Summary
Follicular helper T (Tfh) cells are specialized providers of T cell help to B cells, and are essential for germinal center formation, affinity maturation, and the development of most high affinity antibodies and memory B cells. Tfh cell differentiation is a multi-stage, multi-factorial process involving B cell lymphoma 6 (Bcl6) and other transcription factors. This article reviews understanding of Tfh cell biology, including their differentiation, migration, transcriptional regulation, and B cell help functions. Tfh cells are critical components of many protective immune responses against pathogens. As such, there is strong interest in harnessing Tfh cells to improve vaccination strategies. Tfh cells also have roles in a range of other diseases, particularly autoimmune diseases. Overall, there have been dramatic advances in this young field, but there is much to be learned about Tfh cell biology in the interest of applying that knowledge to biomedical needs.
Introduction
There has been a great deal of recent activity in the study of T follicular helper (Tfh) cells. While the first evidence of Tfh cells was reported in human lymphoid tissue more than a decade ago, much of the interest in Tfh cells traces its origins to the identification of Bcl6 as an essential transcription factor in CD4+ T cells for Tfh cell differentiation and the development of germinal centers (GCs) (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). The field of Tfh cell biology has now exploded with activity, examining everything from the biochemistry of transcription factors involved in programming Tfh cell differentiation to the cellular biology of Tfh cell-mediated selection of germinal center B cells, and examining important roles of Tfh cells in biological processes as diverse as vaccine elicited immune responses to chronic autoimmune diseases and even to roles of Tfh cells in protective immunity in human cancers. This article reviews our understanding of Tfh cell differentiation, molecular biology, and function, and discusses the most recent advances in these areas as well as the complexities of Tfh cell biology. In addition, a new conceptual model is introduced to explain the relationship between Tfh cell and other CD4+ T cell differentiation programs. For an oral presentation of the review see supplemental video 1.
Stages of Tfh Cell Differentiation
Tfh cell differentiation is a multi-stage, multi-factorial process. There is no single event that defines Tfh cell differentiation, unlike Th1 cell differentiation for instance, which can be fully induced by interleukin-12 (IL-12) exposure in vitro or in vivo. Instead, Tfh cell differentiation is a multistep, multisignal process that also accommodates a significant amount of heterogeneity. The canonical Tfh cell differentiation process starts at initial dendritic cell (DC) priming of a naive CD4+ T cell (Goenka et al., 2011) (Fig. 1A). The CD4+ T cell undergoes a cell fate decision within the first few rounds of cell division (Choi et al., 2011; 2013b). If the chemokine receptor CXCR5 is expressed, the early Tfh cell will migrate to the border of the B cell follicle and undergo further Tfh cell differentiation. If the cell instead receives Th1, Th2, or Th17 cell signals (Fig. 1) the CD4+ T cell follows a Th1, Th2, or Th17 cell differentiation program, including upregulation of chemokine receptors for inflammatory chemokines that will drive the effector cell to exit the lymphoid tissue and traffic to the site of infection or inflammation.
Figure 1. Overview of Tfh cell differentiation.
(a) Stages of Tfh cell differentiation, highlighting roles of migration-associated molecules. (b) Signals in CD4 T cell differentiation. A simplified model of CD4 T cell differentiation pathways, showing transcription factors and inducing factors, highlighting apparent differences between human and mouse Tfh cell differentiation.
Early Tfh cell differentiation (the DC priming phase) is regulated by IL-6, inducible costimulator (ICOS), IL-2, and T cell receptor (TCR) signal strength in mouse models. TCR signal strength can bias T cell differentiation in vivo (Tubo et al., 2013), but a single naive mature T cell can give rise to multiple different differentiated effector cell types upon stimulation and proliferation, demonstrating that non-TCR and TCR signals combine to determine T cell differentiation fates. CD4+ T cells possessing TCRs with high affinity preferentially differentiated into Tfh cells in a pigeon cytochrome C (PCC) model (Fazilleau et al., 2009), but not a Friend virus infection (Ploquin et al., 2011). Utilizing a range of systems it was found that TCR: major histocompatibility complex-II (MHCII) dwell time is a more accurate predictor of cell fate preference, with a nonlinear relationship (Tubo et al., 2013), such that there was no simple relationship between TCR signal strength and Tfh cell differentiation. IL-6 is the earliest non-TCR signal involved in initiation of Tfh cell differentiation. IL-6 signaling through IL-6 receptor (IL-6R - gp130) transiently induces Bcl6 expression by newly activated CD4+ T cells (Nurieva et al., 2009). Bcl6 is necessary for early CXCR5 expression in multiple models (Choi et al., 2011; 2013a; Pepper et al., 2011). In the absence of IL-6 an early defect in Tfh cell differentiation is observed (Choi et al., 2013a). The DC type responsible for initiating Tfh cell differentiation is unknown. Most likely there are multiple Tfh cell differentiation pathways and there is no single DC type responsible for priming Tfh cells. Instead, multiple DC and monocyte types can prime Tfh cell differentiation in different conditions (Ballesteros-Tato and Randall, 2014). Many DC types are robust producers of IL-6. Prdm1−/− DCs are hyperactive producers of IL-6, resulting in spontaneous Tfh cell and GC development in vivo (Kim et al., 2011). IL-6 can also be a signal for Th17 cell differentiation, therefore it is assumed that IL-6 in combination with different signals are involved in Tfh cell versus Th17 cell differentiation. Interestingly, no increase in Th17 cells was seen in hyper IL-6 producing mice, in contrast to the increase in Tfh cells. IL-1 is an important driver of Th17 differentiation, while ICOS is important for Tfh differentiation (Choi et al., 2011; Nurieva et al., 2008). ICOS has roles in both Tfh cell differentiation and migration, and there are data supporting a synergistic role of ICOS and IL-6. The importance of ICOS is highlighted by the multiple ways in which ICOS signaling is regulated. Roquin inhibits ICOS, and combined loss of Roquin1 and Roquin2 results in spontaneous Tfh cell and GC development (Pratama et al., 2013; Vogel et al., 2013). In addition, the miR-19~72 complex is necessary for Tfh cell differentiation, and it works, in part, via dampening the PI(3)K inactivating phosphatases PHLPP2 and PTEN, which are inhibitors of ICOS signaling (Baumjohann et al., 2013; Kang et al., 2013). IL-2 signaling is another major regulator of Tfh cell differentiation. IL-2 is a potent inhibitor of Tfh cell differentiation (Ballesteros-Tato et al., 2012; Johnston et al., 2012), and can act very early during T cell priming (Johnston et al., 2012). Thus, the interplay between IL-6, ICOS, IL-2, and TCR signaling orchestrates early induction of mouse Tfh cell differentiation during DC priming via control of CXCR5, Bcl6, and other targets.
The second stage of Tfh cell differentiation occurs when the T cell interacts with antigen-specific B cells in the follicle, interfollicular zone, or the T-B border. Much of Tfh cell differentiation and function is tightly interconnected with the microanatomical geography of the T and B zones of the lymph node (LN) and spleen. The early Tfh cells co-localize with B cells because they express CXCR5, downregulate C-C chemokine receptor type 7 (CCR7) (the primary chemotactic receptor for the T zone), and downregulate P-selectin glycoprotein ligand 1 (PSGL1), which is thought to anchor T cells to CCL19 and CCL21 decorating the T zone extracellular matrix (Fig. 1A). Tfh cells have a highly symbiotic relationship with B cells, and B cells are required for Tfh cell development under almost all conditions (Crotty, 2011). ICOS is a costimulatory molecule, but it has been recently demonstrated that ICOS-ICOS ligand (ICOSL) binding also induces directional migration of CD4+ T cells, which can play an important role in proper localization of the early Tfh cells to the B cell follicle (Xu et al., 2013). B cells serve both as antigen-presenting cells (APCs) and as a source of ICOSL (Choi et al., 2011; Haynes et al., 2007; Nurieva et al., 2008). B cells rapidly become the primary APCs available in a LN during an acute infection or immunization because mature DCs last for only a few days before dying, whereas the antigen-specific B cells undergo geometric replication. Antigen presentation is critical, because unlike effector CD8 T cells, antigen-specific CD4 T cells require antigen recognition for virtually every cell division (Choi et al., 2013b; Obst et al., 2005; Yarke et al., 2008).
The third stage of Tfh cell differentiation involves the GC (Fig. 1A). The GC is a distinct structure consisting of GC Tfh cells, GC B cells, follicular dendritic cells (FDCs), macrophages, and stroma. The majority of GC Tfh cells can be observed to possess a canonical Tfh cell differentiation program. The majority of GC Tfh cells are CXCR5hiPD1hiBcl6hiMafhiSAPhi. They are also PSGL1loCD200+BTLAhiCCR7lo. The canonical secreted Tfh cell molecules are C-X-C motif chemokine 13 (CXCL13), IL-21, and IL-4 (Crotty, 2011; Kroenke et al., 2012; Liang et al., 2012; Linterman et al., 2011). These GC Tfh cell surface proteins, transcription factors, and secreted molecules are well conserved across in vivo conditions and species. GC Tfh cells can be readily identified in mice, humans, and non-human primates as CXCR5hiPD1hiBcl6hi CD4 T cells. The biology of GC Tfh cells is strongly associated with changes in several chemokine receptors and related molecules. GC Tfh cells have very high expression of CXCR5, low expression of CCR7, elevated C-X-C chemokine receptor type 4 (CXCR4), low sphingosine 1-phosphate 1 receptor (S1P1R), and very low amounts of PSGL1. Loss of Epstein-Barr virus-induced G-protein coupled receptor 2 (EBI2) expression is notable because the chemoattractant ligand for EBI2 is present in the B cell follicle but not the GC environment. Reduction of EBI2 expression by both GC B cells and GC Tfh cells is important for their proper localization to GCs (Hannedouche et al., 2011). In addition, adhesion molecules play an important role on GC Tfh cells, regulating their interaction with GC B cells and their localization. The signaling lymphocyte activation molecule (SLAM) family receptors SLAMF6 (also known as Ly108 and NTB-A), CD84, and SLAM are all self-ligands differentially expressed on GC Tfh cells and/or GC B cells. SLAM-associated protein (SAP), the product of the Sh2d1a gene, is an SH2-domain adaptor protein that binds to the cytoplasmic tails of SLAM family receptors and is specifically upregulated in GC Tfh cells. SAP expression is essential for GC Tfh cell development, GC development, and the generation of the majority of memory B cells and memory plasma cells (Hu et al., 2013). In the absence of SAP, Tfh cells have defective adhesion to GC B cells and fail to be retained in GCs (Qi et al., 2008); as a result, insufficient help is provided by SAP-deficient Tfh cells to B cells. The functions of SAP are central to Tfh cell biology, as loss of SAP is one of the few genetic mutations in CD4+ T cells that results in a complete loss of GC Tfh cells and GC B cells in virtually all experimental settings. Surprisingly, much of the importance of SAP is due to a requirement for SAP to prevent powerful inhibitory signaling through SLAMF6 (Kageyama et al., 2012). SAP competes with the phosphatase SHP-1 for binding to SLAMF6. With SAP bound, SLAMF6 transmits positive signals within the Tfh cells, supporting adhesion and help functions. With SHP-1 bound, SLAMF6 transmits potent negative signals that truncate Tfh:B cell adhesion. Important roles for SLAMF6 as a rheostat of cell:cell adhesion for lymphocytes have been shown for Tfh, natural killer T (NKT), CD8 T, and NK cells. This also suggests that other adhesion molecules are likely to be regulators of both GC Tfh cell differentiation and function.
