyco enriched, mixed-mode separated Part 4 PXD000654 – CSF, mixed-mode separated, depleted fraction Part 5 PXD000655 – CSF, mixed-mode separated, bound fraction Part 6 PXD000656 – Plasma, mixed-mode separated, bound fraction Part 7 PXD000657 – Plasma, mixed-mode separated, depleted fraction.
All data about proteins and peptides identified in this study, including accession numbers, number of distinct peptides for identified proteins, precursor charges, modifications, score, confidence etc. is available through CSF-PR at http://probe.uib.no/csf-pr and through PRIDE. The Supplemental Files (S1A–S1C) contain detailed descriptions for some of the methods. Supplemental File S1A and S1B gives a description of in-gel and in-solution digestion, respectively, and Supplemental File S1C gives a detailed description of the analysis of the glycopeptide data. Supplemental Table S1 includes information about the 21 subjects who donated the CSF and plasma used in this study. Supplemental Table S2 lists all the peptides where new glycosylation sites have been identified. Supplemental Table S3 lists the 24 most widespread proteins in the gel in the depleted and gel separated experiment. Supplemental Table S4 lists the 18 proteins which were identified only in the glycopeptide enrichment experiment. Supplemental Table S5 lists the 148 peptides which were identified both by the global and the glyco approach (true glycopeptides), indicating partial occupancy of glycosylation sites.
1 The abbreviations used are:
- BBB
- blood-brain barrier
- CID
- collision-induced dissociation
- CNS
- central nervous system
- CSF
- cerebrospinal fluid
- FA
- formic acid
- FDR
- false discovery rate
- IAA
- iodoacetamide
- MARS
- multiple affinity removal system
- MPS
- maximal protein set
- MM (RP-AX)
- mixed mode reversed phase-anion exchange chromatography
- NOG
- N-octyl-β-D-glucopyranoside
- PNGase F
- Peptide-N-Glycosidase F
- PTMs
- post-translational modifications
- SPEG
- solid-phase extraction of N-linked glycopeptides.
REFERENCES
- 1. Segal M. B. (1993) Extracellular and cerebrospinal fluids. J. Inherit. Metab. Dis. 16, 617–638 [DOI] [PubMed] [Google Scholar]
- 2. Kroksveen A. C., Opsahl J. A., Aye T. T., Ulvik R. J., Berven F. S. (2011) Proteomics of human cerebrospinal fluid: discovery and verification of biomarker candidates in neurodegenerative diseases using quantitative proteomics. J. Proteomics 74, 371–388 [DOI] [PubMed] [Google Scholar]
- 3. McComb J. G. (1983) Recent research into the nature of cerebrospinal fluid formation and absorption. J. Neurosurg. 59, 369–383 [DOI] [PubMed] [Google Scholar]
- 4. Regeniter A., Kuhle J., Mehling M., Moller H., Wurster U., Freidank H., Siede W. H. (2009) A modern approach to CSF analysis: pathophysiology, clinical application, proof of concept, and laboratory reporting. Clin. Neurol. Neurosurg. 111, 313–318 [DOI] [PubMed] [Google Scholar]
- 5. Schutzer S. E., Liu T., Natelson B. H., Angel T. E., Schepmoes A. A., Purvine S. O., Hixson K. K., Lipton M. S., Camp D. G., Coyle P. K., Smith R. D., Bergquist J. (2010) Establishing the proteome of normal human cerebrospinal fluid. PLoS One 5, e10980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zougman A., Pilch B., Podtelejnikov A., Kiehntopf M., Schnabel C., Kumar C., Mann M. (2008) Integrated analysis of the cerebrospinal fluid peptidome and proteome. J. Proteome Res. 7, 386–399 [DOI] [PubMed] [Google Scholar]
- 7. Yuan X., Desiderio D. M. (2005) Proteomics analysis of prefractionated human lumbar cerebrospinal fluid. Proteomics 5, 541–550 [DOI] [PubMed] [Google Scholar]
