SUMMARY
Hippo signaling is a tumor suppressor pathway involved in organ size control and tumorigenesis, through the inhibition of YAP and TAZ. Here we show that energy stress induces YAP cytoplasmic retention and Ser127 phosphorylation and inhibits YAP transcriptional activity and YAP-dependent transformation. These effects require the central metabolic sensor AMP-activated protein kinase (AMPK), and the upstream Hippo pathway components Lats1/2 and Angiomotin-like 1 (AMOTL1). We further show that AMPK directly phosphorylates Ser793 of AMOTL1. AMPK activation stabilizes and increases AMOTL1 steady-state protein levels, contributing to YAP inhibition. The phosphorylation-deficient Ser793Ala mutant of AMOTL1 showed a shorter half-life and conferred resistance to energy stress-induced YAP inhibition. Our findings link energy sensing to the Hippo-YAP pathway, and suggest that YAP may integrate spatial (contact inhibition), mechanical and metabolic signals to control cellular proliferation and survival.
INTRODUCTION
Hippo signaling has been implicated in organ size control by restricting the transcriptional co-activators YAP/TAZ(Dong et al., 2007; Harvey and Tapon, 2007; Harvey et al., 2013; Pan, 2007). The core components of the pathway construct a kinase cascade, in which Mst1/2 in complex with SAV1, phosphorylate and activate Lats1/2 kinases. Subsequently, Lats1/2 kinases in complex with Mob, phosphorylate YAP/TAZ, leading to their cytoplasmic retention and inhibition (Harvey et al., 2003; Udan et al., 2003; Wu et al., 2003). Inhibition of this kinase cascade leads to dephosphorylation of YAP/TAZ, and their accumulation in the nuclei. Nuclear YAP/TAZ bind to TEA domain transcription factors (TEAD), and promote the expression of target genes, and modulate diverse cellular functions, including proliferation, apoptosis, migration, and differentiation(Harvey et al., 2013; Hong and Guan, 2012). Consistently, loss-of-function mutations of upstream Hippo pathway components, or overexpression of YAP, lead to tissue expansion and tumorigenesis in many tissues (Pan, 2010; Schlegelmilch et al., 2011; Zhou et al., 2009; Zhou et al., 2011).
Previously, it has been shown that multiple upstream signals, such as cell-cell contact(Zhao et al., 2007), mechanical forces and cytoskeletal reorganization(Dupont et al., 2011; Zhao et al., 2012), and serum lipids and their receptors(Miller et al., 2012; Yu et al., 2012), could modulate YAP localization and Ser127 phosphorylation through Hippo pathway kinases dependent or independent mechanisms. To identify novel modulators of the Hippo-YAP pathway, we developed a high-content imaging assay using HEK293A cells to directly visualize the nuclear localization of endogenous YAP. YAP nuclear localization can be quantified by the Pearson’s correlation coefficient with the nuclear staining, providing a sensitive and robust cellular assay to study the regulation of YAP (Figure S1A). Using such a system, we have discovered that energy stress and inhibition of glucose metabolism could inhibit YAP, providing new molecular mechanisms linking cellular metabolism to tumorigenesis.
RESULTS
Energy stress induces YAP cytoplasmic retention, Ser127 phosphorylation and inhibits its transcriptional activity
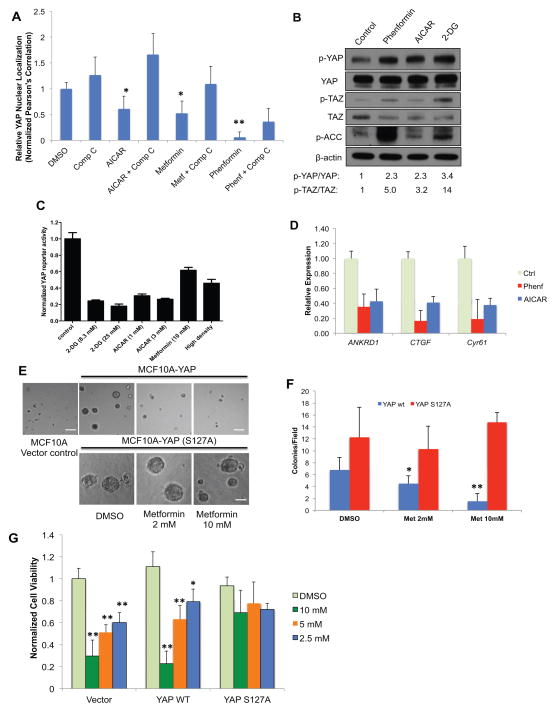
Previously, we and others have discovered that serum deprivation significantly induces YAP cytoplasmic retention through serum lipids. To further study whether other nutrient and energy stress signals could modulate YAP, we screened a set of small molecule compounds known to modulate nutrient and energy sensing pathways, including inhibitors of glucose metabolism and ATP production, PI3K/AKT/mTOR signaling, and growth factor signaling, in the YAP translocation assay. In serum stimulated, confluent HEK293A cells, we observed that treatment of the mitochondrial complex I inhibitor metformin (Glucophage) and its more potent analogue phenformin, inhibits YAP nuclear localization within 3 hr (Figure 1A, and Figure S1B). Metformin and phenformin lower cellular ATP levels, increase AMP to ATP ratio in the cell, and activate AMP-activated protein kinase (AMPK), a central cellular metabolic sensor(Hardie, 2007; Mihaylova and Shaw, 2011). Consistently, we also found that treatment with 5-aminoimidazole-4-carboxamide-1-β-riboside (AICAR), a precursor of ZMP which acts as an AMP mimetic and direct activator of AMPK(Shackelford and Shaw, 2009), has similar effects on YAP cytoplasmic retention. AICAR also potently inhibits YAP nuclear localization in cells cultured at low density (Figure S1C). In HaCaT keratinocyte cells, treatment of phenformin (1mM) and AICAR (1mM) for 6h also elevates p-YAP (S127) and p-TAZ (S89) levels (Figure 1b). Phosphorylation of a well-known AMPK substrate, acetyl-CoA carboxylase (ACC) indicates the activation of AMPK. Similarly, treatment of HEK293 cells with the specific AMPK activator A-769662, induces p-YAP (S127) (Cool et al., 2006)(Figure S1D). Furthermore, energy stressors inhibit YAP-dependent transcription using a TEAD-binding element driven luciferase reporter (MCAT-YAP-Luc) and the expression of direct YAP target genes (ANKRD1, CTGF and Cyr61) by qRT-PCR (Figure 1C, D).
Figure 1. Energy stress inhibits YAP.