Once Tfh cells have differentiated into GC Tfh cells and provided help to GC B cells, they are not confined to the GC. GC Tfh cells can exit GCs (Fig. 1A). GC B cells are strictly confined to a single GC, and the majority of GC B cells within a GC represent oligoclonal antigen-specific B cell clones undergoing hypermutation and selection by Tfh cells. In contrast, the GC Tfh cells can readily exit a GC and (a) transit to a neighboring follicle and enter a different GC (Shulman et al., 2013), or (b) temporarily reside in the adjacent B cell follicle before re-entering the same GC (Fig. 1A), or (c) exit a GC and downregulate Bcl6 and develop into a memory Tfh cell (Kitano et al., 2011; Shulman et al., 2013) (Fig. 2. Memory is discussed further in a section below.). Therefore, a CXCR5+ Tfh cell outside of a GC may have already been a GC Tfh cell and is in the process of transiting to a new GC, or it may be a newly activated Tfh cell on its way to becoming a GC Tfh cell, or it may have another fate, including (but not limited to) memory formation or being a Tfh cell destined to provide help primarily outside of a GC.
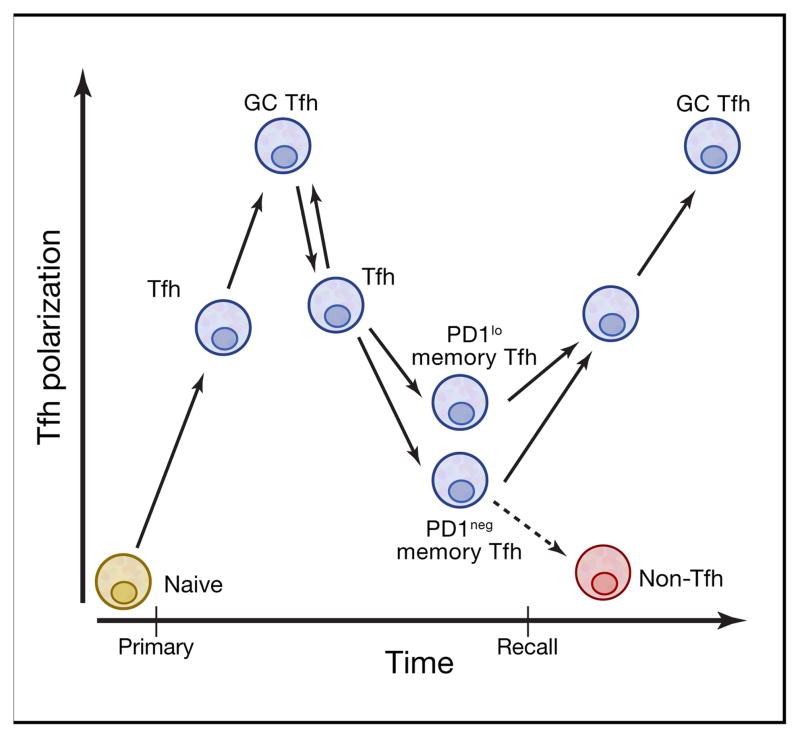
Figure 2. Tfh cell memory development.
Memory Tfh cells develop over time and appear to develop from either Tfh cells or GC Tfh cells. Memory Tfh cells exhibit phenotypic heterogeneity. Memory Tfh cells that retain stable expression of low amounts of PD-1 (PD-1lo or PD-1+) are more polarized and highly functional memory Tfh cells, when compared to PD-1neg memory Tfh cells. Upon reactivation, memory Tfh cells predominantly become Tfh cells and can go on to become GC Tfh cells, although some memory Tfh cells can go on to become non-Tfh cells in a recall response.
A canonical Tfh cell differentiation pathway was described above, involving multiple signals, a multi-stage process, and two different APCs. Alternative Tfh cell differentiation processes exist. This is expected. There is not a single immutable Tfh cell phenotype. It is quite clear that CD4+ T cells have enormous intrinsic heterogeneity. This is an important aspect of CD4+ T cell biology, allowing the cells to adapt to a variety of environmental conditions, locations, and needs. Some of this variability is almost certainly stochastic, and valuable for preventing pathogen evasion by virtue of its randomness and diversity. Th1, Th2, Th17, and Th9 responses are selectively valuable for responses to specific categories of pathogens (e.g. Th1 cells in response to viral infections), and are induced by pathogen-associated molecular patterns (PAMPs) associated with a pathogen category (e.g. viral RNA triggering of TLR7 and TLR8 causes IL-12 production by DCs to instruct Th1 cell differentiation). That conceptual framework does not hold true for Tfh cells. Antibody responses are valuable against almost all pathogens, irrespective of whether they are viral, bacterial, fungal, or multicellular parasites. Consequently, it is critical that the immune system trigger Tfh cell inductive signals whenever any form of pathogen is detected.
As a result of the need to trigger Tfh cell inductive signals in the context of a wide range of potential pathogen assaults, multiple redundant signals are likely involved. IL-6 is an example of this. IL-6 is produced by DCs, macrophages, B cells, and a variety of other cell types in response to a range of external and internal PAMPs and damage-associated molecular patterns (DAMPs). In the absence of IL-6 an early defect in murine Tfh cell differentiation is observed, most likely due to a failure of IL-6 production by the DCs priming naive T cells. Nevertheless, that defect is rapidly compensated for by IL-21 or IL-27 in most cases (Batten et al., 2010; Choi et al., 2013a; Eto et al., 2011; Harker et al., 2013; Karnowski et al., 2012), though late roles of IL-6 for Tfh cells can also be dramatic (Harker et al., 2011). A second example of multiple alternative Tfh cell differentiation pathways involves ICOS (Weinstein et al., 2014; Xu et al., 2013). ICOSL was found to not be essential on antigen-specific B cells (Xu et al., 2013), but a follow-up study found that ICOSL is normally required on antigen-specific B cells, but that can be overcome when large numbers of antigen-specific B cells are transferred, or in the presence of large amounts of antigen (Weinstein et al., 2014).
Given the importance of Tfh cells, one concept is that Tfh cell differentiation is a default pathway for newly activated CD4 T cells. That concept appears to be incorrect. In vitro activated CD4+ T cells in unbiased conditions fail to acquire any of the key features of Tfh cells such as CXCR5, Bcl6, SAP, or IL-21 expression (Eto et al., 2011).
While much is now understood about the multiple stages of Tfh cell differentiation and signals involved in the process, critical knowledge is still lacking. Most importantly, understanding of Tfh cell differentiation is still insufficient to establish a defined and reproducible in vitro Tfh cell differentiation condition. This is the single most serious knowledge gap in the field of Tfh cell biology. Features of some partial aspects of Tfh cell differentiation have emerged from in vitro studies. It is well established that IL-6 is a potent inducer of IL-21 expression by activated murine CD4+ T cells. IL-6 also induces transient Bcl6 expression (Eto et al., 2011; Nurieva et al., 2009). However, while IL-6 was initially reported to drive CXCR5 expression (Nurieva et al., 2009), IL-6 does not induce significant amounts of CXCR5 mRNA or protein in most conditions (Eto et al., 2011; Liu et al., 2014). Furthermore, IL-6 is not a good inducer of IL-21, Bcl6, or CXCR5 expression by activated human CD4 T cells (Ma et al., 2009; Schmitt et al., 2009). In contrast, IL-12 is a potent inducer of IL-21 expression by human CD4 T cells, but not murine CD4 T cells (Ma et al., 2009; Schmitt et al., 2009). Interestingly, IL-6 deficiency results in a severe reduction in CXCR5+Bcl6+ early Tfh cells in vivo, with effectively no CXCR5 or Bcl6 expressing cells present 72 hours after an acute viral infection (Choi et al., 2013a). Nevertheless, constitutive Bcl6 expression in activated murine CD4+ T cells is not sufficient to induce CXCR5 expression in vitro (Liu et al., 2014). In contrast, constitutive expression of Bcl6 in previously activated (CD45RO+) human CD4+ T cells does induce elevated CXCR5 in vitro (Kroenke et al., 2012). A recent paper reports the most successful conditions yet for human Tfh cell differentiation in vitro, with successful short-term induction of CXCR5, Bcl6, and IL-21 in the presence of TGF-β and IL-12 or IL-23 (Schmitt et al., 2014). A contribution by TGF-β is surprising. Importantly, these same conditions were not effective at inducing murine CD4+ T cells to differentiate into Tfh cells in vitro, implying a difference between the species. In vitro TGF-β + IL-12 or TGF-β + IL-23 generated human Tfh cells possessed enhanced B cell help activity, indicating that TGF-β is important for human Tfh cell differentiation and function.
Clues regarding human Tfh cell differentiation are also being provided by analysis of humans with genetic deficiencies. The importance of SAP and ICOS in human Tfh cell differentiation and function was recognized a number of years ago (Crotty, 2011). Now it is also clear that IL-21R and STAT3, as well as IL-12R and STAT4 are two pairs of proteins for which genetic mutations are associated with loss of Tfh cells in humans (Ma et al., 2012a; Schmitt et al., 2013). Importantly, it remains unclear whether murine and human Tfh cell differentiation are regulated by the same cytokines (Fig 1B), and therefore not all lessons from one species may be applicable to the other, even though the cell gene expression program and Tfh cell functions are highly conserved between the species.