- 8. Pan S., Wang Y., Quinn J. F., Peskind E. R., Waichunas D., Wimberger J. T., Jin J., Li J. G., Zhu D., Pan C., Zhang J. (2006) Identification of glycoproteins in human cerebrospinal fluid with a complementary proteomic approach. J. Proteome Res. 5, 2769–2779 [DOI] [PubMed] [Google Scholar]
- 9. Noben J. P., Dumont D., Kwasnikowska N., Verhaert P., Somers V., Hupperts R., Stinissen P., Robben J. (2006) Lumbar cerebrospinal fluid proteome in multiple sclerosis: characterization by ultrafiltration, liquid chromatography, and mass spectrometry. J. Proteome Res. 5, 1647–1657 [DOI] [PubMed] [Google Scholar]
- 10. Sickmann A., Dormeyer W., Wortelkamp S., Woitalla D., Kuhn W., Meyer H. E. (2000) Identification of proteins from human cerebrospinal fluid, separated by two-dimensional polyacrylamide gel electrophoresis. Electrophoresis 21, 2721–2728 [DOI] [PubMed] [Google Scholar]
- 11. Maccarrone G., Milfay D., Birg I., Rosenhagen M., Holsboer F., Grimm R., Bailey J., Zolotarjova N., Turck C. W. (2004) Mining the human cerebrospinal fluid proteome by immunodepletion and shotgun mass spectrometry. Electrophoresis 25, 2402–2412 [DOI] [PubMed] [Google Scholar]
- 12. Davidsson P., Folkesson S., Christiansson M., Lindbjer M., Dellheden B., Blennow K., Westman-Brinkmalm A. (2002) Identification of proteins in human cerebrospinal fluid using liquid-phase isoelectric focusing as a prefractionation step followed by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 16, 2083–2088 [DOI] [PubMed] [Google Scholar]
- 13. Ogata Y., Charlesworth M. C., Muddiman D. C. (2005) Evaluation of protein depletion methods for the analysis of total-, phospho- and glycoproteins in lumbar cerebrospinal fluid. J. Proteome Res. 4, 837–845 [DOI] [PubMed] [Google Scholar]
- 14. Wenner B. R., Lovell M. A., Lynn B. C. (2004) Proteomic analysis of human ventricular cerebrospinal fluid from neurologically normal, elderly subjects using two-dimensional LC-MS/MS. J. Proteome Res. 3, 97–103 [DOI] [PubMed] [Google Scholar]
- 15. Xu J., Chen J., Peskind E. R., Jin J., Eng J., Pan C., Montine T. J., Goodlett D. R., Zhang J. (2006) Characterization of proteome of human cerebrospinal fluid. Int. Rev. Neurobiol. 73, 29–98 [DOI] [PubMed] [Google Scholar]
- 16. Pan S., Zhu D., Quinn J. F., Peskind E. R., Montine T. J., Lin B., Goodlett D. R., Taylor G., Eng J., Zhang J. (2007) A combined dataset of human cerebrospinal fluid proteins identified by multi-dimensional chromatography and tandem mass spectrometry. Proteomics 7, 469–473 [DOI] [PubMed] [Google Scholar]
- 17. Li S. J., Peng M., Li H., Liu B. S., Wang C., Wu J. R., Li Y. X., Zeng R. (2009) Sys-BodyFluid: a systematical database for human body fluid proteome research. Nucleic Acids Res. 37, D907–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., Apweiler R. (2005) PRIDE: the proteomics identifications database. Proteomics 5, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 19. Berger M. S., Locher G. W., Saurer S., Gullick W. J., Waterfield M. D., Groner B., Hynes N. E. (1988) Correlation of c-erbB-2 gene amplification and protein expression in human breast carcinoma with nodal status and nuclear grading. Cancer Res. 48, 1238–1243 [PubMed] [Google Scholar]
