Abstract
In this paper, it is demonstrated how Raman spectroscopy can be used to detect different carotenoids as possible biomarkers in various groups of microorganisms. The question which arose from previous studies concerns the level of unambiguity of discriminating carotenoids using common Raman microspectrometers. A series of laboratory-grown microorganisms of different taxonomic affiliation was investigated, such as halophilic heterotrophic bacteria, cyanobacteria, the anoxygenic phototrophs, the non-halophilic heterotrophs as well as eukaryotes (Ochrophyta, Rhodophyta and Chlorophyta). The data presented show that Raman spectroscopy is a suitable tool to assess the presence of carotenoids of these organisms in cultures. Comparison is made with the high-performance liquid chromatography approach of analysing pigments in extracts. Direct measurements on cultures provide fast and reliable identification of the pigments. Some of the carotenoids studied are proposed as tracers for halophiles, in contrast with others which can be considered as biomarkers of other genera. The limits of application of Raman spectroscopy are discussed for a few cases where the current Raman spectroscopic approach does not allow discriminating structurally very similar carotenoids. The database reported can be used for applications in geobiology and exobiology for the detection of pigment signals in natural settings.
Keywords: detecting carotenoids, bacteria, cyanobacteria, algae, Raman spectroscopy, high-performance liquid chromatography
1. Introduction
In the geological record and also for astrobiology, carotenoids as well as other photosynthetic or protective pigments can be considered as biomarkers. Understanding the mechanisms of their conservation in terrestrial or extraterrestrial rocks, as well as the mode how to detect them unambiguously, are key questions in organic geochemistry and in astrobiology. To propose solutions in these areas, we need to improve our knowledge of the formation and distribution of pigments as well as their unambiguous identification in living or decayed biota.
For planetary research, including astrobiology, the possibility of using high-performance but complex and heavy instrumentation is excluded. For this reason, Raman spectrometry, with high potential of miniaturization [1], is considered as a very useful tool of detecting biomarkers.
Pigments of different nature are widely used as biomarkers for the classification of microorganisms. Different classes of pigments are found in the prokaryotes. They include magnesium-containing chlorophylls and bacteriochlorophylls, phaeophytins and the phycobiliproteins characteristic especially for cyanobacteria, retinal-containing pigments such as bacteriorhodopsin, halorhodopsin and proteorhodopsin, carotenoids, flexirubins and many others [2,3]. Especially within the carotenoids, a great structural variety is found [4]. Carotenoids, prominently present in a large number of prokaryotes and also in eukaryotes, represent a very diversified group of chemicals having a polyene skeletal backbone. Examples of carotenoids are given in figure 1. The vital roles they play include participation in energy-harvesting complexes, UV protection and in the repair of cellular damage [5].
Figure 1.
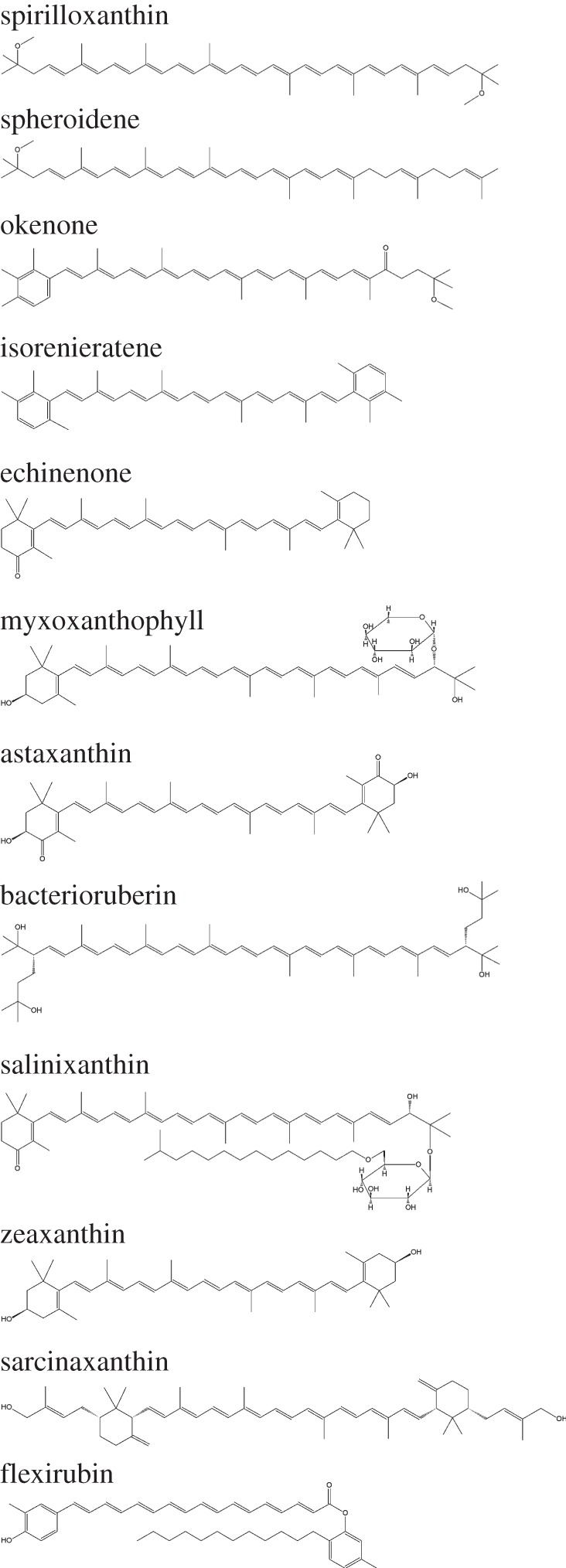
Structural formulas of carotenoids.
Some carotenoids are very common in many groups of microorganisms (e.g. β-carotene); others are only synthesized by a limited number of species, e.g. salinixanthin in the bacterium Salinibacter ruber.
For extremely halophilic heterotrophs, the specific carotenoids bacterioruberin and salinixanthin are excellent biomarkers. Bacterioruberin, the main carotenoid component responsible for the colour of the red Archaea of the family Halobacteriaceae, has a distinct molecular structure in that it has a primary conjugated isoprenoid chain length of 13 C=C units with no subsidiary conjugation arising from terminal groups, which contain four (or less) OH group functionalities. In salinixanthin from Salinibacter ruber the C40 conjugated chain with 13 double bonds and a fatty acid chain are linked to a β-d-glycoside [2,3].
Cyanobacteria often contribute to the colour of hypersaline waters and surface sediments. Unicellular cyanobacteria, designated Aphanothece halophytica, Cyanothece or Halotheceare frequently found in the upper layer of the saltern sediments which may be consequently coloured yellow to bright orange due to the abundance of different carotenoid pigments, mainly myxoxanthophylls and echinenone [6].