Tfh Cell Memory
While Tfh cell memory was controversial (Ma et al., 2012b; Marshall et al., 2011; Pepper et al., 2011), there are now a series of clear studies demonstrating Tfh cell memory in both mice (Choi et al., 2013b; Hale et al., 2013; Liu et al., 2012; Weber et al., 2012) and humans (Bentebibel et al., 2013; Locci et al., 2013), building upon earlier observations in both species (Chevalier et al., 2011; Lüthje et al., 2012; Morita et al., 2011). Memory Tfh cells can be long-lived (Hale et al., 2013; Locci et al., 2013), and are transferrable (Hale et al., 2013). Some of the hesitancy regarding Tfh cell memory was based on an incorrect assumption that GC Tfh cells were terminally differentiated and could not leave GCs. In fact, GC Tfh cells regularly exit from GCs, as discussed above. Upon leaving a GC, the Tfh cell acquires a less activated, less polarized Tfh phenotype and can upregulate IL-7Rα and develop into resting memory Tfh cells (Choi et al., 2013b; Hale et al., 2013; Kitano et al., 2011; Liu et al., 2012; Shulman et al., 2013; Yusuf et al., 2010) (Fig. 1B). In addition, it is not required that a Tfh progress through a GC Tfh state to become a memory Tfh cell (He et al., 2013) (Fig. 2). Memory Tfh cells have a central memory phenotype and predominantly reside in spleen, LNs, and bone marrow, and have the capacity to recirculate in blood (Chevalier et al., 2011; Hale et al., 2013). Approximately 20% of all human central memory CD4+ T cells are CXCR5+, demonstrating that memory Tfh cells are a major component of human T cell memory. Memory Tfh cells preferentially become Tfh cells and GC Tfh cells upon reactivation (Hale et al., 2013) (Fig. 2). In humans, memory Tfh cells are heterogeneous in phenotype, at least in blood (Schmitt and Ueno, 2013), including a significant fraction of resting memory Tfh cells that express low amounts of programmed cell death-1 (PD-1). These PD-1+ memory Tfh cells are the most polarized and functional memory Tfh cells, as measured by gene expression profiling and B cell help (Locci et al., 2013). As a result, one model is that those memory Tfh cells are the most likely to retain their Tfh differentiation program upon reactivation (Fig. 2), but this requires additional studies.
Bcl6 expression is not stable, it requires continuous reinforcement. As such, when a GC Tfh cell leaves a GC and transitions to a non-GC Tfh cell state (a Tfh cell) Bcl6 expression is reduced (Kitano et al., 2011; Yusuf et al., 2010). Bcl6 expression is further reduced as the cell transitions to a fully resting state, becoming a memory Tfh cell (Choi et al., 2013b; Hale et al., 2013; Liu et al., 2012). This is not unlike the phenotype of other central memory CD4+ T cell subsets, which frequently exhibit relatively low amounts of canonical master regulator transcription factors. Interestingly, activated memory B cells induce rapid re-expression of Bcl6 by memory Tfh cells (Ise et al., 2014), reinforcing the concept that many features of Tfh cells are highly intertwined with those of its partners, the B cells.
The Transcription Factor Network Driving Tfh Cell Differentiation
Bcl6 is essential for Tfh differentiation and is frequently referred to as the Tfh master regulator transcription factor. However, the master regulator transcription factor concept is an oversimplification of lymphocyte biology. More that one transcription factor is critical for any CD4 T cell differentiation program. As such, the transcription factors Bcl6, RORγt, Tbet, GATA3, and Foxp3 are now more frequently referred to as “lineage defining” transcription factors for Tfh, Th17, Th1, Th2, and pTreg cells respectively. Even this nomenclature is imperfect, given the evidence of reversible plasticity by most CD4+ T cell types (O’Shea and Paul, 2010). It appears that most cell types can be distinguished by differential expression of a set of 4–6 TFs (Ravasi et al., 2010), and this concept applies to CD4+ T cell differentiation (Ciofani et al., 2012). For Tfh cells, several transcription factors are required in addition to Bcl6. These include Maf, Interferon regulatory factor 4 (IRF4), the activator protein 1 (AP-1) family member ‘basic leucine zipper transcription factor’ (Batf), STATs, and E proteins.
IRF4 and Batf are both essential for early Tfh cell differentiation (Bollig et al., 2012; Ise et al., 2011), but they are required for multiple different CD4+ T cell programs and can be considered early T cell activation-associated transcription factors that enable expression and function of downstream cell fate determining transcription factors (Vahedi et al., 2013). This may occur via cell type specific super enhancer regulation. For example, Batf is a positive regulator of the Bcl6 gene, and combined expression of Bcl6 and Maf is required to obtain CXCR5 expression in vivo in the absence of Batf (Ise et al., 2011). Maf is highly expressed in Tfh cells and is associated with CXCR5, IL-21, and IL-4 expression (Bauquet et al., 2009; Hiramatsu et al., 2010; Ise et al., 2011; Kroenke et al., 2012).
The E protein Ascl2 is a newly recognized contributor to Tfh differentiation. Constitutive expression of Ascl2 in murine CD4+ T cells induces CXCR5 expression by some CD4+ T cells in vitro (Liu et al., 2014). Ascl2fl/fl Cd4Cre mice had normal Tfh cell differentiation and CXCR5 expression, which indicates that Ascl2 is not a unique regulator of CXCR5 and there is redundancy among the several E proteins expressed in T cells and multiple E proteins can enhance CXCR5 expression by binding enhancer regions (Liu et al., 2014; Miyazaki et al., 2011). The signals inducing Ascl2 expression or other E-box proteins remain undefined. Additionally, E protein function is heavily regulated by Inhibitor of DNA-binding/differentiation 2 (Id2) and Id3 (Yang et al., 2011a). Foxp1 and Foxo1 represent transcription factor examples of the converse biology. Foxp1 and Foxo1 are expressed in resting naïve CD4+ T cells and are required for quiescence and homing of naïve CD4+ T cells (Hedrick et al., 2012). Eliminating expression of either of these two genes—by genetic ablation or, at least in the case of Foxo1, ubiquitination and degradation by Itch—is critical for Tfh differentiation (Stone et al., 2013; Wang et al., 2014; Xiao et al., 2014). These transcription factors appear to repress Bcl6 and Tfh cell-associated migration genes, among others, though conflicting Foxo1 data have been reported (Oestreich et al., 2012). As such, there are Tfh transcription factors upstream of Bcl6, as well as transcription factors that coordinate with Bcl6 or are downstream.
Signal transducers and activators of transcription (STATs) are also deeply entwined with Tfh differentiation, as they are with all CD4+ T cell differentiation pathways (Vahedi et al., 2013). Roles of STAT proteins in Tfh cells are complex and overlapping. STAT3 is the most important STAT in murine Tfh differentiation (Ma et al., 2012a; Ray et al., 2014), while STAT1 and STAT4 can also contribute (Choi et al., 2013a; Nakayamada et al., 2011; Schmitt et al., 2013). This includes a critical role for STAT3 in IL-21 expression by murine CD4+ T cells (Suto et al., 2008), whereas both STAT4 and STAT3 regulate IL-21 expression by human CD4+ T cells (Ma et al., 2009; 2012a; Schmitt et al., 2009; 2013). Interestingly, STAT1 can either enhance (Choi et al., 2013a; Nakayamada et al., 2014) or inhibit (Ray et al., 2014) Bcl6 expression and Tfh cell differentiation. Regarding murine CXCR5, it is feasible that IL6-driven STAT3 (and STAT1) binds at the CXCR5 promoter in conjunction with Maf and Batf and drives CXCR5 expression in an E protein-dependent manner (Ise et al., 2011). This remains to be tested. In contrast to mice, STAT3 and STAT4 may be equally important in human Tfh differentiation (Ma et al., 2012a; Schmitt et al., 2013) (Fig. 1B). In contrast, STAT5 represses Tfh differentiation (Johnston et al., 2012). The opposing roles of STAT3 and STAT5 in Tfh differentiation appears similar to the antagonistic roles of STAT3 and STAT5 in Th17 differentiation (Yang et al., 2011b). These multifaceted regulators of Tfh differentiation also highlight the need to always consider the complex features of CD4+ T cell gene regulation.
The mechanisms by which Bcl6 controls CD4+ T cells have remained only partially elucidated. Bcl6 is a DNA binding transcription factor, and it contains three main domains, the BTB (broad complex, tramtrack, bic-a-brac), RDII (repressor domain II, or ‘middle domain’), and the Zn finger DNA binding domain (Fig. 3). However, within each of those domains Bcl6 has the capacity to interact with a range of proteins, particularly transcription factors and chromatin modifiers (Fig. 3). The Bcl6 BTB domain is known to bind multiple other BTB-containing and non-BTB containing proteins. The Bcl6 Zn finger domain actually consists of three different Zn finger pairs, one of which is involved in DNA binding, while the others are involved in protein-protein interactions (Fig. 3). Together, these features allow for vast combinatorial possibilities for Bcl6 control of gene expression.
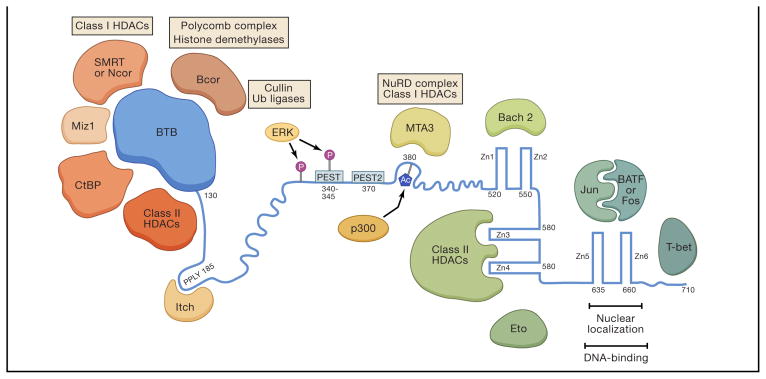
Figure 3. Bcl6 protein domain structure and interacting proteins.