- 20. Hudziak R. M., Schlessinger J., Ullrich A. (1987) Increased expression of the putative growth factor receptor p185HER2 causes transformation and tumorigenesis of NIH 3T3 cells. Proc. Natl. Acad. Sci. U. S. A. 84, 7159–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vogelzang N. J., Lange P. H., Goldman A., Vessela R. H., Fraley E. E., Kennedy B. J. (1982) Acute changes of alpha-fetoprotein and human chorionic gonadotropin during induction chemotherapy of germ cell tumors. Cancer Res. 42, 4855–4861 [PubMed] [Google Scholar]
- 22. Bosl G. J., Lange P. H., Fraley E. E., Goldman A., Nochomovitz L. E., Rosai J., Waldmann T. A., Johnson K., Kennedy B. J. (1981) Human chorionic gonadotropin and alphafetoprotein in the staging of nonseminomatous testicular cancer. Cancer 47, 328–332 [DOI] [PubMed] [Google Scholar]
- 23. Thompson D. K., Haddow J. E. (1979) Serial monitoring of serum alpha-fetoprotein and chorionic gonadotropin in males with germ cell tumors. Cancer 43, 1820–1829 [DOI] [PubMed] [Google Scholar]
- 24. Catalona W. J., Richie J. P., Ahmann F. R., Hudson M. A., Scardino P. T., Flanigan R. C., Dekernion J. B., Ratliff T. L., Kavoussi L. R., Dalkin B. L., Waters W.B., MacFarlane M. T., Southwick P. C. (1994) Comparison of digital rectal examination and serum prostate specific antigen in the early detection of prostate cancer: results of a multicenter clinical trial of 6630 men. J. Urol. 151, 1283–1290 [DOI] [PubMed] [Google Scholar]
- 25. Canney P. A., Moore M., Wilkinson P. M., James R. D. (1984) Ovarian cancer antigen CA125: a prospective clinical assessment of its role as a tumour marker. Br. J. Cancer 50, 765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Roth J. (2002) Protein N-glycosylation along the secretory pathway: relationship to organelle topography and function, protein quality control, and cell interactions. Chem. Rev. 102, 285–303 [DOI] [PubMed] [Google Scholar]
- 27. Sato Y., Endo T. (2010) Alteration of brain glycoproteins during aging. Geriatr. Gerontol. Int. 1, S32–40 [DOI] [PubMed] [Google Scholar]
- 28. Nilsson J., Ruetschi U., Halim A., Hesse C., Carlsohn E., Brinkmalm G., Larson G. (2009) Enrichment of glycopeptides for glycan structure and attachment site identification. Nat. Methods 6, 809–811 [DOI] [PubMed] [Google Scholar]
- 29. Teunissen C., Menge T., Altintas A., Alvarez-Cermeno J. C., Bertolotto A., Berven F. S., Brundin L., Comabella M., Degn M., Deisenhammer F., Fazekas F., Franciotta D., Frederiksen J. L., Galimberti D., Gnanapavan S., Hegen H., Hemmer B., Hintzen R., Hughes S., Iacobaeus E., Kroksveen A. C., Kuhle J., Richert J., Tumani H., Villar L. M., Drulovic J., Dujmovic I., Khalil M., Bartos A. (2013) Consensus definitions and application guidelines for control groups in cerebrospinal fluid biomarker studies in multiple sclerosis. Mult. Scler. [DOI] [PubMed] [Google Scholar]
- 30. Teunissen C. E., Petzold A., Bennett J. L., Berven F. S., Brundin L., Comabella M., Franciotta D., Frederiksen J. L., Fleming J. O., Furlan R., Hintzen R. Q., Hughes S. G., Johnson M. H., Krasulova E., Kuhle J., Magnone M. C., Rajda C., Rejdak K., Schmidt H. K., van Pesch V., Waubant E., Wolf C., Giovannoni G., Hemmer B., Tumani H., Deisenhammer F. (2009) A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology 73, 1914–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kroksveen A. C., Aasebo E., Vethe H., Van Pesch V., Franciotta D., Teunissen C. E., Ulvik R. J., Vedeler C., Myhr K. M., Barsnes H., Berven F. S. (2012) Discovery and initial verification of differentially abundant proteins between multiple sclerosis patients and controls using iTRAQ and SID-SRM. J. Proteomics 78, 312–325 [DOI] [PubMed] [Google Scholar]