The chemical properties of carotenoids originating from biota as well as from sediments in organic solvent extracts are known in depth. However, few studies have been undertaken of carotenoids in vivo [7–9]. Classical techniques of pigment identification and discrimination are absorption spectroscopy and high-performance liquid chromatography (HPLC). Mass spectrometry and nuclear magnetic resonance are largely used for precise conformation and structural studies. Raman spectroscopy has several advantages for carotenoid work: it is non-destructive and provides spectroscopic molecular and structural information. In Raman microspectrometry, Raman spectral information can be obtained from optically observed regions of samples commonly at the micrometric level, which is advantageous for the direct in situ interrogation of biological inclusions in mineralogical matrices. For carotenoid work, advantageous enhancement of spectroscopic features can be obtained by employing Resonance Raman microspectrometry. For biological molecules absorbing light in the visible portion of the electromagnetic spectrum, new possibilities were discovered for Raman spectroscopy using the resonance Raman effect [10]. Tuning the excitation wavelength to the electronic absorption spectrum can produce selective enhancement of certain Raman bands [10,11].
Previous studies have focused on the identification of carotenoids in pure cultures of different microorganisms using the resonance Raman effect. In this way, trace pigment detection is sometimes possible using especially the excitation wavelengths 514.5 and 532 nm [12–14]. Carotenoids can also be identified in native samples from surface or subsurface colonizations of rocks or sediments using Raman spectroscopy.
Interesting new possibilities have become available by the use of portable instruments that allow obtaining Raman spectra under field conditions. Other studies have used Raman spectroscopy to detect carotenoids in native samples from surface or subsurface colonizations of rocks or sediments. These studies have high relevance for geobiology and exobiology [15–23]. In fact, Raman spectrometer in a miniaturized version was proposed to be included onboard a rover to be part of the forthcoming projects of astrobiological priority on Mars [1,17]. Biosignatures of preexisting life, as well as mineral associations confirming the presence of water on Mars in the past, are the main objects of interest to be detected by Raman spectroscopic instrumentation. As biomarkers, pigments and especially carotenoids are of highest relevance.
In this paper, Raman spectra of different carotenoids from representative species of archaea, bacteria, cyanobacteria and algae were obtained. The Raman spectroscopic approach was compared with HPLC of extracts. It was shown that carotenoids can very easily be detected within lyophilized biomass. However, the ability of conventional Raman spectrometry to detect individual carotenoids when present in complex microbiological matrices is limited.
Here, we shall focus on halophiles and a few microorganisms from other groups and provide an estimation of the potential of commercially available Raman spectrometers for the unambiguous detection of their carotenoid biomarkers. We will deal with microorganisms of different taxonomic affiliation (heterotrophic bacteria, cyanobacteria, anoxygenic phototrophs, non-halophilic heterotrophs, as well as eukaryotes: Chromista, Rhodophyta, Chlorophyta) and provide an estimation of the potential of commercially available Raman spectrometers for the unambiguous detection of their carotenoid biomarkers. It is shown that reliable results can be obtained on simple lyophilized aggregates of microbes as well as on acetone/methanol extracts. The data presented can be considered as a basis for the refinement of Raman spectroscopic pigment detection in complex samples from natural environments.
2. Material and methods
(a). Samples
In total, samples of 20 organisms were investigated, belonging to the three domains of life.
The following halophilic Archaea were included in the study for comparative reasons: Halobacteriumstrain NRC-1 (American-type culture collection (ATCC 700922)), Halobacterium salinarum R1 (DSM 671), Haloarcula vallismortis (ATCC 29715T), Halorubrum sodomense (ATCC 33755T) and Haloquadratum walsbyi.
Bacterial strains used were the cyanobacteria: Chamaesiphon polymorphus(Culture Collection of Autotrophic Organism (CCALA 37)), Chroococcussp. (CCALA 701),Chroococcussp. (CCALA 702),Microcoleuscf. vaginatus(CCALA 154), Oscillatoria limnetica (halophilic, from Solar Lake, Egypt), Oscillatoria sancta(CCALA 135), anoxygenic phototrophs: Ectothiorhodospira marismortui (DSM 4180T) (halophilic), Rhodospirillum rubrum(DSM 107) (non-halophilic), the extremely halophilic heterotroph Salinibacter ruber (DSM 13855T) and the non-halophilic heterotrophsMicrococus luteus (ATCC 4698T).
Eukaryotes studied belong to the Ochrophyta (Diadesmis gallicaCCALA 766,Botrydiopsis alpinaCCALA 217), the Rhodophyta (Balbiania investiensSAG 50.93,Porphyridium cruentum CCALA 415) and the Chlorophyta (Dunaliella salina,Dunaliella parva,Haematococcus pluvialis CCALA 357).
Most strains of cyanobacteria and eukaryotes were obtained from the CCALAs of the Institute of Botany AS CR in Třeboň, Czech Republic. The other bacterial strains and most of the Archaea were obtained either from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH or from the ATCC.
(b). Culture conditions
Dunaliella parva, Dunaliella salina and Oscillatoria limnetica were grown at 30°C and continuous irradiance of approximately 100 μmol m−2 s−1 in Chu-11 medium prepared in double strength artificial seawater (‘Turks Island Salts’; the Merck Index). Balbiania investienswas grown in WC medium [24], Porphyridium cruentumwas grown as described by Brody & Emerson [25], and all other oxygenic phototrophs were cultivated at room temperature in Bold's Basal Medium [26,27]. The medium and growth conditions for Ectothiorhodospira marismortui were described by Oren et al. [28]. Rhodospirillum rubrum was grown in Rhodospirillaceae medium (DSM medium 27; (http://www.dsmz.de/microorganisms/medium/pdf/DSMZ.Medium27.pdf)). The medium and culture conditions for Salinibacter ruber were described by Anton et al. [29].Halobacteriumstrains and Haloarcula vallismortis were grown as described by Oren [30]. The medium for Halorubrum sodomense was derived from Oren [31]. Micrococcus luteus was grown at 30°C on Nutrient Agar plates. Cells from liquid cultures were harvested in late exponential phase by centrifugation (10 min, 5000g); when solid media were used, cell material was scraped off the plates. The collected material was dried by lyophilization.
(c). Extractions
Simple acetone/methanol extractions were carried out following Eimhjellen & Liaaen-Jensen [32].
