Abstract
AMPylation (adenylylation) is a recently discovered mechanism employed by infectious bacteria to regulate host cell signaling. However, despite significant effort, only a few host targets have been identified, limiting our understanding of how these pathogens exploit this mechanism to control host cells. Accordingly, we developed a novel nonradioactive AMPylation screening platform using high-density cell-free protein microarrays displaying human proteins produced by human translational machinery. We screened 10,000 unique human proteins with Vibrio parahaemolyticus VopS and Histophilus somni IbpAFic2, and identified many new AMPylation substrates. Two of these, Rac2, and Rac3, were confirmed in vivo as bona fide substrates during infection with Vibrio parahaemolyticus. We also mapped the site of AMPylation of a non-GTPase substrate, LyGDI, to threonine 51, in a region regulated by Src kinase, and demonstrated that AMPylation prevented its phosphorylation by Src. Our results greatly expanded the repertoire of potential host substrates for bacterial AMPylators, determined their recognition motif, and revealed the first pathogen-host interaction AMPylation network. This approach can be extended to identify novel substrates of AMPylators with different domains or in different species and readily adapted for other post-translational modifications.
Protein AMPylation (adenylylation) was recently discovered in bacteria-host interactions where virulence factors catalyze AMPylation using either a conserved Fic domain (e.g., VopS, Vibrio parahaemolyticus (V. para) and IbpA, Histophilus somni) or an adenylyl transferase domain (e.g., DrrA, Legionella pneumophila). These bacterial AMPylation enzymes, or AMPylators, are secreted into the host cells by bacterial secretion systems and transfer AMP from ATP to Tyr or Thr residues of their respective substrates (1–3). In the case of VopS and IbpA, several Rho family GTPases (Rac1, RhoA, and Cdc42) are known substrates and AMPylation disrupts the binding of the GTPase to its downstream effectors, for example, PAK1 (2–6). Considering the conservation of AMPylation domains in both prokaryotic and eukaryotic organisms, we expect that AMPylation plays an important role in a wide range of cellular processes (2, 4, 5, 7–9). Nevertheless, our understanding of this post-translational modification (PTM) is still limited to only a handful of known eukaryotic AMPylation substrates, exclusively belonging to the Rho and Rab GTPase families(10–14). Determining the repertoire of substrates modified by AMPylators will help illuminate both the functional consequences of AMPylation and the mechanistic strategies of pathogens that employ them (6).
Significant effort has been devoted to identifying AMPylation substrates. Li et al. systematically investigated the fragmentation patterns of chemically synthesized peptides with Thr, Ser, and Tyr AMPylation using matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). They detected AMPylation sites with high confidence and selectively scanned AMPylated peptides in protein mixtures (10). Hao et al. produced a polyclonal antibody that specifically recognized proteins with AMPylation at threonine residues (11). Grammel et al. synthesized an ATP analog, N6pATP (N6-propargyl adenosine-5′-triphophate), which allows the labeling of AMPylated proteins with azide-functionalized fluorescein or a cleavable biotin enrichment tag (ortho-hydroxy-azidoethoxy-azobiotin) based on copper-catalyzed azide-alkyne cycloaddition (CuAAC)1. The identification of new substrates for VopS in HeLa cell lysates was explored by a combination of AMP-specific pull-down and LC-MS (12). Using the same approach, Lewallen et al. tried to identify the substrates of VopS in MCF7 cell extracts by employing a commercial N6-(6-amino)hexyl-ATP-5-carboxyl-fluorescein (F1-ATP) and anti-fluorescein antibody(13). With these efforts combined, four potential new VopS substrates have been identified (SCCA2, NAGK, NME1, and PFKP), though not yet confirmed. These approaches might miss substrates because of temporal and spatial expression or low abundance in cell lysate, poor recognition by the capture molecules or loss during pull-down procedures (12, 14).
Protein microarrays offer a promising approach to identify candidate substrates because they display thousands of unique proteins in a high-throughput and reproducible format (15–17). However, producing arrays with consistent levels of well-folded proteins is challenging because of limitations of protein production, purification, and storage, particularly for mammalian proteins (18).
To circumvent these limitations, cell-free protein arrays, which do not require protein purification, have been developed over the past decade (19–22). These methods provide rapid and economical approaches of fabricating protein arrays in terms of cost, shelf life, and storage (23, 24). In cell-free protein arrays, a nucleotide template is printed on the slide and used to produce proteins in vitro with cell-free expression systems from several organisms such as E. coli, wheat germ, and rabbit reticulocyte lysate, etc. (24, 25). These proteins can be engineered to contain fusion tags that enable their capture to the array surface with an appropriate agent. Of these cell-free protein array methods, the Nucleic Acid Programmable Protein Array (NAPPA) is the most advanced, having achieved both high-density and high content containing ∼2300–8000 proteins per slide (20, 26, 27). In NAPPA, a plasmid-based cDNA configured to include an epitope tag is printed on a microscope slide along with the corresponding tag-specific binding reagent, such as an anti-tag antibody, and stored. At the time of experimentation, the cDNA is transcribed/translated into recombinant protein and captured/displayed in situ by the binding reagent. Using a rabbit reticulocyte lysate-based cell-free expression system, NAPPA has been applied toward the identification of novel protein-protein interactions and disease-related antibody biomarkers (20, 26, 28, 29). However, cell-free protein arrays have yet to be employed in the study of PTMs.
In this work, we established a novel, nonradioactive unbiased AMPylation screening platform by developing a novel click chemistry-based detection assay for use on high-density cell-free protein microarrays displaying human proteins. Labeling AMP-modified substrates covalently with a fluorophore coupled with the use of human ribosomal machinery and chaperones to produce proteins achieved much higher sensitivity and signal to noise (S/N) ratio compared with previous studies. We screened 10,000 human proteins with two bacterial pathogen AMPylators, VopS and IbpAFic2, identifying more than twenty new substrates each. Two novel Rho GTPases (Rac2 and Rac3) were validated in vivo as substrates of the virulence factor VopS in HEK293T cells during V. para infection. Using mass spectrometry, we verified that a non-GTPase protein, ARHGDIB/LyGDI, was AMPylated by VopS on its threonine 51, which is located in a highly regulated part of this protein. This modification inhibited phosphorylation of LyGDI by Src kinase in vitro. Finally, the identification of these new targets allowed us to build the first bacteria-host interaction AMPylation network and may reveal signaling interactions that could potentially be important for bacterial pathogenesis in the future functional studies.
EXPERIMENTAL PROCEDURES
Preparation of Bacterial Fic AMPylators and Click Reagents
VopS NΔ30 and Rac1 proteins were generated as previously described (2). IbpAFic2 proteins were expressed as a His-SUMO fusion in the pE-SUMO vector in BL21 (DE3) E. coli and captured with His-Select Nickel Affinity gel (Sigma, St. Louis, MO), washed with 25 column volumes of lysis buffer and eluted with lysis buffer plus 250 mm Imidazole. They were further purified on mono Q column and Superdex 75 PG columns. Proteins were concentrated, adjusted to 10% glycerol, flash frozen, and stored at −80 °C. The click reagents, including N6pATP and az-rho, were generated as previously described (12).
Preparation of Plasmid DNA and Fabrication of NAPPA Arrays
All sequence-verified, full-length human genes in T7-based mammalian expression vectors (pANT7-cGST and pLDNT7_nFLAG) were obtained from DNASU (http://dnasu.asu.edu/DNASU/). The preparation of DNA plasmids and NAPPA arrays were performed as previously reported (26) and described in supplemental Materials and Methods.