The functional domains of Bcl6 are shown with regions of interest indicated. Bcl6 interacting proteins are also shown with the Bcl6 regions with which they interact, if known. Numbers indicate amino acid positions of human Bcl6. Bcl6 is shown in blue. ‘BTB’ is the broad-complex, tramtrack, and bric-à-brac domain. Bcl6 self dimerizes via the BTB domain, but the self-dimerization is not shown, for simplicity. ‘HDAC’, histone deacetylase complex. ‘Ub’, ubiquitin. ‘PEST’ indicates a PEST (proline, glutamic acid, serine, threonine-rich) ubiquitination domain (1 and 2) in Bcl6. Ⓟ symbolizes phosphorylation. Ac symbolizes acetylation, and the acetylation is mediated by p300. The Zn fingers are numbered 1–6 (‘Zn1’). NuRD, nucleosome-remodeling and histone deacetylation complex.
Bcl6 has only been described as a repressor. In GC B cells there is no clear evidence of Bcl6 directly binding and activating any gene, though it clearly results in indirect upregulation of many genes (Hatzi et al., 2013). In Tfh cells Bcl6 appears to participate in control of at least four major categories of genes: cell migration, repression of alternative fates, Tfh differentiation, and Tfh products (Fig. 4A). Bcl6 expression is closely associated with CXCR5 expression in vivo (Baumjohann et al., 2011; Choi et al., 2011; 2013b; Johnston et al., 2009; Pepper et al., 2011). Nevertheless, CXCR5 expression can be initiated independently of Bcl6 (Liu et al., 2012). Furthermore, memory Tfh cells continue to express CXCR5 without evidence of elevated Bcl6 expression (Chevalier et al., 2011; Hale et al., 2013; Locci et al., 2013). Genome-wide Bcl6 occupancy in GC Tfh cells has revealed that Bcl6 binds over 3,000 genes, with several key patterns emerging (Hatzi et al.). Bcl6 binds many migration-associated genes, apparently inhibiting GC Tfh cells from mislocalizing to the T cell zone or sites of inflammation. Bcl6 expression enhances CXCR5, CXCR4, and PD-1 expression (Kroenke et al., 2012), but Bcl6 does not appear to regulate them via direct binding and induction of gene expression (Hatzi et al.). Furthermore, Bcl6 is an important repressor of alternative cell fates (Fig. 4A and B), which is discussed separately in the section below.
Figure 4. An architectural blueprint conceptual model of CD4 T+ cell differentiation programs.
(a) Regulation of Tfh differentiation by Bcl6 and cooperating Tfh transcription factors (‘TFs’). Tfh cell biology can be divided into four categories, indicated in different colors, with representative genes shown for each category. All differentiation and product genes shown are upregulated in Tfh cells. All alternative fates genes shown are downregulated, and are grouped in subcategories (Th1, Th2, IL-2 and Blimp1, Th17, and forkhead box gene regulation). Genes in the location category include genes that are upregulated (+) or downregulated (−).
(b) An architectural blueprint model of Tfh cell differentiation and how it relates to other CD4+ T cell differentiation programs, within a single cell. The Tfh cell differentiation program (shown in 4a) is projected as a flat plane, as if it were the architectural blueprint for a floor of a multistory building, but in this case it is the blueprint within a CD4+ T cell instead of a building. The Th1 cell program is projected as the blueprint of another floor. Bcl6 prevents activity of the Th1 cell program in a Tfh cell by blocking the expression of genes central to the Th1 cell blueprint. Extracellular signals can enter as inputs from the surface of the cell, such as through IL2R or IFNγR, shown at the edge of the Th1 level. Proteins whose genes are inhibited by Bcl6 are indicated by a red —|. The Th17 cell program is projected as another blueprint, which is also inhibited by Bcl6. The Th2 and Treg cell programs are not shown due to space constraints but are conceptually analogous to the Th1 and Th17 cell program blueprints.
Layers of Differentiation
As noted above, the vast majority of GC Tfh cells have highly conserved gene expression. However, GC Tfh cells interconvert with Tfh cells, therefore the GC Tfh cell gene expression program is neither terminal nor immutable. Tfh outside of GCs have more heterogeneous gene expression profiles, perhaps related to their lower expression of Bcl6, resulting in less repression of alternative cell fate programs. Heterogeneity among Tfh cells is not unlike Th17, Th1, Th2, or even Treg cells, where substantial heterogeneity is observed. For example, it is well established that Th17 cells can be converted to Th1 or Treg cells. IL17+IFNγ+ CD4+ T cells are observed under a number of conditions. There have been longstanding data indicating interconversion of Th1 and Th2 cells. It is quite clear that epigenetic marks require continuous maintenance and can be changed when cells enter different environments or experience new external signals (Mukasa et al., 2010). The behavior of the Tfh differentiation program is like that of other CD4+ T cell differentiation programs in this regard.
One way to conceptualize this biology is with architectural blueprints showing layers of differentiation, as if a cell is a multistory building, and a single floor represents a differentiation program (Figure 4B). Each differentiation program has a specific blueprint, which is the gene expression program controlled by a core transcription factor network and external signals. In the case of the Tfh program, the core transcription factors are Bcl6, Maf, and STAT3 (STAT3 and STAT4 in humans), with supporting contributions by additional transcription factors. These transcription factors control four general aspects of Tfh cell biology, including location, function, and differentiation/positive reinforcement (Fig 4A). The fourth important attribute of Tfh biology is repression of alternative cell fate differentiation programs. One obvious way this could be accomplished is by downregulating cytokine receptors necessary for Th1 [IL-12R, interferon-γ receptor-1 (IFNGR1)], Th2 (IL-4R), pTreg (TGF-βR), or Th17 (IL-23R) cell differentiation. Indeed, Bcl6 directly represses each of these genes (Hatzi et al.), thereby cutting off alternative inductive signals. Bcl6 also represses Blimp-1 (Johnston et al., 2009), which is strongly associated with non-Tfh cell fates (Fig. 4). Nevertheless, external cues can still induce Th1, Th2, or Th17 differentiation signals in the cell through the residual cytokine receptor expression. This can result in expression of some amount of Th1, Th2, or Th17 genes. Thus, Bcl6 inhibits the Th1, Th17, Treg, and Th2 blueprints by also directly targeting central Th1, Th17, Treg, and Th2 transcription factors and cytokine genes for repression (Hatzi et al.; Nurieva et al., 2009) (Fig. 4B). IFN-γ, IL-17, or IL-5 and IL-13 expression is uncommon in human GC Tfh cells (Kroenke et al., 2012; Ma et al., 2009), indicating that the Bcl6 amounts in GC Tfh are sufficient to block Th1, Th2, and Th17 gene expression in the majority of in vivo conditions. However, mouse models have demonstrated that, under conditions of intense polarization, expression of other differentiation programs occurs within GC Tfh cells. For example, in a strongly Th1 systemic LCMV infection GC Tfh cells express IFN-γ (Yusuf et al., 2010). In the systemic autoimmunity prone BXD2 mice, spontaneous GC development is associated with IL17+ RORγt+ GC Tfh cells (Ding et al., 2013). The canonical Tfh, Th1, Th2, and Th17 blueprints can also interact in unexpected ways, further adding to the biological complexity, such as enhancement of Tfh cell frequencies in the presence of excessive IFN-γ (Lee et al., 2012). In summary, the majority of GC Tfh cells exhibit a canonical gene expression profile, which is conserved across species and predominantly excludes expression of Th1, Th17, Th2, Th9 and Treg cell-associated genes. Nevertheless, substantial heterogeneity of GC Tfh (and Tfh) phenotypes are possible depending on environmental cues.
Complexities of Tfh cell differentiation and memory
An alternative model to consider is that Tfh differentiation is a secondary program, such that there are Th1 cell-type Tfh (Tfh1), Th2 cell-type Tfh (Tfh2), and Th17 cell-type Tfh (Tfh17) cells (Figure S1) (Crotty, 2011). In this conceptual framework, there are Th1 effector cells and Tfh1 cells in response to a viral infection, for example. In support of this model, Tfh cells in the context of viral infection express some Tbet and IFN-γ (Johnston et al., 2009; Yusuf et al., 2010), and there are rare IFN-γ+ Tfh in L. major infected mice (Reinhardt et al., 2009). There are also examples of IL-17+ Tfh cells in autoimmune prone BXD2 mice (Ding et al., 2013). IL-5 or IL-13 Th2 cytokine-expressing Tfh cells were reported (Zaretsky et al., 2009). However, a more recent extensive study concluded that Tfh cells do not express IL-13 or IL-5 (Liang et al., 2012). Transfer experiments are challenging to interpret, since the outcome can be the result of outgrowth of a small number of cells that did not share the biology of the majority of the cells. Cell transfers appear to show that Th2 cells can convert into Tfh cells (Zaretsky et al., 2009), and Th17 cells can convert into Tfh cells (Hirota et al., 2013). However, in other studies Tfh, Th1, and Th17 cells have largely retained their identities after transfer (Choi et al., 2013b; Hale et al., 2013; Weber et al., 2012). When faced with the human biology, the interpretations are more complicated. While very few GC Tfh cells express non-Tfh cell cytokines in lymphoid tissue (Kroenke et al., 2012; Ma et al., 2009), when observing memory Tfh cells in human blood a high percentage of the memory Tfh cells express chemokine receptors generally affiliated with human Th1, Th17, or Th2 cell memory cells (Morita et al., 2011). However, conventional chemokine receptor-based definitions of human blood central memory Th17 and Th2 cells are flawed, as only a small percentage of those cells actually produce Th17 or Th2 cell-associated cytokines upon restimulation. Additional complexities to understanding CD4+ T cell differentiation programs include overlapping biology, including the observation that Tbet can form a complex with Bcl6 and both inhibit major functions for Bcl6 and enhance other functions (Oestreich et al., 2012). Finally, there are a great deal of data not in support of the secondary program conceptualization of Tfh cell differentiation, many of which were stated earlier in this review. In particular, Tfh cell differentiation can start at DC priming, and can be distinguished from Th1 cells within the first two cell divisions in vivo during an acute infection (Choi et al., 2011). Second, Tfh cell differentiation is independent in that it does not require Th1, Th2, or Th17 cell programming (Nurieva et al., 2008).