- 32. Phillips H. L., Williamson J. C., van Elburg K. A., Snijders A. P., Wright P. C., Dickman M. J. (2010) Shotgun proteome analysis utilizing mixed mode (reversed phase-anion exchange chromatography) in conjunction with reversed phase liquid chromatography mass spectrometry analysis. Proteomics 10, 2950–2960 [DOI] [PubMed] [Google Scholar]
- 33. Tian Y., Zhou Y., Elliott S., Aebersold R., Zhang H. (2007) Solid-phase extraction of N-linked glycopeptides. Nat. Protoc. 2, 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Berven F. S., Ahmad R., Clauser K. R., Carr S. A. (2010) Optimizing performance of glycopeptide capture for plasma proteomics. J. Proteome Res. 9, 1706–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vaudel M., Barsnes H., Berven F. S., Sickmann A., Martens L. (2011) SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics 11, 996–999 [DOI] [PubMed] [Google Scholar]
- 36. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 37. Fenyo D., Beavis R. C. (2003) A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal. Chem. 75, 768–774 [DOI] [PubMed] [Google Scholar]
- 38. Barsnes H., Vaudel M., Colaert N., Helsens K., Sickmann A., Berven F. S., Martens L. (2011) Compomics-utilities: an open-source Java library for computational proteomics. BMC Bioinformatics 12, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nesvizhskii A. I. (2010) A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. J. Proteomics 73, 2092–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mi H., Muruganujan A., Thomas P. D. (2013) PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Beausoleil S. A., Villen J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 43. Vaudel M., Breiter D., Beck F., Rahnenfuhrer J., Martens L., Zahedi R. P. (2013) D-score: a search engine independent MD-score. Proteomics 13, 1036–1041 [DOI] [PubMed] [Google Scholar]
- 44. Frank M., Kemler R. (2002) Protocadherins. Curr. Opin. Cell Biol. 14, 557–562 [DOI] [PubMed] [Google Scholar]
- 45. Kroksveen A. C., Guldbrandsen A., Vedeler C., Myhr K. M., Opsahl J. A., Berven F. S. (2012) Cerebrospinal fluid proteome comparison between multiple sclerosis patients and controls. Acta Neurol. Scand. 126, Supplement s195, 90–96 [DOI] [PubMed] [Google Scholar]
- 46. Ottervald J., Franzén B., Nilsson K., Andersson L., Khademi M., Eriksson B., Kjellström S., Marko-Varga G., Végvári A., Harris R., Laurell T., Miliotis T., Matusevicius D., Salter H., Ferm M., Olsson T. (2010) Multiple sclerosis: identification and clinical evaluation of novel CSF biomarkers. J. Proteomics 73, 1117–1132 [DOI] [PubMed] [Google Scholar]
- 47. Abdi F., Quinn J. F., Jankovic J., McIntosh M., Leverenz J. B., Peskind E., Nixon R., Nutt J., Chung K., Zabetian C., Samii A., Lin M., Hattan S., Pan C., Wang Y., Jin J., Zhu D., Li G. J., Liu Y., Waichunas D., Montine T. J., Zhang J. (2006) Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J. Alzheimer's Dis. 9, 293–348 [DOI] [PubMed] [Google Scholar]
- 48. Simonsen A. H., McGuire J., Podust V. N., Davies H., Minthon L., Skoog I., Andreasen N., Wallin A., Waldemar G., Blennow K. (2008) Identification of a novel panel of cerebrospinal fluid biomarkers for Alzheimer's disease. Neurobiol. Aging 29, 961–968 [DOI] [PubMed] [Google Scholar]
- 49. Davidsson P., Westman-Brinkmalm A., Nilsson C. L., Lindbjer M., Paulson L., Andreasen N., Sjogren M., Blennow K. (2002) Proteome analysis of cerebrospinal fluid proteins in Alzheimer patients. Neuroreport 13, 611–615 [DOI] [PubMed] [Google Scholar]
