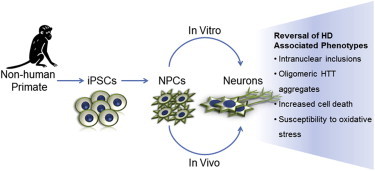
Summary
Huntington’s disease (HD) is a dominant neurodegenerative disorder caused by the expansion of glutamine residues in the N-terminal region of the huntingtin (HTT) protein. The disease results in progressive neuronal loss, leading to motor, cognitive, and psychiatric impairment. Here, we report the establishment of neural progenitor cell (NPC) lines derived from induced pluripotent stem cells (iPSCs) of transgenic HD monkeys. Upon differentiation to neurons, HD neural cells develop cellular features of HD, including the formation of nuclear inclusions and oligomeric mutant HTT (mHTT) aggregates, as well as increased apoptosis. These phenotypes are rescued by genetic suppression of HTT and pharmacological treatment, demonstrating the ability of our HD cell model to respond to therapeutic treatment. The development and reversal of HD-associated phenotypes in neural cells from HD monkeys provides a unique nonhuman primate (NHP) model for exploring HD pathogenesis and evaluating therapeutics that could be assessed further in HD monkeys.
Graphical Abstract
Highlights
-
•
Neural progenitor cell lines were derived from HD monkey iPSCs
-
•
HD monkey neural progenitor cells are capable of differentiating into GABA+ neurons
-
•
HD monkey neural cells develop HD-associated cell phenotypes
-
•
HD monkey neural cells respond to genetic correction and pharmacological treatments
In this article, Chan and colleagues report the establishment of neural progenitor cells derived from pluripotent stem cells of transgenic Huntington’s disease (HD) monkeys. Differentiated HD neural cells develop cellular features of HD that can be rescued by genetic suppression of the huntingtin gene and pharmacological treatment, thus demonstrating disease modeling and drug screening possibilities for HD stem cell-based therapy.
Introduction
Huntington’s disease (HD) is an autosomal dominant neurodegenerative disorder caused by the expansion of CAG repeats in the coding region of the huntingtin (HTT) gene IT15 (MacDonald et al., 1993). The mutation produces an extended stretch of glutamine residues spanning the N terminus of the HTT protein. Aggregation of oligomeric mHTT fragments and the formation of nuclear inclusions have been identified as hallmarks of the disease (DiFiglia et al., 1997); however, the role of mHTT aggregation in neurodegeneration remains unclear (Arrasate et al., 2004). Advances in cellular reprogramming techniques have led to broad applications for induced pluripotent stem cells (iPSCs) derived from patients as a model to recapitulate disease conditions because they share identical genetic defects with patients (Grskovic et al., 2011). Recent studies on iPSCs derived from HD patients have shown promise as possible HD models (An et al., 2012; Camnasio et al., 2012; Cheng et al., 2013; HD iPSC Consortium, 2012; Zhang et al., 2010). As these studies reveal the potential for pluripotent HD stem cell models as a tool for drug discovery and therapeutic development, the need arises for a preclinical nonhuman primate (NHP) model of HD that will be ready to assess the safety and efficacy of new therapeutics (Emborg et al., 2013; Perrier and Peschanski, 2012). Because safety concerns limit the direct translation of findings from human HD iPSC studies, HD monkey models provide a unique platform for long-term in vivo assessments, especially in the case of genetic correction and cell therapies. Here, we report the derivation of NPCs from HD monkey iPSCs. The resulting HD-NPCs are capable of differentiating into neural cells and develop classic HD cellular phenotypes. Furthermore, both genetic and pharmacologic treatments could reverse HD phenotypes in HD neural cells. HD monkeys and their derivative iPSCs provide an unprecedented opportunity for assessing personal medicine in higher primates, and HD-NPCs are a unique cellular platform for studying HD pathogenesis and evaluating drug efficacy.
Results
Monkey NPCs Generate GABA+ Neurons following In Vitro and In Vivo Neural Differentiation
HD monkey dental pulp stromal cells (DPSCs) and fibroblasts were reprogrammed by introducing rhesus genes encoding OCT4, SOX2, and KLF4 through retroviral gene transfer following protocols described in our previous report (Chan et al., 2010; Snyder et al., 2011). HD-iPSC lines (HD-3 and HD-14) were generated from two HD monkeys. HD-2 was embryonic stem cells (ESCs) derived from transgenic embryos from an HD monkey (Putkhao et al., 2013) (Table S1 available online). HD cell lines express exon 1 of the human HTT and green fluorescent protein (GFP) genes under the control of the human polyubiquitin-C promoter (UBC) (Figure S1). HD-2, HD-3, and HD-14 lines carried CAG repeat sizes of 29, 72, and 27/65, respectively. Wild-type (WT) cells lines (WT-2, WT-14, and WT-28) were generated from nontransgenic wild-type control monkeys derived from ESCs (WT-2 and WT-28) or by cellular reprogramming (WT-14) (Table S1). Stem cell lines expressed pluripotent stem cell markers, including OCT4, SSEA4, Tra-1-60, and alkaline phosphatase (Figure S1). In addition, cytogenetic analysis revealed a normal diploid karyotype matching gender to that of the HD monkey donor (Figure S1) (Chan et al., 2010; Putkhao et al., 2013).