A, Confluent and serum stimulated HEK293A cells were treated with DMSO control, AICAR (1mM), metformin (10mM) or phenformin (1mM), with or without Compound C (10 μM) for 3h. YAP nuclear localization is quantified by determining the Pearson’s correlation coefficient of nuclear staining. Data are represented as mean, S.D. n=3. (*, p<0.03; **, p<0.001). B, Small molecule energy stressors induce YAP S127 and TAZ Ser89 phosphorylation in HaCaT cells. HaCaT cells were treated with DMSO (control), 1 mM phenformin, 1 mM AICAR or 25 mM 2-DG for 6h. C, Inhibition of YAP-reporter activity. HEK293 YAP reporter cells were treated with DMSO control, 2-DG (8.3 or 25 mM), AICAR (1 or 3 mM) or Metformin (10 mM). (data are represented as mean, S.D. n=3). D, Energy stressors suppress YAP target gene expression. HEK293A cells were treated with DMSO (ctrl), 1mM phenformin, or 1mM AICAR for 16 hours. Relative mRNA levels are normalized to DMSO (ctrl). E, MCF10A cells transduced with a vector control, YAP wild type or YAP S127A mutant were grown on Matrigel without EGF. Cells were treated with metformin (2mM or 10mM) or DMSO control. Scale bar: 100 μm. F, The colony numbers per field are quantified by Image J, and shown in bar graph (data are represented as mean, SD, n=4; *, p<0.05; **, p<0.0005 by comparing to the S127A mutant samples with the same treatment). G, Metformin dose dependently inhibits the proliferation of primary mouse hepatocellular carcinoma (HCC) cell line JF001 (Mst1−/−Mst2−/−). JF001 cells transduced with vector, wild-type YAP, or YAP S127A mutant were treated as indicated. (Data are represented as mean, SD, n=3; *, p<0.05; **, p<0.001 by comparing to the DMSO control). See also Figure S1.
In addition, deprivation of glucose from the culture media and inhibition of glucose metabolism by 2-deoxy-D-glucose (2-DG) elevated p-YAP (S127) and inhibited YAP-dependent reporter (Figure 1B, C). Consistently, glucose deprivation inhibits, and adding back glucose largely rescued YAP reporter activity (Figure S1E, F). Taken together, these data indicate that energy stress and direct activation of AMPK could inhibit YAP. Similar effects were observed in other cell types, although less significantly in fibroblast cells such as NIH3T3 (Figure S1G). These effects are independent from inhibition of cell growth, as other anti-proliferative compounds, including inhibitors of PI3K, AKT, mTOR and CDK4 (wortmannin, LY294002, rapamycin and roscovitine) have little effect on p-YAP induction under similar conditions (Figure S1H, I). Taken together, these results suggest that inhibition of YAP activity by energy stress could be a conserved and direct signaling event.
Energy stress inhibits YAP-dependent transformation and cancer cell proliferation
Expression of YAP in MCF10A cells disrupts the normal acinar structures and promotes EGF-independent growth in Matrigel(Overholtzer et al., 2006; Zhang et al., 2009). To evaluate whether energy stress could inhibit YAP oncogenic activity, we tested metformin in MCF10A cells transduced with YAP, constitutively active YAP S127A mutant and a vector control. Treatment with metformin (2mM and 10mM) showed a significant and dose dependent reduction in colony number in cells expressing wild type YAP (Figure 1E, F). Although the mean colony size is decreased upon metformin treatment, this effect was not statistically significant due to high variation of the size measurement in 3-D culture (Figure S1J). The significant colony number inhibition likely reflects the inhibition of YAP-mediated cell survival rather than cell growth. In contrast, metformin did not inhibit colony formation in cells expressing YAP S127A mutant, suggesting that the effects of metformin are specifically through the inhibition of YAP nuclear localization. Consistently, the expression of YAP target genes (CTGF and Cyr61) is inhibited by metformin in wild type YAP-expressing cells, while expression of YAP S127A confers resistance (Figure S1K).
Mice lacking Mst1/2 in the liver develop aggressive YAP-dependent hepatocellular carcinoma (HCC). We tested whether energy stress would block the proliferation of HCC cells isolated from liver specific Mst1−/−Mst2−/− mice (Zhou et al., 2009). Indeed, metformin dose dependently inhibits the proliferation of an HCC cell line (JF001). Expression of YAP S127A mutant confers resistance to the compound treatment, confirming that metformin inhibits HCC proliferation through the inhibition of YAP (Figure 1G). Taken together, our results illustrate that energy stress can block YAP-dependent transformation and cancer cell proliferation.
Energy stress inhibits YAP involving AMPK, Lats1/2 kinases and the tight junction protein AMOTL1
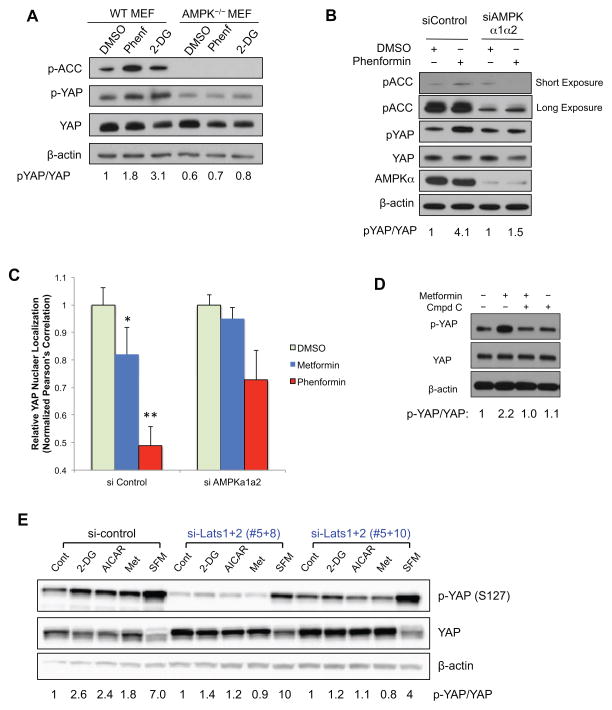
To further explore the mechanisms, we treated wild type and AMPK-null (AMPKα1−/−α2−/−) mouse embryonic fibroblasts (MEFs) with 2-DG (25mM) or phenformin (1mM) for 12h. 2-DG and phenformin increased p- ACC, as well as p-YAP (S127) in wild type MEFs, albeit to a lesser extent than in epithelial cells. In AMPK-null MEFs, p-YAP induced by phenformin and 2-DG were largely inhibited, suggesting that AMPK activity is necessary for p-YAP induction (Figure 2A). AMPK-null MEFs also have lower basal p-YAP levels compared to wild type MEFs, suggesting that loss of AMPK might lead to YAP activation. Moreover, we found that a small molecule AMPK inhibitor, Compound C, blocks AICAR, metformin and phenformin-induced YAP cytoplasmic retention (Figure 1A, and Figure S1B, C). As Compound C might also inhibit other protein kinases(Bain et al., 2007), we used siRNAs targeting AMPKα1 and α2 to confirm the effects. The siRNAs (~90% knock-down efficiency) decreased p-ACC, and greatly inhibited p-YAP induced by phenformin, suggesting that AMPK is indeed required to mediate the regulation of YAP (Figure 2B). Consistently, we also observed that siRNAs targeting AMPKα1 and α2 rescued the effects of metformin and phenformin on YAP nuclear localization (Figure 2C, Figure S2A). Taken together, these data showed that the cellular energy sensor, AMPK is required to mediate YAP regulation in response to metabolic and energy stress.
