Abstract
Background
Risk stratification, detection of minimal residual disease (MRD), and implementation of novel therapeutic agents have improved outcome in acute lymphoblastic leukemia (ALL), but survival of adult patients with T-cell acute lymphoblastic leukemia (T-ALL) remains unsatisfactory. Thus, novel molecular insights and therapeutic approaches are urgently needed.
Methods
We studied the impact of B-cell CLL/lymphoma 11b (BCL11b), a key regulator in normal T-cell development, in T-ALL patients enrolled into the German Multicenter Acute Lymphoblastic Leukemia Study Group trials (GMALL; n = 169). The mutational status (exon 4) of BCL11b was analyzed by Sanger sequencing and mRNA expression levels were determined by quantitative real-time PCR. In addition gene expression profiles generated on the Human Genome U133 Plus 2.0 Array (affymetrix) were used to investigate BCL11b low and high expressing T-ALL patients.
Results
We demonstrate that BCL11b is aberrantly expressed in T-ALL and gene expression profiles reveal an association of low BCL11b expression with up-regulation of immature markers. T-ALL patients characterized by low BCL11b expression exhibit an adverse prognosis [5-year overall survival (OS): low 35% (n = 40) vs. high 53% (n = 129), P = 0.02]. Within the standard risk group of thymic T-ALL (n = 102), low BCL11b expression identified patients with an unexpected poor outcome compared to those with high expression (5-year OS: 20%, n = 18 versus 62%, n = 84, P < 0.01). In addition, sequencing of exon 4 revealed a high mutation rate (14%) of BCL11b.
Conclusions
In summary, our data of a large adult T-ALL patient cohort show that low BCL11b expression was associated with poor prognosis; particularly in the standard risk group of thymic T-ALL. These findings can be utilized for improved risk prediction in a significant proportion of adult T-ALL patients, which carry a high risk of standard therapy failure despite a favorable immunophenotype.
Keywords: Adult T-cell acute lymphoblastic leukemia, BCL11b, Outcome, Standard risk, Expression, Mutation
Background
Improved treatment strategies, integrating risk stratification and minimal residual disease (MRD) monitoring, have improved survival of adult patients with acute lymphoblastic leukemia (ALL) over the last decades [[1],[2]]. Nevertheless, overall survival (OS) remains unsatisfactory with about 40-70%, depending on protocol and age group. Thus far, novel therapy approaches have mainly been developed in B-lineage ALL, where new targeted therapies with monoclonal antibodies like Rituximab or small molecule tyrosine kinase inhibitors (TKI) such as imatinib for Philadelphia chromosome/BCR-ABL-positive patients have been established [[3]–[6]]. In T-cell acute lymphoblastic leukemia (T-ALL), less progress has been made despite the introduction of nelarabine in relapsed and refractory disease [[7],[8]]. Other molecular driven approaches, including γ-secretase inhibitors, have until now been less successful [[9]].
In the German Multicenter Study Group for Adult ALL (GMALL), immunologic subtypes are routinely used as prognostic factors for the risk stratification of T-ALL. Within the high risk group of early T-ALL patients the recently identified subgroup of Early T-cell precursor (ETP-) ALL has been characterized by an immature immunophenotype with a high rate of FLT3-mutations, suggesting a potential role for TKI in the treatment for these high risk patients [[10]–[12]]. In contrast, T-ALL patients with a thymic phenotype have a more favorable prognosis [[1]] and are therefore classified as standard risk. Nevertheless, this large group (56% of adult T-ALL) [[3]] of standard risk T-ALL contains a molecularly and clinically heterogeneous group of patients. Thus, novel insights into the molecular stratification will further aid in refining treatment options.
The B-cell CLL/lymphoma 11b (BCL11b) gene, a Krüppel-like C2H2 zinkfinger transcription factor located on chromosome 14q32.2, is a key player in physiologic T-cell development with potential impact on T-ALL leukemogenesis. In normal hematopoiesis, the onset of BCL11b expression in T-cell progenitors occurs during the onset of DN2 phase and is maintained throughout subsequent differential stages (Figure 1) [[13],[14]]. For several target genes BCL11b acts as repressor (p21, p57), for others as activator of transcription (IL-2) [[15]–[17]]. In vitro studies demonstrated that knockdown inhibited proliferation and led to apoptosis in human T-ALL cell lines [[18],[19]] and a chemo-protective effect of BCL11b overexpression was also observed [[20]].
Figure 1.
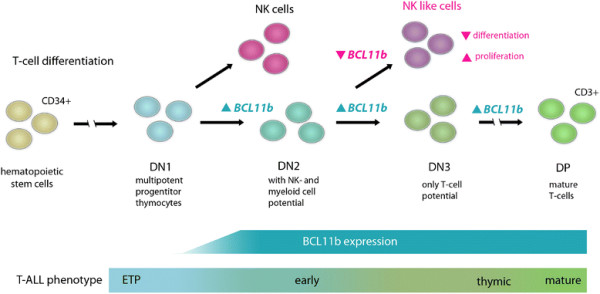
BCL11b expression in the stages of normal T-cell development. In the murine model BCL11b expression begins during the transition of DN1 to DN2 and continues throughout further differentiation steps and in mature T-cells [[39]]. Knockout at DN2 stage leads to a NK like phenotype with differentiation arrest and a high proliferative potential [[13]]. The corresponding human T-ALL immunophenotypes are indicated. DN = Double Negative, DP = Double Positive, NK = Natural Killer cells, ETP = Early T-cell progenitors, ▲up-regulation, ▼down-regulation.
In addition, BCL11b is proposed to act as haploinsufficient tumor-suppressor, as its disruption was found to be associated with lymphomagenesis in mice [[21],[22]]. In humans, chromosomal translocations involving the BCL11b gene locus were identified in patients with acute myeloid leukemia (AML), T-ALL and T/myeloid acute bilineage leukemia [[23]–[28]]. Likewise, deletions and missense mutations, disrupting gene function, were reported in 9 to 16% of pediatric T-ALL patients [[29],[30]]. One study found BCL11b more frequently mutated in adult patients compared to children (20% vs. 5.3%) [[31]], with a lower mutation rate in early immature (3.6%) and a higher rate (12%) in cortical/mature adult T-ALL [[32]]. Studies on the prognostic impact of BCL11b mutations gave diverging results: a small study reported a favorable outcome for adult T-ALL patients with BCL11b mutations (n = 4) [[32]], however, studies in pediatric patients reported no prognostic effect of mutations [[29],[30]].
We hypothesized that deregulated expression of BCL11b, which is tightly regulated in normal T-cell differentiation, and BCL11b mutations play an important role in T-ALL. Therefore, we analyzed BCL11b mRNA expression levels in a large cohort of adult T-ALL patients and screened for mutations in the zinc finger region.