(d). Instrumental analyses
(i). Raman spectroscopy
Micro-Raman analyses of lyophilizates were performed on a multichannel Renishaw In Via Reflex spectrometer coupled with a Peltier-cooled CCD detector. Excitation was generally provided by a 514.5 nm Ar laser. To achieve enhanced signal-to-noise ratios, 10–30 scans were accumulated, each of 20 s exposure time with laser power ranging between 30 and 100 mW. Spectra were recorded at a spectral resolution of 2 cm−1 between 100 and 1800 cm−1. Other excitation wavelengths were used to complement the measurement especially where the spectra obtained with argon-ion excitation were of insufficient quality. Several Raman spectra were recorded also with 488 and 844 nm excitation (tunable Ar+ laser, SpectraPhysics) (600 lines mm−1 grating) of a Horiba Jobin Yvon microspectrometer. FT-Raman data were obtained using a Bruker RFS 100 spectrometer (Nd3+/YAG laser operating at 1064 nm excitation) with 4 cm−1 spectral resolution. Five to 15 measurements were obtained on pellets. Polystyrene, sulfur, ϵ-caprolactone, acetonitrile/toluene (50/50 v/v), cyclohexane, stearic acid and glycerol were used to check the spectral wavenumber calibration. Raman spectra were exported into the Galactic *.SPC format. Spectra were then compared using GRAMS AI (V. 8.0, Thermo Electron Corp., Waltham, MA, USA). Raman spectra were not subjected to any data manipulation or processing techniques and are reported as collected, except for baseline correction performed in a few cases.
(ii). High-performance liquid chromatography
Pigment analyses of selected microorganisms were carried out using the Agilent 1100 Series HPLC system (Agilent Technologies Inc., Palo Alto, CA, USA). The instrument was equipped with the UV-VIS diode-array detector (Agilent DAD 61315B). Pigments were separated using a modified method of Van Heukelem and Thomas (2001) on the thermostated (35°C) Phenomenex Luna 3 μm C8(2) 100 Å column with binary solvent system (0 min 100% A, 20 min 100% B, 25 min 100% B, 27 min 100% A, 30 min 100% A; A: 70% methanol+28 mM ammonium acetate, B: methanol). The solvent flow rate was 0.8 ml min−1. The peak assignment was based on the acquired absorption spectra.
3. Results
Carotenoids are exceptional pigments in Raman spectroscopic analysis of microbial communities, because they have an extremely strong Raman signal in a non-resonant mode and are widespread in microbial communities and are present in all phototrophic microorganisms. Jehlička & Oren [13] and Oliveira et al. [33] reported Raman spectra of pure carotenoids astaxanthin, neoxanthin, myxoxanthophyll, zeaxanthin, lutein, fucoxanthin, canthaxanthin and echinenone. These data are helpful when starting evaluating the Raman spectra obtained on an unknown biogeological, geological or biological sample. However, in biological materials the mode of binding of carotenoids in the functional parts of organisms represents one of the potentially significant origins of shifts of the Raman band positions [33]. Caution when interpreting minor shifts is therefore recommended.
Tables 1–4 report the observed main carotenoid Raman skeletal features: C=C stretching in the region of around 1520 cm−1, C−C stretching around 1150 cm−1 and C=CH bending around 1000 cm−1 of a series of microorganisms investigated as well as complementary information on their detection using HPLC. Tables including all the Raman bands are given in the electronic supplementary material, table S1. Examples of Raman spectra obtained on lyophilized sample of microorganisms are reported in figures 2–4.
Table 1.
Main carotenoid Raman skeletal features observed—halophilic Archaea.
| halophilic Archaea, lyophilized samples/extracts |
||||
|---|---|---|---|---|
| 514.5 nm |
||||
| Halobacterium NRC1 | Halobacterium R1 | Haloarcula vallismortis | Halorubrum sodomense | Haloquadratum walsbyi |
| 1001 m/1001 m | 1001 m/1002 m | 1000 w/1002 m | 1001 m/1002 m | 1000 m |
| 1152 s/1151 s | 1150 s/1152 s | 1151 m/1154 m | 1152 s/1153 s | 1152 s |
| 1506 vs/1506 vs | 1505 vs/1509 vs | 1506 s/1508 s | 1506 vs/1507 vs | 1508 s |
| bacterioruberin | bacterioruberin | bacterioruberin | bacterioruberin | bacterioruberin |
Table 4.
Main carotenoid Raman skeletal features observed—eukaryotes.
| eukaryotes—lyophilized samples/extracts |
|||||
|---|---|---|---|---|---|
| 514.5 nm |
1064 nm |
514.5 nm |
|||
| Diadesmis gallica | Botrydiopsis alpina | Balbiania investiens | Porphyridium cruentum | Dunaliella parva | Haematococcus pluvialis |
| 1007 m/1009 m | 1007 m | 1005 m/1005 m | 1004 m | 1005 m/1007 m | 1003 m/1006 m |
| 1158 s/1159 s | 1159 s | 1158 vs/1158 s | 1157 vs | 1157 vs/1156 s | 1154/1157 s |
| 1527 s/1530 s | 1527 vs | 1527 vs/1523 s | 1525 vs/1530 s | 1525 s/1519 s | |
| 1524 w | |||||
| β-carotene | β-carotene | β-carotene | β-carotene | β-carotene | β-carotene |
| lutein | lutein | zeaxanthin | zeaxanthin | lutein, | lutein, |
| chlorophyll a, | chlorophyll a, | cis-zeaxanthin | cis-zeaxanthin | violaxanthin, | violaxanthin, |
| fucoxanthin | myxoxanthophyll | neoxanthin | neoxanthin, | ||
| (expected) | antheraxanthin | ||||
Figure 2.