Preparation of NAPPA Protein Arrays
The NAPPA array was blocked with Superblock solution (Pierce, Rockford, IL) for 1 h at room temperature, followed by incubation with 160 μl expression solution (HeLa lysates, Accessory proteins, Reaction mix, and Nuclease-free Water) for 1.5 h at 30 °C and 0.5 h at 15 °C.
After washing with PBST (PBS, 0.2%Tween) three times, the plasmid DNA on the microarray spots was removed by incubating the array with 200 μl DNase (Sigma, St. Louis, MO) for 20 min at room temperature with a coverslip. Then the resulting protein array was blocked with 1%BSA solution for 1 h at room temperature.
To load the GTPase proteins, the array was treated with 2.5 ml 50 μm GTPγS solution (50 mm Tris-HCl and 7.5 mm EDTA, pH 7.5) for 15 min at 30 °C, followed with the addition of 12.5 μl 1 m MgCl2 for additional 5min to stop the reaction and washing two times with washing solution (50 μm HEPES and 5 mm MgCl2, pH 7.4)(2).
NAPPA AMPylation Assay
The AMPylation reaction was performed by incubation of the prepared NAPPA protein array with 40 μg/ml AMPylators and 250 μm N6pATP in 160 μl AMPylation solution (20 mm HEPES pH 7.4, 100 mm NaCl, 5 mm MgCl2, 0.1 mg/ml BSA, and 1 mm DTT) for 1 h at 30 °C. Following by washing three times with PBST, the detection was performed with the 160 μl click reagents containing 250 μm azido-rhodamine (az-rho), 1 mm TCEP, 0.1 mm TBTA, and 1 mm CuSO4, as previously described (12) for 1 h at room temperature. After washing overnight, the microarray was scanned with a Tecan's PowerScanner (Männedorf, Switzerland). The fluorescent signal was quantitated using Array-Pro Analyzer, version 6.3 (Media Cybernetics, Bethesda, MD).
Bead-based AMPylation Assay
The proteins with GST or Flag tag was expressed using the expression solution and captured to the anti-GST or anti-Flag antibody coupled Dynabeads (Invitrogen, Carlsbad, CA) (supplemental Materials and Methods). 10 μl GST-proteins conjugated beads were used for each reaction. The supernatant of GST-protein conjugated magnetic beads was removed using a magnetic stand and GTPase conjugated beads were loaded with GTPγS solution as in NAPPA. After washing with GTP washing buffer, 15 μl AMPylation solution (20 mm HEPES pH 7.4, 100 mm NaCl, 5 mm MgCl2, 0.1 mg/ml BSA, and 1 mm DTT) containing 40 μg/ml AMPylators and 250 μm N6pATP was added and the incubation was performed at 30 °C for 1 h. After washing again, 20 μl click reagents were added (250 μm az-rho, 1 mm TCEP, 0.1 mm TBTA, and 1 mm CuSO4), and the incubation was carried out for 1 h at room temperature. The reaction was stopped by addition of 20 μl 1×SDS loading buffer containing 10% 2-mercaptoethanol. Samples were boiled 5 min and analyzed in a 4–15% Tris-Criterion™ Precast Gel (Bio-Rad, Hercules, CA).
Protein gel images were obtained using an Amersham Bioscience Typhoon 9400 variable mode imager (excitation 532 nm, 580 nm filter, 30 nm band-pass). All image adjustments were performed using Adobe® Photoshop® CS4 software (Adobe Systems, San Jose, CA).
The protein expression was examined in the same gel followed with Western blot using 1:3000 dilution of mouse anti-GST antibody (Cell Signaling Technology, Danvers, MA) and HRP conjugated sheep anti-mouse antibody (Jackson ImmunoResearch, Labs, West Grove, PA) separately. The Chemiluminescence detection was performed with SuperSignal West Femto Luminol/Enhancer Solution (Pierce, Rockford, IL).
Data Analysis
Before statistical analysis, all the fluorescent microarray images were examined for the spot shape, dust, and nonspecific binding to remove false positive signals. We then normalized the raw signal intensity to decrease the variation caused by different slides' background. The normalization was performed by subtracting the background signal attributable to nonspecific binding of AMPylators or fluorescent molecules, which was estimated by the first quartile of the printing buffer-only control. The normalized value was calculated by dividing the result for each feature by the median background-adjusted value of all proteins on the array. Proteins with a normalized value 20% above the median were considered positive signals.
To choose potential substrates of VopS and IbpAFic2, we set a cut-off ratio of 1.2-fold, which was calculated by dividing the signal caused by AMPylator by its buffer control. Moreover, each experiment was repeated three times on independent days and only the protein showing a positive ratio in all three experiments were selected as candidates. With this cut-off, we identified and confirmed six of the seven expected targets in a preliminary test of VopS (supplemental Fig. S5).
Bioinformatics Analysis
The protein annotation was performed manually with The Universal Protein Resource (UniProt) databases and PANTHER (Protein ANalysis THrough Evolutionary Relationships) Classification System. The networks of pathogen-host AMPylation interaction and biological processes were constructed with Cytoscape (v2.6.3, available on http://www.cytoscape.org/) using BiNGO, a Java-based tool that is implemented as a plug-in for Cytoscape was utilized to check the overrepresented gene ontology terms. The GO Biological process analysis using our NAPPA 10,000 human genes as reference was executed based on the hypergeometric test and Benjamini & Hochberg False Discovery Rate (FDR) correction with the p value of 0.05. The detail of biological processes analyzed are shown in supplemental Tables S5, S6.
Probing of Proteins for AMPylation using anti-AMP Antibodies
The recombinant proteins for RhoGDI1, LyGDI, and LyGDI T51A were produced using bacterial coexpression experiments (supplemental Materials and Methods). To probe the proteins for AMPylation, 20 μl of precapture lysate or 100 ng of each eluted protein was loaded onto miniProtean SDS-PAGE gels (Bio-Rad, Hercules, CA) and transferred to PVDF membranes. Membranes were incubated for 1 h in blocking buffer (TBS-T containing 5% milk), washed three times in TBS-T, and then incubated with primary antibodies in blocking buffer for 2 h. Membranes containing eluted proteins were probed with 1:1000 dilution of anti-His (Life Technology, Carlsbad, CA), anti-T-AMP (11), or anti-Y-AMP, whereas precapture lysates were blotted with anti-VopS (peptide antibody generated at UTSW antibody core, 1:1000). Membranes were again washed three-times, probed for 1 h with either anti-rabbit or anti-mouse HRP (1:20,000), washed three-times, incubated in homemade luminol substrate and exposed to film. Rabbit anti-Y-AMP antibody was made in house using a chymotrypsin-digested peptide, TTNAFPGEY(AMP)IPTV, from IbpA AMPylated Rac1 through Invitrogen.
Intact Mass Measurement
Intact mass measurements of His-LyGDI proteins expressed with empty vector or VopS in solution were obtained by the UT Southwestern Proteomics Core using an Agilent 6540 UHD Accurate-Mass QTOF machine. A mass shift of 328.05 was observed for His-LyGDI coexpressed with VopS (supplementary Fig. S6).