To overcome technical hurdles associated with long-term stem cell culture, we derived stable NPC lines from HD monkey pluripotent stem cells (Figure 1A). NPCs displayed ectodermal rosette-like structures and exhibited characteristic NPC morphology after monolayer culture (Figure 1B). NPC populations were expandable for over 30 passages and could be frozen and thawed with high viability and maintain neural differentiation competency. NPCs expressed canonical neural precursor markers, including Nestin, PAX6, Musashi 1 (MSI1), and SOX2, confirmed by immunocytochemistry and quantitative RT-PCR (qRT-PCR) (Figures 1B and S2). Homogeneity of NPCs was confirmed by fluorescence-activated cell sorting (FACS) analysis (Figure 1C and Table S2). These results showed the successful establishment of HD- and WT-NPCs at high homogeneity that can be used for in vitro derivation of neural cells.
Figure 1.
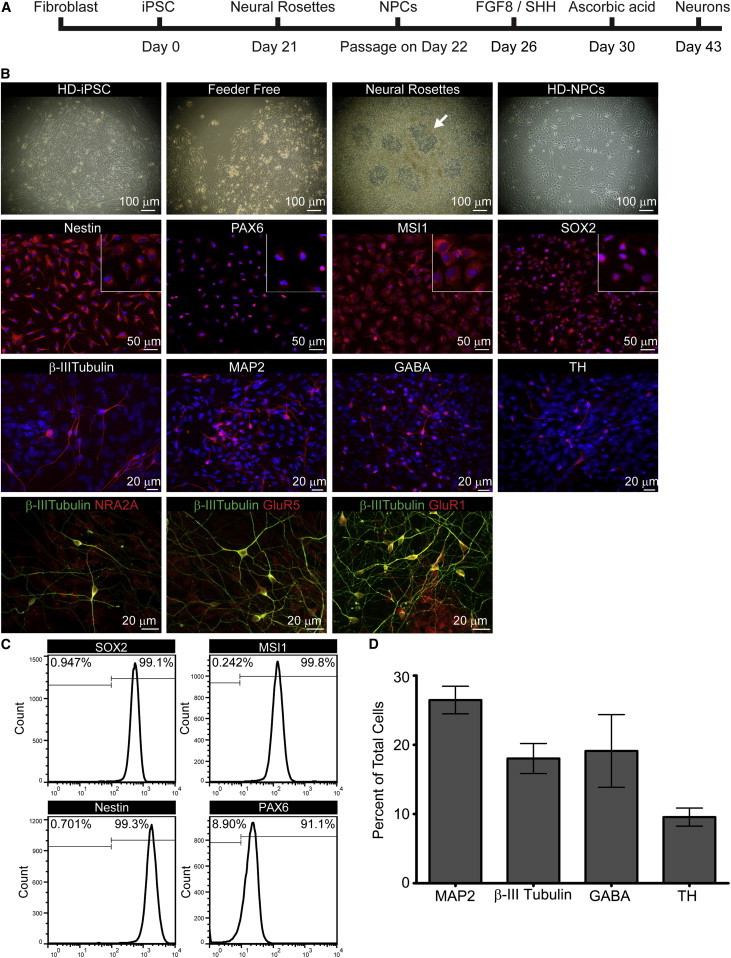
Derivation of HD-NPCs from iPSCs and In Vitro Neural Differentiation of HD-NPCs
(A) Schematic diagram depicting NPC and neuron derivation from rhesus macaque iPSCs.
(B) Bright-field image of HD-14 NPCs at progressing stages of differentiation. Arrows indicate neural rosettes (first row). Immunocytochemistry of HD-14 NPCs stained for Nestin, PAX6, MSI1, and SOX2 (red; second row). HD-14 in vitro differentiated neurons stained for β-III tubulin, MAP2, GABA, and TH (red; third row). Neural receptor subunits are expressed in HD neural cells; NR2A (red), GluR1 (red), and mGlur5 (red) colabeled with β-III tubulin (green) (fourth row). Nuclear staining is shown using Hoechst (blue).
(C) FACS analysis of HD-14 NPCs using antibodies for NPC markers. FACS data for all cell lines are listed in Table S2.
(D) Quantification of neurons derived from in vitro differentiated NPCs. Error bars indicate the SEM. Data represent three biological replicates.
See also Figures S1 and S2 and Table S2.
To determine that HD-NPCs were capable of differentiating into neuronal cell types, HD-NPCs were cultured for 21 days on polyornithine/laminin (P/L)-coated glass slides with neural differentiation medium (Figure 1A). Neurons derived from HD-NPCs expressed structural neural markers, microtubule-associated protein (MAP2, 27%), and β-III tubulin (18%), as well as neurotransmitter markers γ-aminobutyric acid (GABA, 21%) and tyrosine hydroxylase (TH, 10%), as demonstrated by immunostaining using specific antibodies (Figures 1B and 1D). β-III tubulin-positive neurons derived from HD-NPCs also stained positive for neural receptor subunits, including NR2A, GluR1, and mGluR5 (Figure 1B).
