Abstract
Background
The sepals, petals and stamens of Arabidopsis flowers detach via abscission zones formed at their boundaries with the underlying receptacle. The ASYMMETRIC LEAVES1 (AS1) MYB transcription factor plays a critical role in setting boundaries between newly formed leaf primordia and the shoot meristem. By repressing expression of a set of KNOTTED1-LIKE HOMEODOMAIN (KNOX) genes from developing leaf primordia, AS1 and its partner ASYMMETRIC LEAVES2 allow the patterning and differentiation of leaves to proceed. Here we show a unique role for AS1 in establishing the positions of the sepal and petal abscission zones in Arabidopsis flowers.
Results
In as1 mutant flowers, the sepal abscission zones are displaced into inverted V-shaped positions, leaving behind triangular stubs of tissue when the organs abscise. Movement of the petal abscission zones is also apparent. Abscission of the medial sepals is delayed in as1 flowers; loss of chlorophyll in the senescing sepals contrasts with proximal zones that remain green. AS1 has previously been shown to restrict expression of the KNOX gene, BREVIPEDICELLUS (BP), from the sepals. We show here that loss of BP activity in as1 flowers is sufficient to restore the positions of the sepal and petal abscission zones, the sepal-receptacle boundary of the medial sepals and the timing of their abscission.
Conclusions
Our results indicate that AS1 activity is critical for the proper placement of the floral organ abscission zones, and influences the timing of organ shedding.
Keywords: Abscission, Floral organ shedding, Organ boundary, Flower development, AS1, BP
Background
The process of organ abscission allows plants to detach leaves, floral organs, fruit and seeds at specific points in their life cycles or in response to environmental cues. Genetic analysis in Arabidopsis has been particularly productive in revealing factors that activate the cell separation phase of floral organ abscission [1],[2]. Less is known about the regulatory circuits that control abscission zone differentiation at the boundaries between the outer floral organs and underlying receptacle [3],[4].
Several genes that regulate lateral organ and/or inter-organ boundaries also affect floral organ abscission. The transcriptional activators BLADE-ON-PETIOLE1 (BOP1) and BOP2 redundantly control patterning of the proximal regions of developing leaves and floral organs [5]–[7]. Anatomical evidence of abscission zone differentiation cannot be detected in bop1 bop2 flowers and the sepals, petals and stamens remain strongly attached [8]. The BELL-type homeodomain transcription factor, ARABIDOPSIS THALIANA HOMEOBOX GENE1 (ATH1), represses growth in regions that will become the floral organ-receptacle boundaries and is required for stamen abscission zone formation [9]. HAWAIIAN SKIRT (HWS), an F-box protein, prevents the fusion of adjacent sepals and influences the timing of their abscission [10].
Five mutant alleles of ASYMMETRIC LEAVES1 (AS1), a key determinant of polarity and cell fate in lateral organs [11], were identified through a screen for floral organ shedding mutants [12] (Additional file 1: Table S1). AS1, a MYB transcription factor, acts in conjunction with the AS2 LATERAL ORGAN BOUNDARIES domain (LBD) transcription factor to repress expression of a set of meristem-promoting KNOTTED1-LIKE HOMEODOMAIN (KNOX) genes during leaf development [13]–[15]. Ectopic activity of the BREVIPEDICELLUS (BP, also known as KNAT1), KNAT2 and KNAT6 homeodomain transcription factors has been genetically linked to the reduced size, shorter petioles and rumpled appearance of as1 mutant leaves [13],[16]. The AS1-AS2 repressor complex recruits components of the Polycomb-repressive complex2 to the promoters of BP and KNAT2, to establish an inactive chromatin state in leaf cells [17]–[19].
AS1 also restricts expression of BP and KNAT2 within developing flower primordia [13]. BP is expressed in the pedicel, receptacle, and organ boundaries of wild-type flowers, while KNAT2 is primarily restricted to the floral organ boundaries and receptacle [13],[20]–[22]. In as1 mutant flowers, which open prematurely due to their smaller sepals and petals, expression of BP and KNAT2 expands into the sepals [13],[23]. Genetic analysis has shown that the combined loss of BP, KNAT2 and KNAT6 activity is sufficient to rescue the reduced organ size and open bud defects of as1 mutant flowers [16].
BP directs growth of the receptacle, an expanded region of the pedicel [20]. bp flowers have slender, abbreviated pedicels due to radial constriction of the receptacle and reduced cell division [20],[23],[24]. Due to increased constriction on the abaxial side of the pedicel, bp pedicels bend down rather than pointing up [20]. BP also functions during floral organ abscission to prevent premature shedding and inhibit expansion of abscission zone cells [21],[25]. The abscission zones of bp flowers are notably enlarged, similar to those of flowers with constitutive expression of INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) [26]. Signaling by the IDA peptide through the HAESA (HAE) and HAESA-LIKE2 receptor-like kinases has been proposed to activate organ abscission by inhibiting BP activity [25],[27],[28].
Here we show that AS1 controls the placement of the sepal and petal organ abscission zones in Arabidopsis flowers. The medial sepals of as1 mutant flowers are most affected: delayed organ abscission occurs along the edges of a triangular-shaped proximal domain with altered identity. We further show that loss of BP activity is sufficient to rescue the abscission-related defects of as1 mutant flowers.
Methods
Plants
Five as1 (formerly known as bibb) mutants were previously identified through a genetic screen for organ shedding mutants [12] (Additional file 1: Table S1); all mutants are of the Landsberg erecta (Ler) ecotype. The F1 progeny of a cross between the as1-21 and as1-20 mutants showed abscission defects, consistent with an allelic relationship between the mutations. as1-1 (Ler; CS146) and bp-1 (Ler; CS30) seeds were obtained from ABRC (Columbus, OH). Plants homozygous for as1-20 were distinguished with a CAPS (cleaved amplified polymorphic sequence) [29] marker based on a BccI site present in the wild-type allele of AS1. The oligos used to amplify this region of the AS1 gene are described in Additional file 2: Table S2. Plants were grown at 21°C with 50% humidity and a 16 hour photoperiod.