Results
BCL11b is heterogeneously expressed in adult T-ALL
BCL11b mRNA expression levels were not detectable in CD34 positive hematopoietic progenitor cells or unselected bone marrow (BM) samples from healthy donors, whereas high expression levels were found in CD3 positive mature T-cells (median: 2.5, Figure 2). In contrast, diagnostic BM samples of adult T-ALL patients (n = 195) showed an aberrant and highly heterogeneously BCL11b expression pattern (median: 0.5 range = 0-12.3; Figure 2).
Figure 2.
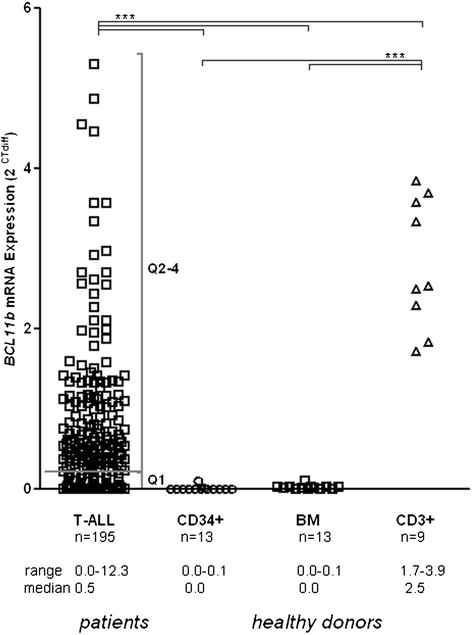
BCL11bmRNA expression in T-ALL patients and healthy donors. A heterogeneous expression pattern was observed within the T-ALL patient samples. Samples from healthy donors had a significantly higher expression of BCL11b in CD3 positive mature T-cells than in CD34 positive progenitor cells or unselected bone marrow (BM) samples. T-ALL patients were split into BCL11b expression quartiles and patients with low or lacking expression were combined in Q1 and patients with high expression in Q2-4. Outliers (n = 4 T-ALL patients, BCL11b expression values: 6.3, 7.6, 9.7, 12.3) were omitted from the diagram but included in the statistical analysis. BM = bone marrow; *** P < 0.01.
As explorative approach, we divided the samples into BCL11b-low and BCL11b-high expression groups by quartiles. Expression levels in the lowest quartile Q1 (≤0.2) were regarded as BCL11b-low, and samples with levels in quartiles Q2-4 (>0.2) as BCL11b-high.
BCL11b associated global gene expression profile
To explore the underlying transcriptional profile associated with aberrant BCL11b expression in T-ALL, we analyzed microarray expression data of an independent cohort of 86 adult T-ALL patients [[33]]. Samples were classified into a low and a high BCL11b expression group as described in the material and methods section. We identified 229 up-regulated probe sets (corresponding to 183 unique genes, hypothetical genes/proteins and open reading frames) and 200 down-regulated probe sets (corresponding to 166 genes, hypothetical genes/proteins and open reading frames) in the BCL11b-low group compared to the BCL11b-high group (Figure 3A, Additional file 1: Table S2 and Additional file 1: Table S3). In the BCL11b-low group, genes reported to be suppressed by BCL11b were highly expressed: cyclin-dependent kinase inhibitor 1A (p21) and cyclin-dependent kinase inhibitor 1C (p57) [[15],[17]]. Interestingly, genes associated with an immature stem cell-like phenotype were up-regulated in the BCL11b-low cohort including IGFBP7, BAALC, CD34, and FLT3 [[34]–[36]]. In contrast, the BCL11b-high group showed co-expression of markers of a mature T-cell phenotype including several T-cell receptor genes as well as CD8 and CD6. This was further underscored in gene set enrichment analysis (GSEA), in which gene sets associated with physiological T-cell development were enriched in BCL11b-high (P < 0.01) and genes down regulated in normal T-cells were enriched in BCL11b-low group (P = 0.01; Figure 3B) [[37]].
Figure 3.
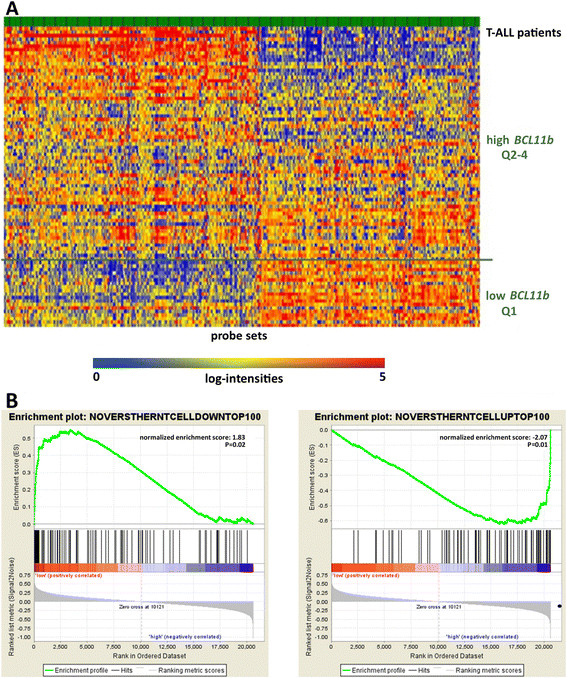
Gene expression profiles reveal an immature phenotype inBCL11b-lowgroup. A: Heat map of two-fold or greater differentially expressed genes between BCL11b quartile 1 (Q1) and quartiles 2-4 (Q2-4). B: Gene set enrichment analysis for differentially regulated genes in physiological T-cell development according to their BCL11b expression. On the left side the top hundred down-regulated genes are shown, on the right side the top hundred up-regulated genes. The lists were taken from Noversthern et al. [[37]].
BCL11b expression with respect to molecular and clinical characteristics
We further explored molecular characteristics associated with BCL11b expression in the T-ALL GMALL patient cohort by quantitative RT-PCR. Samples in the lowest expression quartile (Q1; n = 49; BCL11b expression range = 0-0.2) were compared to samples with aberrantly high BCL11b expression levels defined as Q2-4 (n = 146; BCL11b expression range >0.2-12.3, Additional file 1: Table S4). In concordance with the gene expression profiles (GEP) data, these molecular studies by RT-PCR confirmed IGFBP7 to be overexpressed in the BCL11b-low compared to the BCL11b-high group. No significant difference between BCL11b-low and BCL11b-high patients was observed for the previously described negative prognostic factors including BAALC, ERG, and WT1 [[34],[38]]. GATA3, a transcription factor up-regulated in DN1 phase of normal T-cell development [[39]] was significantly lower expressed in BCL11b-low (median: 2.1 vs. 5.7, P < 0.01) compared to BCL11b-high patients. The frequency of TCR rearrangements was significantly lower (50% vs. 80%, P = 0.01) in BCL11b-low compared to the BCL11b-high group. The BCL11b low and the high expressing groups did not differ regarding NOTCH1 or WT1 mutation frequencies (Additional file 1: Table S4).