We further assessed neural differentiation potential for HD-NPCs in vivo. WT-2 and HD-14 NPCs were transplanted into the striatum of 10-week-old SCID mice. Although HD-NPCs carry both mHTT and GFP transgenes, WT-NPCs were introduced with lentivirus expressing GFP gene under the control of the UBC promoter for identification. At 12.5, 15, and 16 weeks posttransplantation, the brains of the transplanted mice were sectioned and stained for neuronal markers. Confocal imaging revealed that GFP-positive WT-2 and HD-14 grafts were also positive for neuronal markers DCX (Figures 2A and 2B), GABA (Figures 2C and 2D), and striatal neuron marker DARPP-32 (Figures 2E and 2F). At 12.5 weeks, WT-2 neurons showed positive staining for neuronal marker NeuN (Figure 2I). NeuN-positive HD-14 neurons were seen after 16 weeks (Figure 2J), suggesting that neural differentiation and maturation may possibly be impacted by mHTT in HD cells.
Figure 2.
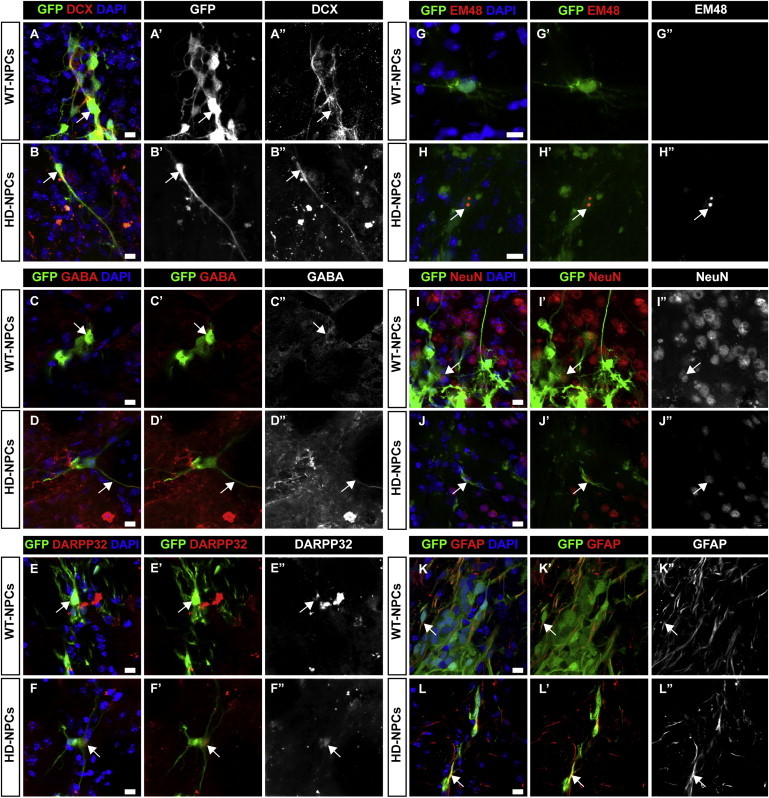
Characterization of Neuron Differentiation in Grafted NPCs
(A and B) Twelve and a half weeks posttransplantation, WT and HD cell grafts were positive for DCX (red).
(C and D) Fifteen weeks posttransplantation, WT and HD cell grafts were positive for GABA (red).
(E and F) Fifteen weeks posttransplantation, WT and HD cell grafts were positive for DARPP32 (red).
(G and H) Sixteen weeks posttransplantation, WT cell grafts were negative for mEM48; HD cell grafts show colocalization of GFP (green) and mEM48-positive nuclear inclusions (red).
(I and J) Twelve and a half weeks posttransplantation, WT cell grafts express neuronal marker NeuN (red). NeuN-positive HD cell grafts were observed after 16 weeks grafting (red).
(K and L) WT and HD cell grafts differentiate to glial cells indicated by positive GFAP staining (red).
Arrows indicate marker-positive transplanted neurons. Scale bars, 10 μm.
Immunostaining with mEM48, an antibody reactive to mHTT with expanded glutamine repeats (Chan et al., 2010), revealed grafted cells positive for mHTT inclusions (Figures 2G and 2H). WT and HD-NPCs were also capable of generating glial cells as indicated by positive GFAP staining (Figures 2K and 2L). These results show that iPSC-derived HD-NPCs survived after xenograft in the striatum and were capable of differentiating into striatal GABA+ neurons both in vitro and in vivo.