Microscopy and marker analyses
Wild-type and mutant flowers were fixed as previously described [30], and prepared for scanning electron microscopy using a Tousimis Samdri-790 critical point dryer (Tousimis, Rockville, MD) and EMS 550 sputter coater equipped with a film thickness monitor (EMS, Hatfield, PA). Samples were examined using an accelerating voltage of 10 kV in a FEI XL-30 scanning electron microscope (FEI, Hillsboro, OR). Young flowers (stages 8–12) were dissected and reimaged to determine their developmental stage [31].
A transgenic HAE::GUS line [32] was crossed to the as1-20 and as1-20 bp-1 mutants to generate single and double mutants carrying this abscission zone marker. For β-glucuronidase assays, wild-type and mutant flowers were fixed and stained as described [33] with minor modifications. Digital images were taken with a Stemi SV11 dissecting microscope and Axiocam HR camera (Carl Zeiss, Germany) or PowerShot SX160 IS (Canon, Melville, NY). Image brightness and contrast were adjusted with Photoshop CS6 (Adobe, Mountain View, CA). Abscission zone displacement was measured using NIH ImageJ [34].
Molecular biology
Genomic DNA samples were prepared from wildtype (Ler) and two independent as1-21 mutant plants using the DNeasy Plant Mini Kit (Qiagen, Venlo, Netherlands). Regions of the AS1 gene were PCR amplified as described in Additional file 2: Table S2. PCR products were purified using the Zymoclean Gel DNA recovery kit (Irvine, CA) and sequenced (MacrogenUSA, Rockville, MD). Sequences were analyzed using Geneious R6.1 software (Biomatters, Auckland, NZ).
Results
Identification of a new mutation in the AS1 MYB domain
AS1 is one of 126 proteins in Arabidopsis with a MYB domain composed of R2 and R3 repeats [35],[36]. Each repeat consists of three helices, of which the second and third form a helix-turn-helix motif that binds DNA (Figure 1A). To characterize the role of AS1 in organ abscission, we selected as1-20 (formerly known as bibb-1) [12] and as1-1[11],[23],[37] as reference alleles (Additional file 1: Table S1). The associated nonsense mutation for as1-20 is in the third helix of the AS1 R2 repeat (Figure 1A) [12]. The as1-21 mutant (formerly known as bibb-2), was found to introduce a nonsense mutation in the first helix of the AS1 R2 repeat (Figure 1A).
Figure 1.
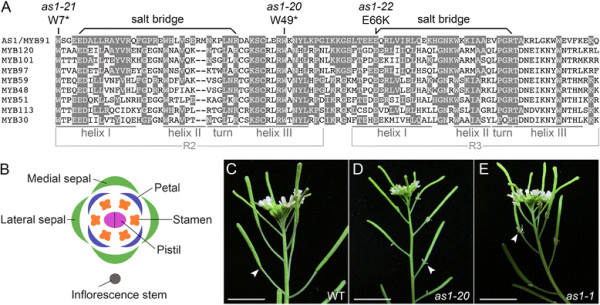
Mutations inAS1delay sepal abscission. A: Sequence alignment of the DNA-binding domain of AS1 and related Arabidopsis R2R3 MYB proteins. Conserved amino acid residues are shaded. The pairs of acidic glutamic acid (E) and basic arginine (R) residues that form salt bridges in the R2 and R3 repeats are linked by bars. The positions of three as1 mutations that alter organ abscission are indicated. The as1-20 (reference allele for this study) and as1-21 mutations introduce stop codons in place of codons specifying tryptophan. The as1-22 mutation replaces a conserved glutamic acid with a lysine; the affected residue is an integral part of the salt bridge formed in the R3 repeat of AS1. B: Diagram of a wild-type flower illustrating the orientation of the medial and lateral sepals with respect to the inflorescence stem. C: Wild-type inflorescence. Flowers shed their outer organs (stage 16), leaving the developing fruit behind. Scale bar, 1 cm. D/E: as1-20(D) and as1-1(E) mutant inflorescences. Mutant flowers typically retain their medial sepals until after the fruit is fully elongated (stage 17). Scale bars, 1 cm.
The as1-22 mutation disrupts a salt bridge in the DNA binding domain
Within each repeat of eukaryotic R2R3 MYB proteins, an acidic amino acid residue in the first helix forms a salt bridge with a basic amino acid residue in the turn adjacent to the third helix [38] (Figure 1A). The glutamic acid and arginine residues that form salt bridges in R2R3 MYB proteins are conserved in more than 97% of the Arabidopsis family members surveyed [35] (Figure 1A). Of the as1 alleles that introduce point mutations [12],[37], as1-22 (formerly known as bibb-5) [12] represents the only missense mutation identified to date. Replacement of the affected glutamic acid with lysine, a basic residue, would prevent salt bridge formation in the R3 repeat of the as1-22 mutant protein (Figure 1A). An alanine substitution of the corresponding glutamic acid within the R3 repeat of the c-Myb transcription factor abolishes both DNA binding and transcriptional activation [38].
Mutations in AS1 delay shedding of the medial sepals
Arabidopsis flowers contain pairs of medial and lateral sepals, four petals and six stamens, which are shed shortly after fertilization (stage 16) (Figure 1B, C) [31]. In each of the as1 mutants known to alter organ abscission (Figure 1D, E; Additional file 1: Table S1; Additional file 3: Figure S1), shedding of the medial sepals is delayed until after the fruit is fully elongated (early stage 17). When as1 flowers are touched during the normal period of abscission (stage 16), the medial sepals remain firmly attached. Of 30 as1-1 flowers (early to mid stage 17) surveyed after touching, 80% (48/60) of the medial sepals remained attached compared to 0% (0/20) for 10 wild-type flowers. Abscission of the lateral sepals is also delayed in some as1-21, as1-22 and as1-23 flowers (Additional file 3: Figure S1B, E).