- 50. Simonsen A. H., McGuire J., Hansson O., Zetterberg H., Podust V. N., Davies H. A., Waldemar G., Minthon L., Blennow K. (2007) Novel panel of cerebrospinal fluid biomarkers for the prediction of progression to Alzheimer dementia in patients with mild cognitive impairment. Arch. Neurol. 64, 366–370 [DOI] [PubMed] [Google Scholar]
- 51. Kroksveen A. C., Opsahl J. A., Aye T. T., Ulvik R. J., Berven F. S. (2011) Proteomics of human cerebrospinal fluid: discovery and verification of biomarker candidates in neurodegenerative diseases using quantitative proteomics. J. Proteomics 74, 371–388 [DOI] [PubMed] [Google Scholar]
- 52. Kroksveen A. C., Aasebo E., Vethe H., Van Pesch V., Franciotta D., Teunissen C. E., Ulvik R. J., Vedeler C., Myhr K. M., Barsnes H., Berven F. S. (2013) Discovery and initial verification of differentially abundant proteins between multiple sclerosis patients and controls using iTRAQ and SID-SRM. J. Proteomics 78, 312–325 [DOI] [PubMed] [Google Scholar]
- 53. Benjannet S., Leduc R., Adrouche N., Falgueyret J. P., Marcinkiewicz M., Seidah N. G., Mbikay M., Lazure C., Chretien M. (1987) Chromogranin B (secretogranin I), a putative precursor of two novel pituitary peptides through processing at paired basic residues. FEBS Lett. 224, 142–148 [DOI] [PubMed] [Google Scholar]
- 54. Helle K. (2010) Chromogranins A and B and secretogranin II as prohormones for regulatory peptides from the diffuse neuroendocrine system. Results Probl. Cell Differ. 50, 21–44 [DOI] [PubMed] [Google Scholar]
- 55. Haddad J. G. (1995) Plasma vitamin D-binding protein (Gc-globulin): multiple tasks. J. Steroid Biochem. Mol. Biol. 53, 579–582 [DOI] [PubMed] [Google Scholar]
- 56. Kim J. S., Monroe M. E., Camp D. G., 2nd, Smith R. D., Qian W. J. (2013) In-source fragmentation and the sources of partially tryptic peptides in shotgun proteomics. J. Proteome Res. 12, 910–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wein S. P., Cote R. G., Dumousseau M., Reisinger F., Hermjakob H., Vizcaino J. A. (2012) Improvements in the Protein Identifier Cross-Reference service. Nucleic Acids Res. 40, W276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Martins-de-Souza D., Maccarrone G., Wobrock T., Zerr I., Gormanns P., Reckow S., Falkai P., Schmitt A., Turck C. W. (2010) Proteome analysis of the thalamus and cerebrospinal fluid reveals glycolysis dysfunction and potential biomarkers candidates for schizophrenia. J. Psychiatr. Res. 44, 1176–1189 [DOI] [PubMed] [Google Scholar]
- 59. Constantinescu R., Andreasson U., Li S., Podust V. N., Mattsson N., Anckarsater R., Anckarsater H., Rosengren L., Holmberg B., Blennow K., Wikkelso C., Ruetschi U., Zetterberg H. (2010) Proteomic profiling of cerebrospinal fluid in parkinsonian disorders. Parkinsonism Relat. Disord. 16, 545–549 [DOI] [PubMed] [Google Scholar]
- 60. Wright B. L., Lai J. T., Sinclair A. J. (2012) Cerebrospinal fluid and lumbar puncture: a practical review. J. Neurol. 259, 1530–1545 [DOI] [PubMed] [Google Scholar]
- 61. Aasebo E., Opsahl J. A., Bjorlykke Y., Myhr K. M., Kroksveen A. C., Berven F. S. (2014) Effects of blood contamination and the rostro-caudal gradient on the human cerebrospinal fluid proteome. PLoS One 9, e90429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nesvizhskii A. I., Aebersold R. (2005) Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics 4, 1419–1440 [DOI] [PubMed] [Google Scholar]
- 63. Vizcaino J. A., Cote R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Perez-Riverol Y., Reisinger F., Rios D., Wang R., Hermjakob H. (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.