Expression of mHTT and Formation of Intranuclear Inclusions during Neural Differentiation
The aggregation of mHTT and the formation of intranuclear inclusions are classic HD-associated neuronal phenotypes (DiFiglia et al., 1997) that have been described in humans as well as animal models (Wang et al., 2008; Yang et al., 2008). Although HTT expresses in all cell types, neuronal cell types are most susceptible to the toxicity caused by mHTT (Johnson and Davidson, 2010). qRT-PCR analysis of HD-NPCs during neural differentiation revealed elevated expression of total HTT transcripts in HD neural cells compared to WT neurons (Figure 3A). Western blot analysis of HD and WT cells at different stages of differentiation revealed increased accumulation of oligomeric mHTT in HD-3 and HD-14 (Figure 3B). Interestingly, the increased oligomeric aggregates are only seen in HD neural cells and are undetectable in HD-iPSCs. Immunostaining using mEM48 revealed the development of HTT aggregates and intranuclear inclusions in differentiated HD neurons colabeled with neuronal markers DCX, MAP2, and GABA (Figure 3C).
Figure 3.
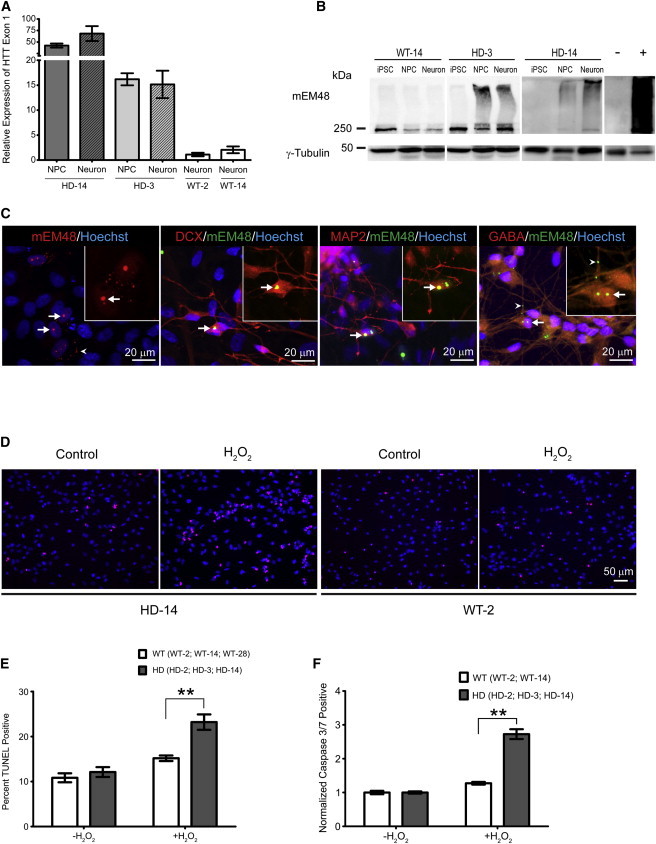
Elevated Expression and Aggregation of mHTT Accompany Increased Vulnerability to Oxidative Stress in HD Cells
(A) qRT-PCR analysis of HTT exon 1 during in vitro neural differentiation. Expression levels are compared to WT-2 neuron control line. Results were from three biological replicates. qRT-PCR samples run in duplicate. Data and error bars are represented as mean ± SEM.
(B) Western blot analysis revealed an increase in oligomeric mHTT that correlates with neuronal differentiation stage. Notice aggregate accumulation in the stacking gel (above 250 kDa). Negative (−) and positive (+) lanes are frontal cortex samples from nontransgenic and HD monkeys, respectively.
(C) Left: staining of in vitro differentiated HD-NPCs with mEM48 (red) reveals distinct intranuclear inclusions and cytosolic aggregates (arrowhead) of mHTT. Middle left: mHTT inclusions (green) colabeled with DCX (red). Middle right: mHTT inclusions (green) colabeled with MAP2 (right; red). Right: mHTT inclusions and neuropil aggregates (arrowhead; green) colabeled with GABA (red). Staining using Hoechst (blue). Arrows indicate intranuclear inclusions.
(D) Images represent TUNEL staining of WT-2 and HD-14 NPCs after 24 hr H2O2 challenge. H2O2 TUNEL-positive cells (red). Nuclear staining is shown using Hoechst (blue).
(E) Quantification of TUNEL assay on HD- and WT-NPCs following H2O2 exposure. TUNEL-positive cell (percentage) was averaged for three HD lines (HD-2, HD-3, HD-14) and three WT lines (WT-2, WT-14, WT-28). Percentage of TUNEL-positive cells was calculated from total number of cells from three biological replicates of each cell line. Data are represented as mean ± SEM (∗∗p < 0.001, ANOVA).
(F) Quantification of caspase 3/7-positive HD-NPCs compared to WT-NPCs following H2O2 exposure. Apoptotic population percentage was averaged for three HD lines (HD-2, HD-3, HD-14) and two WT lines (WT-2, WT-14). Values represent fold change from untreated sample. Results are shown from three biological replicates of each cell line. Data and error bars are represented as mean ± SEM (∗∗p < 0.001, ANOVA).