The sepal and petal abscission zones of as1 flowers are displaced
After abscission (stage 17), the positions of the floral organ abscission zones can be easily visualized in wild-type flowers (Figure 2A, E; Figure 3A, E). Discrete domains of the abscission zone cells that remain with the plant body are present for the petals and stamens, along with a band of sepal abscission zone cells that encircles the receptacle. Placement of the sepal abscission zones is altered in as1 flowers (stage 17). Instead of developing at the sepal base, abscission zones are usually formed in an inverted V-shape within the proximal regions of as1 medial and lateral sepals (Figure 2B-D, F-H; Figure 3B; Additional file 3: Figure S1). When the sepals are shed, triangular regions of tissue remain attached at the bases of as1 flowers (Figure 2D, F, H; Figure 3C, F, G; Additional file 3: Figure S1F). Displacement of the medial sepal abscission zones (Figure 2D; Figure 3B-D) is more pronounced relative to that of the lateral sepal abscission zones (Figure 2F; Figure 3F-H). The distalmost position of as1-20 medial abscission zones is 279 ± 15 μm (n = 5) from the sepal base, compared to 133 ± 13 μm (n = 4) for as1-20 lateral abscission zones. In as1-1 flowers, the medial sepal abscission zones were displaced 268 ± 34 μm (n = 7) compared to 107 ± 11 μm (n = 6) for the lateral abscission zones.
Figure 2.
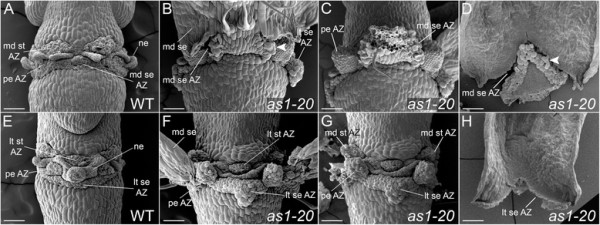
The positions of the sepal and petal abscission zones are altered inas1mutant flowers. Scanning electron micrographs of wild-type and mutant flowers (stage 17). Medial (A-D) and lateral (E-H) views are shown. A/E: After the outer organs are shed in wild-type flowers, the sepal (se) abscission zone (AZ) appears as a continuous ring adjacent to the pedicel. Individual petal (pe) and stamen (st) AZ regions can be distinguished. The remaining AZ cells have a rounded appearance. Nectary (ne) tissue is sandwiched between the stamen and petal AZs. B: Abscission of the medial (md) sepals is delayed in as1 flowers. Cell separation first occurs at the proximal edges of an inverted V-shaped region. Cell expansion is apparent in the sepal AZ cells that remain associated with this region (see arrowhead). C: Flower shown in (B) with medial sepal removed. Stubs of tissue are visible at the petal attachment sites. D: Inner view of an as1 medial sepal. Expansion of cells within an inverted V-shaped AZ is apparent (see arrowhead). F: Rounded cells are present at the tips of the stubs found at the petal attachment sites. Triangular-shaped regions of tissue are present at the lateral (lt) sepal attachment sites. G: Flower shown in (F) with the medial sepals removed. Positioning of the lateral stamen AZs in as1 flowers is not detectably altered; the medial stamen AZs appear to be slightly displaced. H: Inner view of an as1 lateral sepal (stage 16). An inverted V-shaped AZ is present. Scale bars, 100 μm.
Figure 3.

Mutations inAS1alter the proximal domains of the medial sepals. Medial (A-D, I-L) and lateral (E-H) views of wild-type and as1 mutant flowers (stage 17). A/E: Wild-type flowers. B-D: Detachment of the medial sepals of as1-20(B) and as1-1(C) flowers usually occurs at inverted V-shaped abscission zones (AZ). The triangular-shaped regions of proximal zone (PZ) tissue that remain behind do not senesce. In some as1 flowers (D), the sides of the sepals abscise along the edges of larger proximal-distal zones (PDZ) that remain green. F-H: Shedding of the lateral sepals and petals of as1-20(F/G) and as1-1(H) flowers leaves behind tissue stubs that remain green. I/J: The altered PZs of as1-20(I) and as1-1(J) medial sepals can be visualized during the extended senescence that occurs prior to organ abscission. The constriction of growth associated with the sepal-receptacle boundary in wild-type flowers is located at the base of the altered PZs (see arrowhead, I). K/L: Substantially larger, hourglass-shaped PDZs are observed in some as1-20(K) and as1-1(L) medial sepals. Scale bars, 200 μm.
The positions of the petal abscission zones are also shifted in as1 flowers (Figure 2C, F; Figure 3F-H; Additional file 3: Figure S1). After abscission, the tissue stubs that remain at the petal attachment sites are 126 ± 18 μm (n = 8) for as1-20 flowers and 92 ± 8 μm (n = 8) for as1-1 flowers. While placement of the lateral stamen abscission zones appears to be unaffected in as1 flowers (Figure 2F), small tissue projections are sometimes observed at the attachment sites for the medial stamens (Figure 2G).
The proximal regions of as1 medial sepals show an altered identity
The outer organs of wild-type flowers begin to wither and senesce shortly before abscission (stage 16), but are still turgid when they detach [39]. In as1 flowers (stage 17), due to the delay in abscission of the medial sepals, two distinctive patterns of chlorophyll loss are observed after the extended senescence period (Figure 3B, D, I-L). The primary pattern is that chlorophyll is retained in a triangular-shaped proximal zone and lost elsewhere in as1 medial sepals (Figure 3I, J). Larger proximal-distal green zones are also observed in some as1 medial sepals (Figure 3K, L). Of 31 as1-1 medial sepals surveyed, 19 (61%) had green proximal zones with a triangular shape (Figure 3I, J). The other 12 (39%) had green proximal-distal zones with an hourglass shape (Figure 3K, L). If the neck of the altered proximal-distal zone is narrow, only the triangular-shaped proximal region remains attached after abscission (data not shown). If it is wide, the sides of the medial sepal begin to abscise, starting from the proximal edges (Figure 3D).