Figure 2. Energy stress inhibits YAP through AMP-activated protein kinase (AMPK) and Lats1/2.
A, AMPK is required for YAP inhibition. Wild type or AMPK−/− (AMPKα1α2 double knockout) MEFs were treated with DMSO control, 2-DG (25 mM) or phenformin (1 mM) for 12h. Cells were then harvested for western blot analysis. B, HaCaT cells were transfected with siRNAs targeting AMPKα1 and α2, and then treated for 16h with DMSO or phenformin (1mM). C, siRNAs targeting AMPKα1 and α2 blocks YAP cytoplasmic retention. Cells were transfected with siRNAs targeting AMPK, and then treated for 16h with DMSO, metformin (10mM) or phenformin (1mM). YAP nuclear localization is quantified by determining the Pearson’s correlation coefficient between YAP positive areas with the nuclear staining. Data are represented as mean, S.D. n=3. (*, p<0.05; **, p<0.001 by comparing to the DMSO control). D, HCCs were isolated from tumors derived from liver specific Mst1−/−Mst2−/− mice. Cells are treated with metformin (10mM) or co-treated with Compound C (10 μM) for 8h. p-YAP levels were evaluated by western blot. E, HEK293A cells were transfected with siRNA control or siRNA targeting Lats1 and Lats2 (#5+8, or #5+10). Cells were treated with AICAR (1mM) or Metformin (10mM) for 8h, serum free media (SFM, 3h) or 2-DG (25mM, 3h). Cells were then harvested for western blot analysis of p-YAP (Ser127). See also Figure S2.
To probe whether AMPK-mediated YAP inhibition requires upstream core Hippo pathway kinases, we first tested the effects in Mst1−/−Mst2−/− HCCs. Interestingly, metformin treatment induces and co-treatment of Compound C inhibits p-YAP (Ser127) in these cells, suggesting that Mst1/2 kinases are dispensable to inhibit YAP (Figure 2D). This is also consistent with our data that metformin blocks the proliferation of Mst1−/−Mst2−/− HCCs (Figure 1H). We then used siRNAs to silence Lats1 and Lats2 (~ 90% knockdown) (Figure S2B), which decreased the basal level of p-YAP and completely blocked the effects of energy stressors (Figure 2E), suggesting that energy stress-mediated YAP phosphorylation requires Lats1/2 kinases.
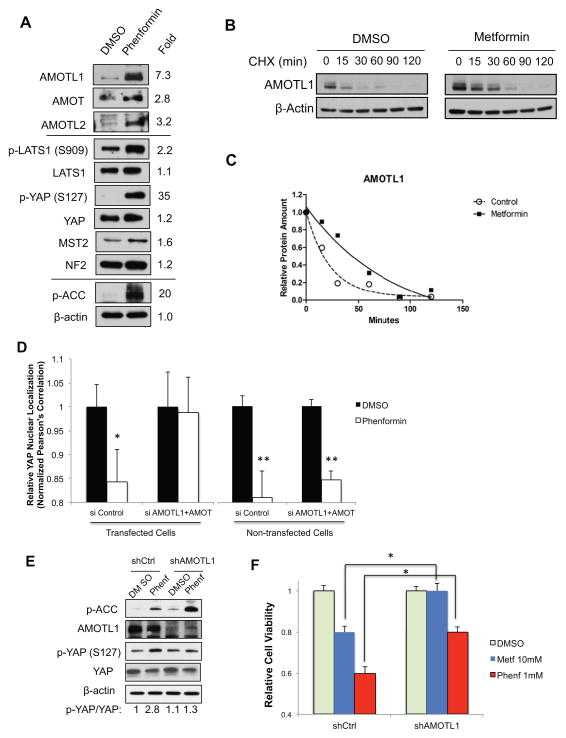
Next, we examined the changes of known Hippo pathway regulators upon energy stress. We found that phenformin treatment significantly increased the protein levels of angiomotin like 1 (AMOTL1) by 7.3-fold. The protein levels of other motin family proteins (AMOT and AMOTL2) also increased, although less significantly (Figure 3A). In total, the levels of motin proteins increased more than 12-fold upon phenformin treatment. AMOT, AMOTL1 and AMOTL2 are tight junction proteins that inhibit YAP through three mechanisms: 1) binding to and sequestering YAP out of nuclei (Yi et al., 2011; Zhao et al., 2011); 2) binding to and enhancing Lats1/2 activation(Paramasivam et al., 2011); 3) reducing the stability of YAP by promoting its ubiquitination (Adler et al., 2013a). This is consistent with our observation that, upon metabolic stress, changes in YAP translocation are more quickly detected than its Ser127 phosphorylation, suggesting that direct sequestration and enhanced Lats1/2 activity both play roles in the regulation. Indeed, we observed moderate activation of Lats1 (p-Lats1 Ser909) by 2.2 fold with phenformin treatment. Other upstream Hippo pathway components, such as NF2 and MST2 were not significantly changed. We then evaluated the mRNA levels of motin family members by qRT-PCR. We observed that phenformin treatment did not significantly increase the mRNA levels of AMOTL1 and AMOTL2, and AMOT mRNA levels increase by ~2-fold (Figure S3A). We chose to focus our studies on AMOTL1 in energy stress mediated YAP inhibition, as its changes are the most significant.
Figure 3. Endogenous angiomotin like-1 (AMOTL1) is stabilized in response to energy stress and is required to mediate AMPK-induced YAP-cytoplasmic retention and Ser127 phosphorylation.