Tandem LC-MS/MS
An SDS-PAGE excised band containing His-LyGDI protein coexpressed with VopS was digested with trypsin and run on the Q-Exactive mass-spectrometer for LC-MS/MS analysis at the UT Southwestern Medical Center Proteomics Core. Resulting data file was searched using in house Central Proteomics Facilities Pipeline and phosphoadenosine (AMPylation) definitions present in the Unimod database were used as variable modifications in the searches. GDIR2 Human sequence was identified with 740 spectra/41 unique peptides, yielding 97.5% sequence coverage. AMPylation of Threonine T51 was confidently identified. 34 identifications of the TLLGDGPVVTDPK peptide or mis-cleaved variants were found with AMPylation present on the threonine 51. Identifications were made from charge states 2+, 3+, 4+, and characteristic AMPylation reporter ions from Li et al. are visible in spectra (10) (supplementary Fig. S7 and supplementary Table S7).
In Vitro Kinase Assays
20 μl radioactive kinase assays were prepared using a 4x Src kinase buffer (100 mm Tris pH 7.2, 125 mm MgCl2, 25 mm MnCl2, 8 mm EGTA, 250 μm NaVO4, and 2 mm DTT), 100 mm ATP, 5 μCi [γ-32P] ATP (Perkin Elmer 6000Ci/mmol), 4 μg recombinant LyGDI protein, and 3 units Src p60c-src (Millipore cat #14–117, Billerica, MA). Prior to assay setup, His-LyGDI proteins were cleaved with thrombin as the His tag was found to be inhibitory to phosphorylation compared with GST tagged wild type LyGDI. Proteins were incubated at 30 °C for 45 min and reactions were stopped by the addition of Laemmli buffer and boiled 2 min. Samples were loaded onto miniProtean SDS-PAGE gels (Bio-Rad, Hercules, CA),transferred to PVDF membranes, and exposed to film (Phenix, Candler, NC) for between 2–24 h until an exposure in the linear range was obtained. Densitometry was calculated using ImageJ, and phosphorylation bands were normalized to the coomassie stained PVDF bands. A student's 1 tailed t test with unequal variance (p = 0.034) was used to compare relative phosphorylation of LyGDI WT and AMPylated proteins from three independent experiments.
RESULTS
Development of an AMPylation Assay on NAPPA Arrays
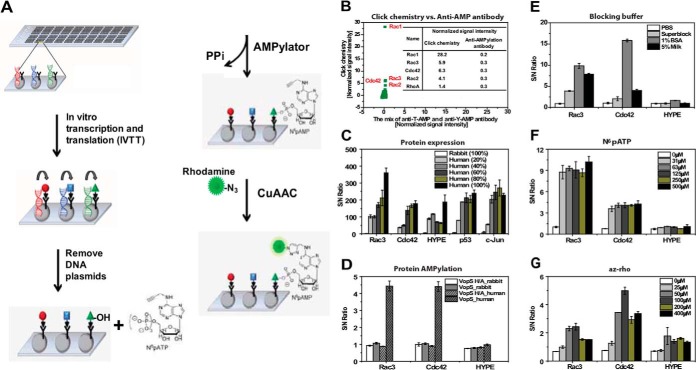
The principle of our click chemistry-based AMPylation assay on NAPPA is shown in Fig. 1A. Briefly, the protein expression plasmids were printed on slides and stored anhydrously at room temperature. At the time of the AMPylation experiment, we produced proteins by mammalian lysate-based cell-free expression system. The slides were then treated with DNase to eliminate potential cross-reaction of detection agents with the DNA. After incubation with N6pATP and AMPylators for 1h at 30 °C to allow the transfer of AMP to the substrates, the AMPylated proteins on the NAPPA slides were detected with az-rho following CuAAC labeling (12).
Fig. 1.
Development of AMPylation screening platform on NAPPA arrays. A, The principle of AMPylation assay on NAPPA arrays. B, Comparison of AMPylation assay on NAPPA arrays using click chemistry and anti-AMPylation antibody. The experiment was performed by using VopS enzyme and NAPPA GST2 arrays comprising 1823 human proteins. 1:50 dilution of anti-T-AMP antibody and anti-Y-AMP antibody were mixed and used as the primary antibody, and 1:100 dilution of Alex647 chick anti-rabbit IgG antibody was used as the secondary detection antibody. X-axis and Y-axis represent the normalized signal intensity produced by taking the signal intensity for each feature divided by the median background-adjusted value for all the proteins on the array as described in the methods. C–G, Optimization of the assay parameters for AMPylation assay on NAPPA arrays, including the cell-free expression system C, D, blocking buffer E, N6pATP F, and az-rho G, respectively. The signal to noise ratio (S/N) ratio was calculated using MSP-1, which expresses a 25 kDa protein with the HA tag and is not captured to the anti-GST coated array surface, as a negative control. CuAAC, copper-catalyzed azide-alkyne cycloaddition.
We initially compared the ability to detect AMP modification by VopS between this click chemistry and a mix of anti-T-AMP and anti-Y-AMP antibodies using a NAPPA array displaying ∼2000 human proteins produced from human HeLa lysate based cell-free expression system (Fig. 1B). In contrast to the antibody-based assay, where we observed high background noise presumably because of nonspecific binding, we readily identified three known VopS substrates (Rac1, RhoA, and Cdc42) and two new RhoGTPase substrates (Rac2 and Rac3) using N6pATP. By comparing the normalized signal intensity, which was calculated by using the signal intensity of each feature divided by the median background-adjusted value for all the proteins on the array, we found the click chemistry had much higher sensitivity, specificity, and a wider dynamic range than the anti-AMPylation antibodies in the detection of proteins with AMPylation on NAPPA (Fig. 1B).
Using VopS and its inactive H348A mutant as a control, we systematically optimized the experimental conditions of the AMPylation assay on NAPPA. First, we tested the influence of DNase and RNase treatment on the AMPylation assay. Treatments of either DNase alone or combined with RNase removed nearly all DNA when estimated by PicoGreen staining (Fig. 2A and supplemental Fig. S1A). The pilot assay showed that the AMPylation of VopS targets (Rac2, Rac3, and Cdc42) could be detected at high S/N ratios only after removal of DNA molecules from the slides. However, no additional benefit was observed after the addition of RNase (supplemental Fig. S1B, S1C). Second, we tested the effect of various time lengths for incubation and washing steps, and found that an overnight washing step after the az-rho treatment improved the signal and S/N ratio of the AMPylation assay, presumably by reducing nonspecific binding of fluorescent dyes (supplemental Fig. S2). Third, we achieved a significant improvement in candidate substrate display by using a new cell-free expression system based on human HeLa cell lysate, which has a theoretical advantage of using human ribosomes and chaperones to produce human proteins in their native forms, compared with the initially used rabbit reticulocyte lysate system(25, 30). Indeed, we found that both protein yields and the S/N ratio of the AMPylation assay were markedly improved with the human HeLa system (Fig. 1C, 1D). Finally, further optimizations were performed using different blocking buffers (PBS, Superblock, 1%BSA, and 5%Milk), different concentrations of N6pATP (0, 31, 63, 125, 250, and 500 μm) and different concentrations of az-rho (0, 25, 50, 100, 200, and 400 μm). These combined experiments yielded optimized conditions using 1% BSA, N6pATP at 63 μm, and az-rho at 100 μm (Fig. 1E–1G).
Fig. 2.