Establishment of the medial sepal-receptacle boundary is altered in as1 flowers
In wild-type flowers, a crease marking the future sepal-receptacle boundary is apparent at the bases of the sepal primordia by stage 8 (Figure 4A; stages assigned according to [31]). As development continues (stage 9), constriction of growth in this region compared to the cells found in the sepal primordia above and floral receptacle below (Figure 4C) results in the formation of a clearly defined boundary (Figure 4E, stage 12) [20],[40]. In as1 mutant flowers (stage 8), the position of the initial crease is distally displaced at the midpoints of the medial sepal primordia (Figure 4B). Although some growth suppression is evident (Figure 4D, stage 9), the medial sepal-receptacle boundary regions of as1 flowers (Figure 4F, stage 12) are disorganized and less pronounced than those of wild-type (Figure 4E). As as1 flowers mature (stage 15), the slight displacement of this boundary in the medial sepals can still be detected (Additional file 4: Figure S2B) compared to wild-type (Additional file 4: Figure S2A). The lateral sepal-receptacle boundary regions of as1 flowers (stage 15) resemble those of wild-type flowers (Additional file 4: Figure S2E, F).
Figure 4.

Mutations inAS1affect the medial sepal-receptacle boundary. Scanning electron micrographs of wild-type and mutant sepal primordia. Boundary regions between the medial sepal (md se) primordia and underlying flower pedicels (pd) are indicated by arrowheads. A: In wild-type flowers (stage 8), a crease marks the site of the future sepal-receptacle (distal pedicel) boundary. B: This incipient boundary is shifted in as1 medial sepal primordia (stage 8). C: As wild-type flowers (stage 9) develop, the surface of the sepal is uniformly rounded. D: The central, proximal region of as1 sepal primordia (stage 9) protrudes from the surface. E: A well-defined boundary between the sepals and receptacle is present in wild-type flowers (stage 12). Trichomes are not observed in the proximal regions of sepals. F: The boundary between the medial sepals and pedicel is altered in as1 flowers (stage 12). Trichomes (see arrow, also in (C)) are frequently observed in the central, proximal regions of as1 medial sepals. Scale bars, 100 μm.
Whereas wild-type sepal primordia are rounded (Figure 4C, E), the central-proximal domains of as1 medial sepal primordia become ridged (Figure 4D, F). Trichomes are frequently present in these regions of as1 sepals (Figure 4B, F; Figure 3J, L; Figure 2B), and are not observed in the corresponding areas of wild-type sepals (Figure 4A, C, E). The altered appearance of the proximal domains of as1 medial sepals corresponds with the regions that retain chlorophyll as the rest of the sepal senesces (Figure 3I-J). Abscission occurs along the distal edges of these proximal zones (Figure 3B; Figure 2D) rather than coinciding with the regions where growth is constricted at their bases (Figure 2B; Figure 3I).
The abscission zone defects of as1 flowers are BP-dependent
To determine whether BP activity contributes to the displaced abscission zones of as1 flowers, we generated the as1-20 bp-1 double mutant. bp-1 is a null allele due to a deletion that spans the entire locus [20],[24]. As previously reported for the as1-1 bp-2 double mutant [23], the overall appearance of the as1-20 bp-1 inflorescences is largely additive with respect to those of the single mutants. Like the bp-1 mutant (Figure 5A, E, I) [24], as1-20 bp-1 mutant inflorescences have abbreviated internodes and slender, short, downward-pointing pedicels (Figure 5B, F, J). As in the as1-20 mutant (Figure 1D), the reduced size of the sepals and petals of as1-20 bp-1 flowers results in premature bud opening and exposure of the developing stamens and carpels (Figure 5B).
Figure 5.
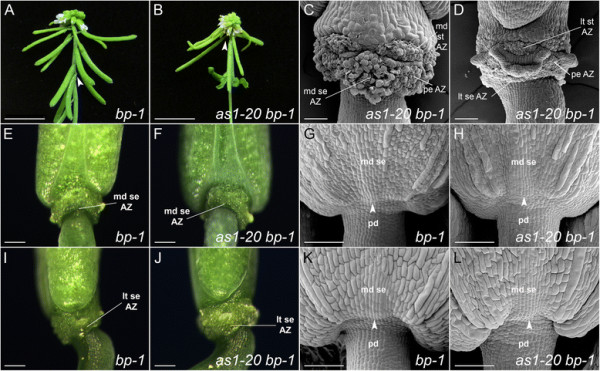
Loss ofBPrestores abscission zone placement and the position of the medial sepal-receptacle boundary inas1mutant flowers. A: bp inflorescence. The flower pedicels are short, slender and point downward. B: as1 bp inflorescence. Abscission of the medial sepals is not delayed. C/D: Scanning electron micrographs of as1 bp flowers (stage 17) showing medial (C) and lateral (D) views. Placement of the abscission zones in as1 bp flowers (D) is similar to that of wild-type (see Figure 2A, E). The abscission zones of as1 bp flowers (C), as previously reported for bp-101, bp-3 and bp-10 flowers [21],[25], show increased cell expansion during fruit development. E/F/I/J: Medial (E/F) and lateral (I/J) views of bp and as1 bp flowers (stage 17). The positions of the enlarged abscission zones of bp(E/I) and as1 bp(F/J) flowers are similar to those of wild-type flowers (see Figure 3A, E). G/H/K/L: Scanning electron micrographs of bp and as1 bp medial sepal primordia. Boundary regions between the medial sepal primordia and underlying flower pedicels are indicated by arrowheads. G: In bp flowers (stage 9), the pedicels are slender and the receptacle region beneath the floral organs fails to expand. H: Displacement of the sepal-receptacle boundary is not evident in as1 bp flowers (stage 9) or bp flowers (G). K/L: Due to radial constriction of the pedicel, the boundary between the sepals and receptacle is less distinct in bp(K) and as1 bp(L) flowers (stage 11). Scale bars: 1 cm (A/B); 100 μm (C/D/G/H/K/L); 200 μm (E/F/I/J).
Loss of BP activity is, however, sufficient to restore the positions of the sepal and petal abscission zones in as1 flowers (Figure 5C, D, F, J) as well as the timing of medial sepal abscission (Figure 5B). As in wild-type flowers, distinct abscission zones are initially observed in as1 bp flowers after organ shedding (Figure 5D; Figure 2A, E). As fruit maturation progresses (stage 17), the sepal abscission zones of as1 bp flowers become enlarged and have a disorganized appearance (Figure 5C). This result is consistent with the abscission zone enlargement that is characteristic of bp flowers [21],[25].