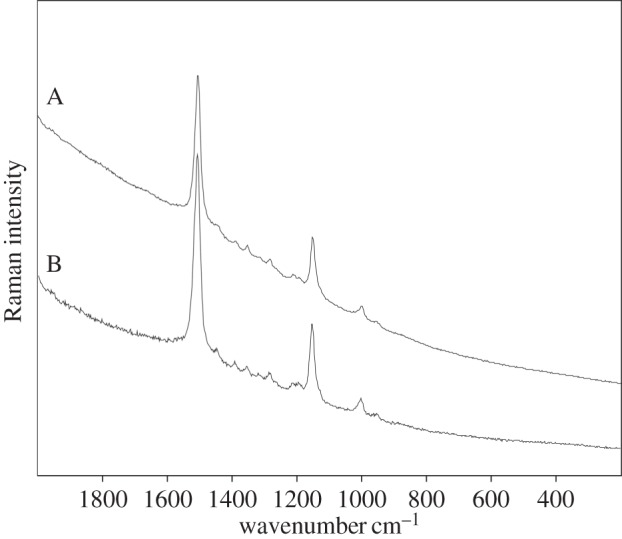
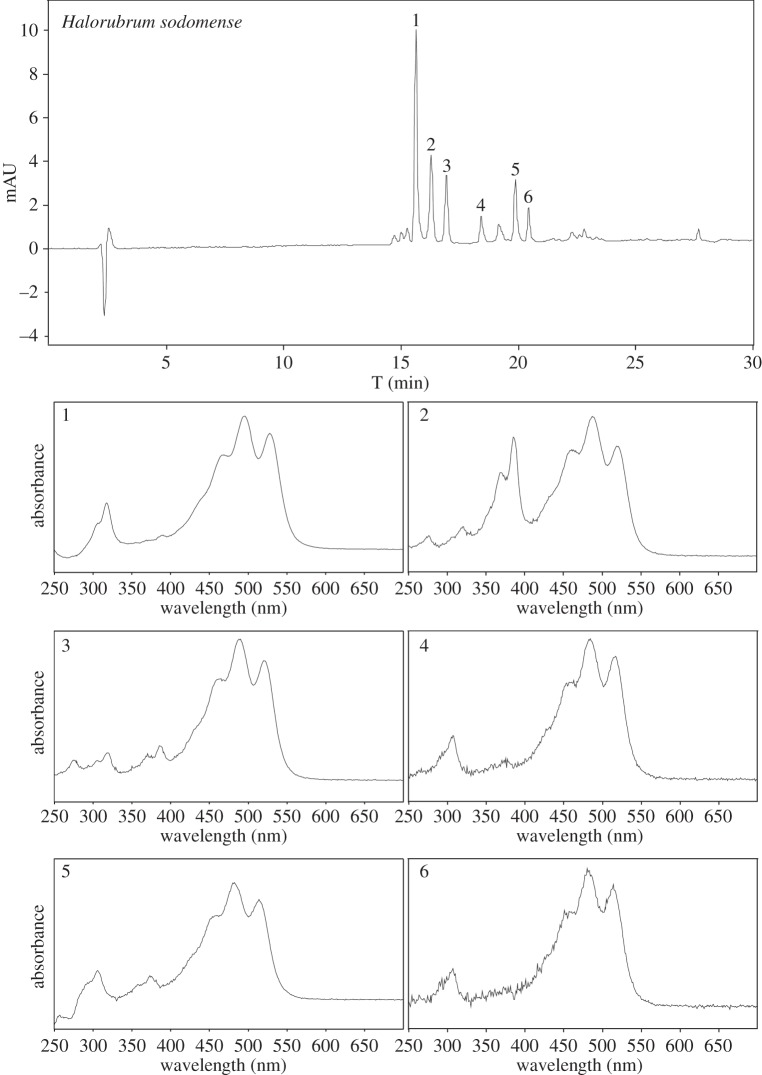
Example of Raman spectra (532 nm) on an example of the Archeae—Halorubrum sodomense: (A) lyophilized material and (B) extract.
Figure 4.
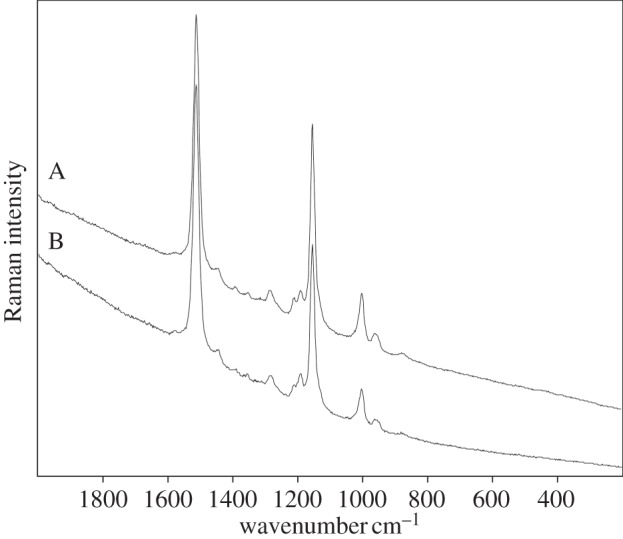
Example of Raman spectra (532 nm) on an example of the extremely halophilic heterotroph Salinibacter ruber: (A) lyophilized material and (B) extract.
Bacterioruberin, the main carotenoid component responsible for the colour of the red Archaea of the family Halobacteriaceae, has a rather different molecular structure in that it has a primary conjugated isoprenoid chain length of 13 C=C units with no subsidiary conjugation arising from terminal groups, which contain only four OH group functionalities. The main Raman bands (figure 2) and their tentative assignments to α-bacterioruberin, the main carotenoid found in Halobacterium salinarum,Halorubrum sodomense and Haloarcula vallismortis are given in the first three columns of table 1. The observed Raman band at 1506 cm−1 can be attributed to the C=C stretching band of bacterioruberin, and the bands at 1152 and 1001 cm−1 belong to the corresponding C–C and CCH, respectively.
In the case of cyanobacteria, frequently the 1064 nm excitation allowed to record acceptable Raman spectra without strong fluorescence (figure 3 and table 2). All the samples of representatives of Synechococcales contain four carotenoids (β-carotene, lutein, cantxanthin, echinenone, as documented by HPLC, Chroococcus sp. seems to contain myxoxanthophyll). However, the Raman spectra contain only a set of features corresponding to only one carotenoid: β-carotene.
Figure 3.
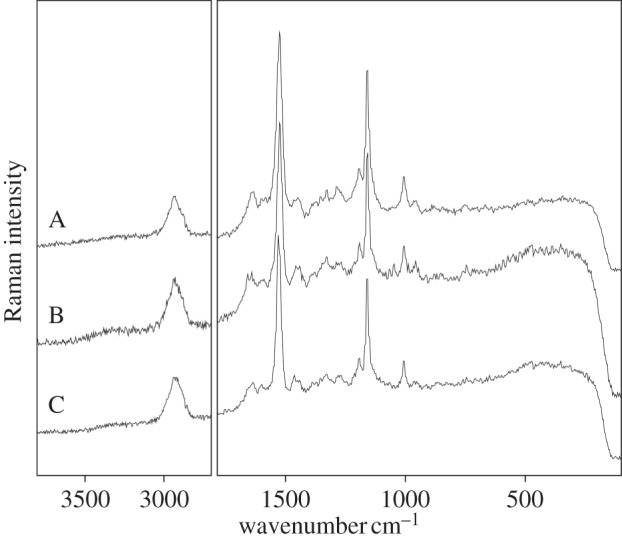
Examples of Raman spectra obtained using an FT Raman spectrometer (1064 nm) on different microorganisms ((A) Phormidium cf. subfuscum, (B) Porphyridium cruentum and (C) Balbiania investens). Note good resolution of the spectra, presence of a broad band in the second order.
Table 2.