Global identification of substrates for VopS and IbpAFic2 using high-density NAPPA arrays. A, Representative images show the quality control of fabricated high-density NAPPA arrays. Expression clones (n = 1823) encoding the target proteins fused to a C-terminal GST tag were printed along with a polyclonal anti-GST antibody in single spot on the array surface. DNA capture was confirmed by PicoGreen staining (Fig. 2A, left). The plasmid DNA was removed using DNase after expression (Fig. 2A, middle), and the protein displayed in situ was assessed by a monoclonal anti-GST antibody (Fig. 2A, right) (GST color code: red>orange>yellow>green>blue). B, The PicoGreen signal was quantified on the slide and displayed as a box (25th–75th percentiles) and whisker plot both before (Green) and after (Black) DNase treatment. MFI is the abbreviation of mean of fluorescence intensity. C, The correlation coefficient of GST signal between two NAPPA protein arrays is 0.93. D, Representative fluorescent images show human targets with strong fluorescent signals. E, F, Histograms show the distribution of the molecular weights of identified substrates for VopS and IbpAFic2 as well as the entire human 10k protein collection respectively. X-axis is the molecular weight and y axis is the number of substrate proteins. IVTT, in vitro transcription and translation.
In addition to the signals from candidate substrates, we reproducibly observed signal from a human AMPylator, HYPE, detected even in the absence of the AMPylators, presumably because of auto-AMPylation (Fig. 1F–1G and supplemental Fig. S3). The wild type HYPE showed extremely low auto-AMPylation activity in previous studies with autoradiography, presumably because an inhibitory α-helix containing a conserved (S/T)XXXE(G/N) motif before the Fic domain prevents the binding of ATP to Fic (3, 9). Mutation of this inhibitory motif (E→G) increased HYPE auto-AMPylation and it's AMPylation of other substrates significantly (9). This result demonstrates our method has the potential to identify novel mammalian AMPylators with auto-AMPylation regardless of Fic and adenylyl transferase motifs (6, 7, 9, 31).
Global Identification of Substrates for Bacterial Fic AMPylators with Human 10K NAPPA Arrays
To execute a high-throughput screen for AMPylation substrates, we fabricated high-density NAPPA arrays containing 10,000 human proteins on five arrays, four with C-terminal GST-tagged proteins and one with N-terminal Flag-tagged proteins (Fig. 2A). DNA quantity before expression and after DNase treatment was measured to ensure the quality of newly fabricated NAPPA arrays (Fig. 2A, 2B). In addition, we probed several arrays in each printing batch with either a monoclonal anti-GST antibody or an anti-Flag antibody to assess the amount of protein displayed (Fig. 2A, right). Using the mean plus two standard deviations of the signal for negative control features (MSP-1 protein, which lacks the GST tag) as a cutoff, about 95% of the human proteins were successfully displayed (supplemental Fig. S4), consistent with previous studies (26, 32). The correlation of GST signals (i.e., protein levels) between arrays was r = 0.93, indicating a high reproducibility in the fabrication of NAPPA protein arrays (Fig. 2C).
We then performed high-throughput screens using the Fic AMPylators (VopS or IbpAFic2) with the N6pATP substrate (Fig. 2D). Two known substrates (Rac1 and RhoA) for VopS and IbpAFic2 gave strong fluorescent signals, which were not observed on NAPPA with buffer alone. We also observed several features with fluorescent signals that corresponded to novel candidate substrates (Fig. 2D).
Before statistical analysis, all fluorescent microarray images were visually examined to ensure their quality to exclude false positive signals. Normalized signals showing a 1.2-fold increase over the buffer-only control were considered positive. Only the features showing a positive ratio in all three experiments on different days were selected as final candidates. With these criteria, a total of 20 and 21 substrates for VopS and IbpAFic2 were discovered from the screening, respectively (supplemental Tables S1, S2). The distribution of these candidate substrates' molecular weights agreed with previous findings using gel-based AMPylation assays (Fig. 2E, 2F) (2, 3, 12, 14). The annotations showed that many of the VopS and IbpAFic2 substrates were RhoGTPases, 8 and 7, respectively. In addition, we found dozens of novel potential non-GTPase substrate proteins.
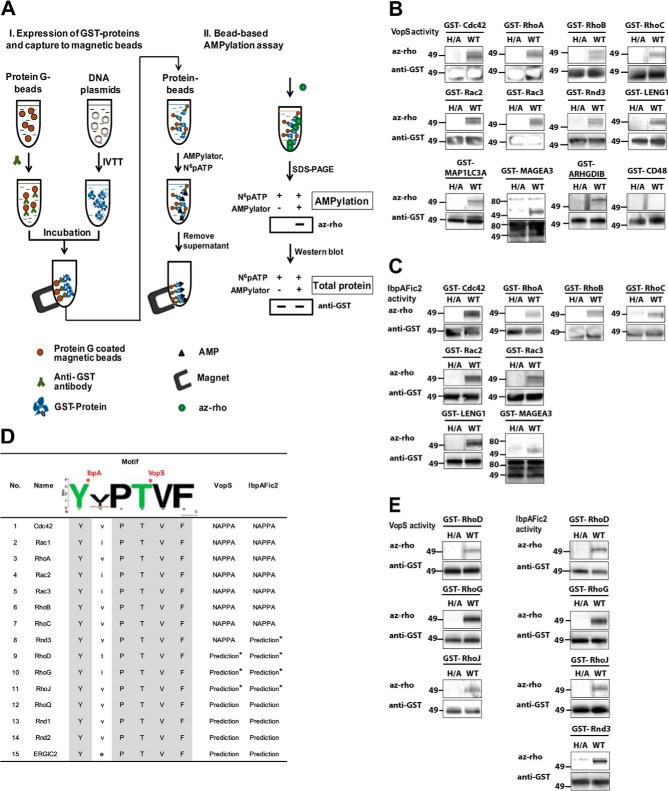
Validation of Candidate Substrates of AMPylators with a Bead-based Assay
To further assess the AMPylation substrates, we developed an independent AMPylation assay based on a magnetic bead platform (Fig. 3A). Briefly, a GST-tagged target protein was expressed in solution with the HeLa cell-free expression system and then captured by a polyclonal anti-GST antibody coupled to magnetic beads. Next, N6pATP and either active or inactive mutant AMPylators were added and incubated for an hour at 30 °C. After addition of az-rho reagents, fluorescently labeled proteins were visualized and protein levels were confirmed by Western blotting on the same gel using anti-GST antibody. Of the 20 and 21 candidates for VopS and IbpAFic2 identified by the NAPPA screening, 11 and 8, respectively, were validated with the bead-based AMPylation assays (Fig. 3B, 3C). Interestingly, the confirmed in vitro substrates included several non-GTPase proteins, ARHGDIB/LyGDI, MAP1LC3A, LENG1, and MAGEA3.
Fig. 3.
Validation of identified human substrates from NAPPA screens. A, Schematic illustration of bead-based AMPylation assays. B, C, Validation of identified human substrates for VopS and IbpAFic2, respectively. VopS H348A and IbpAFic2 H359A, both inactive enzymes, were used as their corresponding negative controls. CD48 is an example of a substrate candidate that did not validate in the bead-based assay. D, Prediction of new substrates for VopS and IbpAFic2 by using motif analysis. In Rac1, RhoA, and Cdc42, the conserved Y and T indicated were reported as the AMPylation substrate of IbpA and VopS respectively (2, 49). Additional candidate substrates with the motif were searched in the UniProtKB/Swiss-Prot human database (http://prosite.expasy.org/scanprosite/). E, Validation of predicted substrates using bead-based AMPylation assays. Each experiment was repeated three times on independent days. az-rho, azide-rhodamine; WB, Western blot.