A, HEK293A cells were treated with DMSO control or phenformin (1 mM) for 16h. Protein levels of Hippo pathway components (AMOTL1, AMOT, AMOTL2, p-YAP, YAP, p-Lats1, Lats1, MST2, and NF2) were analyzed by western blots. B, Cells were treated with metformin (10mM) for 4h and with cycloheximide (CHX) with indicated time. Protein levels of endogenous AMOTL1 were analyzed by western blots. C, Protein levels from B were quantified by densitometry of the bands. Projected degradation curve of AMOTL1 was plotted and fitted using Prism software. D, HEK293A cells were transfected with siRNAs targeting AMOT and AMOTL1, with GFP to mark transfected cells. Cells were treated with phenformin (1mM) or DMSO. YAP nuclear localization is quantified by determining the Pearson’s correlation coefficient, in GFP positive (transfected) or GFP negative (non-transfected) cells. Data are represented as mean, S.D. n=3. (*, p<0.02; **, p<0.001 by comparing to the DMSO controls). E. HaCaT cells were transfected with shRNA targeting AMOTL1. Cells were treated with DMSO control or phenformin (1mM) and p-YAP level is analyzed. F. Cells were transfected shRNA targeting AMOTL1, and then treated with DMSO, metformin (10mM) or phenformin (1mM) for 48 hours. The cell proliferation is determined by measuring the cell viability (*, p<0.01). See also Figure S3.
To test whether metabolic stress could stabilize AMOTL1, we treated cells with metformin or DMSO control, together with protein synthesis inhibitor cycloheximide, and analyzed the protein levels of endogenous AMOTL1 by western blotting at different time points (Figure 3B). We observed that the half-life of endogenous AMOTL1 increases from 15.4 min to 56.2 min with metformin treatment (Figure 3C). Therefore, energy stress might stabilize the AMOTL1 protein and thus increase its steady state protein levels, leading to YAP inhibition.
To confirm that Motin proteins are required to mediate the effects, we transfected siRNAs targeting AMOT and AMOTL1 with GFP into HEK293A cells, and treated the cells with 1mM phenformin or DMSO. The siRNAs effectively knock down AMOT and AMOTL1 (Figure S3B). In siRNA-transfected cells (GFP+), we observed that knocking down AMOT/AMOTL1 blocked phenformin effects. In the non-transfected cells (GFP-), phenformin remains effective, serving as a good internal control (Figure 3D, Figure S3C). In HaCaT cells, AMOTL1 shRNA alone could sufficiently block phenformin-induced p-YAP (Figure 3E). In addition, when AMOTL1 is silenced by shRNA, inhibition of YAP target genes expression (CTGF, Cyr61 and ANKD1) by phenformin could be significantly rescued (Figure S3D). Most importantly, the growth inhibition of these cells by phenformin and metformin could also be significantly rescued when AMOTL1 is silenced (Figure 3F). Taken together, these results suggest that the motin family proteins, particularly AMOTL1, are involved in the regulation of YAP in response to energy stress.
AMPK directly phosphorylates AMOTL1 at Ser793
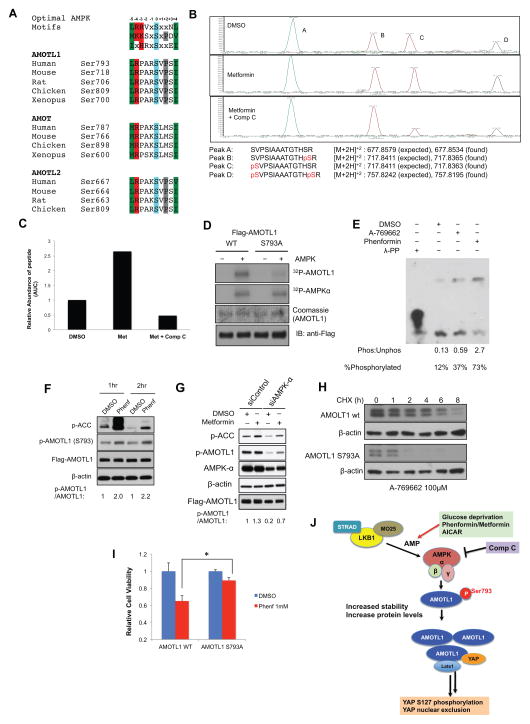
We then set out to study how AMPK regulates AMOTL1. To test whether motin proteins could be direct substrates of AMPK, we carried out a bioinformatics search for conserved AMPK substrate motifs (Dale et al., 1995; Gwinn et al., 2008; Scott et al., 2002). We found that all motin proteins contain AMPK substrate motif (AMOTL1 Ser793, AMOT Ser787 and AMOTL2 Ser667), all of which are evolutionarily conserved among vertebrates (Figure 4A). We then performed co-immunoprecipitation (IP) experiments, and found that the AMPKα subunit directly bind to AMOTL1 (Figure S4A). To confirm that AMOTL1 could be phosphorylated upon AMPK activation in cells, we carried out phosphoproteomic studies using Flag-AMOTL1 isolated from cells treated with DMSO control, metformin, or co-treated with Compound C. We detected non-phosphorylated peptides (Peak A), peptides phosphorylated at an adjacent site Ser805 (Peak B), peptides phosphorylated at the predicted AMPK site Ser793 (Peak C) and peptides with double phosphorylation at Ser793 and Ser805 (Peak D, Figure 4B) by LC-MS/MS methods (Figure S4B). We then quantified their relative abundance by calculating the area under the curve (AUC). Treatment with metformin increased the abundance of peptide fragments containing p-Ser793 (Peak C and D) and co-treatment with Compound C dramatically decreased p-Ser793 peptides abundance, suggesting that activation of AMPK leads to increased phosphorylation of AMOTL1 at Ser793 in cells (Figure 4C). In addition, the phosphorylation levels of the adjacent Ser805 site are independent of AMPK activation and inhibition, serving as an ideal internal control (Figure 4B).
Figure 4. AMPK directly phosphorylates AMOTL1 Ser793 leading to its stabilization and YAP inhibition.