In addition to the development of classic HD cellular phenotypes, we also investigated whether HD-NPCs were more susceptible to oxidative stress compared to WT-NPCs, as previously shown in human HD-iPSCs and neurons (An et al., 2012; HD iPSC Consortium, 2012). Hydrogen peroxide (H2O2) was supplemented into culture medium to induce oxidative stress. TUNEL assay following H2O2 incubation demonstrated a significant increase in cell death in HD-NPCs compared to WT-NPCs (Figures 3D and 3E; p < 0.01). This observation was further supported by flow cytometric analysis of caspase 3/7 activity, which revealed a significant increase in apoptotic cells in HD-NPCs compared to WT-NPCs following exposure to H2O2 (Figure 3F; p < 0.01). Together, these findings show that HD neural cells mirror key cellular phenotypes associated with mHTT.
Reversal of HD-Associated Phenotypes by RNAi and Memantine
Following the characterization of HD phenotypes in neural cells derived from HD monkeys, we investigated whether these phenotypes could be rescued by therapeutic treatment. RNAi-mediated reduction of HTT in HD models has shown promise in a number of studies (Boudreau et al., 2009; McBride et al., 2011). We postulated that reducing the expression of HTT in HD-NPCs would confer a therapeutic benefit. HD-NPCs were infected with a lentivirus expressing small hairpin RNA (shRNA) targeting the HTT transcript. After Zeocin selection to establish stable transformed NPC lines, qRT-PCR analysis showed that HTT expression in shRNA-treated NPCs (shHD-3 and shHD-14) was reduced by approximately 50% (Figure 4A; p < 0.05). Furthermore, oligomeric HTT aggregation was reduced in shHD-3 and shHD-14 as revealed by western blot using mEM48 antibody (Figure 4B; top panel). Slight reduction of soluble HTT detected by 1C2 antibody was also observed in cells treated with shRNA-HTT (Figure 4B; lower panel). We next determined whether the reduction of HTT could reduce apoptosis in HD-NPCs. shHD-3 and HD-3 NPCs were challenged with H2O2 to induce oxidative stress. As demonstrated by caspase 3/7 assay, the percentage of apoptotic cells was significantly reduced in shHD-3 NPCs in comparison to HD-3 NPCs (Figure 4C; p < 0.01). Next, we performed TUNEL assay to assess cell death in shHD-3 and shHD-14 neural cells following differentiation. In correlation with our caspase assay, we observed a significant reduction of apoptotic neural cells that had reduced HTT expression compared to HD neural cells not expressing shRNA (Figure 4D; p < 0.05).
Figure 4.
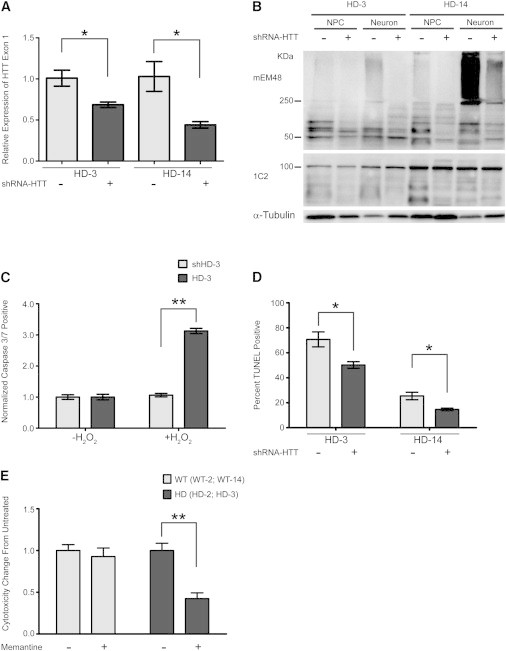
Reversal of HD-Associated Phenotypes Using shRNA and Memantine
(A) qRT-PCR analysis of HTT exon 1 expression in HD-3 and HD-14 NPCs after infection with lentivirus expressing shRNA-HTT. Results shown from three biological replicates, qRT-PCR samples run in duplicate. Data and error bars are represented as mean ± SEM (∗p < 0.05, ANOVA).
(B) Western blot analysis of mHTT aggregation in HD-3 and HD-14 NPCs and in vitro differentiated neural cells with (+) or without (−) expressing shRNA-HTT.
(C) Quantification of caspase 3/7-positive HD-3 and shHD-3 NPCs following H2O2 exposure. Values represent fold change from untreated sample. Results are shown from three biological replicates. Data and error bars are represented as mean ± SEM (∗∗p < 0.001, ANOVA).
(D) Quantification of TUNEL assay on HD-3 and HD-14 neurons after in vitro differentiation with or without expressing shRNA-HTT. Percentage of TUNEL-positive cells calculated from total number of cells identified by Hoechst staining. Results are shown from three biological replicates. Data and error bars are represented as mean ± SEM (∗p < 0.05, ANOVA).
(E) Cytotoxicity profile of neurons treated with 10 μM memantine drug measured by G6PD assay. Values represent mean fold change in cytotoxicity from untreated cells. Cytotoxicity values averaged from two HD lines (HD-2, HD-3) and two WT lines (WT-2, WT-28) from three repeated experiments. Data and error bars are represented as mean ± SEM (∗p < 0.05, ANOVA).