Universal AMPylation of the Rho GTPase Family by Bacterial Fic AMPylators
As the NAPPA set covered about half of all predicted human proteins, we performed bioinformatics analyses to predict additional potential substrates of AMPylators based on the sequences of the hits we identified. As previously shown, Rho GTPases share a homologous sequence within the switch I region (3) where conserved tyrosine and threonine residues are AMPylated by IbpAFic2 and VopS, respectively (2, 3). Sequence alignment of all eight GTPases identified as targets for IbpAFic2 and VopS in the NAPPA screening revealed a highly conserved motif of YxPTVF (Fig. 3D). A motif scan using ScanProsite (http://prosite.expasy.org/scanprosite/) identified seven more proteins with the motif, including six GTPases and a non-GTPase, ERGIC2. Among these potential substrates, expression clones for RhoD, RhoG, and RhoJ were available and their AMPylation by VopS and IbpAFic2 was confirmed (Fig. 3E). In addition, we showed AMPylation of Rnd3 by IbpAFic2, which also shares the motif and was not detected in the initial NAPPA screening. Taken together, these data show that bacterial AMPylators could potentially regulate a wide range of Rho GTPases by targeting the highly conserved switch I regulatory region.
To determine the range of Rho GTPase targets for VopS in cells during an infection, we developed an assay in which candidate substrates are transiently expressed in HEK293T cells, followed by infection with V. para and immunoprecipitation of the target substrate (Fig. 4A). Using a strain that expresses and secretes VopS as the only known T3SS effector (CabS), we found that VopS AMPylated the known substrate Rac1 and two new GTPases, Rac2, and Rac3 (Fig. 4B). This indicates that VopS can AMPylate most Rho GTPase family members during infection and serve as a general inhibitor of all Rho GTPase activity. We also tested AMPylation of three in vitro confirmed non-GTPase substrates (ARHGDIB/LyGDI (LyGDI), LC3, and LENG1) in vivo, but did not observe the modification under these conditions. AMPylation of these substrates may be transient or prevented by other PTMs (such as phosphorylation) and we may have failed to evaluate the cells at the relevant time or condition to detect these changes (33). In addition, this measurement is technically challenging because the robust activity of VopS and IbpA for Rho GTPases can mask subtle cellular consequences (3, 9, 12, 13). As a result, we sought to confirm a non-GTPase substrate and elucidate the effect of AMPylation on its potential function in vitro.
Fig. 4.
VopS AMPylates all Rac proteins during infection. A, Schematic of the V. para infection assay with identified substrates. Cells are transfected with substrate candidate and infected with V. para. Candidate substrates are immunoprecipitated from the lysate and blotted for AMPylation. WT is V. para CAB strain which was generated from POR1 (RIMD 2210633 ΔtdhAS). The CabS strain contains a deletion for the transcriptional regulator VtrA to prevent expression of the second type three secretion system, as well as deletions of the effectors VopQ, VopR, and VPA0450 to leave VopS as the only known T3SS effector expressed and secreted. Δ and + are the CabSΔS strain with the deletion of VopS and complemented with a pBAD-VopS expression plasmid, respectively. B, Transfected 3xHA tagged Rac1, Rac2, and Rac3 were immunoprecipitated from infected cells and immunoblotted for AMPylation.
Characterization of In Vitro AMPylation of LyGDI AMPylation by VopS
LyGDI was chosen for further study because it represents a new class of substrate that nevertheless plays important roles in GTPase regulation for innate immunity, including T-cell activation, phagocytosis of bacteria by macrophages, and reactive oxygen species production for microbial killing(34–36). LyGDI is primarily expressed in hematopoietic cells and some cancers, and belongs to the RhoGDI family of which there are three members: ARHGDIA/RhoGDI, LyGDI, and ARHGDIG/RhoGDI3. Unlike the GTPase substrates, LyGDI does not have a YxPTVF motif, suggesting that if LyGDI were AMPylated, modification must occur at a novel recognition sequence.
To test these non-GTPase substrates for VopS modification, we co-expressed either LyGDI or RhoGDI with VopS, as previously described (2). Using antibodies that recognize AMPylated threonine (T-AMP) or tyrosine (Y-AMP) we observed that purified His-LyGDI co-expressed with VopS, but not empty vector, was strongly recognized by the T-AMP antibody (Fig. 5A). Interestingly, despite strong homology between the RhoGDI family members, RhoGDI was not AMPylated by VopS. Comparison of the apparent mass of His-LyGDI co-expressed with GST-VopS revealed the appearance of a protein population with an increase in molecular weight of 328 Daltons over the LyGDI coexpressed with empty vector (supplemental Fig. S6), indicative of a protein modified with AMP.
Fig. 5.
AMPylation of LyGDI on residue threonine 51 inhibits phosphorylation by Src. A, Validation of LyGDI AMPylation using anti-threonine and anti-tyrosine AMPylation antibodies. His-RhoGDI and His-LyGDI proteins were expressed in E. coli with GST-NΔ30VopS or empty vector and blotted for total protein (anti-His), threonine AMPylation (anti-T-AMP), tyrosine AMPylation (anti-Y-AMP), and VopS expression (anti-VopS, total lysate). Lane 1 (*) is tyrosine AMPylated Cdc42. B, AMPylated or unAMPylated LyGDI incubated with Src kinase, 100 μm ATP and 5 μCi [γ-32P] ATP for 45 min. CB, coomassie staining. C, Relative phosphorylation of AMPylated versus unAMPylated LyGDI over three experiments. p = 0.034.
To identify the AMPylated residue(s) on LyGDI, we used LC-MS/MS as previously described (2). Only one residue, threonine 51, was identified in the peptide T51LLGDGPVVTDPK63 to be modified with AMP (supplemental Fig. S7) (10). Analysis of the reporter ions eliminated threonine 60 as the possible site for AMPylation (supplemental Fig. S7). Mutation of threonine 51 to alanine abrogated AMPylation of LyGDI in an identical co-expression experiment, demonstrating that threonine 51 is a specific AMPylation site (Fig. 5A). The targeting of this specific residue in LyGDI is reminiscent of the way in which VopS precisely targets the threonine in the Switch I region of Rho GTPases to inhibit their interaction with downstream effectors (2).
Sequence analysis of RhoGDIs reveals that the site of AMPylation is present only in LyGDI, not RhoGDI or RhoGDI3 (supplemental Fig. S8A), explaining its specificity. The tertiary structure of RhoGDIs can be divided into two parts: the prenyl binding, immunoglobulin-like C terminus and the dual helix containing N terminus. The latter is the site of multiple PTMs and may be considered the regulatory region of the protein (37, 38) (supplemental Fig. S8B, C). Interestingly, the site of AMPylation of LyGDI by VopS is located in the second N-terminal alpha helix, which is near the location of several PTMs, including phosphorylation by Src kinase (39–42). The diversity of regulation on and around the N-terminal helical region of LyGDI presents multiple avenues by which AMPylation by VopS could disrupt its function.
Inhibition of Src Phosphorylation of LyGDI by VopS AMPylation
To test if AMPylation by VopS might have potential as a competitive PTM, we assessed the ability of AMPylated LyGDI to be phosphorylated by Src. We observed that unmodified LyGDI is readily phosphorylated by Src, but LyGDI previously AMPylated by VopS is phosphorylated at a significantly lower (58%) level than unmodified LyGDI (Fig. 5B, 5C), indicating that AMPylation of LyGDI may be competitive with Src phosphorylation and potentially other sites of modification in the N terminus of LyGDI. A complete loss of phosphorylation was not expected, as the AMPylated-LyGDI sample is a mixed population of modified and unmodified LyGDI, although the relative amounts cannot be deduced from unquantitative mass spectrometry (supplemental Fig. S6). It is possible but not certain the portion resistant to Src phosphorylation also represents the portion of AMPylated LyGDI.