Both conceptual models do have strengths. While it is appealing to have a simple shared nomenclature, this is inherently problematic for cells as heterogeneous as CD4+ T cells are. From a semantic perspective, this is in part a question of the perceived primary biology of the cell in question. Is it primarily a Tfh cell, helping B cells? Or is it primarily a Th1 cell, inducing inflammation in an infected tissue? Treg cells are an illustrative example here. While there are clearly Tbet+ or RORγt+ Tregs (Th1-related and Th17-related), these cells are clearly still Tregs, as their primary biology is to dampen/regulate immune responses. A similar perspective is applicable to Tfh cells. While there can be Tbet+ or RORγt+ Tfh cells, these cells are clearly still Tfh cells, as their primary biology is to help B cells. Hence, the conceptual frameworks discussed thoroughly earlier in the review and shown in Figures 1B and 4B seem most relevant. Th1, Th2, or Th17 cell-associated cytokines produced by Tfh cells or GC Tfh cells can be instructive for class switch recombination and are therefore not irrelevant. However, it must be noted that the importance of this cytokine biology appears to be exacerbated in mice, as the difference in IgG class switch recombination between Th1 and Th2 conditions is dramatic in mice (murine IgG1 and murine IgG2a/b/c have very different functions), while human IgG1 (the dominant isotype) has broad functionality. In addition, while IFN-γ is a major class switch recombination factor in mice, its role in human IgG class switching appears to be trivial (Holland and Casanova, 2006). In summary, Tfh cells can exhibit features consistent with the Tfh1/2/17 cell conceptual framework, but on balance the majority of the data support Tfh cells as a distinct cell type as the more instructive conceptual framework.
Within the context of categorizing cells and differentiation programs, T follicular regulatory (Tfr) cells are a separate issue, as these cells are thymic Treg (tTreg or ‘natural Treg’) cells that also express Bcl6 and CXCR5 (Chung et al., 2011; Linterman et al., 2011; Sage et al., 2013; Wollenberg et al., 2011). They are analogous to type-specific Treg cells, such as Th1 cell-specific Treg cells, which express Tbet as a mechanism to express CXCR3. Expression of the appropriate chemokine receptor is a necessary part of Treg cell biology, so that they migrate to the same location of the effector cells (Tfh or Th1 cells in these examples) such that they can dampen immune responses at that site. Bcl6+CXCR5+ induced pTreg cells have yet to be identified.
Bcl6 does have some role in the development of T cell memory, which also impacts interpretations of Tfh cell biology at different time points. While Bcl6 has a role in development of memory CD8 T cells, the cell intrinsic effect of Bcl6 deficiency is modest (Cui et al., 2011). In CD4+ T cells, the role of Bcl6 in memory requires further investigation. Early experiments with Bcl6-deficient CD4+ T cells lacked key controls for cellular rejection (Ichii et al., 2007), while more recent studies did not examine Bcl6-deficient CD4+ T cells at memory time points (Hale et al., 2013; He et al., 2013; Pepper et al., 2011). An additional challenge to studying Tfh cell memory is that active Tfh cell responses continue for much longer than Th1 or Th2 cell responses after an acute immunization or infection. Tfh cells are active for the duration of the GCs, with continuous exposure to antigens, which frequently last for 60 or more day. Therefore, to stringently study resting Tfh cell memory it is likely necessary to wait more than 90 days, depending on the model system, while resting Th1 or Th2 cell memory develops by day 30 after an acute antigen exposure.
Tfh Cell Function
The most prominent role of Tfh cells is their requirement for GC development and function. The GC is the primary site of B cell affinity maturation. The extraordinary process of Ig gene somatic hypermutation and selection is one of the miracles of immunology. A GC reaction is effectively evolution in miniature. Regulation of Tfh cell help is central for achieving the goal of GC responses, which is to generate and select GC B cells with higher affinity for the pathogen (Victora and Nussenzweig, 2012). In GCs, B cells circulate through two regions: the light zone (LZ) and the dark zone (DZ). In the LZ, GC B cells bind to antigen and present antigen peptide:MHC complexes to Tfh cells that, in turn, provide help signals to GC B cells that are essential for their survival and proliferation. GC B cells that receive survival signals then migrate to the DZ, where they undergo proliferation and somatic hypermutation (SHM), allowing for the generation of BCRs with a spectrum of affinities to antigen. These mutated GC B cells then move back to the LZ, where the highest affinity B cells are selected again by the Tfh cells for another round of proliferation and mutation. Multiple studies have revealed the roles of Tfh cells in regulating GCs (Crotty, 2011; Victora and Nussenzweig, 2012). Tfh cells regulate GC size (Hams et al., 2011; Johnston et al., 2009; Rolf et al., 2010), restrict low-affinity B cell entry into the GC, support high affinity B cell occupancy of the GC (Schwickert et al., 2011), and select high affinity B cells during affinity maturation (Good-Jacobson et al., 2010; Victora et al., 2010). In addition, most GC B cells cannot trigger BCR signaling (Khalil et al., 2012). Therefore the GC B cells are dependent on help signals from the Tfh cells to discriminate which GC B cells proliferate. Tfh cells selectively provide help to the B cells with the most antigen peptides, which are the high affinity B cells that bound and endocytosed the most antigen. Surprisingly, the amount of help provided by the Tfh cell directly translates to the number of cell divisions—and mutations—a GC B cell will undergo in the DZ in a single selection cycle (Gitlin et al., 2014). Therefore, Tfh cells regulate the mutation diversity of BCRs in a GC. Strong Tfh help to B cells can result in GC B cell accumulation of multiple mutations in a single round of selection, which may be important for generating rare multiple mutation combinations, particularly if a single mutation happens to be deleterious on its own. For example, development of broadly neutralizing antibodies against HIV is very difficult and requires extensive somatic hypermutation (Burton et al., 2012). Some of these antibodies develop new disulfide bonds that stabilize long CDR loops (Doria-Rose et al., 2014). Single cysteines are likely to be deleterious, and are unlikely to be positively selected. Simultaneous generation of two cysteine mutations in a single selection cycle may be an example of how strong Tfh cell help to GC B cells controls the mutation spectrum of GC B cells and may allow some B cells to overcome large hurdles in the evolutionarily landscape to develop high affinity for their target antigen.
The help signals provided by Tfh cells to GC B cells consist of both cytokines and cell surface receptors. While the help signals are incompletely characterized, CD40L, IL-21, and IL-4 are major ‘help’ molecules produced by GC Tfh cells to keep GC B cells alive and induce their proliferation (Crotty, 2011). IL-21 and IL-4 are also potent inducers of IgG1 class switch recombination for human B cells (Avery et al., 2008), and IgG1 is the most prevalent class switched immunoglobulin. The help factors IL-21 and IL-4 are not simply produced by Tfh cells upon TCR engagement; they are regulated by additional cell surface molecules that effectively communicate additional dialogue between the Tfh cell (the provider of help) and the GC B cell soliciting help. IL-4 is produced by GC Tfh cells in a SLAM-dependent manner (Yusuf et al., 2010), and SLAM is selectively upregulated on LZ GC B cells (Victora et al., 2012). Separately, ICOS triggering is important for Tfh cell production of IL-21 (Morita et al., 2011). Manipulating the amount of Tfh cell help given to B cells dramatically alters affinity maturation. Indiscriminate Tfh cell help to both high and low affinity B cells leads to the generation of lower affinity antibodies over time (Victora et al., 2010). Therefore, the outcome of the GC B cell response depends on the proper regulation of Tfh cell help to high affinity versus low affinity B cells. For this reason, negative signaling through SLAMF6 may serve a critical purpose in limiting Tfh cell help (Kageyama et al., 2012), thereby enhancing selective pressure.
Inhibitory signals to Tfh cells may limit Tfh cell help signals or Tfh cell proliferation, or both. One of the central challenges of Tfh cells in GCs is that they are constantly exposed to antigen and must retain sensitive TCR signaling—so as to distinguish between GC B cells with modest differences in their numbers of p:MHC complexes—but the Tfh cell needs to not respond by proliferating, in contrast to most effector CD4+ T cells. Instead, the Tfh cell needs to respond to that TCR signaling by only providing transient help to the cognate B cells. After all, the purpose of the GC is for the GC B cells to rapidly proliferate, mutate, and evolve, not the Tfh cell. As such, control of multiple components of TCR signaling and downstream pathways [including regulation of transcription elongation by Cyclin T1 (Chen et al, 2014)] may be essential for striking the balance in Tfh cells between sensitive sensing of p:MHC and unwanted proliferation. The extremely high amount of PD-1 expressed by GC Tfh cells critically contributes to limiting GC Tfh cell proliferation in GCs by dampening TCR signaling. Nevertheless, excessive availability of the PD-1 ligand PD-L1 on GC B cells can occur and is associated with defective Tfh cell function (Cubas et al., 2013). High amounts of PD-L1 on GC B cells results in reduced ICOS and IL-21 expression, resulting in minimal help to the PD-L1hi GC B cells (Cubas et al., 2013). In addition to PD-1 and SLAMF6, Tfh cells express multiple additional inhibitory receptors, which may control both Tfh cell proliferation and function. Immunotherapy targeting Tfh cells by blocking PD-L1 and LAG-3 in a mouse malaria model led to an increase in Tfh cell and GC B cell numbers, followed by rapid development of protective antibodies and clearance of the Plasmodium (Butler et al., 2012). In summary, Tfh cell help to GC B cells is provided via a combination of secreted and surface bound help molecules, which are counter regulated by inhibitory molecules that can critically affect the amount of help provided, the duration of the T:B interaction (which is related to the amount of help provided), or the cocktail of molecules expressed by the Tfh cell.
Negative regulation of the GC is also accomplished by Tfr cells, introduced earlier in this review. Tfr cells can have potent inhibitory roles in GCs, and it is thought that the ratio between the Tfh and Tfr cells is an important determinant of the GC reaction. Nevertheless, there are many unanswered questions about Tfr cells. It is unclear whether the Tfr cells primarily function by interacting with T cells (Tfh cells in this case), which is the primary mechanism by which tTregs cells act, or alternatively, the Tfr cells may mainly act directly on the GC B cells. There is currently no evidence for Tfr cells regulating immune responses in an antigen-specific manner, and it may be that their primary role is to eliminate autoreactive B cells that arise via mutation in the GC (Linterman et al., 2011).
It is worth noting that while the most important and well-studied role of Tfh cells is their requirement in GCs, Tfh cells also have critical roles outside the GC. This is an area that is understudied. In addition, there are GC independent memory B cells, which form early during responses and generally have no mutations. These memory B cells are Tfh cell-independent (Kaji et al., 2012).