Main carotenoid Raman skeletal features observed—cyanobacteria.
| cyanobacteria, lyophilized samples |
|||
|---|---|---|---|
| 1064 nm |
|||
| Chamaesiphon polymorphus | Chroococcus sp. green | Chroococcus sp. | Microcoleus cf. vaginatus |
| 1005 m | 1003 m | 1001 m | 1005 m |
| 1158 s | 1156 s | 1154 s | 1157 vs |
| 1523 s | 1521 m | 1528 w | 1525 m |
| β-carotene | β-carotene | β-carotene | β-carotene |
| zeaxanthin | zeaxanthin | zeaxanthin | zeaxanthin |
| myxoxanthophyll | cis-zeaxanthin | myxoxanthophyll | |
| echinenone | echinenone | ||
The main carotenoid Raman bands of the halophilic cyanobacterium Oscilatoria limnetica samples are 1517, 1156 and 1003 cm−1 falling in the characteristic area of β-carotene. However, HPLC analysis allowed to detect zeaxanthin as an additional pigment. Raman spectra of Oscilatoria sancta as well as Microcoleuscf.vaginatus (table 3) document only the presence of β-carotene in contrast to HPLC which showed also traces of chlorophyll a, α-carotene and zeaxanthin. Raman spectra of Diadesmis gallica and Botrydiopsis alpina display Raman features 1527, 1159 and 1009 cm−1. HPLC permitted to detect chlorophyll, α-carotene, β-carotene, lutein and myxoxanthophyll.
Table 3.
Main carotenoid Raman skeletal features observed—bacteria.
| bacteria, lyophilized samples/extracts |
||||
|---|---|---|---|---|
| 514.5 nm |
1064 nm | |||
| Oscillatoria limnetica | Ectothiorhodospira | Rhodospirillum rubrum | Salinibacter M31 | Micrococcus luteus |
| 1003 m | 1004 m | 1000 m | 1003 m | 1004 w |
| 1156 vs/1156 s | 1153 s/1153 s | 1147 s/1147 s | 1155 vs | 1158 s |
| 1517 vs/1516 | 1514 vs/1514 | 1504 vs | 1512 vs | 1529 s |
| β-carotene | spirilloxanthin | salinixanthin | sarcinaxanthin, | |
| zeaxanthin | anhydrorhodovibrin | sarcinaxanthin | ||
| bacteriophaeophytin | monoglucoside, | |||
| sarcinaxanthin | ||||
| diglucoside | ||||
Raman spectra of Ectothiorhodospira and Halochromatium-like purple sulfur bacteria, especially spirilloxanthin-like carotenoids are expected to occur. The main Raman bands recorded on Ectothiorhodospira marismortui are 1514, 1152 and 1000 cm−1. Raman bands obtained on samples of Rhodospirilum rubrum: 1504, 1147, 1000 cm−1 could correspond to somewhat different carotenoid (table 3). Rhodovibrin and spirilloxanthin are the compounds expected here. Strong carotenoid signals were observed in Salinibacter ruber at 1512, 1155 and 1003 cm−1 (figure 4). The signals correspond to salinixanthin a carotenoid with 11 C=C in the primary isoprenoid chain conjugation plus one more in a terminal alicyclic ring.
The yellow-coloured Micrococcus luteus exhibits, strong Raman bands at 1530, 1158 and 1005 cm−1 (FT-Raman spectroscopy, 1064 nm; table 3), which could be assigned to C=C stretching, C–C stretching and C–CH3 deformation modes of carotenoid sarcinaxanthin. Moreover, other medium and strong features at 1666 and 1452 cm−1 are present in the spectrum, which probably correspond to a protein signal [34].
In the case of Rhodophyta, HPLC analysis permitted to detect chlorophyll a, β-carotene as well as zeaxanthin. Raman spectra of Balbiania investiens contain bands at 1527, 1158, 1005 cm−1, and those of Porphyridium cruentum 1524, 1157, 1004 cm−1 (table 4).
(a). Comparison of Raman spectroscopy and high-performance liquidchromatography/UV-VIS
It is well known that pigments, namely carotenoids, occur in microorganisms associated in specific parts/organelles of the cells. Previous studies on pigments of biological tissues were carried out using HPLC—an appropriate (or even optimal) technique for studying mixtures in extracts. Clearly, this technique allows good separation of pigments from mixtures of organic compounds in solution (in extracts) and powerful detection of their minor contents using UV/visible detectors. Examples of these studies are investigations of pollen carotenoids [35] or a recent study on Cyclotella diatoms [14]. In the frame of the present critical evaluation of conventional Raman spectrometry, a comparison of detecting individual pigments was made for a few examples of microorganisms. Identification of the carotenoids in the samples was based on chromatography and characteristics of the UV-VIS spectra. Figures 5 and 6 as well as electronic supplementary material, figures S1and S2 show HPLC chromatograms and UV-VIS spectra of the compounds corresponding to the main bands
Figure 5.
HPLC chromatogram of a pigment extract of Halorubrum sodomense and UV-VIS spectra of the components.
Figure 6.
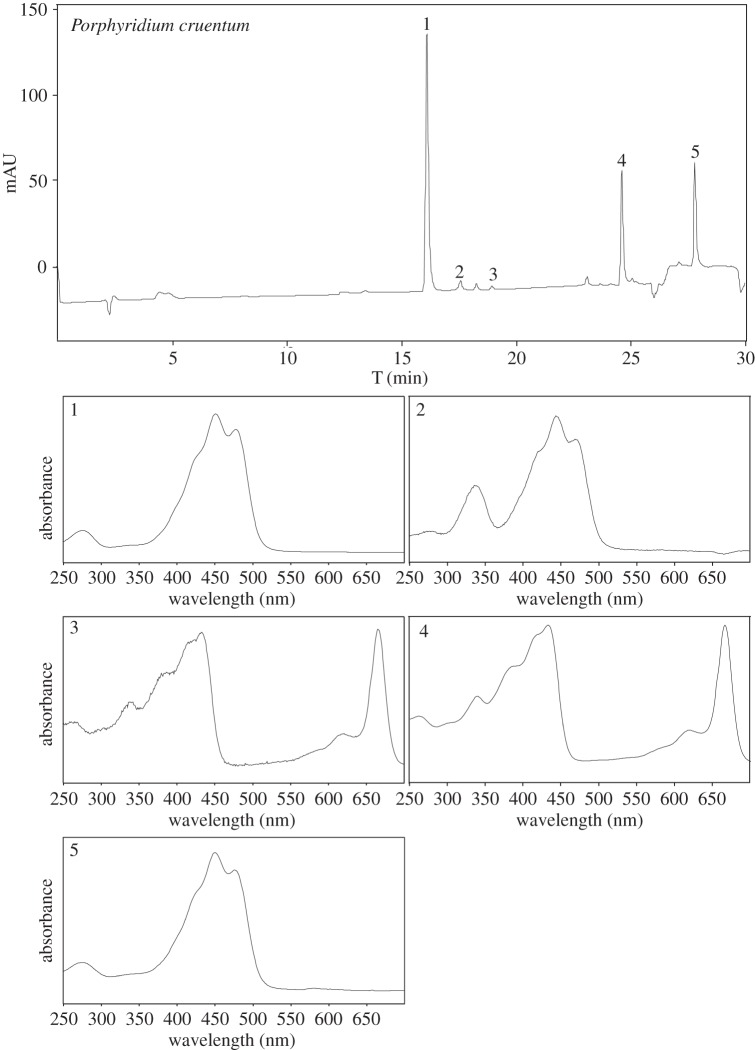
HPLC chromatogram of a pigment extract of Porphyridium cruentumand UV-VIS spectra of the components.