A, Alignment of the conserved AMPK substrate motifs in angiomotin family proteins (AMOTL1, AMOT and AMOTL2) of different species. B, LC-MS/MS studies of phosphorylation of AMOTL1 Ser793. Flag-AMOTL1 was purified from HEK293A cells treated with metformin (10mM) or metformin and C ompound C (10μM). Extracted ion chromatographs of peptides containing Ser793 were shown. Peak A (unphosphoryated peptide), Peak B (peptide with p-Ser805), Peak C (peptide with p-Ser793), Peak D (peptide with p-Ser793 and p-Ser805). C, The abundance of the peptides was quantified by measuring the area under the curve (AUC) in the spectra. D, Recombinant AMPK phosphorylates AMOTL1 wild type, but not S793A mutant in vitro. E, HEK239T cells were treated for 8 hours with DMSO (control), 1μM A-769662, or 1mM Phenformin. Protein lysates were then analyzed by Phos-Tag SDS-PAGE followed by Western blotting with an anti-AMOTL1 antibody. F, Phenformin treatment increased AMOTL1 S793 phosphorylation. An antibody recognizing p-AMOTL1 (Ser793) was used in western blot. HaCaT cells were transfected with Flag-AMOTL1, and then treated with DMSO, or phenformin (1mM) for 1hr or 2hr. G, Knock-down of AMPK decreased the levels of p-AMOTL1. Cells were transfected with siRNAs targeting AMPKα1 and α2, and Flag-AMOTL1. Cells were then treated with metformin (10mM) or DMSO control. p-AMOTL1 levels were determined by using a phosphospecific antibody recognizing AMOTL1 Ser793. H, AMOTL1 S793A has a shorter half-life than wild type AMOTL1. HEK293T cells transfected with either wild type or S793A mutant AMOTL1 were treated for 16 hours with A-769662 then treated for the indicated time with cycloheximide (CHX). I. Cells were transfected with AMOTL1 wild type or S793A mutant, and then treated with DMSO control, or phenformin (1mM) for 48 hours. The cell viability is determined by CellTiter Glo (*, p<0.01). J. A proposed model of energy stress-mediated YAP inhibition. See also Figure S4.
To confirm that Ser793 is directly phosphorylated by AMPK, we generated an AMOTL1 (S793A) mutant construct. Indeed, purified AMPK kinase could phosphorylate AMOTL1 wild type, but not the S793A mutant in vitro (Figure 4D). In addition, we quantified the radioactivity of the phosphorylated AMOTL1 in this assay. We observed the phosphorylation stoichiometry of 0.78mole of phosphate incorporation per mole of AMOTL1 (78% phosphorylated). These results suggest that Ser793 is indeed an AMPK substrate site. Furthermore, we utilized a Phos-tag gel to visualize phosphorylated and non-phosphorylated species of AMOTL1. In DMSO treated cells, 12% of the AMOTL1 protein is phosphorylated. Upon treatment with A-769662 or phenformin, the percentage of phosphorylated AMOTL1 increases to 37% and 73%, respectively (Figure 4E). Thus AMPK activation increases the amount of phosphorylated AMOTL1 in cells. Next, we generated a phosphospecific antibody against p-Ser793 of AMOTL1. Although the antibody is too weak to reliably detect endogenous AMOTL1 phosphorylation, it showed good specificity and sensitivity toward exogenous wild type but not Ser793 mutants (Figure S4C). We transfected HaCaT cells with Flag-AMOTL1, and treated the cells with phenformin, or DMSO control for 1hr or 2 hr. We observed that the AMOTL1 p-Ser793 increased within 1hr (Figure 4F), suggesting that its phosphorylation is indeed enhanced by AMPK activation. We also observed that exogenous Flag-AMOTL1 has high basal Ser793 phosphorylation levels, possibly resulting from the basal AMPK activity in the cells (Figure 4F, G). Consistently, when AMPK α1/α2 subunits are silenced by siRNAs (with knockdown efficiency of ~50%) in HEK293A cells, we observed a significant decrease in the basal p-AMOTL1 levels (Figure 4G), thus suggesting that inhibition of AMPK could inhibit p-AMOTL1. With the decrease of basal p-AMOTL1 levels, the fold induction of p-AMOTL1 by metformin treatment is more significant, suggesting that the phosphorylation of exogenous AMOTL1 at Ser793 indeed could be regulated by AMPK (Figure 4G). Collectively, these data suggest that AMOTL1 is indeed an AMPK substrate, and AMOTL1 Ser793 could be phosphorylated by AMPK directly.
AMPK-mediated AMOTL1 Ser793 phosphorylation leads to increased protein stability
We then probed the functional consequences of AMOTL1 Ser793 phosphorylation. We transfected HEK293T cell with either wild type or Ser793A AMOTL1 and treated cells with AMPK activator A-769662 (100μM). We then measured the protein half-life of wild type and S793A mutant of AMOTL1 by cycloheximide chase followed by western blotting (Figure 4H). We found that the half-life of the S793A mutant is markedly shorter than the wild type, suggesting that loss of Ser793 phosphorylation destabilizes AMOTL1. Furthermore, dephosphorylation of AMOTL1 by Compound C treatment also dramatically decreased the half-life of endogenous AMOTL1 (Figure S4D). As Lats1/2 phosphorylation of AMOT at Ser175 has been implicated in regulation of its localization and stability(Adler et al., 2013b; Dai et al., 2013), we tested whether Lats1/2 are required to stabilize AMOTL1 upon energy stress. We found that knocking down Lats1/2 decreased basal AMOTL1 protein levels, however had no effects on AMPK activator-mediated increase of AMOTL1 protein levels (Figure S4E). Further studies are needed to elucidate the cooperation between the Lats1/2 and AMPK phosphorylation sites on motin proteins.
To test whether AMOTL1 S793 phosphorylation is functionally involved in YAP inhibition, we knocked down endogenous AMOTL1 using shRNA and then overexpress the shRNA-resistant version of AMOTL1 wild type or AMOTL1 S793A mutant. Interestingly, we found that in cells expressing wild type AMOTL1, phenformin could significantly inhibit YAP target gene expression. However, in cells expressing AMOTL1 S793A mutant, phenformin failed to inhibit YAP target genes (Figure S4F). Furthermore, in cells expressing wild type AMOTL1, phenformin could potently inhibit cell proliferation, while expressing AMOTL1 S793A mutant significantly rescues the inhibitory effects of phenformin (Figure 4I). These results suggest that AMOTL1 S793 phosphorylation could modulate YAP functions in response to energy stress, and could be one of the mechanisms leading to YAP inhibition.
DISCUSSION
Here we propose a new signaling mechanism (Figure 4J), in which cellular energy level is an upstream regulator of Hippo signaling. We found that energy stress activates AMPK, stabilizes AMOTL1, and leads to YAP inhibition. Although glucose deprivation and metformin treatment could activate AMPK relatively quickly (in 1hr) (Nguyen et al., 2012), sustained AMPK activation (~2–6h) was required to inhibit YAP. This is consistent with our proposed mechanism in which phosphorylation of AMOTL1 by AMPK leads to AMOTL1 protein accumulation due to its increased half-life. However, other possible AMPK substrates (Kibra, ZO-2 and YAP) and other motin family members could also be involved in the regulation of YAP. Therefore, further studies to characterize them as AMPK substrates would shed new insights into the mechanisms of YAP regulation. In addition, it is also possible that other AMPK related kinases (such as SIK or MARK) could mediate the basal phosphorylation of AMOTL1, and AMPK activation boosts the existing phosphorylation in response to metabolic stress, which is not uncommon in AMPK-regulated substrates(Shackelford and Shaw, 2009).