In younger as1 bp and bp flowers (stage 9), the positions of the sepal-receptacle boundaries are not distally displaced (Figure 5G, H) as they are in as1 flowers (Figure 2B, D). In addition, the central-proximal regions of as1 bp medial sepals are not ridged, nor do they contain ectopic trichomes (Figure 5H, L). These results suggest that the altered identity of the proximal zone and the shifted sepal-pedicel boundary in as1 flowers are also BP-dependent.
Due to radial constriction of the receptacle, the creases at the bases of bp and as1 bp sepal primordia (stage 11) are notably less distinct (Figure 5K, L) than those of wild-type flowers (Figure 2E). By promoting expansion of the receptacle in wild-type flowers, BP activity influences the definition of the sepal-pedicel boundary.
Expression of the HAE::GUS abscission zone marker is altered in as1 flowers
We have presented morphological evidence that the placement of sepal and petal abscission zones is shifted in as1 mutant flowers (Figures 2 and 3; Additional file 3: Figure S1). To examine the profile of a known marker of floral organ abscission zones in as1 flowers, we crossed the as1-20 mutant to a transgenic plant carrying a fusion of the HAE promoter to the β-Glucuronidase (GUS) reporter gene [32],[41]. In wild-type HAE::GUS flowers (stage 17), expression of GUS persists in the remaining floral organ abscission zone cells in discrete domains that mark the sepal, petal and stamen attachment sites (Figure 6A, E). In as1-20 flowers (stage 17), HAE regulatory regions usually direct GUS expression in an inverted V-shape in the proximal zone of the medial sepals (Figure 6B). Like the delayed progression of cell separation (Figure 2B, D; Figure 3B, D), expression of this marker originates at the proximal edges of as1-20 medial sepals (Additional file 5: Figure S3A, B) and usually expands distally until the lines intersect at the sepal midpoint (Additional file 5: Figure S3C-F; Figure 6B). GUS expression is also found in the tissue remnants that remain at the petal and lateral sepal attachment sites of as1-20 flowers (Figure 6B, F).
Figure 6.
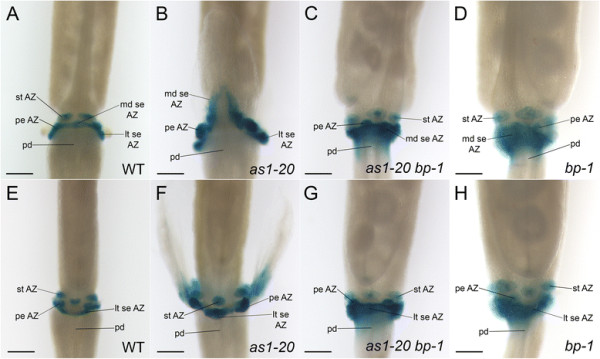
Expression of theHAE::GUSabscission zone marker is displaced inas1medial sepals. Images of wild-type and mutant flowers (stage 17) histochemically stained for β-Glucuronidase (GUS) activity. Medial (A-D) and lateral (E-H) views are shown. A/E: In wild-type flowers, HAE regulatory regions direct expression of GUS in discrete domains of the remaining sepal (se), petal (pe) and stamen (st) abscission zone cells. B/F: In as1-20 flowers, GUS is expressed in an inverted V-shaped proximal region of the medial sepals (B), and in the remaining petal and lateral sepal stubs (B/F). C/D/G/H: Expression of GUS expands beyond the enlarged sepal abscission zones of as1-20 bp-1(C/G) and bp-1(D/H) flowers into the distal pedicels. Scale bars: 200 μm.
We have shown that loss of BP activity is sufficient to restore the positions of the sepal and petal abscission zones in as1-20 flowers. To determine whether the expression profile of the HAE::GUS abscission zone marker is also rescued in as1-20 bp-1 flowers, mutants carrying this marker were generated. We discovered that, as in wild-type flowers (Figure 6A, E), GUS is expressed in the remaining sepal and petal abscission zone cells at the bases of as1-20 bp-1 (Figure 6C, G) as well as bp-1 (Figure 6D, H) flowers (stage 17). Interestingly, the band of GUS expression corresponding to the sepal abscission zone cells is conspicuously wider and diffuses into the distal pedicel of both as1-20 bp-1 (Figure 6C, G) and bp-1 flowers (Figure 6D, H), in correspondence with the enlarged abscission zones of these mutant flowers.
Taken together, these results provide molecular evidence that sepal and petal abscission zones are distally displaced in as1 flowers, and that this displacement is BP-dependent.
Discussion
We report here a novel role for the AS1 MYB transcription factor in establishing the positions of the sepal and petal abscission zones. Three aspects of the altered pattern of sepal abscission in as1 flowers are noteworthy. First, maximal abscission zone displacement occurs at the midpoint of each as1 sepal (Figures 2 and 3). Second, the abscission zone midpoints of the medial sepals are shifted distally about twice as far as those of the lateral sepals (Figure 2B, F). Third, expression of the HAE::GUS abscission zone marker is not only displaced but shows a late onset in as1 medial sepals (Additional file 5: Figure S3), which is mirrored by a significant delay in abscission (Figure 1D, E). These results suggest that the loss of AS1 activity primarily affects the position of abscission zone cell specification, and that the degree of displacement is influenced by orientation with respect to the inter-sepal boundary regions as well as the medial-lateral axis of the flower (Figure 1B). The delay in detachment of as1 medial sepals may represent an alteration in either the timing of abscission zone specification or of the subsequent activation of cell separation.
Our analysis of as1 flowers reveals that proximal zones within the medial sepals have an altered identity. As the attached organs senesce (stage 17), a distinct demarcation appears between proximal regions that remain green and the rest of the organ that loses its color as chlorophyll is catabolized (Figure 3I-L). Trichomes are frequently observed in the central, ridged proximal zones of as1 medial sepals (Figure 2B; Figure 4B, F) and are not found within the corresponding rounded areas of wild-type sepals (Figure 4 A, C, E). The displaced abscission zones of as1 medial sepals outline the distal edges of these altered proximal zones (Figure 3B). The region of constricted growth that defines the medial sepal-receptacle boundary of wild-type flowers [40],[42] is disorganized and slightly shifted at the bases of as1 medial sepals (Figure 4; Additional file 4: Figure S2; Figure 2B). It is intriguing that the growth suppression associated with this boundary can be uncoupled from abscission zone differentiation in as1 flowers. These results suggest that the sepal-receptacle boundary is disrupted in as1 flowers but not completely dismantled.