There was no difference with respect to age or sex of T-ALL patients enrolled on the GMALL 06/99 and 07/03 trials, but within the BCL11b-low group significantly more patients had a low white blood cell count (WBC; 62% vs. 18% <30.000 × 109/l WBC; P = 0.01; Table 1). Patients with early T-ALL showed a significantly lower BCL11b expression (median: 0.3) compared to patients with mature (median: 0.6, P = 0.03) or thymic (median: 0.6, P = 0.01) T-ALL (Additional file 1: Figure S1).
Table 1.
Clinical characteristics of GMALL T-ALL patients with respect to BCL11b expression
|
Characteristics |
BCL11b
-low |
BCL11b-
high |
|
|---|---|---|---|
| Q1 | Q2-4 | P-value | |
| n |
40 |
129 |
|
|
BCL11b expression |
|
|
|
| Median |
0.1 |
0.8 |
|
| Range |
0-0.2 |
0.2-12.3 |
|
| Age |
|
|
n.s. |
| 15-35 yrs |
27 |
70 |
|
| 36-55 yrs |
13 |
48 |
|
| 56-65 yrs |
0 |
11 |
|
| Sex |
|
|
n.s. |
| Female % |
20% |
26% |
|
| WBC, × 109/l |
|
0.01 |
|
| <30,000 |
24 |
42 |
|
| 30-100,000 |
10 |
53 |
|
| >100,000 |
5 |
30 |
|
| T-ALL subtype |
|
0.01 |
|
| Early n (%) |
17 (42%) |
23 (18%) |
|
| Mature n (%) |
5 (13%) |
22 (17%) |
|
| Thymic n (%) |
18 (45%) |
84 (65%) |
|
| Response to induction therapy |
n.s. |
||
| CR n (%) |
37 (95%) |
115 (94%) |
|
| ED n (%) |
1 (3%) |
3 (2%) |
|
| Failure n (%) | 1 (3%) | 5 (4%) | |
Abbreviations: CR = complete remission, ED = early death, WBC = white blood cell count.
BCL11b expression is associated with outcome in adult T-ALL
While there was no difference between BCL11b-low and BCL11b-high within the overall GMALL cohort of T-ALL patients with respect to the response to induction therapy (Table 1), the overall survival probability of BCL11b-low patients was significantly lower compared to BCL11b-high patients (Q1: n = 40, 35% at 5 yrs; Q2-4: n = 129, 53% at 5 yrs; P = 0.02; Figure 4A). When outcomes were analyzed separately for each quartile, T-ALL patients with BCL11b-low (Q1) showed an inferior outcome (Q1: n = 40; 35% OS at 5 yrs) compared to patients in the remaining quartile groups (Q2: n = 42; 52% OS at 5 yrs; Q3: n = 42; 52% OS at 5 yrs; Q4: n = 45; 55% OS at 5 yrs; Additional file 1: Figure S2A).
Figure 4.
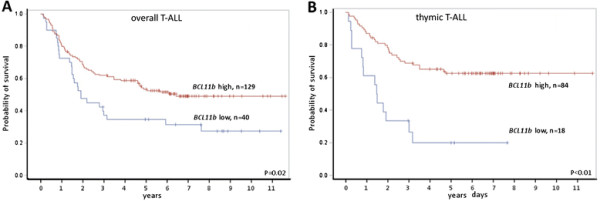
Kaplan–Meier analyses of overall survival (OS) and remission duration in T-ALL with respect toBCL11bmRNA expression. A: T-ALL patients with low BCL11b expression showed significant inferior OS than patients with higher expression (low expression quartile Q1 vs. high Q2-4 expression quartiles; P = 0.02; log-rank test). B: The BCL11b low expressing thymic T-ALL patients had a highly significant inferior OS (P < 0.01).
Low BCL11b defines high risk patients within the standard risk group of thymic T-ALL
The identification of novel prognostic markers is in particular important for the largest subgroup of standard risk thymic T-ALL. As these patients are regarded as standard risk, allogeneic stem cell transplantation is not recommended in first complete remission within GMALL trials. In thymic T-ALL, BCL11b expression groups did not differ in the expression levels of molecular markers including BAALC, ERG, IGFBP7 and WT1. The BCL11b-low group showed significantly lower expression of T-cell regulator GATA3 compared to BCL11b-high (median Q1: 1.6, median Q2-4: 5.1, P < 0.01). Significantly fewer thymic BCL11b-low patients remained in continuous complete remission at five years compared to BCL11b-high thymic T-ALL patients (at 5 yrs: Q1: n = 15, 38%; Q2-4: n = 78, 72%; P = 0.02; Additional file 1: Figure S2B). Moreover, BCL11b-low thymic T-ALL had a significantly inferior OS compared to BCL11b-high thymic T-ALL patients: while only 20% (n = 18) of BCL11b-low patients were alive at 5 years, the 5 year OS of the BCL11b-high group was 62% (n = 84; P < 0.01, Figure 4B). Similar to the entire cohort, patients with thymic T-ALL BCL11b-low (Q1) showed a significantly inferior outcome compared to higher expression quartiles (Q1: n = 18, 20% at 5 yrs; Q2 n = 26, 67% at 5 yrs¸ Q3 n = 18, 50% at 5 yrs; Q4 n = 30, 70% at 5 yrs; P = 0.01; Additional file 1: Figure S2C).
High frequency of BCL11b mutations in adult T-ALL
We sequenced BCL11b exon 4 and identified in 14% (24/178) of the T-ALL patients protein modifying alterations (Figure 5, Additional file 1: Table S6). A single T-ALL patient carried two mutations. Sixteen of the mutations were point mutations with single base pair exchanges leading to amino acid exchanges in 13 and a translation stop in three cases. Seven patients carried deletions and two insertions causing frame shifts. 23 of 24 mutations were either located within zinc finger domains or had an impact on these domains through frame shift or stop of translation, and thus have a likely impact on protein function. Nearly all patients with BCL11b mutations were in the BCL11b-high group (n = 22/23, P = 0.01). The presence of BCL11b mutations was associated with the maturation stage of the T-ALL. Of the 21 patients with BCL11b mutations, 18 were classified as thymic T-ALL (85.5%; P = 0.03; Additional file 1: Table S7). WT1, a negative prognostic factor, was significantly lower expressed in the BCL11b mutated group and other known oncogenes or molecular factors were not associated with BCL11b mutation status (not shown). However, outcome analyses showed no significant impact of BCL11b mutations and deletions on overall survival (Additional file 1: Figure S3).