Among the motin family proteins, AMOTL1 might be the major mediator of the effects. We could not detect phosphorylation of the predicted AMPK motif (Ser787) in AMOT by mass spectrometry. Other non-AMPK sites (Ser97, Ser714) are phosphorylated upon metformin treatment in mass spectrometry studies (Figure S4G). These results suggest that other kinases downstream of AMPK or less stringent AMPK substrate sites might be involved. Therefore, AMOT might be regulated differently than AMOTL1 and future studies are needed to elucidate the regulation of AMOT and AMOTL2 in response to energy stress, and their roles in YAP regulation.
LKB1/STK11 is a known tumor suppressor(Shackelford and Shaw, 2009) and a major upstream regulator of AMPK. Our results suggest that loss of LKB1 and AMPK activities might contribute to tumorigenesis through destabilizing AMOTL1, leading to hyperactivation of YAP. A recent study has showed that LKB1 acts through the microtubule affinity-regulating kinase (MARK) family to regulate the localization of Scribble(Mohseni et al., 2014). Our results and the results of Mohseni et al suggest that there are multiple pathways downstream of LKB1 to regulate YAP activity. This could have important implications for the treatment of LKB1-dependent tumors. Recent work by the Cancer Genome Atlas Project has identified mutations of AMOTL1 at the AMPK recognition motif (R789C, P790I, and R792H) in multiple cancers. Although at low frequency, such mutations might lead to loss of AMPK-mediated Ser793 phosphorylation, and promote YAP activation in these cancers. In summary, our studies have shown that cellular energy sensor AMPK could regulate YAP by directly phosphorylating and stabilizing tight junction protein AMOTL1, connecting energy sensing to the regulation of Hippo pathway.
METHODS
Immunofluorescence staining, YAP translocation assay and imaging analysis
Cells were fixed and then stained with primary antibody (anti-YAP). The images were acquired by high throughput confocal microscopy (Opera® High Content Screening System, Waltham, MA). 4 images/well were captured using a 20X objective at a resolution of ~0.65 μm/pixels. The images of YAP immunostaining and nuclear Hoechest were analyzed using a custom Acapella (PerkinElmer) script as previously reported or with CellProfiler (Carpenter et al., 2006). YAP nuclear/cytoplasmic translocation was defined using the Pearson’s correlation coefficient (R), between the YAP and the Hoechst fluorescence channels, across each pixel of the cellular object detected.
Supplementary Material
Figure S1. High content imaging assay revealed energy stress inhibits YAP. Related to Figure 1.
A. Development of a high content imaging assay of YAP nuclear localization. HEK293A cells are cultures in sparse or dense conditions. Cells are fixed and stained with anti-YAP antibody and imaged under high content microscope. An algorithm measuring the correlation of YAP staining vs. nuclear staining is developed to quantify YAP nuclear localization. The Pearson’s correlation between YAP positive area and nuclear staining was calculated to quantify YAP nuclear localization. R=1 (YAP in nuclei), R=-1 (YAP in cytoplasm); Density-dependent YAP localization validated the method and showed good dynamic window of the assay.
B. Metabolic stressors induce YAP cytoplasmic retention, and Comp C inhibits their effects. The images were associated with the graph in Figure 1a. Normalized YAP/nuclear Pearson’s correlation is listed to quantify the level of YAP nuclear localization. Scale bar: 10μm.
C. HEK293A cells at low density culture were treated with DMSO control, AICAR (0.5 mM), or AICAR (0.5mM) and Compound C (10 μM) for 12h. Cells were subsequently fixed and immunostained for YAP (green) and the nuclei (Hoechst, blue). Scale bar: 5 μm.
D. A specific AMPK activator (A-768662) has similar effects on induction of p-YAP compared with energy stressor phenformin.
E. Glucose deprivation induces YAP Ser127 phosphorylation. Cells were cultured in normal medium (control, 4.5g/L glucose) or glucose free medium (Glucose −) for 12h. Cells were harvested for western blot analysis of p-YAP (Ser127) and total YAP. Protein levels were quantified by densitometry of the bands.
F. HEK293A cells were transfected with a YAP reporter plasmid (TEAD-binding element driven firefly luciferase) along with a p-CMV-Renilla-Luc plasmid. Cells at medium density (8000 cells/well) are cultured in normal media (no media change), glucose free media (Glucose −), adding back glucose (Glucose +) or serum free media (SFM, positive control). Firefly luciferase activities were read 14h after media change, and normalized to renilla luciferase controls (data are represented as mean, S.D. n=3, *, p<0.01, comparing to the Glucose + condition).
G. Energy stressors induce p-YAP (Ser127) in NIH3T3, OVCAR8 and HaCaT cells.
H–I. Anti-proliferative compounds such as inhibitors of PI3K, AKT, mTOR and CDK4 do not induce p-YAP. Cells were treated with DMSO control, Wortmannin (1μM), LY294002 (10 μM), rapamycin (100nM) for overnight, and p-YAP levels were analyzed by western blot (H). CDK inhibitor roscovitine was used at 10 μM to treat HEK293 cells at different time points (I).
I. Quantification of the colony sizes in YAP transformed MCF10A cells. The pixel area of each colony was measured by ImageJ.
J. Metformin (10mM) treatment inhibits YAP target genes expression (CTGF and Cyr61) in wild type YAP transduced MCF-10A cells. YAP (S127A) mutant transduced cells are largely resistant to metformin. Relative RNA levels were normalized to vector transduced MCF-10A cells (data are represented as mean, S.D. n=3).
Figure S2. AMPK and Lats1/2 are required to inhibit YAP. Related to Figure 2.
A. siRNA targeting AMPK α1α2 blocked YAP translocation induced by metformin and phenformin in HEK293A cells. Scale bar: 10 μm.
B. Knock-down efficiency of siRNAs targeting Lats1 and Lats2 as shown in Fig. 2e.
Figure S3. AMOTL1 is required to mediate the inhibition of YAP. Related to Figure 3.