Displacement of the sepal and petal abscission zones in as1 flowers is BP-dependent (Figures 5 and 6). In wild-type flowers, the AS1-AS2 repressor complex restricts the BP expression domain to the pedicel, receptacle and organ boundary regions of the floral organ primordia [13]. Although the combined loss of BP, KNAT2 and KNAT6 activity is required to restore normal sepal and petal size in as1 flowers [16], loss of BP alone is sufficient to rescue the shifted position of the sepal-receptacle boundary and the altered features of the proximal zone associated with the displaced abscission zones of as1 medial sepals (Figure 5).
An interesting parallel to our results is the AS1-mediated regulation of BP expression during fruit patterning. Arabidopsis fruit are composed of two carpel valves attached to a medial replum [43]. Seed dispersal is facilitated by dehiscence zones that form at the valve-replum boundaries [30]. Within the developing gynoecium, AS1 restricts expression of a BP::GUS marker to the region that will become the replum [44]. Compared to wild-type fruit, as1 mutant fruit contain a wider replum and narrower valves, and show expansion of the replum domain of BP expression as well as ectopic expression of BP in the valves [44]. Loss of BP activity was found to partially restore the respective sizes of the repla and valves in as1 bp fruit [44]. Through its regulation of the valve-replum boundary, AS1 controls the position of the fruit dehiscence zones. In as1 fruit, the dehiscence zones are shifted laterally due to expansion of the replum [44]. We propose that AS1 similarly regulates placement of the floral organ-receptacle boundaries and thereby the positions of the sepal and petal organ abscission zones.
BP plays dual roles in regulating organ abscission. In addition to influencing the specification sites of abscission zone cells, BP prevents premature abscission by inhibiting the cell separation phase of abscission [25]. Signaling through the IDA-HAE/HSL2 pathway has been proposed to relieve the BP-mediated repression of organ abscission [25]. bp and as1 bp flowers develop enlarged, disorganized sepal abscission zones (Figure 5C, E) that resemble those of flowers constitutively expressing IDA [21],[25],[26]. We have found that expression of a HAE::GUS marker expands into the receptacles of bp and as1 bp flowers (Figure 6C, D, G, H). These results suggest that BP may control sepal abscission zone size by restricting HAE expression from the receptacle.
Our study provides fresh evidence to support the links between organ boundary formation and abscission zone development. Unlike ATH1 and BOP1/BOP2, which are required for some or all of the floral organ abscission zones to form [8],[9], AS1 and BP appear to play a more indirect role in abscission zone specification by regulating their positions. Our observation that displacement of the sepal abscission zones in as1 flowers is less affected at the sepal margins than at the sepal midpoint (Figure 2D, H) suggests that factors active at the inter-sepal boundaries also influence abscission zone differentiation. The growth suppression that occurs at inter-organ boundaries has been previously found to affect the establishment of lateral organ boundaries. Flowers with mutations in the CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 genes have fused sepals, but their sepal-receptacle boundaries are not altered [45]. However, reduction of CUC activity further obscures the definition of the sepal-receptacle boundaries of ath1 flowers [9]. Further dissection of the complex regulatory networks that establish lateral and inter-organ boundaries is expected to uncover critical connections between the definition and positioning of the receptacle boundary, proximal-distal organ patterning, and floral organ abscission zone development.
Conclusions
We have identified a novel role for AS1 in establishing the positions of the sepal and petal abscission zones. In as1 mutant flowers, the sepal and petal abscission zones are displaced distally, detachment of the medial sepals is significantly delayed, and proximal domains within the medial sepals show an altered identity. Loss of BP activity rescues the abscission zone and proximal domain defects of as1 flowers. Our results suggest that further advances in understanding the process of floral organ abscission zone development can be made by analyzing the network of genes known to control organ boundaries and proximal-distal patterning in Arabidopsis leaves.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SL conceived and coordinated this study. CG, MC, WG and SL performed SEMs. CG photographed plants and carried out marker analyses. DW and WG sequenced the as1-21 allele. CG, MC, WG and DW prepared figures; SL wrote the manuscript. All authors read and approved the final manuscript.
Additional files
Supplementary Material
Alleles of ASYMMETRIC LEAVES1 (AS1) that alter organ abscission [12],[37].
AS1 oligos used in this study.
Shedding of the lateral sepals is delayed in some as1 flowers. Medial (A/B/E-H) and lateral (C/D) views of mutant flowers (stage 17). Shedding of the medial sepals is delayed in as1-21(A/B), as1-101(E), as1-23(F), and as1-22(G/H) flowers. Shedding of the lateral sepals is delayed in some as1-21(B/D), as1-23(F), and as1-22 flowers. Of 10 flowers (early to mid stage 17) surveyed per genotype after light touching, all of the medial sepals (20 of 20) remained attached in as1-21 and as1-23 flowers, while 75% (15 of 20) and 70% (14 of 20) remained in as1-22 and as1-101 flowers, respectively. None of the lateral sepals remained attached in as1-101 (0 of 20) or as1-1 (0 of 60) flowers; 20% (4 of 20), 15% (3 of 20), and 15% (3 of 20) remained in as1-21, as1-22, and as1-23 flowers, respectively. Scale bars, 200 μm.
Displacement of the receptacle boundary is not detected in the lateral sepals of as1-20 flowers. Medial (A, B) and lateral (C, D) views of wild-type and as1-20 flowers (stage 15). In comparison with wild-type flowers (A/C), placement of the sepal-pedicel boundary is affected in the medial (B, see arrow) but not the lateral (D) sepals of as1-20 flowers. Scale bars, 200 μm.