Figure 5.
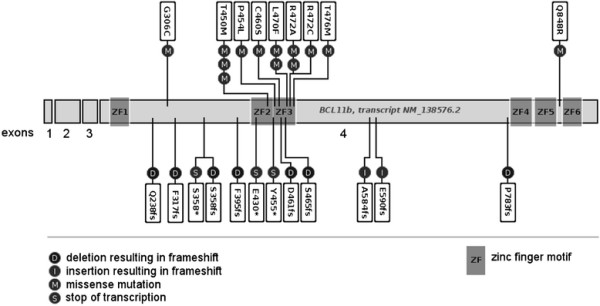
Mutations found in exon 4 ofBCL11bin T-ALL. Mutation analysis was performed by Sanger Sequencing in 178 T-ALL patients, 14% of patients were revealed to harbor protein changing mutations in exon 4 of BCL11b.
Discussion
During normal T-cell development, T-cell progenitors pass through separate differentiation stages and this process is tightly regulated by lineage specific transcription factors. While onset of GATA3 and TCF-1 expression characterize the early DN1 (ETP) phase in T-cell differentiation, BCL11b is expressed at the later DN2 stage [[39]]. As a gatekeeper of T-cell development, it is essential for the correct αβ T-cell development [[3],[40]]. Loss of BCL11b at various stages was shown to induce a natural killer cell-like phenotype with a differentiation arrest and was associated with a high proliferative potential [[14],[40]]. In addition, BCL11b is necessary for T-cell identity maintenance. Alteration of different stage-specific transcription factors and key regulators of T-cell differentiation by silencing, ectopic expression or mutations have shown to contribute in oncogenic transformation in T-ALL. For example, aberrant NOTCH1 signaling through activating mutations found in 50-60% of T-ALL cases [[41],[42]], is a prominent example of a potent driver event in T-cell leukemogenesis. For TAL1, which plays a key role in hematopoietic stem cell development, rearrangements and aberrant expression were found in T-ALL patients [[43]]. In this case, TAL1 expression was positively correlated with survival in pediatric patients [[44]]. Likewise for the homeobox transcription factor TXL-1 deregulation and gene locus abnormalities had been associated with improved outcome for TXL-1-high T-ALL patients [[45]]. Understanding of the molecular processes involved in T-ALL pathology offers the possibility to refine prognosis and stratify therapeutic algorithms.
For BCL11b aberrant expression levels, deletions and mutations have been reported in T-ALL [[29]–[32],[46],[47]]. Here we comprehensively investigated the implications of altered BCL11b expression and loss of function mutations in a large cohort of adult T-ALL patients (n = 169). While CD34+ hematopoietic progenitor cells and unselected BM samples of healthy donors lack BCL11b expression, T-ALL patients showed an aberrant and highly heterogonous BCL11b expression pattern. Similar to normal T-cell differentiation, the expression of BCL11b reflected the maturation stage in T-ALL and thus was significantly lower in the early compared to thymic and mature T-ALL subgroups (Figure 1, Additional file 1: Figure S1). Analysis of microarray expression data confirmed this observation revealing up-regulation of genes associated with an immature phenotype in BCL11b-low and an enrichment of markers of a mature T-cell phenotype in BCL11b-high T-ALL.
While the combined patient data suggested that BCL11b expression reflects the differentiation arrest of leukemic cells, expression was heterogeneously distributed and patients that lacked or had very low BCL11b expression were found in all immunophenotypic subgroups. This suggests that BCL11b is not just a mere marker of genetically more differentiated blasts, but may act as a maturation dependent tumor suppressor, which is supported in other studies [[29],[30],[47]]. If deregulated during differentiation, disruption of normal BCL11b function may contribute to malignant transformation.
In this study the cohort of uniformly treated adult T-ALL patients, the BCL11b-low subgroup had a significantly inferior OS compared to the BCL11b-high patients. In particular, in the standard risk group of thymic T-ALL, BCL11b was of relevant prognostic impact: 62% of BCL11b-high patients were alive after 5 years, whereas survival was only 20% at 5 years in the BCL11b-low subgroup. Remission duration was also significantly shorter for patients within the BCL11b-low subgroup. This contrasts a study in pediatric T-ALL patients, which showed that BCL11b expression had no impact on OS [[30]], although the difference may be due to the smaller sample size of the study and that patients were not classified into immunophenotypic subgroups. Nevertheless, low BCL11b expression was associated with chemotherapy induction failure in the same study.
While immunophenotypic classification of T-ALL has improved outcome prediction, a relevant percentage of patients classified as standard risk based on their thymic immunophenotype, fail conventional chemotherapy. As thymic T-ALL on the molecular and clinical levels compromises a highly heterogeneous cohort, it remains essential to identify patients with a high risk of relapse to adjust treatment stratification. Our results indicate that thymic BCL11b-low T-ALL patients represent a high risk subgroup which would benefit from intensified MRD monitoring and treatment stratification including allogeneic stem cell transplantation. Since lack of BCL11b expression proved to indicate inferior survival, we investigated disruption of the gene’s function on the mutational level. BCL11b mutations in pediatric and adult patients had been reported in T-ALL in the zinc finger structures of exon 4 [[29],[30],[32],[46]]. In agreement with these studies, we discovered a high rate (14%) of BCL11b mutations in this large cohort of T-ALL patients. We found an association of immunophenotype and mutation frequency: only 2% mutational rate in early T-ALL patients and 19% mutational rate in thymic T-ALL patients. Our results support a recent study in which adult T-ALL patients characterized as “early/immature” had fewer BCL11b mutations [[32]]. The number of BCL11b mutations in thymic T-ALL samples in this report was low, limiting the statistical significance regarding outcome. Also, gene expression studies may be more sensitive to identify patients with the specific outcome-associated phenotype caused by BCL11b down-regulation.
Further studies are needed to fully understand the biological relevance of BCL11b mutations, and to explore potential directed therapies.
In summary, we identified BCL11b expression as a potent prognostic factor in the overall cohort and in particular in the standard risk subgroup of thymic T-ALL. These findings will help to identify patients with an enhanced risk of failure to standard therapy, however, standardized detection analyses of aberrant gene expression levels in a routine diagnostic setting remains challenging. More importantly, alterations in critical transcription factors contribute to leukemogenesis and may be regarded as ideal candidates for differentiation directed therapies in the future.