AMPylation of LyGDI during an infection was not observed in preliminary experiments, but further studies will be required to rule it out as a naturally occurring modification. Nevertheless, the ability of this in vitro AMPylation to compete with naturally occurring PTMs presents a tool to understand the regulation of this protein in processes like immunity and cancer.
DISCUSSION
Over 40 years ago, Stadtman and colleagues identified the modification of a protein with AMP as a mechanism to reversibly modulate the activity of glutamine synthetase (43). This modification has been implicated as a fundamental mechanism to regulate protein-protein interactions of modified proteins with downstream signaling machinery. Undoubtedly, the molecular mechanism of AMPylation-mediated cellular changes has not been fully elucidated, because of a limited knowledge of the repertoire of enzymes and substrates (6, 44). Advancement in the identification of novel substrates has been stalled by the dependence on anti-AMPylation antibodies and radioactive detection to find substrates, which have only yielded a handful of substrates since the discovery of AMPylation (2, 3, 33). To start to address these questions, we established a high-throughput discovery platform by developing a click chemistry-based assay that selectively labels enzyme substrates and works on a cell free protein microarray platform. The alkynyl chemical reporter, N6pATP, allows nonradioactive and robust detection of AMPylated proteins with high sensitivity and specificity (Fig. 1B, 1E–1G, Fig. 2, and supplemental Figs. S1–3)(12). Moreover, the covalent bond formed by alkynyl and azido moieties enabled us to examine both the AMPylation status and the expression of target proteins in the same gel using the bead-based AMPylation assay followed by Western blotting (Fig. 3).
Including both the screen and the targeted testing, a total of 27 and 29 potential substrates were identified for VopS and IbpAFic2, respectively, after screening 10,000 human proteins by NAPPA and the following prediction analysis. Among them, 14 and 12 hits showed strong detection using the bead-based AMPylation assay for VopS and IbpAFic2, respectively. The discrepancy of the results between NAPPA and bead-based assays might be caused by the differential sensitivity, resolution and the amount of protein required for signal by these two methods. False positives are also inherent in any screen and careful interpretation of unvalidated hits is required. In addition, although we did not observe any difference for the detection of protein AMPylation between N6pATP analog and native ATP with the substrates we tested, false negatives might still exist in click chemistry based screening in which the alkynyl group on N6pATP may cause steric hindrance and block the transfer of AMP to some potential substrates.
We identified a highly conserved motif (YxPTVF) by sequence alignment of all GTPases targets of IbpAFic2 and VopS (Fig. 3D). Further searching of human protein sequence databases with ScanProsite identified six GTPases and a non-GTPase, ERGIC2. ERGIC2 is an ER-Golgi transmembrane protein with possible functions in transport between ER and Golgi (45). Altogether, these hits expanded the repertoire of host substrates for the bacterial AMPylators to the entire Rho GTPase family and potentially beyond (12, 13).
This motif appears to be specific for GTPase substrates, as it was not present in any of the non-GTPase substrates. The proximal sequence at the documented modification site in LyGDI was KYKKT*LLGDGP. Using the Eukaryotic Linear Motif tool, we could not find a related sequence in the other non-GTPases that had been confirmed in the bead-based assay. Notably, the modified region in LyGDI is alpha helical and the modified threonine residue is facing directly away from the protein, perhaps enabling accessibility to VopS. Nonetheless, there is likely to be additional specificity required for VopS targeting, as there were less than two dozen observed substrates from among the 10,000 candidates.
A number of other proteins were identified as potential substrates for VopS and IbpA (Fig. 6), covering a diverse range of protein classes in addition to small GTPases, including enzyme modulators, signaling molecules, receptors, and enzymes. These potential substrates localize to diverse subcellular compartments, such as cytoplasm, cell membrane, and the nucleus, and can be lipid-anchored or secreted. VopS has recently been shown to contain a conserved phosphoinositide binding domain (BPD) that localizes it to the cell membrane. However, a mutation of the BPD that renders VopS cytosolic had no appreciable effect on cell rounding during infection (46). Proteins accessible both in the cytoplasm and on the cell membrane are likely to be possible substrates of VopS. The molecular function analysis of the potential substrates reveals a variety of binding activities that might change through AMPylation, for example, GBD domain binding, Wnt receptor binding, BH3 domain binding and thioesterase binding, etc. (supplemental Fig. S9 and supplemental Tables S3, S4). It is important to avoid over-interpretation of the results of a screen without thorough in vivo validation.
Fig. 6.
Bacteria-human interactions through AMPylation. A, Bacteria-human interaction AMPylation network. B, C, Distribution of PANTHER protein classes and sub-cellular location of VopS and IbpAFic2 substrates.
LyGDI was confirmed as an in vitro substrate and was found to be specifically AMPylated on threonine 51 by VopS in the highly regulated N terminus of the protein. LyGDI appears to be involved in several processes of the innate immune system and is also implicated as a metastatic factor in several cancers (35, 42, 47) Although we have been unable to confirm LyGDI as an in vivo substrate, its AMPylation may prove to be a competitive PTM (Fig. 5) and could be a powerful tool for understanding its complex roles in immunity and cancer. AvrAC provides a previous precedent for phosphorylation-competitive modifications by Fic proteins, as addition of UMP to the plant kinases Bik1 and RipK by AvrAC masks their phosphorylation sites (33, 48).
We analyzed the biological processes represented by the substrates identified in the screen using BINGO with p value of 0.05, which shows that most of the proteins selected are Rho GTPases (Fig. 7, supplemental Figs. S10, S11 and supplemental Tables S5, S6). The results indicate that the actin cytoskeleton and Rho signaling are the main targets of VopS and IbpA enzymes, which is in accord with previous studies in which the ectopic expression of the bacterial AMPylators induced host cell rounding because of cytoskeletal collapse (2, 3).
Fig. 7.
Biological processes of VopS and IbpAFic2 involved in the human host by AMPylation of their substrates. In both panels, GO biological processes are represented in a tree format that shows the hierarchy of GO terms. The adjusted p value from enrichment tests is represented by the color from yellow (5 × 10−2) to orange (5 × 10−7) (details in Methods, full-scale images in Supplemental Fig. S10 and S11 with details in supplemental Tables S5, S6) of identified substrates for each biological process.
Finally, we believe this work contributes major conceptual and methodological advances of cell-free protein arrays, including array fabrication, assay novelty and application in the detection of a novel PTM. We have developed a high-throughput approach for the identification and characterization of proteins modified by AMPylation using a combination of multi-disciplinary technologies that covers proteomics, chemistry, bioinformatics and molecular biology. Furthermore, this study validated the high-density NAPPA based cell-free protein arrays as a powerful method of characterizing PTMs, beginning with the assay development of a fluorescent reporter screen for an enzyme, identification of substrates, analysis of the interaction web of the enzyme, and ending in the validation and preliminary characterization of unexpected and novel substrates. The breadth of information gained about an enigmatic modification like AMPylation demonstrates the great potential of cell-free protein arrays in unbiased, discovery-based research on PTMs.
Supplementary Material
Acknowledgments
We thank the members of our lab for their helpful discussions and kindly assistance. Thanks to the UT Southwestern Proteomics core for assistance and advice.
Footnotes
Author contributions: X.Y., A.W., K.O., and J.L. designed research; X.Y. and A.W. performed research; P.L., Y.H., M.G., N.W., J.W., X.B., G.D., and H.H. contributed new reagents or analytic tools; X.Y., A.W., J.P., K.O., and J.L. analyzed data; X.Y., A.W., K.O., and J.L. wrote the paper.