Expression of the HAE::GUS marker progresses distally from the proximal edges of as1 medial sepals. Medial views of as1-20 mutant flowers (stage 17) histochemically stained for β-Glucuronidase (GUS) activity. Expression of GUS initiates at the proximal margins of the medial sepals (A, B). Stripes of GUS expression extend from each origin in a distal direction (B, C) until they intersect (D) to outline the edges of an inverted V-shaped proximal domain. In some as1-20 medial sepals (E, F), stripes of GUS expression expand distally toward the sepal tip without intersecting in a proximal region.
Contributor Information
Catherine M Gubert, Email: cmgubert@olemiss.edu.
Megan E Christy, Email: mechris2@go.olemiss.edu.
Denise L Ward, Email: deniseward1963@gmail.com.
William D Groner, Email: wdgroner@gmail.com.
Sarah J Liljegren, Email: liljegren@olemiss.edu.
Acknowledgments
We thank Brad Jones and Sara Patterson for helpful discussions, Lou Boykins at the University of Memphis Integrated Microscopy Center for assistance with scanning electron microscopy, Charles McCrory for technical assistance, and the Arabidopsis Biological Resource Center for providing seeds. This research was supported by University of Mississippi startup funds and National Science Foundation grants to SL (IOS-1239311) and the Mississippi EPSCoR program (EPS-0903787). MC was supported by the Mississippi EPSCoR community college research program; DW was supported by the Ronald E. McNair Scholars program at the University of Mississippi.
References
- Niederhuth CE, Cho SK, Seitz K, Walker JC. Letting go is never easy: abscission and receptor-like protein kinases. JIPB. 2013;55:1251–1263. doi: 10.1111/jipb.12116. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ. Organ abscission: exit strategies require signals and moving traffic. Curr Opin Plant Biol. 2012;15:670–676. doi: 10.1016/j.pbi.2012.09.012. [DOI] [PubMed] [Google Scholar]
- Van Nocker S. Development of the abscission zone. Stewart Postharvest Rev. 2009;1:5. [Google Scholar]
- Estornell LH, Agusti J, Merelo P, Talon M, Tadeo FR. Elucidating mechanisms underlying organ abscission. Plant Sci. 2013;199–200:48–60. doi: 10.1016/j.plantsci.2012.10.008. [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Zhang Y, McKim S, Li X, Haughn GW. BLADE-ON-PETIOLE-dependent signaling controls leaf and floral patterning in Arabidopsis. Plant Cell. 2005;17:1434–1448. doi: 10.1105/tpc.104.030536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norberg M, Holmlund M, Nilsson O. The BLADE ON PETIOLE genes act redundantly to control the growth and development of lateral organs. Development. 2005;132:2203–2213. doi: 10.1242/dev.01815. [DOI] [PubMed] [Google Scholar]
- Jun JH, Ma CM, Fletcher JC. BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell. 2010;22:62–76. doi: 10.1105/tpc.109.070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim SM, Stenvik GE, Butenko MA, Kristiansen W, Cho SK, Hepworth SR, Aalen RB, Haughn GW. The BLADE-ON-PETIOLE genes are essential for abscission zone formation in Arabidopsis. Development. 2008;135:1537–1546. doi: 10.1242/dev.012807. [DOI] [PubMed] [Google Scholar]
- Gomez-Mena C, Sablowski R. ARABIDOPSIS THALIANA HOMEOBOX GENE1 establishes the basal boundaries of shoot organs and controls stem growth. Plant Cell. 2008;20:2059–2072. doi: 10.1105/tpc.108.059188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Carranza ZH, Rompa U, Peters JL, Bhatt AM, Wagstaff C, Stead AD, Roberts JA. HAWAIIAN SKIRT: an F-box gene that regulates organ fusion and growth in Arabidopsis. Plant Physiol. 2007;144:1370–1382. doi: 10.1104/pp.106.092288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature. 2000;408:967–971. doi: 10.1038/35050091. [DOI] [PubMed] [Google Scholar]
- Hazen SP, Borevitz JO, Harmon FG, Pruneda-Paz JL, Schultz TF, Yanovsky M, Liljegren SJ, Ecker JR, Kay SA. Rapid array mapping of circadian clock and developmental mutations in Arabidopsis. Plant Physiol. 2005;138:990–997. doi: 10.1104/pp.105.061408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development. 2000;127:5523–5532. doi: 10.1242/dev.127.24.5523. [DOI] [PubMed] [Google Scholar]
- Guo M, Thomas J, Collins G, Timmermans MCP. Direct repression of KNOX loci by the ASYMMETRIC LEAVES1 complex in Arabidopsis. Plant Cell. 2008;20:48–58. doi: 10.1105/tpc.107.056127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Li Z, Zhu Y, Wang H, Ma H, Dong A, Huang H. Arabidopsis genes AS1, AS2, and JAG negatively regulate boundary-specifying genes to promote sepal and petal development. Plant Physiol. 2008;146:566–575. doi: 10.1104/pp.107.113787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezaki M, Kojima M, Sakakibara H, Kojima S, Ueno Y, Machida C, Machida Y. Genetic networks regulated by ASYMMETRIC LEAVES1 (AS1) and AS2 in leaf development in Arabidopsis thaliana: KNOX genes control five morphological events. Plant J. 2009;61:70–82. doi: 10.1111/j.1365-313X.2009.04033.x. [DOI] [PubMed] [Google Scholar]
- Phelps-Durr TL, Thomas J, Vahab P, Timmermans MC. Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell. 2005;17:2886–2898. doi: 10.1105/tpc.105.035477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Yu C-W, Chen F-F, Zhao L, Tian G, Liu X, Cui Y, Yang J-Y, Wu K. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in Arabidopsis. PLoS Genet. 2012;8:e1003114. doi: 10.1371/journal.pgen.1003114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodha M, Marco CF, Timmermans MCP. The ASYMMETRIC LEAVES complex maintains repression of KNOX homeobox genes via direct recruitment of Polycomb-repressive complex2. Genesx Dev. 2013;27:596–601. doi: 10.1101/gad.211425.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SJ, Riggs CD. Pedicel development in Arabidopsis thaliana: contribution of vascular positioning and the role of the BREVIPEDICELLUS and ERECTA genes. Dev Biol. 2005;284:451–463. doi: 10.1016/j.ydbio.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Wang X-Q, Xu W-H, Ma L-G, Fu Z-M, Deng X-W, Li J-Y, Wang Y-H. Requirement of KNAT1/BP for the development of abscission zones in Arabidopsis thaliana. JIPB. 2006;48:15–26. [Google Scholar]