Methods
Patients
We analyzed diagnostic BM material of 195 adult T-ALL patients sent to the GMALL reference laboratory [[48]]. Immunophenotyping of the samples was centrally performed in the GMALL reference laboratory at the Charité, University Hospital Berlin, Germany, as previously described [[49],[50]], and classified into the T-ALL subgroups: early (n = 50), mature (n = 33) and thymic (n = 112). Of these, 169 T-ALL patients were consecutively enrolled in the GMALL trials 06/99 and 07/2003 [[51]] with available clinical follow-up data. Additionally, samples of healthy adult donors were used, 13 BM samples, 13 CD34+ progenitors and nine CD3+ selected T-cell samples. Written informed consent according to the declaration of Helsinki had been given. Studies were approved by the ethics board and registered in a public registry (clincaltrials.gov NCT00199056, NCT0098991).
Sample preparation and qRT-PCR
Total RNA and DNA extractions were performed using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. To analyze BCL11b expression, complementary DNA was synthesized and quantitative real-time polymerase chain reaction (qRT-PCR) was performed, using Glucose-6-Phosphate Isomerase (GPI) as internal control, as previously described [[52]]. For BCL11b amplification forward primer AACCCGCAGCACTTGTCC, reverse-primer ATTTGACACTGGCCACAGGT and probe FAM-CTCATCACCCCAGAGGCTGACCAT-BHQ1 spanning exons 1 and 2 were used. Expression levels of BCL11b were calculated using the mean of ΔCT from two replicates and expressed as 2μ(ΔCT). The mRNA expression levels for IGFBP7, WT1, BAALC, ERG, and GATA3 by qRT-PCR, as well as mutation status of WT1 and NOTCH1 and TCR-rearrangements had been determined in previous studies [[12],[34],[35],[52]–[54]].
Gene expression profiles
BCL11b-associated GEP of an independent set of 86 adult T-ALL samples were generated from raw data obtained from the Microarrays Innovations in Leukemia (MILE) multicenter study (HG-U133 Plus 2.0; Affymetrix, Santa Clara, CA, USA) [[33]]. For the GEP-analysis, samples were divided into quartiles (Q) according to BCL11b expression [median of the two probe sets (219528_s_at, 222895_s_at)]. To identify BCL11b-associated GEP signatures, the lowest expression quartile (Q1) was compared to quartiles 2 to 4 united (Q2-4). Lists of genes with a 2-fold under- or over-expression were generated. Statistical significance was calculated by the non-parametric t-test with a P-value ≤0.05. The data analyses were carried out with GeneSpring software version 4.2 (Silicon Genetics, Redwood City, CA, USA).
Mutational analysis
Quantity and quality of genomic DNA was sufficient for the mutational analysis of BCL11b in 178 T-ALL samples. As previous studies had detected only very few mutations outside exon 4, which harbors all six of the gene’s zinc finger domains, we focused on this region [[29],[30]]. Primer pairs were newly designed or used as previously published for bidirectional Sanger sequencing of exon 4 (complete list see Additional file 1: Table S1) [[29]]. Geneious version 5.4.3 software (Biomatters Ltd., Auckland, NZ) was used for analysis.
Abbreviations
ALL: Acute lymphoblastic leukemia
AML: Acute myeloid leukemia
BM: Bone marrow
DN: Double negative
ETP-ALL: Early t-cell precursor ALL
GEP: Gene expression profile
GSEA: Gene set enrichment analysis
GMALL: German multicenter acute lymphoblastic leukemia study group
MILE: Microarrays innovations in leukemia
MRD: Minimal residual disease
OS: Overall survival
qRT-PCR: Quantitative real-time polymerase chain reaction
T-ALL: T-cell acute lymphoblastic leukemia
TKI: Tyrosine kinase inhibitors
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
IB performed laboratory work, data analysis and wrote the manuscript. NG supervised the GMALL study center, performed statistical analysis and reviewed the manuscript. CS performed laboratory work for this study. SH helped to design the study. LF provided expression data. SS, RS, KSE, MS, AR and DH recruited the study patients and performed the study procedures. CDB coordinate the research and reviewed the manuscript. MN performed statistical analysis and reviewed the manuscript. All authors read and approved the final manuscript.
Additional file
Supplementary Material
Statistical analysis. Figure S1. BCL11b mRNA expression in T-ALL immunophenotypic subtypes. Figure S2. Kaplan–Meier analysis of overall survival (OS) in T-ALL with respect to BCL11b mRNA expression. Figure S3. Kaplan–Meier analysis of overall survival (OS) in T-ALL with respect to BCL11b mutation status. Table S1. Primer sets designed for human BCL11b exon 4 (NP_612808.1). Table S2. Probe sets up-regulated in the BCL11b-low group of T-ALL patients. Table S3. Probe sets up-regulated in the BCL11b-high group of T-ALL patients. Table S4. Molecular characteristics of T-ALL patients with respect to BCL11b expression. Table S5. Molecular characteristics of thymic GMALL T-ALL patients in respect to BCL11b expression. Table S6. BCL11b exon 4 mutations in T-ALL. Table S7. Clinical characteristics of GMALL T-ALL patients in respect to BCL11b mutations. Table S8. Members of the German Multicenter Study Group for Adult ALL Supplemental Methods: Statistics. References.
Contributor Information
Isabelle Bartram, Email: isabelle.bartram@charite.de.
Nicola Gökbuget, Email: goekbuget@em.uni-frankfurt.de.
Cornelia Schlee, Email: cornelia.schlee@charite.de.
Sandra Heesch, Email: SaHee@gmx.de.
Lars Fransecky, Email: Lars.Fransecky@charite.de.
Stefan Schwartz, Email: stefan.schwartz@charite.de.
Reingard Stuhlmann, Email: r.stuhlmann@asklepios.com.
Kerstin Schäfer-Eckhart, Email: kerstin.schaefer-eckart@klinikum-nuernberg.de.
Michael Starck, Email: michael.starck@klinikum-muenchen.de.
Albrecht Reichle, Email: albrecht.reichle@ukr.de.
Dieter Hoelzer, Email: hoelzer@em.uni-frankfurt.de.
Claudia D Baldus, Email: claudia.baldus@charite.de.
Martin Neumann, Email: martin.neumann@charite.de.