* This work is supported by NIH Allergy and Infectious Disease (R01-AI056404 and R01-AI087808 to K.O), the Welch Foundation (I-1561 to K.O.), the Leukemia & Lymphoma Society (5210-14 to N.W.) and Early Detection Research Network (5U01CA117374 to J.L.). K.O. is a Burroughs Wellcome Investigator in Pathogenesis of Infectious Disease and a W.W. Caruth Jr. Biomedical Scholar.
 This article contains supplemental Figs. S1 to S11 and Tables S1 to S7.
This article contains supplemental Figs. S1 to S11 and Tables S1 to S7.
1 The abbreviations used are:
- AMP
- Adenosine monophosphate
- az-rho
- azide-rhodamine
- CB
- coomassie staining
- CuAAC
- copper-catalyzed azide–alkyne cycloaddition
- Fic
- filamentation induced by cAMP
- GO
- Gene ontology
- GST
- Glutathione S-transferase
- HRP
- horse radish peroxidase
- IVTT
- in vitro transcription and translation
- NAPPA
- Nucleic Acid Programmable Protein Array
- N6pATP
- N6-propargyl adenosine-5′-triphophate
- PANTHER
- Protein ANalysis THrough Evolutionary Relationships
- SUMO
- Small ubiquitin-related modifier
- TBTA
- Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine
- TCEP
- Tris(2-carboxyethyl)phosphine hydrochloride
- UniProt
- The Universal Protein Resource
- VopS
- Vibrio outer protein S.
REFERENCES
- 1. Muller M. P., Peters H., Blumer J., Blankenfeldt W., Goody R. S., Itzen A. (2010) The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329, 946–949 [DOI] [PubMed] [Google Scholar]
- 2. Yarbrough M. L., Li Y., Kinch L. N., Grishin N. V., Ball H. L., Orth K. (2009) AMPylation of Rho GTPases by Vibrio VopS disrupts effector binding and downstream signaling. Science 323, 269–272 [DOI] [PubMed] [Google Scholar]
- 3. Worby C. A., Mattoo S., Kruger R. P., Corbeil L. B., Koller A., Mendez J. C., Zekarias B., Lazar C., Dixon J. E. (2009) The fic domain: regulation of cell signaling by adenylylation. Mol. Cell 34, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neunuebel M. R., Chen Y., Gaspar A. H., Backlund P. S., Jr., Yergey A., Machner M. P. (2011) De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science 333, 453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tan Y., Luo Z. Q. (2011) Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature 475, 506–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Woolery A. R., Luong P., Broberg C. A., Orth K. (2010) AMPylation: something old is new again. Front. Microbiol. 1, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kinch L. N., Yarbrough M. L., Orth K., Grishin N. V. (2009) Fido, a novel AMPylation domain common to fic, doc, and AvrB. PLoS One 4, e5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rahman M., Ham H., Liu X., Sugiura Y., Orth K., Kramer H. (2012) Visual neurotransmission in Drosophila requires expression of Fic in glial capitate projections. Nat. Neurosci. 15, 871–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Engel P., Goepfert A., Stanger F. V., Harms A., Schmidt A., Schirmer T., Dehio C. (2012) Adenylylation control by intra- or intermolecular active-site obstruction in Fic proteins. Nature 482, 107–110 [DOI] [PubMed] [Google Scholar]
- 10. Li Y., Al-Eryani R., Yarbrough M. L., Orth K., Ball H. L. (2011) Characterization of AMPylation on threonine, serine, and tyrosine using mass spectrometry. J. Am. Soc. Mass Spectrom. 22, 752–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hao Y. H., Chuang T., Ball H. L., Luong P., Li Y., Flores-Saaib R. D., Orth K. (2011) Characterization of a rabbit polyclonal antibody against threonine-AMPylation. J. Biotechnol. 151, 251–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grammel M., Luong P., Orth K., Hang H. C. (2011) A chemical reporter for protein AMPylation. J. Am. Chem. Soc. 133, 17103–17105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lewallen D. M., Steckler C. J., Knuckley B., Chalmers M. J., Thompson P. R. (2012) Probing adenylation: using a fluorescently labelled ATP probe to directly label and immunoprecipitate VopS substrates. Mol. Biosyst. 8, 1701–1706 [DOI] [PubMed] [Google Scholar]
- 14. Broncel M., Serwa R. A., Tate E. W. (2012) A new chemical handle for protein AMPylation at the host-pathogen interface. Chembiochem. 13, 183–185 [DOI] [PubMed] [Google Scholar]
- 15. Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., McCartney R. R., Schmidt M. C., Rachidi N., Lee S. J., Mah A. S., Meng L., Stark M. J., Stern D. F., De Virgilio C., Tyers M., Andrews B., Gerstein M., Schweitzer B., Predki P. F., Snyder M. (2005) Global analysis of protein phosphorylation in yeast. Nature 438, 679–684 [DOI] [PubMed] [Google Scholar]
- 16. Foster M. W., Forrester M. T., Stamler J. S. (2009) A protein microarray-based analysis of S-nitrosylation. Proc. Natl. Acad. Sci. U. S. A. 106, 18948–18953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu J. Y., Lin Y. Y., Sheu J. C., Wu J. T., Lee F. J., Chen Y., Lin M. I., Chiang F. T., Tai T. Y., Berger S. L., Zhao Y., Tsai K. S., Zhu H., Chuang L. M., Boeke J. D. (2011) Acetylation of yeast AMPK controls intrinsic aging independently of caloric restriction. Cell 146, 969–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Popescu S. C., Popescu G. V., Bachan S., Zhang Z., Seay M., Gerstein M., Snyder M., Dinesh-Kumar S. P. (2007) Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. U. S. A. 104, 4730–4735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He M., Taussig M. J. (2001) Single step generation of protein arrays from DNA by cell-free expression and in situ immobilization (PISA method). Nucleic Acids Res. 29, E73–E73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ramachandran N., Hainsworth E., Bhullar B., Eisenstein S., Rosen B., Lau A. Y., Walter J. C., LaBaer J. (2004) Self-assembling protein microarrays. Science 305, 86–90 [DOI] [PubMed] [Google Scholar]
- 21. He M., Stoevesandt O., Palmer E. A., Khan F., Ericsson O., Taussig M. J. (2008) Printing protein arrays from DNA arrays. Nat. Methods 5, 175–177 [DOI] [PubMed] [Google Scholar]
- 22. Tao S. C., Zhu H. (2006) Protein chip fabrication by capture of nascent polypeptides. Nat. Biotechnol. 24, 1253–1254 [DOI] [PubMed] [Google Scholar]
- 23. He M., Stoevesandt O., Taussig M. J. (2008) In situ synthesis of protein arrays. Curr. Opin. Biotechnol. 19, 4–9 [DOI] [PubMed] [Google Scholar]
- 24. Chandra H., Srivastava S. (2010) Cell-free synthesis-based protein microarrays and their applications. Proteomics 10, 717–730 [DOI] [PubMed] [Google Scholar]