In addition to mHTT expression, neurodegeneration has been attributed to N-methyl-D-aspartate receptor (NMDAR)-mediated cytotoxicity in HD (Marco et al., 2013). Reports of using memantine, a well-characterized NMDAR antagonist, have shown some promise as a potential therapy for HD (Milnerwood et al., 2010). To test whether treatment with memantine could rescue the cell death observed in HD neurons, 10 μM memantine was supplemented into neural differentiation medium 24 hr prior to assessing cytotoxicity. The cytotoxicity of HD neurons treated with memantine was significantly reduced compared to untreated HD neurons, whereas there was no significant change in cytotoxicity in WT neurons (Figure 4E; p < 0.01). Together, these results show that HD monkey neural cells respond to potential therapeutic treatments, suggesting our model will be useful as a platform for drug assessment.
Discussion
This study reports the development of an HD-NPC model derived from the iPSCs of HD NHPs. We established stable and expandable NPC lines from HD monkeys that are capable of neural differentiation in vitro and in vivo. HD neural cells develop classic cellular phenotypes of HD that could be reversed using both genetic and pharmacological treatments. Though efforts to differentiate iPSCs or directly differentiate somatic cells to target cells of interest are ongoing, establishing potent NPC lines from iPSCs provides an advantage by allowing easy maintenance of a progenitor cell population committed to the neuronal lineage, thus enhancing homogeneity in subsequent neural differentiation (Koch et al., 2009). Such HD-NPC lines are important for in vitro neural differentiation for drug discovery research, whereas competency in in vivo differentiation is important for cell replacement therapy.
In vitro differentiation methods used in this system were similar to protocols described for human iPSC neural differentiation, resulting in neurons positive for GABA, TH, and mature neural markers. NPC grafts after in vivo differentiation were positive for glial marker GFAP and mature neural markers; however, we noticed that survival of NPCs and derivative neural cells at the graft site was more impaired in HD-NPCs compared to the contralateral side implanted with WT-NPCs. A possible explanation for this observation is that only 50,000 NPCs were grafted into the mouse striatum. This number is relatively low compared to other studies that grafted over 100,000 cells (Nicholas et al., 2013; Nori et al., 2011). Furthermore, in this study NPCs were transplanted as a single cell suspension, potentially leading to reduced survival of cell grafts compared to other transplantation studies that graft neurospheres (Nori et al., 2011). Together, these results cohere with our findings that the progressive increase in mHTT expression and the accumulation of oligomeric mHTT in neural differentiating HD-NPCs, combined with reduced tolerance to oxidative stress, result in a higher apoptotic rate and increased cell death.
As a potential platform to model HD cellular pathogenesis, an important goal of this study was to assess whether our cellular model recapitulates disease-associated pathologic events. We found that an increase of intranuclear inclusions and the accumulation of oligomeric, soluble, and insoluble mHTT aggregates were correlated with neuronal differentiation. This observation mirrors a key neural pathology seen in HD patients’ brains, as well as in HD monkey models (DiFiglia et al., 1997; Yang et al., 2008). In addition to mHTT aggregation, HD-NPCs show increased apoptosis induced by exposure to H2O2 as a means of inducing oxidative stress. This disease-dependent difference in vulnerability to exogenous stress recapitulates phenotypes characterized by various studies using human pluripotent stem cells (HD iPSC Consortium, 2012). These findings suggest that our in vitro system is comparable to other previously described cellular models using HD-NPCs, and derivative neural cells develop pronounced HD cellular phenotypes. Furthermore, these HD phenotypes develop independent of treatment with proteasome inhibitors, which in some reports were necessary to induce HD-associated phenotypes (Cheng et al., 2013). We suspect that, unlike human iPSCs, our HD-NPCs express multiple copies of the truncated mHTT gene with an expanded polyglutamine tract regulated by the UBC promoter, which may lead to more dramatic phenotypes and impact neural differentiation.
The capability of HD monkey neural cells to respond to in vitro drug and gene therapy provides a proof of principle that this cellular model of HD could make a powerful resource for the assessment and development of candidate therapeutic strategies. By using an RNAi approach to reduce HTT expression and by treating neurons with memantine, we were able to ameliorate cell death phenotypes. With this result, we show the potential utility of our system for screening therapeutic targets in HD, with the important bonus that our model provides a unique opportunity for future assessments in HD monkeys in vivo. Furthermore, reports describing the successful transplantation and integration of monkey neural progenitors into targeted brain regions of NHPs (Emborg et al., 2013; Hashimoto et al., 2012) point to the feasibility of cell replacement studies in NHP models of neurodegenerative disease. These reports, combined with our recent success in developing HD monkeys (Chan et al., 2014; Kocerha et al., 2013; Yang et al., 2008), lay the foundation for long-term assessment of the safety and efficacy of cell and gene therapies in HD monkey models.