References
- Gokbuget N, Hoelzer D. Treatment of adult acute lymphoblastic leukemia. Semin Hematol. 2009;46:64–75. doi: 10.1053/j.seminhematol.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Hunault M, Harousseau JL, Delain M, Truchan-Graczyk M, Cahn JY, Witz F, Lamy T, Pignon B, Jouet JP, Garidi R, Caillot D, Berthou C, Guyotat D, Sadoun A, Sotto JJ, Lioure B, Casassus P, Solal-Celigny P, Stalnikiewicz L, Audhuy B, Blanchet O, Baranger L, Béné MC, Ifrah N. Better outcome of adult acute lymphoblastic leukemia after early genoidentical allogeneic bone marrow transplantation (BMT) than after late high-dose therapy and autologous BMT: a GOELAMS trial. Blood. 2004;104:3028–3037. doi: 10.1182/blood-2003-10-3560. [DOI] [PubMed] [Google Scholar]
- Bassan R, Hoelzer D. Modern therapy of acute lymphoblastic leukemia. J Clin Oncol. 2011;29:532–543. doi: 10.1200/JCO.2010.30.1382. [DOI] [PubMed] [Google Scholar]
- Hoelzer D. Novel antibody-based therapies for acute lymphoblastic leukemia. Hematol Am Soc Hematol Educ Progr. 2011;2011:243–249. doi: 10.1182/asheducation-2011.1.243. [DOI] [PubMed] [Google Scholar]
- Daver N, O'Brien S. Novel therapeutic strategies in adult acute lymphoblastic leukemia - a focus on emerging monoclonal antibodies. Curr Hematol Malig Rep. 2013;8:123–131. doi: 10.1007/s11899-013-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp MS, Gokbuget N, Zugmaier G, Degenhard E, Goebeler ME, Klinger M, Neumann SA, Horst HA, Raff T, Viardot A, Stelljes M, Schaich M, Köhne-Volland R, Brüggemann M, Ottmann OG, Burmeister T, Baeuerle PA, Nagorsen D, Schmidt M, Einsele H, Riethmüller G, Kneba M, Hoelzer D, Kufer P, Bargou RC. Long-term follow-up of hematologic relapse-free survival in a phase 2 study of blinatumomab in patients with MRD in B-lineage ALL. Blood. 2012;120:5185–5187. doi: 10.1182/blood-2012-07-441030. [DOI] [PubMed] [Google Scholar]
- Berg SL, Blaney SM, Devidas M, Lampkin TA, Murgo A, Bernstein M, Billett A, Kurtzberg J, Reaman G, Gaynon P, Whitlock J, Krailo M, Harris MB. Phase II study of nelarabine (compound 506U78) in children and young adults with refractory T-cell malignancies: a report from the children’s oncology group. J Clin Oncol. 2005;23:3376–3382. doi: 10.1200/JCO.2005.03.426. [DOI] [PubMed] [Google Scholar]
- Robak T. New nucleoside analogs for patients with hematological malignancies. Expert Opin Investig Drugs. 2011;20:343–359. doi: 10.1517/13543784.2011.554822. [DOI] [PubMed] [Google Scholar]
- Palomero T, Ferrando A. Therapeutic targeting of NOTCH1 signaling in T-cell acute lymphoblastic leukemia. Clin Lymphoma Myeloma. 2009;9(Suppl 3):S205–S210. doi: 10.3816/CLM.2009.s.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, Biondi A, Pui CH, Downing JR, Campana D. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijerink JP. Genetic rearrangements in relation to immunophenotype and outcome in T-cell acute lymphoblastic leukaemia. Best Pract Res Clin Haematol. 2010;23:307–318. doi: 10.1016/j.beha.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Neumann M, Heesch S, Gokbuget N, Schwartz S, Schlee C, Benlasfer O, Farhadi-Sartangi N, Thibaut J, Burmeister T, Hoelzer D, Hofmann WK, Thiel E, Baldus CD: Clinical and molecular characterization of early T-cell precursor leukemia: a high-risk subgroup in adult T-ALL with a high frequency of FLT3 mutations.Blood Cancer J 2012, 2:e55. [DOI] [PMC free article] [PubMed]
- Liu P, Li P, Burke S. Critical roles of Bcl11b in T-cell development and maintenance of T-cell identity. Immunol Rev. 2010;238:138–149. doi: 10.1111/j.1600-065X.2010.00953.x. [DOI] [PubMed] [Google Scholar]
- Kominami R. Role of the transcription factor Bcl11b in development and lymphomagenesis. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88:72–87. doi: 10.2183/pjab.88.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier T, Suzanne S, Redel L, Calao M, Marban C, Samah B, Mukerjee R, Schwartz C, Gras G, Sawaya BE, Zeichner SL, Aunis D, Van Lint C, Rohr O. p21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene. 2009;28:3380–3389. doi: 10.1038/onc.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cismasiu VB, Ghanta S, Duque J, Albu DI, Chen HM, Kasturi R, Avram D. BCL11B participates in the activation of IL2 gene expression in CD4+ T lymphocytes. Blood. 2006;108:2695–2702. doi: 10.1182/blood-2006-05-021790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topark-Ngarm A, Golonzhka O, Peterson VJ, Barrett B Jr, Martinez B, Crofoot K, Filtz TM, Leid M. CTIP2 associates with the NuRD complex on the promoter of p57KIP2, a newly identified CTIP2 target gene. J Biol Chem. 2006;281:32272–32283. doi: 10.1074/jbc.M602776200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabarczyk P, Przybylski GK, Depke M, Volker U, Bahr J, Assmus K, Broker BM, Walther R, Schmidt CA. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene. 2007;26:3797–3810. doi: 10.1038/sj.onc.1210152. [DOI] [PubMed] [Google Scholar]
- Karanam NK, Grabarczyk P, Hammer E, Scharf C, Venz S, Gesell-Salazar M, Barthlen W, Przybylski GK, Schmidt CA, Volker U. Proteome analysis reveals new mechanisms of Bcl11b-loss driven apoptosis. J Proteome Res. 2010;9:3799–3811. doi: 10.1021/pr901096u. [DOI] [PubMed] [Google Scholar]
- Grabarczyk P, Nahse V, Delin M, Przybylski G, Depke M, Hildebrandt P, Volker U, Schmidt CA: Increased expression of bcl11b leads to chemoresistance accompanied by G1 accumulation.PLoS One 2010, 5:e12532. [DOI] [PMC free article] [PubMed]