- Ragni L, Belles-Boix E, Günl M, Pautot V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell. 2008;20:888–900. doi: 10.1105/tpc.108.058230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development. 2002;129:1957–1965. doi: 10.1242/dev.129.8.1957. [DOI] [PubMed] [Google Scholar]
- Venglat SP, Dumonceaux T, Rozwadowski K, Parnell L, Babic V, Keller W, Martienssen R, Selvaraj G, Datla R. The homeobox gene BREVIPEDICELLUS is a key regulator of inflorescence architecture in Arabidopsis. PNAS. 2002;99:4730–4735. doi: 10.1073/pnas.072626099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CL, Stenvik GE, Vie AK, Bones AM, Pautot V, Proveniers M, Aalen RB, Butenko MA. Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell. 2011;23:2553–2567. doi: 10.1105/tpc.111.084608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Butenko MA, Urbanowicz BR, Rose JK, Aalen RB. Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell. 2006;18:1467–1476. doi: 10.1105/tpc.106.042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell. 2008;20:1805–1817. doi: 10.1105/tpc.108.059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SK, Larue CT, Chevalier D, Wang H, Jinn TL, Zhang S, Walker JC. Regulation of floral organ abscission in Arabidopsis thaliana. PNAS. 2008;105:15629–15634. doi: 10.1073/pnas.0805539105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313X.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000;404:766–770. doi: 10.1038/35008089. [DOI] [PubMed] [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie ME, Lewis MW, Youn J-Y, Daniels MJ, Liljegren SJ. The EVERSHED receptor-like kinase modulates floral organ shedding in Arabidopsis. Development. 2010;137:467–476. doi: 10.1242/dev.041335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D. LEAFY expression and flower initiation in Arabidopsis. Development. 1997;124:3835–3844. doi: 10.1242/dev.124.19.3835. [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr Opin Plant Biol. 2001;4:447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:1360–1385. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Sun Y, Zhou Q, Zhang W, Fu Y, Huang H. ASYMMETRIC LEAVES1, an Arabidopis gene that is involved in control of cell differentiation in leaves. Planta. 2002;214:694–702. doi: 10.1007/s004250100673. [DOI] [PubMed] [Google Scholar]
- Ko ER, Ko D, Chen C, Lipsick JS. A conserved acidic patch in the Myb domain is required for activation of an endogenous target gene and for chromatin binding. Mol Cancer. 2008;7:77. doi: 10.1186/1476-4598-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S-C, Fernandez DE. Effect of regulated overexpression of the MADS domain factor AGL15 on flower senescence and fruit maturation. Plant Physiol. 2002;130:78–89. doi: 10.1104/pp.004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Tasaka M. Genetic control of shoot organ boundaries. Curr Opin Plant Biol. 2006;9:72–77. doi: 10.1016/j.pbi.2005.11.011. [DOI] [PubMed] [Google Scholar]
- Jinn TL, Stone JM, Walker JC. HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 2000;14:108–117. [PMC free article] [PubMed] [Google Scholar]
- Zádníková P, Simon R. How boundaries control plant development. Curr Opin Plant Biol. 2014;17:116–125. doi: 10.1016/j.pbi.2013.11.013. [DOI] [PubMed] [Google Scholar]
- Roeder AHK, Yanofsky MF. Fruit development in Arabidopsis. Arabidopsis Book. 2006;4:e0075. doi: 10.1199/tab.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Cantabrana H, Ripoll JJ, Ochando I, Vera A, Ferrándiz C, Martínez-Laborda A. Common regulatory networks in leaf and fruit patterning revealed by mutations in the Arabidopsis ASYMMETRIC LEAVES1 gene. Development. 2007;134:2663–2671. doi: 10.1242/dev.02864. [DOI] [PubMed] [Google Scholar]
- Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M. Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell. 1997;9:841–857. doi: 10.1105/tpc.9.6.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Alleles of ASYMMETRIC LEAVES1 (AS1) that alter organ abscission [12],[37].
AS1 oligos used in this study.
Shedding of the lateral sepals is delayed in some as1 flowers. Medial (A/B/E-H) and lateral (C/D) views of mutant flowers (stage 17). Shedding of the medial sepals is delayed in as1-21(A/B), as1-101(E), as1-23(F), and as1-22(G/H) flowers. Shedding of the lateral sepals is delayed in some as1-21(B/D), as1-23(F), and as1-22 flowers. Of 10 flowers (early to mid stage 17) surveyed per genotype after light touching, all of the medial sepals (20 of 20) remained attached in as1-21 and as1-23 flowers, while 75% (15 of 20) and 70% (14 of 20) remained in as1-22 and as1-101 flowers, respectively. None of the lateral sepals remained attached in as1-101 (0 of 20) or as1-1 (0 of 60) flowers; 20% (4 of 20), 15% (3 of 20), and 15% (3 of 20) remained in as1-21, as1-22, and as1-23 flowers, respectively. Scale bars, 200 μm.
Displacement of the receptacle boundary is not detected in the lateral sepals of as1-20 flowers. Medial (A, B) and lateral (C, D) views of wild-type and as1-20 flowers (stage 15). In comparison with wild-type flowers (A/C), placement of the sepal-pedicel boundary is affected in the medial (B, see arrow) but not the lateral (D) sepals of as1-20 flowers. Scale bars, 200 μm.
Expression of the HAE::GUS marker progresses distally from the proximal edges of as1 medial sepals. Medial views of as1-20 mutant flowers (stage 17) histochemically stained for β-Glucuronidase (GUS) activity. Expression of GUS initiates at the proximal margins of the medial sepals (A, B). Stripes of GUS expression extend from each origin in a distal direction (B, C) until they intersect (D) to outline the edges of an inverted V-shaped proximal domain. In some as1-20 medial sepals (E, F), stripes of GUS expression expand distally toward the sepal tip without intersecting in a proximal region.