Experimental Procedures
In Vitro/In Vivo Neuronal Differentiation
NHP NPCs were seeded onto a P/L-coated glass chamber in differentiation medium (DMEM/F12 [Life Technologies], P/S [Invitrogen], 2 mM L-glutamine, 1 × N2 [Invitrogen], and 1 × B27 [Life Technologies]). At 4 days of neural induction, SHH (R&D Systems, 0.2 μg/ml) and FGF8 (R&D, 0.1 μg/ml) were added. At day 8, 200 mM ascorbic acid was added in combination with SHH and FGF8.
All protocols involving animal care and handling were approved by Emory University’s Institutional Animal Care and Use Committee. For in vivo differentiation to the neuronal lineage, WT-2 NPCs and HD-14 NPCs were dissociated and suspended at 50,000 cells/μl in artificial cerebrospinal fluid solution. By stereotactic injection, the cell suspension was infused into the right and left hemispheres of the striatum. Further details are in the Supplemental Experimental Procedures.
Cell Stress and Apoptosis Assays
NPCs were treated with 5 mM H2O2 in culture medium for 24 hr. Caspase 3/7 activity was assessed using Muse Caspase-3/7 Kit (Millipore) according to the manufacturer’s instructions. Cell counts were analyzed using Muse Cell Analyzer (Millipore). Effects of H2O2 were further assessed by TUNEL analysis as described in the Supplemental Experimental Procedures.
Statistical Analysis
For all comparisons one-way ANOVA analysis and Bonferonni post tests were performed using SPSS statistical software (IBM). Statistical significance was determined at ∗p < 0.05 and ∗∗p < 0.001.
Acknowledgments
We thank Xiao-Jiang Li for providing mEM48 antibody, Gary Bassell for providing NR2A antibody, and Kiran Gill for assistance in FACS analysis. We also thank Cheryl Strauss for assistance in editing. YNPRC is supported by the Office of Research and Infrastructure Program (ORIP)/OD P51OD11132. K.P. and R.P. are supported by the Royal Golden Jubilee PhD program of Thailand Research Fund. R.L.C. is partly supported by NIH training grant to the Department of Human Genetics at Emory University (5T32MH879774). This study is supported by a grant awarded by the ORIP/NIH (OD010930), NINDS/NIH (NS084163), and ACTSI to A.W.S.C.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
References
- An M.C., Zhang N., Scott G., Montoro D., Wittkop T., Mooney S., Melov S., Ellerby L.M. Genetic correction of Huntington’s disease phenotypes in induced pluripotent stem cells. Cell Stem Cell. 2012;11:253–263. doi: 10.1016/j.stem.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camnasio S., Delli Carri A., Lombardo A., Grad I., Mariotti C., Castucci A., Rozell B., Lo Riso P., Castiglioni V., Zuccato C. The first reported generation of several induced pluripotent stem cell lines from homozygous and heterozygous Huntington’s disease patients demonstrates mutation related enhanced lysosomal activity. Neurobiol. Dis. 2012;46:41–51. doi: 10.1016/j.nbd.2011.12.042. [DOI] [PubMed] [Google Scholar]
- Chan A.W.S., Cheng P.-H., Neumann A., Yang J.-J. Reprogramming Huntington monkey skin cells into pluripotent stem cells. Cell. Reprogram. 2010;12:509–517. doi: 10.1089/cell.2010.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.W., Xu Y., Jiang J., Rahim T., Zhao D., Kocerha J., Chi T., Moran S., Engelhardt H., Larkin K. A two years longitudinal study of a transgenic Huntington disease monkey. BMC Neurosci. 2014;15:36. doi: 10.1186/1471-2202-15-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng P.-H., Li C.-L., Chang Y.-F., Tsai S.-J., Lai Y.-Y., Chan A.W.S., Chen C.-M., Yang S.-H. miR-196a ameliorates phenotypes of Huntington Disease in cell, transgenic mouse, and induced pluripotent stem cell models. Am. J. Hum. Genet. 2013;93:306–312. doi: 10.1016/j.ajhg.2013.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- Emborg M.E., Liu Y., Xi J., Zhang X., Yin Y., Lu J., Joers V., Swanson C., Holden J.E., Zhang S.-C. Induced pluripotent stem cell-derived neural cells survive and mature in the nonhuman primate brain. Cell Rep. 2013;3:646–650. doi: 10.1016/j.celrep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grskovic M., Javaherian A., Strulovici B., Daley G.Q. Induced pluripotent stem cells—opportunities for disease modelling and drug discovery. Nat. Rev. Drug Discov. 2011;10:915–929. doi: 10.1038/nrd3577. [DOI] [PubMed] [Google Scholar]