- Wakabayashi Y, Inoue J, Takahashi Y, Matsuki A, Kosugi-Okano H, Shinbo T, Mishima Y, Niwa O, Kominami R. Homozygous deletions and point mutations of the Rit1/Bcl11b gene in gamma-ray induced mouse thymic lymphomas. Biochem Biophys Res Commun. 2003;301:598–603. doi: 10.1016/S0006-291X(02)03069-3. [DOI] [PubMed] [Google Scholar]
- Kamimura K, Ohi H, Kubota T, Okazuka K, Yoshikai Y, Wakabayashi Y, Aoyagi Y, Mishima Y, Kominami R. Haploinsufficiency of Bcl11b for suppression of lymphomagenesis and thymocyte development. Biochem Biophys Res Commun. 2007;355:538–542. doi: 10.1016/j.bbrc.2007.02.003. [DOI] [PubMed] [Google Scholar]
- MacLeod RA, Nagel S, Kaufmann M, Janssen JW, Drexler HG. Activation of HOX11L2 by juxtaposition with 3′-BCL11B in an acute lymphoblastic leukemia cell line (HPB-ALL) with t(5;14)(q35;q32.2) Genes Chromosomes Cancer. 2003;37:84–91. doi: 10.1002/gcc.10194. [DOI] [PubMed] [Google Scholar]
- Przybylski GK, Dik WA, Wanzeck J, Grabarczyk P, Majunke S, Martin-Subero JI, Siebert R, Dolken G, Ludwig WD, Verhaaf B, van Dongen JJ, Schmidt CA, Langerak AW. Disruption of the BCL11B gene through inv(14)(q11.2q32.31) results in the expression of BCL11B-TRDC fusion transcripts and is associated with the absence of wild-type BCL11B transcripts in T-ALL. Leukemia. 2005;19:201–208. doi: 10.1038/sj.leu.2403619. [DOI] [PubMed] [Google Scholar]
- Satterwhite E, Sonoki T, Willis TG, Harder L, Nowak R, Arriola EL, Liu H, Price HP, Gesk S, Steinemann D, Schlegelberger B, Oscier DG, Siebert R, Tucker PW, Dyer MJ. The BCL11 gene family: involvement of BCL11A in lymphoid malignancies. Blood. 2001;98:3413–3420. doi: 10.1182/blood.V98.12.3413. [DOI] [PubMed] [Google Scholar]
- Bezrookove V, van Zelderen-Bhola SL, Brink A, Szuhai K, Raap AK, Barge R, Beverstock GC, Rosenberg C. A novel t(6;14)(q25-q27;q32) in acute myelocytic leukemia involves the BCL11B gene. Cancer Genet Cytogenet. 2004;149:72–76. doi: 10.1016/S0165-4608(03)00302-9. [DOI] [PubMed] [Google Scholar]
- Oliveira JL, Kumar R, Khan SP, Law ME, Erickson-Johnson M, Oliveira AM, Ketterling RP, Dogan A. Successful treatment of a child with T/myeloid acute bilineal leukemia associated with TLX3/BCL11B fusion and 9q deletion. Pediatr Blood Cancer. 2011;56:467–469. doi: 10.1002/pbc.22850. [DOI] [PubMed] [Google Scholar]
- Abbas S, Sanders M, Zeilemaker A, Geertsma-Kleinekoort WM, Koenders JE, Kavelaars FG, Abbas ZG, Mahamoud S, Chu IW, Hoogenboezem R, Peeters JK, van Drunen E, van Galen J, Beverloo HB, Löwenberg B, Valk PJ. Integrated genome-wide genotyping and gene expression profiling reveals BCL11B as a putative oncogene in acute myeloid leukemia with 14q32 aberrations. Haematologica. 2014;99:848–57. doi: 10.3324/haematol.2013.095604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Real PJ, Gatta GD, Palomero T, Sulis ML, Tosello V, Van VP, Barnes K, Castillo M, Sole X, Hadler M, Lenz J, Aplan PD, Kelliher M, Kee BL, Pandolfi PP, Kappes D, Gounari F, Petrie H, Van der Meulen J, Speleman F, Paietta E, Racevskis J, Wiernik PH, Rowe JM, Soulier J, Avran D, Cavé H, Dastugue N, Raimondi S. et al. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat Med. 2010;16:1321–1327. doi: 10.1038/nm.2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A, Kentsis A, Sanda T, Holmfeldt L, Chen SC, Zhang J, Protopopov A, Chin L, Dahlberg SE, Neuberg DS, Silverman LB, Winter SS, Hunger SP, Sallan SE, Zha S, Alt FW, Downing JR, Mullighan CG, Look AT. The BCL11B tumor suppressor is mutated across the major molecular subtypes of T-cell acute lymphoblastic leukemia. Blood. 2011;118:4169–4173. doi: 10.1182/blood-2010-11-318873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, Girardi T, Gianfelici V, Geerdens E, Clappier E, Porcu M, Lahortiga I, Lucà R, Yan J, Hulselmans G, Vranckx H, Vandepoel R, Sweron B, Jacobs K, Mentens N, Wlodarska I, Cauwelier B, Cloos J, Soulier J, Uyttebroeck A, Bagni C, Hassan BA, Vandenberghe P, Johnson AW, Aerts S, Cools J. Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nat Genet. 2013;45:186–190. doi: 10.1038/ng.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P, Ambesi-Impiombato A, De Keersmaecker K, Hadler M, Paietta E, Tallman MS, Rowe JM, Forne C, Rue M, Ferrando AA. Prognostic relevance of integrated genetic profiling in adult T-cell acute lymphoblastic leukemia. Blood. 2013;122:74–82. doi: 10.1182/blood-2013-03-491092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Bene MC, De VJ, Hernandez JM, Hofmann WK, Mills KI, Gilkes A, Chiaretti S, Shurtleff SA, Kipps TJ, Rassenti LZ, Yeoh AE, Papenhausen PR, Liu WM, Williams PM, Foà R. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: report from the international microarray innovations in leukemia study group. J Clin Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldus CD, Martus P, Burmeister T, Schwartz S, Gokbuget N, Bloomfield CD, Hoelzer D, Thiel E, Hofmann WK. Low ERG and BAALC expression identifies a new subgroup of adult acute T-lymphoblastic leukemia with a highly favorable outcome. J Clin Oncol. 2007;25:3739–3745. doi: 10.1200/JCO.2007.11.5253. [DOI] [PubMed] [Google Scholar]
- Heesch S, Schlee C, Neumann M, Stroux A, Kuhnl A, Schwartz S, Haferlach T, Goekbuget N, Hoelzer D, Thiel E, Hofmann WK, Baldus CD. BAALC-associated gene expression profiles define IGFBP7 as a novel molecular marker in acute leukemia. Leukemia. 2010;24:1429–1436. doi: 10.1038/leu.2010.130. [DOI] [PubMed] [Google Scholar]