In the first example, a relatively simple situation of the case of Hrr. sodomenseis presented. Here, the main Raman bands observed at 1506 cm−1 (the C=C stretching band, 1152 and 1001 cm−1, C–C and C–CH3, respectively), can be attributed to α-bacterioruberin. Figure 5 shows the chromatogram and absorption spectra obtained from Hrr. sodomense. The pigments α-bacterioruberin (Rt=15.6 min) and its derivatives (anhydrobacterioruberins) were detected as a unique and major pigment.
Another microorganism which produces only one major carotenoid pigment is the red bacterium S. ruber. The Raman spectrum of its unusual pigment salinixanthin was published recently. The C=C stretching band is present at 1512 cm−1, the C–CH3 mode band is observed at 1003 cm−1 and the C–C stretching vibration lies at 1155 cm−1 [36]. Electronic supplementary material, figure S2 shows HPLC chromatograms and UV-VIS spectra of the compounds corresponding to the main bands. The pigments identified in this case were only salinixanthin (Rt=22.5 min) and minor unidentified derivatives.
A more complex situation can be envisaged in the example of M. luteus. This coloured strain exhibits strong Raman bands at 1530, 1158 and 1005 cm−1 (FT-Raman spectroscopy), which could be assigned to C=C stretching, C–C stretching and C–CH3 deformation modes of carotenoid sarcinaxanthin. Moreover, other medium and strong features at 1666 and 1452 cm−1 are present in the spectrum, which probably correspond to a protein signal [34]. The HPLC elution profile of carotenoids extracted from the M. luteus strain show peaks of sarcinaxanthin diglucoside (Rt=17.2 min), sarcinaxanthin monoglucoside (Rt=20.0 min) and sarcinaxanthin (Rt=22.5 min), which together with absorption spectra of these carotenoids are given in the electronic supplementary material, figure S1.
Another example of a microorganism which produces more than one carotenoid is Porphyridium cruentum.Raman spectroscopy shows well resolved only peaks at 1524, 1157 and 1004 cm−1. These features could be assigned to β-carotene, but only with caution (more probably there is a carotenoid mixture). Hence, HPLC and UV-VIS showed the following major pigments (figure 6): zeaxanthin (Rt=16.1 min), cis-zeaxanthin (Rt=17.6 min), chlorophyll a (Rt=24.6 min) and β-carotene (Rt=27.8 min).
Comparing the data obtained by methods stated earlier, we can infer that Raman spectroscopy is at once specific, sensitive and also reproducible. We were also able to demonstrate that Raman spectroscopy strongly correlates with the HPLC analysis of carotenoids, where a single carotenoid is present. However, on the contrary, Raman spectroscopy was not able to unambiguously discriminate between the individual carotenoids in a complex mixture of similar compounds.
4. Discussion
Raman spectroscopy has become a popular tool for detecting carotenoids in different biological materials, including prokaryotic microorganisms, algae and lichens. This is also true for other pigments including chlorophylls and scytonemins. Raman spectroscopy has several obvious advantages: fast analysis, the non-destructive mode of obtaining data, and the possibility to obtain information on pigments within complex organo-mineral matrices. For carotenoids, additionally, resonance Raman enhancement of the signals is very advantageous, allowing their detections in very small concentrations.
Raman spectroscopy is also an excellent technique to detect other pigments of relevance for geoscience, bioscience and astrobiology and to distinguish between carotenoids and non-polyene pigments (e.g. porphyrins, scytonemins, mycosporine-like amino acids) as was repeatedly shown [17,37].
For astrobiology studies, carotenoids, other pigments and few other biomolecules were suggested as biomarkers, compounds that can be detected by Raman spectroscopy in the frame of future Mars missions. For detecting them in the frame of future Mars missions, the application of Raman spectroscopy was proposed [38–40]. In fact, it was shown that on the Earth carotenoids can unambiguously be detected in lichen encrustations [41–44]. More important for the future Mars missions context, pigments were unambiguously detected in desert halite crusts (scytonemin and carotenoids), bottom gypsum layers of halophiles microbial mats (spirilloxanthin), as well as in microplancton inhabiting brines form salterns (bacterioruberin β-carotene) using miniaturized instruments [23,45].
During laboratory-based studies, however, frequently carotenoids were identified (alongside sometimes with other pigments) without the discrimination of the specific ones. Clearly for astrobiology purposes, the potential detection of Raman spectroscopic signs of the presence of a carotenoid would be a big advance even without the precise discrimination. However, some detail on the type of carotenoid is frequently needed in microbiology as well as in other fields of advanced studies, e.g. for studies of environmental effects on synthesis of carotenes or to learn more in-depth about endolithic colonizations. In several scenarios, working with extracted carotenoids (and applying the powerful separation detection HPLC) could be useful. However in another situation important detail can be lost when separating the biomolecule from its molecular environment.
There can be several reasons of potential minor shifts in the position of the Raman bands in spectra of a given carotenoid within biomass. Some of them are related to the nature of the sample, the others can derive from the experimental conditions. Theoretical studies can help in better understanding of some of those effects; however, these frequently use simplified assumptions [46,47]. The mode how a carotenoid is bound in the biomass and what compounds are in the surrounding environment of the molecule, these are assumed to be few of the effects affecting the precise conformation of the molecule and consequently the position of Raman bands in the spectra [33,48]. Additionally, minor shifts are known to be related to the excitation laser wavelength [49,50]. Those effects are mainly described in the case of unmethylated polyenes (originating in pigments of pearls and corals in the case of the cited studies). To address these effects, few studies presented comparisons using laser wavelengths from 458 to 657 nm. In a detailed study of salinixanthin from Salinibacter ruber [36], it was possible to show that in the case of this carotenoid a systematic shift was not observed for ν1(C=C) neither ν2(C–C) stretching vibrations (in this case, a 458, 514.5 and 1064 nm excitation was used).
In this study, Raman spectroscopy was successfully used for detecting β-carotene in the frame of very different microorganisms as well as bacterioruberin as a key carotene of halophilic Archaea. These pigments are structurally different, they differ in the length of the conjugated chain (11 in β-carotene, 13 in bacterioruberin), so that also their Raman spectra differ considerably. Even Raman spectrometers with mean spectral resolution would easily allow their detection and discrimination.
However in other cases reported, microorganisms synthesize other pigments, and investigated samples contained a mixture of them. In these cases, Raman spectra do not contain a series of sharp bands, each of them corresponding to a given carotene. Also, interestingly, no major broadening of the bands was observed. The dominant carotenoid is reflected in the spectra. Unfortunately, it was not possible to use a unique excitation wavelength for the whole series of microorganisms, e.g. 532 nm, owing to the presence of other compounds which cause intense fluorescence. Deeper understanding of the presence of a mixture of carotenoids (or unmethylated polyenes) would be possible comparing simple compound measurements and model mixtures. The effects of individual compounds under slightly differing resonance conditions would perhaps allow to found precise resonance conditions and record intensity enhanced band corresponding to a carotenoid chain under resonance.