Tfh Cells in Disease
Tfh cells are essential for the generation of most isotype switched and affinity matured antibodies, and therefore they have an obvious role in protective immunity against pathogens. Antibodies are necessary for the control of chronic lymphocytic choriomeningitis virus (LCMV) infection, and defects in Tfh cell frequencies result in failure to control LCMV (Fahey et al., 2011; Harker et al., 2011). LCMV has a surface that is very difficult for antibodies to bind, hence substantial T cell help to B cells is necessary to drive the slow generation of B cell responses capable of controlling the viremia. A similar problem exists for HIV, made even worse by the extreme mutability of HIV. Tfh cell frequencies are associated with the amount and quality of antibody responses against SIV (simian immunodeficiency virus, a close relative to HIV) in SIV-infected macaques (Petrovas et al., 2012). Impaired Tfh cell help to B cells is observed in HIV-infected individuals, which appears to exacerbate the difficulty in generating neutralizing antibodies against HIV (Cubas et al., 2013). Conversely, individuals who have unusually elevated frequencies of highly functional memory Tfh cells (PD1loCXCR3−CXCR5+) are more likely to make broadly neutralizing antibodies against HIV (Locci et al., 2013), again consistent with the hypothesis that Tfh cells are a key limiting resource for the development of high affinity B cell responses.
Almost all licensed human vaccines work on the basis of long-term protective antibody responses, and therefore it is reasonable to assume that Tfh cells are mediators of the development of the protective immunity generated by licensed human vaccines. There is evidence that Tfh cell help is a limiting factor in humans for generating antibody responses after immunizations (Bentebibel et al., 2013; Duan et al., 2014; Pallikkuth et al., 2012; Schmitt et al., 2013). Therefore, learning to control Tfh cells would almost certainly enhance development of new or improved vaccines. As described above, boosting Tfh cell numbers with immunotherapy led to dramatically improved antibody responses in mouse malaria (Butler et al., 2012). Therefore, it may also be that vaccines that employ monoclonal antibodies targeting Tfh cell inhibitory pathways could adjuvant vaccine efficacy.
Tfh cells are not only important in control of pathogens, they are also important in control of the commensal microbiota. The predominant antibody isotype at mucosal surfaces is IgA, and the majority of IgA is T-dependent, based on data from MHCII-or T cell-deficient animals. In addition, in mice with normal IgA levels but having a defect in productive GC somatic hypermutation, the microbiota was expanded and increased susceptibility to the intestinal pathogen Yersinia pestis was observed (Wei et al., 2011). Thus, Tfh cells are required for sufficient antibody response quality to control both commensals and pathogens. Nevertheless, intestinal immune responses are characterized in many ways by their balance between protecting against microbes and avoiding undo inflammation. This paradigm has been seen to also hold true for Tfh cells, as a defect in Tfr cells can result in more Tfh cells, larger antibody responses, and less microbiota diversity which, paradoxically, can result in less healthy gut homeostasis (Kawamoto et al., 2014). Interestingly, Tfh cells are also essential for IgE production (Liang et al., 2012), and therefore Tfh cells are important in allergic responses; however, the literature on Tfh cells in allergy is currently limited and this is an area in need of much more investigation.
Tfh cells are central players in a number of autoimmune diseases, and it is hoped that a greater understanding of Tfh cells can result in new therapeutic approaches against major autoimmune diseases. Increased frequencies of Tfh-like cells (CXCR5+ and PD-1hi or ICOShi) in peripheral blood are observed in subsets of patients with Sjogren’s syndrome (Simpson et al., 2010; Szabo et al., 2013), juvenile dermatomyositis (Morita et al., 2011), and systemic lupus erythematosus (He et al., 2013; Simpson et al., 2010). Each of those diseases is associated with extensive autoantibody production. Autoimmune diseases for which autoantibodies play a direct pathogenic role are likely to have Tfh cells as an important component of the disease, for example, granulomatosis with polyangiitis (GPA). Separately, there are functions of Tfh cells that may be unrelated to antibody responses, per se. For autoimmune diseases like SLE, autoantibodies are considered primarily markers of disease, not causes of disease. Interestingly, a common variation of SLE in humans is lupus nephritis, which can result in severe kidney dysfunction associated with inflammation and the accumulation of high concentrations of autoantibodies. Importantly, ectopic clusters of Tfh cells and B cells can be found in the inflamed kidneys of such patients, including GC Tfh cells and GC B cells (Liarski et al., 2014). These and other data suggest that Tfh cells may be major regulators of ectopic follicles in autoimmune diseases (Craft, 2012). Thus, Tfh cells may contribute to autoimmune diseases both by facilitating the aberrant generation of autoantibodies and by facilitating the formation or maintenance of ectopic follicles, which serve as nucleation points for other cells that may be directly pathogenic in the autoimmune disease.
While roles for Tfh cells in infectious diseases, allergy, and autoimmunity were expected, it was not anticipated that Tfh cells would be relevant for cancer immunity. Therefore, it was surprising that a strong positive correlation was observed in breast cancer between a Tfh gene signature in the tumor tissue and long term patient survival (Gu-Trantien et al., 2013). Histology confirmed that a Tfh cell gene signature was associated with infiltration of the tumor margin with Tfh cells and the development of B cell follicles or ectopic lymphoid organ-like structures. More surprising, similar results were found for human colorectal cancer (Bindea et al., 2013). Positive outcomes in controlling the cancers were associated with Tfh cell gene signatures, including CXCL13 and IL-21 expression. Even more intriguing, tumor deletion of the CXCL13 gene was associated with cancer progression (Bindea et al., 2013). Therefore, this invites the speculation that Tfh cells may have shared roles in cancer immunity and autoimmune diseases independently of helping antibody responses. The Tfh cells may facilitate or maintain ectopic B cell-rich lymphoid structures, sustaining a local microenvironment that is nurturing for other immunological cell types with more direct roles in affecting disease progression or regression, whether they be CTL in the context to tumor immunity (Bindea et al., 2013; Gu-Trantien et al., 2013), or Th17 cells in the context of multiple sclerosis (Hauser et al., 2008).
In conclusion, much has been recently discovered about the biology of Tfh cells and germinal centers. While major knowledge gaps remain, and Tfh cell biology is clearly complex, it is nevertheless easy to predict that in the coming years we will see an ever growing impact of the study of Tfh cells as we appreciate more and more that Tfh cells have pivotal roles in a range of diseases.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avery DT, Bryant VL, Ma CS, de Waal Malefyt R, Tangye SG. IL-21-induced isotype switching to IgG and IgA by human naive B cells is differentially regulated by IL-4. J Immunol. 2008;181:1767–1779. doi: 10.4049/jimmunol.181.3.1767. [DOI] [PubMed] [Google Scholar]
- Ballesteros-Tato A, Randall TD. Priming of T follicular helper cells by dendritic cells. Immunol Cell Biol. 2014;92:22–27. doi: 10.1038/icb.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A, León B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batten M, Ramamoorthi N, Kljavin NM, Ma CS, Cox JH, Dengler HS, Danilenko DM, Caplazi P, Wong M, Fulcher DA, et al. IL-27 supports germinal center function by enhancing IL-21 production and the function of T follicular helper cells. J Exp Med. 2010;207:2895–2906. doi: 10.1084/jem.20100064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Kageyama R, Clingan JM, Morar MM, Patel S, de Kouchkovsky D, Bannard O, Bluestone JA, Matloubian M, Ansel KM, et al. The microRNA cluster miR-17~92 promotes TFH cell differentiation and represses subset-inappropriate gene expression. Nat Immunol. 2013;14:840–848. doi: 10.1038/ni.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Okada T, Ansel KM. Cutting Edge: Distinct Waves of BCL6 Expression during T Follicular Helper Cell Development. J Immunol. 2011;187:2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, Kuchroo VK. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat Immunol. 2009;10:167–175. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, Flaño E, Mejias A, Albrecht RA, Blankenship D, et al. Induction of ICOS+CXCR3+CXCR5+ TH Cells Correlates with Antibody Responses to Influenza Vaccination. Sci Transl Med. 2013;5:176ra32. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- Bollig N, Brüstle A, Kellner K, Ackermann W, Abass E, Raifer H, Camara B, Brendel C, Giel G, Bothur E, et al. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc Natl Acad Sci USA. 2012;109:8664–8669. doi: 10.1073/pnas.1205834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler NS, Moebius J, Pewe LL, Traore B, Doumbo OK, Tygrett LT, Waldschmidt TJ, Crompton PD, Harty JT. Therapeutic blockade of PD-L1 and LAG-3 rapidly clears established blood-stage Plasmodium infection. Nat Immunol. 2012;13:188–195. doi: 10.1038/ni.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Belanger S, Frederick MA, Li B, Johnston RJ, Xiao N, et al. In Vivo RNA Interference Screens Identify Regulators of Antiviral CD4(+) and CD8(+) T Cell Differentiation. Immunity. 2014;41(2):325–338. doi: 10.1016/j.immuni.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier N, Jarrossay D, Ho E, Avery DT, Ma CS, Yu D, Sallusto F, Tangye SG, Mackay CR. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. J Immunol. 2011;186:5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- Choi YS, Eto D, Yang JA, Lao C, Crotty S. Cutting Edge: STAT1 Is Required for IL-6-Mediated Bcl6 Induction for Early Follicular Helper Cell Differentiation. J Immunol. 2013a;190:3049–3053. doi: 10.4049/jimmunol.1203032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S. ICOS Receptor Instructs T Follicular Helper Cell versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Yang JA, Yusuf I, Johnston RJ, Greenbaum J, Peters B, Crotty S. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J Immunol. 2013b;190:4014–4026. doi: 10.4049/jimmunol.1202963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nature Medicine. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, Huang W, Parkurst CN, Muratet M, et al. A validated regulatory network for th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craft JE. Follicular helper T cells in immunity and systemic autoimmunity. Nat Rev Rheumatol. 2012;8:337–347. doi: 10.1038/nrrheum.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- Cubas RA, Mudd JC, Savoye AL, Perreau M, Van grevenynghe J, Metcalf T, Connick E, Meditz A, Freeman GJ, Abesada-Terk G, et al. Inadequate T follicular cell help impairs B cell immunity during HIV infection. Nature Medicine. 2013;19:494–499. doi: 10.1038/nm.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W, Liu Y, Weinstein JS, Craft J, Kaech SM. An Interleukin-21- Interleukin-10-STAT3 Pathway Is Critical for Functional Maturation of Memory CD8(+) T Cells. Immunity. 2011;35:792–805. doi: 10.1016/j.immuni.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Li J, Wu Q, Yang P, Luo B, Xie S, Druey KM, Zajac AJ, Hsu HC, Mountz JD. IL-17RA is essential for optimal localization of follicular Th cells in the germinal center light zone to promote autoantibody-producing B cells. J Immunol. 2013;191:1614–1624. doi: 10.4049/jimmunol.1300479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Schramm CA, Gorman J, Moore PL, Bhiman JN, DeKosky BJ, Ernandes MJ, Georgiev IS, Kim HJ, Pancera M, et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature. 2014;509:55–62. doi: 10.1038/nature13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Chen X, Liang Z, Zeng Y, Zhu F, Long L, McCrae MA, Zhuang H, Shen T, Lu F. Genetic polymorphisms of CXCR5 and CXCL13 are associated with non-responsiveness to the hepatitis B vaccine. Vaccine. 2014 doi: 10.1016/j.vaccine.2014.07.064. [DOI] [PubMed] [Google Scholar]
- Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey LM, Wilson EB, Elsaesser H, Fistonich CD, McGavern DB, Brooks DG. Viral persistence redirects CD4 T cell differentiation toward T follicular helper cells. J Exp Med. 2011;208:987–999. doi: 10.1084/jem.20101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N, McHeyzer-Williams LJ, Rosen H, McHeyzer-Williams MG. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat Immunol. 2009;10:375–384. doi: 10.1038/ni.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014;509:637–640. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka R, Barnett LG, Silver JS, O’Neill PJ, Hunter CA, Cancro MP, Laufer TM. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J Immunol. 2011;187:1091–1095. doi: 10.4049/jimmunol.1100853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, Shlomchik MJ. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol. 2010;11:535–542. doi: 10.1038/ni.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu-Trantien C, Loi S, Garaud S, Equeter C, Libin M, de Wind A, Ravoet M, Le Buanec H, Sibille C, Manfouo-Foutsop G, et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J Clin Invest. 2013;123:2873–2892. doi: 10.1172/JCI67428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale JS, Youngblood B, Latner DR, Mohammed AUR, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hams E, McCarron MJ, Amu S, Yagita H, Azuma M, Chen L, Fallon PG. Blockade of B7-H1 (programmed death ligand 1) enhances humoral immunity by positively regulating the generation of T follicular helper cells. J Immunol. 2011;186:5648–5655. doi: 10.4049/jimmunol.1003161. [DOI] [PubMed] [Google Scholar]
- Hannedouche S, Zhang J, Yi T, Shen W, Nguyen D, Pereira JP, Guerini D, Baumgarten BU, Roggo S, Wen B, et al. Oxysterols direct immune cell migration via EBI2. Nature. 2011;475:524–527. doi: 10.1038/nature10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker JA, Dolgoter A, Zuniga EI. Cell-intrinsic IL-27 and gp130 cytokine receptor signaling regulates virus-specific CD4+ T cell responses and viral control during chronic infection. Immunity. 2013;39:548–559. doi: 10.1016/j.immuni.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 2011;334:825–829. doi: 10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, Zumbo P, Kirouac K, Bhaskara S, Polo JM, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep. 2013;4:578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzi K, Nance JP, Haddad EK, Melnick AM, Crotty S. Tfh Bcl6 ChIP-Seq. (unpublished) [Google Scholar]
- Hauser SL, Waubant E, Arnold DL, Vollmer T, Antel J, Fox RJ, Bar-Or A, Panzara M, Sarkar N, Agarwal S, et al. B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N Engl J Med. 2008;358:676–688. doi: 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- Haynes NM, Allen CDC, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J Immunol. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- He J, Tsai LM, Leong YA, Hu X, Ma CS, Chevalier N, Sun X, Vandenberg K, Rockman S, Ding Y, et al. Circulating Precursor CCR7(lo)PD-1(hi) CXCR5(+) CD4(+) T Cells Indicate Tfh Cell Activity and Promote Antibody Responses upon Antigen Reexposure. Immunity. 2013;39:770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- Hedrick SM, Hess Michelini R, Doedens AL, Goldrath AW, Stone EL. FOXO transcription factors throughout T cell biology. Nat Rev Immunol. 2012;12:649–661. doi: 10.1038/nri3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu Y, Suto A, Kashiwakuma D, Kanari H, Kagami SI, Ikeda K, Hirose K, Watanabe N, Grusby MJ, Iwamoto I, et al. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-beta inhibits c-Maf-induced IL-21 production in CD4+ T cells. J Leukoc Biol. 2010;87:703–712. doi: 10.1189/jlb.0909639. [DOI] [PubMed] [Google Scholar]
- Hirota K, Turner JE, Villa M, Duarte JH, Demengeot J, Steinmetz OM, Stockinger B. Plasticity of Th17 cells in Peyer’s patches is responsible for the induction of T cell-dependent IgA responses. Nat Immunol. 2013;14:372–379. doi: 10.1038/ni.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland SM, Casanova J-L. Inherited Disorders of the Interleukin-12/Interferon-Gamma Circuit. In: Ochs HD, editor. Primary Immunodeficiency Diseases : a Molecular & Genetic Approach. Oxford; 2006. pp. 450–467. [Google Scholar]
- Hu J, Havenar-Daughton C, Crotty S. Modulation of SAP dependent T:B cell interactions as a strategy to improve vaccination. Curr Opin Virol. 2013;3:363–370. doi: 10.1016/j.coviro.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichii H, Sakamoto A, Arima M, Hatano M, Kuroda Y, Tokuhisa T. Bcl6 is essential for the generation of long-term memory CD4+ T cells. Int Immunol. 2007;19:427–433. doi: 10.1093/intimm/dxm007. [DOI] [PubMed] [Google Scholar]
- Ise W, Inoue T, McLachlan JB, Kometani K, Kubo M, Okada T, Kurosaki T. Memory B cells contribute to rapid Bcl6 expression by memory follicular helper T cells. Proc Natl Acad Sci USA. 2014 doi: 10.1073/pnas.1404671111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama R, Cannons JL, Zhao F, Yusuf I, Lao C, Locci M, Schwartzberg PL, Crotty S. The receptor Ly108 functions as a SAP adaptor-dependent on-off switch for T cell help to B cells and NKT cell development. Immunity. 2012;36:986–1002. doi: 10.1016/j.immuni.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji T, Ishige A, Hikida M, Taka J, Hijikata A, Kubo M, Nagashima T, Takahashi Y, Kurosaki T, Okada M, et al. Distinct cellular pathways select germline-encoded and somatically mutated antibodies into immunological memory. J Exp Med. 2012;209:2079–2097. doi: 10.1084/jem.20120127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SG, Liu WH, Lu P, Jin HY, Lim HW, Shepherd J, Fremgen D, Verdin E, Oldstone MBA, Qi H, et al. MicroRNAs of the miR-17~92 family are critical regulators of TFH differentiation. Nat Immunol. 2013;14:849–857. doi: 10.1038/ni.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnowski A, Chevrier S, Belz GT, Mount A, Emslie D, D’Costa K, Tarlinton DM, Kallies A, Corcoran LM. B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1. J Exp Med. 2012;209:2049–2064. doi: 10.1084/jem.20111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Maruya M, Kato LM, Suda W, Atarashi K, Doi Y, Tsutsui Y, Qin H, Honda K, Okada T, et al. Foxp3(+) T cells regulate immunoglobulin a selection and facilitate diversification of bacterial species responsible for immune homeostasis. Immunity. 2014;41:152–165. doi: 10.1016/j.immuni.2014.05.016. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Cambier JC, Shlomchik MJ. B cell receptor signal transduction in the GC is short-circuited by high phosphatase activity. Science. 2012;336:1178–1181. doi: 10.1126/science.1213368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Zou YR, Goldstein J, Reizis B, Diamond B. Tolerogenic function of Blimp-1 in dendritic cells. J Exp Med. 2011;208:2193–2199. doi: 10.1084/jem.20110658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, Okada T. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–972. doi: 10.1016/j.immuni.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Silva DG, Martin JL, Pratama A, Hu X, Chang PP, Walters G, Vinuesa CG. Interferon-γ excess leads to pathogenic accumulation of follicular helper T cells and germinal centers. Immunity. 2012;37:880–892. doi: 10.1016/j.immuni.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Liang HE, Reinhardt RL, Bando JK, Sullivan BM, Ho IC, Locksley RM. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat Immunol. 2012;13:58–66. doi: 10.1038/ni.2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liarski VM, Kaverina N, Chang A, Brandt D, Yanez D, Talasnik L, Carlesso G, Herbst R, Utset TO, Labno C, et al. Cell distance mapping identifies functional T follicular helper cells in inflamed human renal tissue. Sci Transl Med. 2014;6:230ra46. doi: 10.1126/scitranslmed.3008146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nature Medicine. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med. 2012;209:1841–1852. doi: 10.1084/jem.20120219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, et al. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity. 2013;39:758–769. doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthje K, Kallies A, Shimohakamada Y, Belz GT, Light A, Tarlinton DM, Nutt SL. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol. 2012;13:491–498. doi: 10.1038/ni.2261. [DOI] [PubMed] [Google Scholar]