- Small D, Levenstein M, Kim E, Carow C, Amin S, Rockwell P, Witte L, Burrow C, Ratajczak MZ, Gewirtz AM, Civin CI. STK-1, the human homolog of Flk-2/Flt-3, is selectively expressed in CD34+ human bone marrow cells and is involved in the proliferation of early progenitor/stem cells. Proc Natl Acad Sci U S A. 1994;91:459–463. doi: 10.1073/pnas.91.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, Frampton GM, Drake AC, Leskov I, Nilsson B, Preffer F, Dombkowski D, Evans JW, Liefeld T, Smutko JS, Chen J, Friedman N, Young RA, Golub TR, Regev A, Ebert BL. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen HD, Renkl HJ, Rodeck U, Maurer J, Notter M, Schwartz S, Reinhardt R, Thiel E. Presence of Wilms' tumor gene (wt1) transcripts and the WT1 nuclear protein in the majority of human acute leukemias. Leukemia. 1995;9:1060–1067. [PubMed] [Google Scholar]
- Rothenberg EV. Transcriptional drivers of the T-cell lineage program. Curr Opin Immunol. 2012;24:132–138. doi: 10.1016/j.coi.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch U, Radtke F. Notch in T-ALL: new players in a complex disease. Trends Immunol. 2011;32:434–442. doi: 10.1016/j.it.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, Blacklow SC, Look AT, Aster JC. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Van VP, Ferrando A. The molecular basis of T cell acute lymphoblastic leukemia. J Clin Invest. 2012;122:3398–3406. doi: 10.1172/JCI61269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi A, Hayashi Y, Kobayashi S, Hanada R, Moriwaki K, Yamamoto K, Fujimoto J, Kaneko Y, Yamamori S. Clinical significance of TAL1 gene alteration in childhood T-cell acute lymphoblastic leukemia and lymphoma. Leukemia. 1993;7:933–938. [PubMed] [Google Scholar]
- Bergeron J, Clappier E, Radford I, Buzyn A, Millien C, Soler G, Ballerini P, Thomas X, Soulier J, Dombret H, Macintyre EA, Asnafi V. Prognostic and oncogenic relevance of TLX1/HOX11 expression level in T-ALLs. Blood. 2007;110:2324–2330. doi: 10.1182/blood-2007-04-079988. [DOI] [PubMed] [Google Scholar]
- Kraszewska MD, Dawidowska M, Kosmalska M, Sedek L, Grzeszczak W, Kowalczyk JR, Szczepanski T, Witt M. BCL11B, FLT3, NOTCH1 and FBXW7 mutation status in T-cell acute lymphoblastic leukemia patients. Blood Cells Mol Dis. 2013;50:33–38. doi: 10.1016/j.bcmd.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Kurosawa N, Fujimoto R, Ozawa T, Itoyama T, Sadamori N, Isobe M: Reduced level of the BCL11B protein is associated with adult T-cell leukemia/lymphoma.PLoS One 2013, 8:e55147. [DOI] [PMC free article] [PubMed]
- Brüggemann M, Raff T, Flohr T, Gokbuget N, Nakao M, Droese J, Luschen S, Pott C, Ritgen M, Scheuring U, Horst HA, Thiel E, Hoelzer D, Bartram CR, Kneba M. Clinical significance of minimal residual disease quantification in adult patients with standard-risk acute lymphoblastic leukemia. Blood. 2006;107:1116–1123. doi: 10.1182/blood-2005-07-2708. [DOI] [PubMed] [Google Scholar]
- Baldus CD, Thibaut J, Goekbuget N, Stroux A, Schlee C, Mossner M, Burmeister T, Schwartz S, Bloomfield CD, Hoelzer D, Thiel E, Hofmann WK. Prognostic implications of NOTCH1 and FBXW7 mutations in adult acute T-lymphoblastic leukemia. Haematologica. 2009;94:1383–1390. doi: 10.3324/haematol.2008.005272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokbuget N, Raff R, Brüggemann M, Flohr T, Scheuring U, Pfeifer H, Bartram CR, Kneba M, Hoelzer D. Risk/MRD adapted GMALL trials in adult ALL. Ann Hematol. 2004;83(Suppl 1):S129–S131. doi: 10.1007/s00277-004-0850-2. [DOI] [PubMed] [Google Scholar]
- http://www.leukemia-net.org/trial/download/public/ALL_GMALL07-03_ShortProtEN.pdf?id=489 European leukemia Net. European leukemia trial registry trial: ALL GMALL 07/2003.. Accessed September 3, 2013. .
- Baldus CD, Burmeister T, Martus P, Schwartz S, Gokbuget N, Bloomfield CD, Hoelzer D, Thiel E, Hofmann WK. High expression of the ETS transcription factor ERG predicts adverse outcome in acute T-lymphoblastic leukemia in adults. J Clin Oncol. 2006;24:4714–4720. doi: 10.1200/JCO.2006.06.1580. [DOI] [PubMed] [Google Scholar]
- Heesch S, Goekbuget N, Stroux A, Tanchez JO, Schlee C, Burmeister T, Schwartz S, Blau O, Keilholz U, Busse A, Hoelzer D, Thiel E, Hofmann WK, Baldus CD. Prognostic implications of mutations and expression of the Wilms tumor 1 (WT1) gene in adult acute T-lymphoblastic leukemia. Haematologica. 2010;95:942–949. doi: 10.3324/haematol.2009.016386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Coskun E, Fransecky L, Mochmann LH, Bartram I, Sartangi NF, Heesch S, Gokbuget N, Schwartz S, Brandts C, Schlee C, Haas R, Dührsen U, Griesshammer M, Döhner H, Ehninger G, Burmeister T, Blau O, Thiel E, Hoelzer D, Hofmann WK, Baldus CD: FLT3 mutations in early T-cell precursor ALL characterize a stem cell like leukemia and imply the clinical use of tyrosine kinase inhibitors.PLoS One 2013, 8:e53190. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistical analysis. Figure S1. BCL11b mRNA expression in T-ALL immunophenotypic subtypes. Figure S2. Kaplan–Meier analysis of overall survival (OS) in T-ALL with respect to BCL11b mRNA expression. Figure S3. Kaplan–Meier analysis of overall survival (OS) in T-ALL with respect to BCL11b mutation status. Table S1. Primer sets designed for human BCL11b exon 4 (NP_612808.1). Table S2. Probe sets up-regulated in the BCL11b-low group of T-ALL patients. Table S3. Probe sets up-regulated in the BCL11b-high group of T-ALL patients. Table S4. Molecular characteristics of T-ALL patients with respect to BCL11b expression. Table S5. Molecular characteristics of thymic GMALL T-ALL patients in respect to BCL11b expression. Table S6. BCL11b exon 4 mutations in T-ALL. Table S7. Clinical characteristics of GMALL T-ALL patients in respect to BCL11b mutations. Table S8. Members of the German Multicenter Study Group for Adult ALL Supplemental Methods: Statistics. References.