On the other hand, HPLC analysis of extracts permits the identification of multiple and even very similar carotenoids. For the advanced task of identifying individual carotenoids in a complex mixture within biological materials, Raman spectroscopy generally cannot allow excellent discrimination like HPLC.
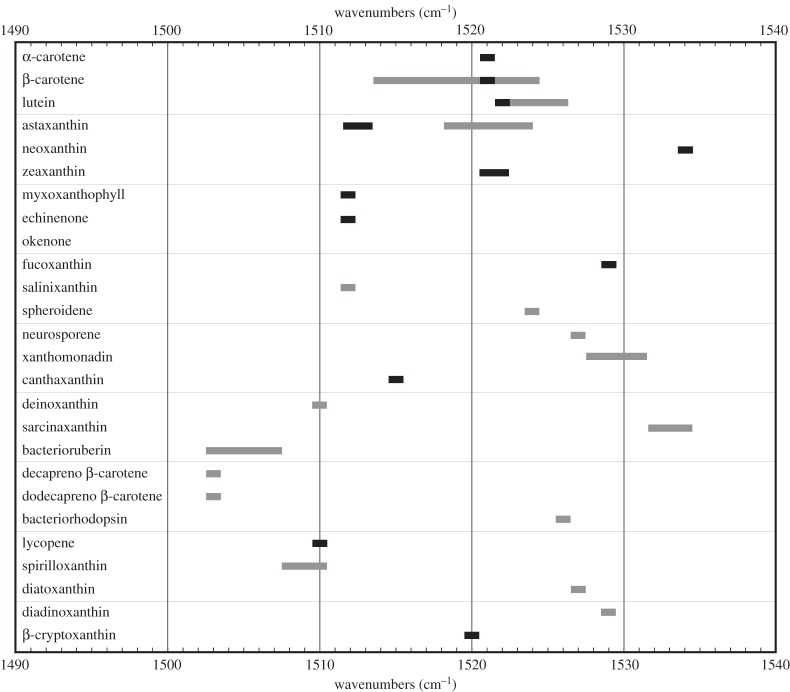
For the practical estimation of the probability to successfully discriminate carotenoids in unknown samples, an identification chart was constructed. In this chart reported as figure 7, the main Raman spectroscopic feature of carotenoids—the band ν1(C=C) in the area of 1505–1530 cm−1 was collected for a series of compounds. The precise frequencies correspond to the length of the conjugated chain. The chart is based on data from this study as well as on those reported in the literature. To simplify, analytical aspects are omitted (excitation wavelength, mode of measurements). Values of pure compounds (when reported) are given as full squares. Some carotenoids can be distinguished from others with high probability, just on the basis of the position of the Raman band reflecting the length of the polyene chain. Lower values of wavenumbers in the Raman spectrum correspond to carotenes with a longer polyene chain (higher number of the conjugated double bonds). In some cases, the length of a pair of carotenes is the same, but they differ by additional functionalities on the extremity(ies) or by additional side chain(s). These features can sometimes, clearly, also affect Raman spectra. This was documented, for example, in the case of salinixanthin of Salinibacter ruber (presence of the bands corresponding to the glycosidic stretching mode of the associated sugar ring (1187 cm−1)) and a COH stretching mode (1041 cm−1) or in the case of polyenic flexirubins (CC stretching of the saturated aliphatic chain 1133/1138 cm−1 in the case of Flavobacterium johnsoniae/Flexibacter elegans [51]). These features are not reported in the present chart and will be collected in the frame of a review prepared for publication.
Figure 7.
Review chart presenting the values of the ν1(C=C) stretching Raman band.
Summarizing, Raman spectroscopy can be used to detect the presence of carotenoids and other pigments in cyanobacteria, bacteria and algae. But the occurrence of coloured compounds in such organism can cause minor to moderate significant variations in intensities and especially in the wavenumber positions of the Raman bands, prohibiting potentially precise identification of a given carotenoid because of shifts in intensities and especially the positions of the Raman bands.
5. Conclusion
— Carotenoids—as a group—are easily detected within cultures of microorganisms using Raman spectroscopy of different excitations without the need of extracting samples or complex treatment. Excellent results are obtained using 514 and 532 nm lasers and due to the advantage of resonance Raman enhancement in some cases.
— Methanol–acetone extracts from cultures confirm the presence of the main carotenoids. The main features in the Raman spectra of carotenoids in extracts generally correspond to those of parent lyophilized biomass. Sometimes non-systematic shifts were observed. Those can be related with the potential modification of the molecular environment as well as the conformation of the carotene molecule.
— Raman spectroscopy permits unambiguous discrimination of structurally differing carotenoids when occurring in a culture in major or moderate concentrations.
— More work is needed to permit unambiguous detection of individual carotenoids in complex natural (biological, geobiological as well as geological) samples. Only careful reading of the spectra can help to unambiguously learn about interesting detail needed for interpretations.
Supplementary Material
Funding statement
This work was supported by grant no. P210/10/0467 from the Grant Agency of the Czech Republic and by institutional support MSM0021620855 from the Ministry of Education of the Czech Republic (to J.J.) and by grant no. 1103/10 from the Israel Science Foundation (to A.O.). We thank Salt of the Earth Eilat Ltd. for allowing access to the Eilat salterns, and the Interuniversity Institute for Marine Sciences of Eilat for logistic support. We thank Adam Culka for the final technical assistance with the manuscript.