A. qRT-PCR to detect the mRNA levels of AMOT, AMOTL1 and AMOTL2 upon metformin and phenformin treatment for 16h. (data are represented as mean, S.D. n=3)
B. Knock-down efficiency of siRNAs targeting AMOT and AMOTL1. Combination of siAMOT and siAMOTL1#3 showed good knock-down effeciecy for both genes, and was used in the experiments in Figure. 3D.
C. Representative images of YAP localization with AMOT and AMOTL1 silencing. The quantification was shown in Figure 4d. The white arrow shows the GFP positive transfected cells used in quantification. The GFP negative population was used as control. Scale bar: 10 μm.
D. shRNA knockdown of AMOTL1 rescues YAP target gene expression inhibition by phenformin.
Figure S4. Phosphorylation studies of AMOT and AMOTL1. Related to Figure 4.
A. Flag-AMOTL1 co-immunoprecipitates with endogenous AMPKα.
B. Mass spectrometry studies of AMOTL1 phosphorylation. Tandem mass spectra of unphosphorylated and phosphorylated peptides confirm the site of the phosphorylation.
C. Phosphospecific antibody against p-AMOTL1 (Ser793) recognized wild type, but not AMOTL1 (Ser793Ala and Ser793Asp) mutants.
D. Compound C treatment decreases the half-life of endogenous AMOTL1 protein.
E. knockdown of Lats1/2 has little effects on AMPK activator (A-769662) induced AMOTL1 stablization.
F. HEK293A cells are infected with shRNA targeting AMOTL1, and then transfected with AMOTL1 wild type or S793A mutant. Cells were treated with DMSO control or phenformin for 24hours. YAP target gene expression is evaluated by qRT-PCR. Data are represented as mean, S.D. n=3. *, p<0.05 by comparing with the DMSO control.
G. Phosphoproteomic studies and in vitro kinase assays of AMOT
Acknowledgments
This work is supported by MGH, HSCI Seed Grant, Children’s Tumor Foundation, American Cancer Society (124929-RSG-13-291-01-TBE), NIH/NCI R01CA181537 (X.W.); and NIH/NCI R01CA166717 (B.Z.). We thank Drs. K.-L, Guan, A. Schmitt, J. Brugge, M. Sudol, D. McCollum, J. Avruch, K. Laderoute and L. Cantley for constructs, antibodies, cell lines or discussions. E.C.P., M.H. and J. L. are employees of Novartis.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures can be found with this article online.
Author contributions J.Y. M.D. J.L. and X.W. conceived the concepts and designed the experiments; E.C.P. and J.A. performed mass spec studies of AMOTL1 and AMOT, respectively; J.Y., M.D. P.C. M.H. S.Z. J.F., N.B, C-H. S. and B.Z. performed the cell biology and biochemistry experiments, and all authors analyzed the data; J.Y., M.D. and X.W. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler JJ, Heller BL, Bringman LR, Ranahan WP, Cocklin RR, Goebl MG, Oh M, Lim HS, Ingham RJ, Wells CD. Amot130 adapts atrophin-1 interacting protein 4 to inhibit yes-associated protein signaling and cell growth. J Biol Chem. 2013a;288:15181–15193. doi: 10.1074/jbc.M112.446534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler JJ, Johnson DE, Heller BL, Bringman LR, Ranahan WP, Conwell MD, Sun Y, Hudmon A, Wells CD. Serum deprivation inhibits the transcriptional co-activator YAP and cell growth via phosphorylation of the 130-kDa isoform of Angiomotin by the LATS1/2 protein kinases. Proc Natl Acad Sci U S A. 2013b;110:17368–17373. doi: 10.1073/pnas.1308236110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cool B, Zinker B, Chiou W, Kifle L, Cao N, Perham M, Dickinson R, Adler A, Gagne G, Iyengar R, et al. Identification and characterization of a small molecule AMPK activator that treats key components of type 2 diabetes and the metabolic syndrome. Cell Metab. 2006;3:403–416. doi: 10.1016/j.cmet.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Dai X, She P, Chi F, Feng Y, Liu H, Jin D, Zhao Y, Guo X, Jiang D, Guan KL, et al. Phosphorylation of angiomotin by Lats1/2 kinases inhibits F-actin binding, cell migration and angiogenesis. J Biol Chem. 2013 doi: 10.1074/jbc.M113.518019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nature reviews Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Pfleger CM, Hariharan IK. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nature reviews Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Hong W, Guan KL. The YAP and TAZ transcription co-activators: Key downstream effectors of the mammalian Hippo pathway. Semin Cell Dev Biol. 2012 doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E, Yang J, Deran M, Wu C, Su AI, Bonamy GM, Liu J, Peters EC, Wu X. Identification of Serum-Derived Sphingosine-1-Phosphate as a Small Molecule Regulator of YAP. Chem Biol. 2012;19:955–962. doi: 10.1016/j.chembiol.2012.07.005. [DOI] [PubMed] [Google Scholar]
- Mohseni M, Sun J, Lau A, Curtis S, Goldsmith J, Fox VL, Wei C, Frazier M, Samson O, Wong KK, et al. A genetic screen identifies an LKB1-MARK signalling axis controlling the Hippo-YAP pathway. Nat Cell Biol. 2014;16:108–117. doi: 10.1038/ncb2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HB, Babcock JT, Wells CD, Quilliam LA. LKB1 tumor suppressor regulates AMP kinase/mTOR-independent cell growth and proliferation via the phosphorylation of Yap. Oncogene. 2012 doi: 10.1038/onc.2012.1431.. Published online Oct. 3rd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overholtzer M, Zhang J, Smolen GA, Muir B, Li W, Sgroi DC, Deng CX, Brugge JS, Haber DA. Transforming properties of YAP, a candidate oncogene on the chromosome 11q22 amplicon. Proc Natl Acad Sci U S A. 2006;103:12405–12410. doi: 10.1073/pnas.0605579103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paramasivam M, Sarkeshik A, Yates JR, 3rd, Fernandes MJ, McCollum D. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell. 2011;22:3725–3733. doi: 10.1091/mbc.E11-04-0300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, et al. Yap1 acts downstream of alpha-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JW, Norman DG, Hawley SA, Kontogiannis L, Hardie DG. Protein kinase substrate recognition studied using the recombinant catalytic domain of AMP-activated protein kinase and a model substrate. Journal of molecular biology. 2002;317:309–323. doi: 10.1006/jmbi.2001.5316. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nature reviews Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udan RS, Kango-Singh M, Nolo R, Tao C, Halder G. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nature cell biology. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- Wu S, Huang J, Dong J, Pan D. hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- Yi C, Troutman S, Fera D, Stemmer-Rachamimov A, Avila JL, Christian N, Persson NL, Shimono A, Speicher DW, Marmorstein R, et al. A tight junction-associated Merlin-angiomotin complex mediates Merlin’s regulation of mitogenic signaling and tumor suppressive functions. Cancer Cell. 2011;19:527–540. doi: 10.1016/j.ccr.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. Regulation of the Hippo-YAP Pathway by G-Protein-Coupled Receptor Signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ji JY, Yu M, Overholtzer M, Smolen GA, Wang R, Brugge JS, Dyson NJ, Haber DA. YAP-dependent induction of amphiregulin identifies a non-cell-autonomous component of the Hippo pathway. Nat Cell Biol. 2009;11:1444–1450. doi: 10.1038/ncb1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes & development. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Wang L, Wang CY, Yu J, Guan KL. Cell detachment activates the Hippo pathway via cytoskeleton reorganization to induce anoikis. Genes & development. 2012;26:54–68. doi: 10.1101/gad.173435.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes & development. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell. 2009;16:425–438. doi: 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Zhang Y, Wu H, Barry E, Yin Y, Lawrence E, Dawson D, Willis JE, Markowitz SD, Camargo FD, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci U S A. 2011;108:E1312–1320. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. High content imaging assay revealed energy stress inhibits YAP. Related to Figure 1.