- Hashimoto K., Shimada H., Okada Y., Ibata K., Ebise H., Ota S.-i., Tomioka I., Nomura T., Maeda T., Kohda K. Efficient derivation of multipotent neural stem/progenitor cells from non-human primate embryonic stem cells. PLoS One. 2012;7:e49469. doi: 10.1371/journal.pone.0049469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HD iPSC Consortium Induced pluripotent stem cells from patients with Huntington’s disease show CAG-repeat-expansion-associated phenotypes. Cell Stem Cell. 2012;11:264–278. doi: 10.1016/j.stem.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C.D., Davidson B.L. Huntington’s disease: progress toward effective disease-modifying treatments and a cure. Hum. Mol. Genet. 2010;19(R1):R98–R102. doi: 10.1093/hmg/ddq148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocerha J., Liu Y., Willoughby D., Chidamparam K., Benito J., Nelson K., Xu Y., Chi T., Engelhardt H., Moran S. Longitudinal transcriptomic dysregulation in the peripheral blood of transgenic Huntington’s disease monkeys. BMC Neurosci. 2013;14:88. doi: 10.1186/1471-2202-14-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch P., Opitz T., Steinbeck J.A., Ladewig J., Brüstle O. A rosette-type, self-renewing human ES cell-derived neural stem cell with potential for in vitro instruction and synaptic integration. Proc. Natl. Acad. Sci. USA. 2009;106:3225–3230. doi: 10.1073/pnas.0808387106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald M.E., Ambrose C.M., Duyao M.P., Myers R.H., Lin C., Srinidhi L., Barnes G., Taylor S.A., James M., Groot N., The Huntington’s Disease Collaborative Research Group A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- Marco S., Giralt A., Petrovic M.M., Pouladi M.A., Martínez-Turrillas R., Martínez-Hernández J., Kaltenbach L.S., Torres-Peraza J., Graham R.K., Watanabe M. Suppressing aberrant GluN3A expression rescues synaptic and behavioral impairments in Huntington’s disease models. Nat. Med. 2013;19:1030–1038. doi: 10.1038/nm.3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride J.L., Pitzer M.R., Boudreau R.L., Dufour B., Hobbs T., Ojeda S.R., Davidson B.L. Preclinical safety of RNAi-mediated HTT suppression in the rhesus macaque as a potential therapy for Huntington’s disease. Mol. Ther. 2011;19:2152–2162. doi: 10.1038/mt.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milnerwood A.J., Gladding C.M., Pouladi M.A., Kaufman A.M., Hines R.M., Boyd J.D., Ko R.W.Y., Vasuta O.C., Graham R.K., Hayden M.R. Early increase in extrasynaptic NMDA receptor signaling and expression contributes to phenotype onset in Huntington’s disease mice. Neuron. 2010;65:178–190. doi: 10.1016/j.neuron.2010.01.008. [DOI] [PubMed] [Google Scholar]
- Nicholas C.R., Chen J., Tang Y., Southwell D.G., Chalmers N., Vogt D., Arnold C.M., Chen Y.J., Stanley E.G., Elefanty A.G. Functional maturation of hPSC-derived forebrain interneurons requires an extended timeline and mimics human neural development. Cell Stem Cell. 2013;12:573–586. doi: 10.1016/j.stem.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nori S., Okada Y., Yasuda A., Tsuji O., Takahashi Y., Kobayashi Y., Fujiyoshi K., Koike M., Uchiyama Y., Ikeda E. Grafted human-induced pluripotent stem-cell-derived neurospheres promote motor functional recovery after spinal cord injury in mice. Proc. Natl. Acad. Sci. USA. 2011;108:16825–16830. doi: 10.1073/pnas.1108077108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier A., Peschanski M. How can human pluripotent stem cells help decipher and cure Huntington’s disease? Cell Stem Cell. 2012;11:153–161. doi: 10.1016/j.stem.2012.07.015. [DOI] [PubMed] [Google Scholar]
- Putkhao K., Kocerha J., Cho I.-K., Yang J., Parnpai R., Chan A.W.S. Pathogenic cellular phenotypes are germline transmissible in a transgenic primate model of Huntington’s disease. Stem Cells Dev. 2013;22:1198–1205. doi: 10.1089/scd.2012.0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder B.R., Cheng P.-H., Yang J., Yang S.-H., Huang A.H.C., Chan A.W.S. Characterization of dental pulp stem/stromal cells of Huntington monkey tooth germs. BMC Cell Biol. 2011;12:39. doi: 10.1186/1471-2121-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.E., Tydlacka S., Orr A.L., Yang S.H., Graham R.K., Hayden M.R., Li S., Chan A.W.S., Li X.J. Accumulation of N-terminal mutant huntingtin in mouse and monkey models implicated as a pathogenic mechanism in Huntington’s disease. Hum. Mol. Genet. 2008;17:2738–2751. doi: 10.1093/hmg/ddn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.-H., Cheng P.-H., Banta H., Piotrowska-Nitsche K., Yang J.-J., Cheng E.C.H., Snyder B., Larkin K., Liu J., Orkin J. Towards a transgenic model of Huntington’s disease in a non-human primate. Nature. 2008;453:921–924. doi: 10.1038/nature06975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N., An M.C., Montoro D., Ellerby L.M. Characterization of human Huntington’s disease cell model from induced pluripotent stem cells. PLoS Curr. 2010;2:RRN1193. doi: 10.1371/currents.RRN1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