- 25. Festa F., Rollins S. M., Vattem K., Hathaway M., Lorenz P., Mendoza E. A., Yu X., Qiu J., Kilmer G., Jensen P., Webb B., Ryan E. T., LaBaer J. (2013) Robust microarray production of freshly expressed proteins in a human milieu. Proteomics Clin. Appl. 7, 372–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramachandran N., Raphael J. V., Hainsworth E., Demirkan G., Fuentes M. G., Rolfs A., Hu Y., LaBaer J. (2008) Next-generation high-density self-assembling functional protein arrays. Nat. Methods. 5, 535–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takulapalli B. R., Qiu J., Magee D. M., Kahn P., Brunner A., Barker K., Means S., Miersch S., Bian X., Mendoza A., Festa F., Syal K., Park J. G., LaBaer J., Wiktor P. (2012) High density diffusion-free nanowell arrays. J. Proteome Res. 11, 4382–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anderson K. S., Sibani S., Wallstrom G., Qiu J., Mendoza E. A., Raphael J., Hainsworth E., Montor W. R., Wong J., Park J. G., Lokko N., Logvinenko T., Ramachandran N., Godwin A. K., Marks J., Engstrom P., Labaer J. (2011) Protein microarray signature of autoantibody biomarkers for the early detection of breast cancer. J. Proteome Res. 10, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu X., Bian X., Throop A., Song L., Moral L. D., Park J., Seiler C., Fiacco M., Steel J., Hunter P., Saul J., Wang J., Qiu J., Pipas J. M., LaBaer J. (2014) Exploration of panviral proteome: high-throughput cloning and functional implications in virus-host interactions. Theranostics. 4, 808–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saul J., Petritis B., Sau S., Rauf F., Gaskin M., Ober-Reynolds B., Mineyev I., Magee M., Chaput J., Qiu J., LaBaer J. (2014) Development of a full-length human protein production pipeline. Protein Sci. 10.1002/pro.2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yarbrough M. L., Orth K. (2009) AMPylation is a new post-translational modiFICation. Nat. Chem. Biol. 5, 378–379 [DOI] [PubMed] [Google Scholar]
- 32. Goshima N., Kawamura Y., Fukumoto A., Miura A., Honma R., Satoh R., Wakamatsu A., Yamamoto J., Kimura K., Nishikawa T., Andoh T., Iida Y., Ishikawa K., Ito E., Kagawa N., Kaminaga C., Kanehori K., Kawakami B., Kenmochi K., Kimura R., Kobayashi M., Kuroita T., Kuwayama H., Maruyama Y., Matsuo K., Minami K., Mitsubori M., Mori M., Morishita R., Murase A., Nishikawa A., Nishikawa S., Okamoto T., Sakagami N., Sakamoto Y., Sasaki Y., Seki T., Sono S., Sugiyama A., Sumiya T., Takayama T., Takayama Y., Takeda H., Togashi T., Yahata K., Yamada H., Yanagisawa Y., Endo Y., Imamoto F., Kisu Y., Tanaka S., Isogai T., Imai J., Watanabe S., Nomura N. (2008) Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods 5, 1011–1017 [DOI] [PubMed] [Google Scholar]
- 33. Feng F., Yang F., Rong W., Wu X., Zhang J., Chen S., He C., Zhou J. M. (2012) A Xanthomonas uridine 5′-monophosphate transferase inhibits plant immune kinases. Nature 485, 114–118 [DOI] [PubMed] [Google Scholar]
- 34. Wu W., Hsu Y. M., Bi L., Songyang Z., Lin X. (2009) CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nat. Immunol. 10, 1208–1214 [DOI] [PubMed] [Google Scholar]
- 35. Mehta P., Wavreille A. S., Justiniano S. E., Marsh R. L., Yu J., Burry R. W., Jarjoura D., Eubank T., Caligiuri M. A., Butchar J. P., Tridandapani S. (2011) LyGDI, a novel SHIP-interacting protein, is a negative regulator of FcgammaR-mediated phagocytosis. PLoS One 6, e21175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Groysman M., Hornstein I., Alcover A., Katzav S. (2002) Vav1 and Ly-GDI two regulators of Rho GTPases, function cooperatively as signal transducers in T cell antigen receptor-induced pathways. J. Biol. Chem. 277, 50121–50130 [DOI] [PubMed] [Google Scholar]
- 37. Scheffzek K., Stephan I., Jensen O. N., Illenberger D., Gierschik P. (2000) The Rac-RhoGDI complex and the structural basis for the regulation of Rho proteins by RhoGDI. Nat. Struct. Biol. 7, 122–126 [DOI] [PubMed] [Google Scholar]
- 38. Tnimov Z., Guo Z., Gambin Y., Nguyen U. T., Wu Y. W., Abankwa D., Stigter A., Collins B. M., Waldmann H., Goody R. S., Alexandrov K. (2012) Quantitative analysis of prenylated RhoA interaction with its chaperone, RhoGDI. J. Biol. Chem. 287, 26549–26562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Griner E. M., Churchill M. E., Brautigan D. L., Theodorescu D. (2013) PKCalpha phosphorylation of RhoGDI2 at Ser31 disrupts interactions with Rac1 and decreases GDI activity. Oncogene 32, 1010–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Danley D. E., Chuang T. H., Bokoch G. M. (1996) Defective Rho GTPase regulation by IL-1 beta-converting enzyme-mediated cleavage of D4 GDP dissociation inhibitor. J. Immunol. 157, 500–503 [PubMed] [Google Scholar]
- 41. Choi M. R., Groot M., Drexler H. C. (2007) Functional implications of caspase-mediated RhoGDI2 processing during apoptosis of HL60 and K562 leukemia cells. Apoptosis 12, 2025–2035 [DOI] [PubMed] [Google Scholar]
- 42. Wu Y., Moissoglu K., Wang H., Wang X., Frierson H. F., Schwartz M. A., Theodorescu D. (2009) Src phosphorylation of RhoGDI2 regulates its metastasis suppressor function. Proc. Natl. Acad. Sci. U. S. A. 106, 5807–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stadtman E. R., Ginsburg A., Ciardi J. E., Yeh J., Hennig S. B., Shapiro B. M. (1970) Multiple molecular forms of glutamine synthetase produced by enzyme catalyzed adenylation and deadenylylation reactions. Adv. Enzyme Regul. 8, 99–118 [DOI] [PubMed] [Google Scholar]
- 44. Itzen A., Blankenfeldt W., Goody R. S. (2011) Adenylylation: renaissance of a forgotten post-translational modification. Trends Biochem. Sci. 36, 221–228 [DOI] [PubMed] [Google Scholar]
- 45. Breuza L., Halbeisen R., Jeno P., Otte S., Barlowe C., Hong W., Hauri H. P. (2004) Proteomics of endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membranes from brefeldin A-treated HepG2 cells identifies ERGIC-32, a new cycling protein that interacts with human Erv46. J. Biol. Chem. 279, 47242–47253 [DOI] [PubMed] [Google Scholar]
- 46. Salomon D., Guo Y., Kinch L. N., Grishin N. V., Gardner K. H., Orth K. (2013) Effectors of animal and plant pathogens use a common domain to bind host phosphoinositides. Nat. Commun. 4, 2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Groysman M., Hornstein I., Alcover A., Katzav S. (2002) Vav1 and Ly-GDI two regulators of Rho GTPases, function cooperatively as signal transducers in T cell antigen receptor-induced pathways. J. Biol. Chem. 277, 50121–50130 [DOI] [PubMed] [Google Scholar]
- 48. DerMardirossian C., Rocklin G., Seo J. Y., Bokoch G. M. (2006) Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol. Biol. Cell 17, 4760–4768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Doyle J. M., Gao J., Wang J., Yang M., Potts P. R. (2010) MAGE-RING protein complexes comprise a family of E3 ubiquitin ligases. Mol. Cell 39, 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.