A. Development of a high content imaging assay of YAP nuclear localization. HEK293A cells are cultures in sparse or dense conditions. Cells are fixed and stained with anti-YAP antibody and imaged under high content microscope. An algorithm measuring the correlation of YAP staining vs. nuclear staining is developed to quantify YAP nuclear localization. The Pearson’s correlation between YAP positive area and nuclear staining was calculated to quantify YAP nuclear localization. R=1 (YAP in nuclei), R=-1 (YAP in cytoplasm); Density-dependent YAP localization validated the method and showed good dynamic window of the assay.
B. Metabolic stressors induce YAP cytoplasmic retention, and Comp C inhibits their effects. The images were associated with the graph in Figure 1a. Normalized YAP/nuclear Pearson’s correlation is listed to quantify the level of YAP nuclear localization. Scale bar: 10μm.
C. HEK293A cells at low density culture were treated with DMSO control, AICAR (0.5 mM), or AICAR (0.5mM) and Compound C (10 μM) for 12h. Cells were subsequently fixed and immunostained for YAP (green) and the nuclei (Hoechst, blue). Scale bar: 5 μm.
D. A specific AMPK activator (A-768662) has similar effects on induction of p-YAP compared with energy stressor phenformin.
E. Glucose deprivation induces YAP Ser127 phosphorylation. Cells were cultured in normal medium (control, 4.5g/L glucose) or glucose free medium (Glucose −) for 12h. Cells were harvested for western blot analysis of p-YAP (Ser127) and total YAP. Protein levels were quantified by densitometry of the bands.
F. HEK293A cells were transfected with a YAP reporter plasmid (TEAD-binding element driven firefly luciferase) along with a p-CMV-Renilla-Luc plasmid. Cells at medium density (8000 cells/well) are cultured in normal media (no media change), glucose free media (Glucose −), adding back glucose (Glucose +) or serum free media (SFM, positive control). Firefly luciferase activities were read 14h after media change, and normalized to renilla luciferase controls (data are represented as mean, S.D. n=3, *, p<0.01, comparing to the Glucose + condition).
G. Energy stressors induce p-YAP (Ser127) in NIH3T3, OVCAR8 and HaCaT cells.
H–I. Anti-proliferative compounds such as inhibitors of PI3K, AKT, mTOR and CDK4 do not induce p-YAP. Cells were treated with DMSO control, Wortmannin (1μM), LY294002 (10 μM), rapamycin (100nM) for overnight, and p-YAP levels were analyzed by western blot (H). CDK inhibitor roscovitine was used at 10 μM to treat HEK293 cells at different time points (I).
I. Quantification of the colony sizes in YAP transformed MCF10A cells. The pixel area of each colony was measured by ImageJ.
J. Metformin (10mM) treatment inhibits YAP target genes expression (CTGF and Cyr61) in wild type YAP transduced MCF-10A cells. YAP (S127A) mutant transduced cells are largely resistant to metformin. Relative RNA levels were normalized to vector transduced MCF-10A cells (data are represented as mean, S.D. n=3).
Figure S2. AMPK and Lats1/2 are required to inhibit YAP. Related to Figure 2.
A. siRNA targeting AMPK α1α2 blocked YAP translocation induced by metformin and phenformin in HEK293A cells. Scale bar: 10 μm.
B. Knock-down efficiency of siRNAs targeting Lats1 and Lats2 as shown in Fig. 2e.
Figure S3. AMOTL1 is required to mediate the inhibition of YAP. Related to Figure 3.
A. qRT-PCR to detect the mRNA levels of AMOT, AMOTL1 and AMOTL2 upon metformin and phenformin treatment for 16h. (data are represented as mean, S.D. n=3)
B. Knock-down efficiency of siRNAs targeting AMOT and AMOTL1. Combination of siAMOT and siAMOTL1#3 showed good knock-down effeciecy for both genes, and was used in the experiments in Figure. 3D.
C. Representative images of YAP localization with AMOT and AMOTL1 silencing. The quantification was shown in Figure 4d. The white arrow shows the GFP positive transfected cells used in quantification. The GFP negative population was used as control. Scale bar: 10 μm.
D. shRNA knockdown of AMOTL1 rescues YAP target gene expression inhibition by phenformin.
Figure S4. Phosphorylation studies of AMOT and AMOTL1. Related to Figure 4.
A. Flag-AMOTL1 co-immunoprecipitates with endogenous AMPKα.
B. Mass spectrometry studies of AMOTL1 phosphorylation. Tandem mass spectra of unphosphorylated and phosphorylated peptides confirm the site of the phosphorylation.
C. Phosphospecific antibody against p-AMOTL1 (Ser793) recognized wild type, but not AMOTL1 (Ser793Ala and Ser793Asp) mutants.
D. Compound C treatment decreases the half-life of endogenous AMOTL1 protein.
E. knockdown of Lats1/2 has little effects on AMPK activator (A-769662) induced AMOTL1 stablization.
F. HEK293A cells are infected with shRNA targeting AMOTL1, and then transfected with AMOTL1 wild type or S793A mutant. Cells were treated with DMSO control or phenformin for 24hours. YAP target gene expression is evaluated by qRT-PCR. Data are represented as mean, S.D. n=3. *, p<0.05 by comparing with the DMSO control.
G. Phosphoproteomic studies and in vitro kinase assays of AMOT