- Ma CS, Avery DT, Chan A, Batten M, Bustamante J, Boisson-Dupuis S, Arkwright PD, Kreins AY, Averbuch D, Engelhard D, et al. Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood. 2012a;119:3997–4008. doi: 10.1182/blood-2011-11-392985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CS, Deenick EK, Batten M, Tangye SG. The origins, function, and regulation of T follicular helper cells. J Exp Med. 2012b;209:1241–1253. doi: 10.1084/jem.20120994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner-Nanan B, Deenick EK, Tangye SG. Early commitment of naïve human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol Cell Biol. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- Marshall HD, Chandele A, Jung YW, Meng H, Poholek AC, Parish IA, Rutishauser R, Cui W, Kleinstein SH, Craft J, et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Rivera RR, Miyazaki K, Lin YC, Agata Y, Murre C. The opposing roles of the transcription factor E2A and its antagonist Id3 that orchestrate and enforce the naive fate of T cells. Nat Immunol. 2011;12:992–1001. doi: 10.1038/ni.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukasa R, Balasubramani A, Lee YK, Whitley SK, Weaver BT, Shibata Y, Crawford GE, Hatton RD, Weaver CT. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity. 2010;32:616–627. doi: 10.1016/j.immuni.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun HW, Vahedi G, Hakim O, Handon R, et al. Early Th1 Cell Differentiation Is Marked by a Tfh Cell-like Transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayamada S, Poholek AC, Lu KT, Takahashi H, Kato M, Iwata S, Hirahara K, Cannons JL, Schwartzberg PL, Vahedi G, et al. Type I IFN induces binding of STAT1 to Bcl6: divergent roles of STAT family transcription factors in the T follicular helper cell genetic program. J Immunol. 2014;192:2156–2166. doi: 10.4049/jimmunol.1300675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Hwang D, Yang XO, Kang HS, Ma L, Wang YH, Watowich SS, Jetten AM, Tian Q, et al. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–149. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obst R, van Santen HM, Mathis D, Benoist C. Antigen persistence is required throughout the expansion phase of a CD4(+) T cell response. J Exp Med. 2005;201:1555–1565. doi: 10.1084/jem.20042521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich KJ, Mohn SE, Weinmann AS. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallikkuth S, Parmigiani A, Silva SY, George VK, Fischl M, Pahwa R, Pahwa S. Impaired peripheral blood T-follicular helper cell function in HIV-infected nonresponders to the 2009 H1N1/09 vaccine. Blood. 2012;120:985–993. doi: 10.1182/blood-2011-12-396648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M, Pagán AJ, Igyártó BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovas C, Yamamoto T, Gerner MY, Boswell KL, Wloka K, Smith EC, Ambrozak DR, Sandler NG, Timmer KJ, Sun X, et al. CD4 T follicular helper cell dynamics during SIV infection. J Clin Invest. 2012;122:3281–3294. doi: 10.1172/JCI63039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploquin MJY, Eksmond U, Kassiotis G. B cells and TCR avidity determine distinct functions of CD4+ T cells in retroviral infection. J Immunol. 2011;187:3321–3330. doi: 10.4049/jimmunol.1101006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratama A, Ramiscal RR, Silva DG, Das SK, Athanasopoulos V, Fitch J, Botelho NK, Chang PP, Hu X, Hogan JJ, et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity. 2013;38:669–680. doi: 10.1016/j.immuni.2013.01.011. [DOI] [PubMed] [Google Scholar]
- Qi H, Cannons JL, Klauschen F, Schwartzberg PL, Germain RN. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray JP, Marshall HD, Laidlaw BJ, Staron MM, Kaech SM, Craft J. Transcription factor STAT3 and type I interferons are corepressive insulators for differentiation of follicular helper and T helper 1 cells. Immunity. 2014;40:367–377. doi: 10.1016/j.immuni.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nat Immunol. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolf J, Bell SE, Kovesdi D, Janas ML, Soond DR, Webb LMC, Santinelli S, Saunders T, Hebeis B, Killeen N, et al. SOM-Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J Immunol. 2010;185:4042–4052. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- Sage PT, Francisco LM, Carman CV, Sharpe AH. The receptor PD-1 controls follicular regulatory T cells in the lymph nodes and blood. Nat Immunol. 2013;14:152–161. doi: 10.1038/ni.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N, Ueno H. Blood Tfh Cells Come with Colors. Immunity. 2013;39:629–630. doi: 10.1016/j.immuni.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N, Bustamante J, Bourdery L, Bentebibel SE, Boisson-Dupuis S, Hamlin F, Tran MV, Blankenship D, Pascual V, Savino DA, et al. IL-12 receptor β1 deficiency alters in vivo T follicular helper cell response in humans. Blood. 2013;121:3375–3385. doi: 10.1182/blood-2012-08-448902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N, Liu Y, Bentebibel SE, Munagala I, Bourdery L, Venuprasad K, Banchereau J, Ueno H. The cytokine TGF-β co-opts signaling via STAT3-STAT4 to promote the differentiation of human TFH cells. Nat Immunol. 2014 doi: 10.1038/ni.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt N, Morita R, Bourdery L, Bentebibel SE, Zurawski SM, Banchereau J, Ueno H. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 2009;31:158–169. doi: 10.1016/j.immuni.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwickert TA, Victora GD, Fooksman DR, Kamphorst AO, Mugnier MR, Gitlin AD, Dustin ML, Nussenzweig MC. A dynamic T cell-limited checkpoint regulates affinity-dependent B cell entry into the germinal center. J Exp Med. 2011;208:1243–1252. doi: 10.1084/jem.20102477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman Z, Gitlin AD, Targ S, Jankovic M, Pasqual G, Nussenzweig MC, Victora GD. T follicular helper cell dynamics in germinal centers. Science. 2013;341:673–677. doi: 10.1126/science.1241680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson N, Gatenby PA, Wilson A, Malik S, Fulcher DA, Tangye SG, Manku H, Vyse TJ, Roncador G, Huttley GA, et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010;62:234–244. doi: 10.1002/art.25032. [DOI] [PubMed] [Google Scholar]
- Stone EL, Pepper M, Katayama C, Yang E, Goldrath AW, Crotty S, Hedrick SM. A role for inactivation of the Foxo1 transcription factor in the differentiation of T follicular helper cells (P1131) J Immunol. 2013;190:50–8. [Google Scholar]
- Suto A, Kashiwakuma D, Kagami SI, Hirose K, Watanabe N, Yokote K, Saito Y, Nakayama T, Grusby MJ, Iwamoto I, et al. Development and characterization of IL-21-producing CD4+ T cells. J Exp Med. 2008;205:1369–1379. doi: 10.1084/jem.20072057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo K, Papp G, Barath S, Gyimesi E, Szanto A, Zeher M. Follicular helper T cells may play an important role in the severity of primary Sjögren’s syndrome. Clinical Immunology. 2013;147:95–104. doi: 10.1016/j.clim.2013.02.024. [DOI] [PubMed] [Google Scholar]
- Tubo NJ, Pagán AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, Jenkins MK. Single Naive CD4(+) T Cells from a Diverse Repertoire Produce Different Effector Cell Types during Infection. Cell. 2013;153:785–796. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahedi GC, Poholek A, Hand TW, Laurence A, Kanno Y, O’Shea JJ, Hirahara K. Helper T-cell identity and evolution of differential transcriptomes and epigenomes. Immunol Rev. 2013;252:24–40. doi: 10.1111/imr.12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Victora GD, Dominguez-Sola D, Holmes AB, Deroubaix S, Dalla Favera R, Nussenzweig MC. Identification of human germinal center light and dark zone cells and their relationship to human B-cell lymphomas. Blood. 2012;120:2240–2248. doi: 10.1182/blood-2012-03-415380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel KU, Edelmann SL, Jeltsch KM, Bertossi A, Heger K, Heinz GA, Zöller J, Warth SC, Hoefig KP, Lohs C, et al. Roquin Paralogs 1 and 2 Redundantly Repress the Icos and Ox40 Costimulator mRNAs and Control Follicular Helper T Cell Differentiation. Immunity. 2013;38:655–668. doi: 10.1016/j.immuni.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Wang H, Geng J, Wen X, Bi E, Kossenkov AV, Wolf AI, Tas J, Choi YS, Takata H, Day TJ, et al. The transcription factor Foxp1 is a critical negative regulator of the differentiation of follicular helper T cells. Nat Immunol. 2014;15:667–675. doi: 10.1038/ni.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JP, Fuhrmann F, Hutloff A. T-follicular helper cells survive as long-term memory cells. Eur J Immunol. 2012;42:1981–1988. doi: 10.1002/eji.201242540. [DOI] [PubMed] [Google Scholar]
- Wei M, Shinkura R, Doi Y, Maruya M, Fagarasan S, Honjo T. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat Immunol. 2011;12:264–270. doi: 10.1038/ni.1991. [DOI] [PubMed] [Google Scholar]
- Weinstein JS, Bertino SA, Hernandez SG, Poholek AC, Teplitzky TB, Nowyhed HN, Craft J. B cells in T follicular helper cell development and function: separable roles in delivery of ICOS ligand and antigen. J Immunol. 2014;192:3166–3179. doi: 10.4049/jimmunol.1302617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg I, Agua-Doce A, Hernández A, Almeida C, Oliveira VG, Faro J, Graca L. Regulation of the germinal center reaction by Foxp3+ follicular regulatory T cells. J Immunol. 2011;187:4553–4560. doi: 10.4049/jimmunol.1101328. [DOI] [PubMed] [Google Scholar]
- Xiao N, Eto D, Elly C, Peng G, Crotty S, Liu YC. The E3 ubiquitin ligase Itch is required for the differentiation of follicular helper T cells. Nat Immunol. 2014;15:657–666. doi: 10.1038/ni.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Li X, Liu D, Li J, Zhang X, Chen X, Hou S, Peng L, Xu C, Liu W, et al. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 2013;496:523–527. doi: 10.1038/nature12058. [DOI] [PubMed] [Google Scholar]
- Yang CY, Best JA, Knell J, Yang E, Sheridan AD, Jesionek AK, Li HS, Rivera RR, Lind KC, D’Cruz LM, et al. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat Immunol. 2011a;12:1221–1229. doi: 10.1038/ni.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XP, Ghoreschi K, Steward-Tharp SM, Rodriguez-Canales J, Zhu J, Grainger JR, Hirahara K, Sun HW, Wei L, Vahedi G, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011b;12:247–254. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarke CA, Dalheimer SL, Zhang N, Catron DM, Jenkins MK, Mueller DL. Proliferating CD4+ T cells undergo immediate growth arrest upon cessation of TCR signaling in vivo. J Immunol. 2008;180:156–162. doi: 10.4049/jimmunol.180.1.156. [DOI] [PubMed] [Google Scholar]
- Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Yusuf I, Kageyama R, Monticelli L, Johnston RJ, DiToro D, Hansen K, Barnett B, Crotty S. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150) J Immunol. 2010;185:190–202. doi: 10.4049/jimmunol.0903505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaretsky AG, Taylor JJ, King IL, Marshall FA, Mohrs M, Pearce EJ. T follicular helper cells differentiate from Th2 cells in response to helminth antigens. 2009;206:991–999. doi: 10.1084/jem.20090303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.