References
- 1.Edwards HGM, Hutchinson I, Ingley R. 2012. The Exomars Raman spectrometer and the identification of biogeological spectroscopic signatures using a flight-like prototype. Anal. Bioanal. Chem. 404, 1723–1731. ( 10.1007/s00216-012-6285-z) [DOI] [PubMed] [Google Scholar]
- 2.Oren A. 2011. Characterization of pigments of prokaryotes and their use in taxonomy and classification. In Methods in microbiology, vol. 38 (eds Rainey F, Oren A.), pp. 261–282. New York, NY: Academic Press. [Google Scholar]
- 3.Oren A. 2002. Pigments of halophilic microorganisms. In Halophilic microorganisms and their environments, pp. 173–206. Dordrecht, The Netherlands: Kluwer Scientific Publishers. [Google Scholar]
- 4.Britton G, Liaaen-Jensen S, Pfander H. (eds). 2004. Carotenoids handbook. Basel, Switzerland: Birkhäuser. [Google Scholar]
- 5.Frank HA, Chynwat V, Desamero RZB, Farhoosh R, Erickson J, Bautista J. 1997. On the photophysics and photochemical properties of carotenoids and their role as light-harvesting pigments in photosynthesis. Pure Appl. Chem. 69, 2117–2124. ( 10.1351/pac199769102117) [DOI] [Google Scholar]
- 6.Caumette P. 1993. Ecology and physiology of phototrophic bacteria and sulfate-reducing bacteria in marine salterns. Experientia 49, 473–481. ( 10.1007/bf01955148) [DOI] [Google Scholar]
- 7.Wachsmann-Hogiu S, Weeks T, Huser T. 2009. Chemical analysis in vivo and in vitro by Raman spectroscopy-from single cells to humans. Curr. Opin. Biotechnol. 20, 63–73. ( 10.1016/j.copbio.2009.02.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner WD. 1986. Raman excitation profiles from pigments?. in vivo. J. Raman Spectrosc. 17, 51–53. ( 10.1002/jrs.1250170111) [DOI] [Google Scholar]
- 9.Heraud P, Beardall J, McNaughton D, Wood BR. 2007. In vivo prediction of the nutrient status of individual microalgal cells using Raman microspectroscopy. FEMS Microbiol. Lett. 275, 24–30. ( 10.1111/j.1574-6968.2007.00861.x) [DOI] [PubMed] [Google Scholar]
- 10.Spiro TG. 1987. Biological applications of Raman spectroscopy. New York, NY: John Wiley & Sons. [Google Scholar]
- 11.Carey PR. 1982. Biochemical applications of Raman and resonance Raman spectroscopies. New York, NY: Academic Press. [Google Scholar]
- 12.Marshall CP, Leuko S, Coyle CM, Walter MR, Burns BP, Neilan BA. 2007. Carotenoid analysis of halophilic Archaea by resonance Raman spectroscopy. Astrobiology 7, 631–643. ( 10.1089/ast.2006.0097) [DOI] [PubMed] [Google Scholar]
- 13.Jehlička J, Oren A. 2013. Raman spectroscopy in halophile research. Front. Microbiol. 4, 380 ( 10.3389/fmicb.2013.00380) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alexandre MTA, Gundermann K, Pascal AA, van Grondelle R, Buechel C, Robert B. 2014. Probing the carotenoid content of intact Cyclotella cells by resonance Raman spectroscopy. Photosynth. Res. 119, 273–281. ( 10.1007/s11120-013-9942-y) [DOI] [PubMed] [Google Scholar]
- 15.Edwards HGM, Villar SEJ, Parnell J, Cockell CS, Lee P. 2005. Raman spectroscopic analysis of cyanobacterial gypsum halotrophs and relevance for sulfate deposits on Mars. Analyst 130, 917–923. ( 10.1039/b503533c) [DOI] [PubMed] [Google Scholar]
- 16.Wynn-Williams DD, Edwards HGM. 2002. Environmental UV radiation: biological strategies for protection and avoidance. In Astrobiology (eds Horneck G, Baumstark-Khan Ch.), pp. 245–260. Heidelberg, Germany: Springer. [Google Scholar]
- 17.Edwards HGM, Hutchinson IB, Ingley R, Parnell J, Vítek P, Jehlička J. 2013. Raman spectroscopic analysis of geological and biogeological specimens of relevance to the ExoMars mission. Astrobiology 13, 543–549. ( 10.1089/ast.2012.0872) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards HGM, Hutchinson IB, Ingley R. 2013. A Raman spectral signatures in the biogeological record: An astrobiological challenge. In Habitability of other planets and satellites (eds de Vera J-P, Seckbach J.), pp. 311–330. The Netherlands: Springer. [Google Scholar]
- 19.Marshall CP, Carter EA, Leuko S, Javaux EJ. 2006. Vibrational spectroscopy of extant and fossil microbes: relevance for the astrobiological exploration of Mars. Vib. Spectrosc 41, 182–189. ( 10.1016/j.vibspec.2006.01.008) [DOI] [Google Scholar]
- 20.Jehlička J, Edwards HGM, Vítek P. 2009. Assessment of Raman spectroscopy as a tool for the non-destructive identification of organic minerals and biomolecules for Mars studies. Planet. Space Sci. 57, 606–613. ( 10.1016/j.pss.2008.05.005) [DOI] [Google Scholar]
- 21.Vítek P, Osterrothová K, Jehlička J. 2009. Beta-carotene-a possible biomarker in the Martian evaporitic environment: Raman micro-spectroscopic study. Planet. Space Sci. 57, 454–459. ( 10.1016/j.pss.2008.06.001) [DOI] [Google Scholar]
- 22.Vítek P, Jehlička J, Edwards HGM. 2013. Practical considerations for the field application of miniaturized portable Raman instrumentation for the identification of minerals. Appl. Spectrosc. 67, 767–778. ( 10.1366/12-06774) [DOI] [PubMed] [Google Scholar]
- 23.Vítek P, Jehlička J, Edwards HGM, Hutchinson I, Ascaso C, Wierzchos J. 2012. The miniaturized Raman system and detection of traces of life in halite from the Atacama desert: Some considerations for the search for life signatures on Mars. Astrobiology 12, 1095–1099. ( 10.1089/ast.2012.0879) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guillard RR, Lorenzen CJ. 1972. Yellow-green algae with chlorophyllide c. J. Phycol. 8, 10–14. ( 10.1111/j.0022-3646.1972.00010.x) [DOI] [Google Scholar]
- 25.Brody M, Emerson R. 1959. The quantum yield of photosynthesis in Porphyridium cruentum, and the role of chlorophyll a in the photosynthesis of red algae. J. Gen. Physiol. 43, 251–264. ( 10.1085/jgp.43.2.251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bold HC. 1949. The morphology of Chlamydomonas chlamydogama sp. nov. Bull. Torrey Bot. Club 76, 101–108. ( 10.2307/2482218) [DOI] [Google Scholar]
- 27.Bischoff HW, Bold HC. 1963. Phycological studies. IV. Some soil algae from Enchanted Rock and related algal species. Univ. Texas. Publ. 6318, 1–95. [Google Scholar]
- 28.Oren A, Kessel M, Stackebrandt E. 1989. Ectothiorhodospira marismortui sp. nov., an obligately anaerobic, moderately halophilic purple sulfur bacterium from a hypersaline sulfur spring on the shore of the Dead-Sea. Arch. Microbiol. 151, 524–529. ( 10.1007/bf00454869) [DOI] [Google Scholar]
- 29.Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R. 2002. Salinibacter ruber gen. Nov., sp nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Microbiol. 52, 485–491. ( 10.1099/ijs.0.01913-0) [DOI] [PubMed] [Google Scholar]
- 30.Oren A. 1996. Sensitivity of selected members of the family Halobacteriaceae to quinolone antimicrobial compounds. Arch. Microbiol. 165, 354–358. ( 10.1007/s002030050338) [DOI] [PubMed] [Google Scholar]
- 31.Oren A. 1983. Halobacterium-sodomense sp-nov, a Dead-Sea Halobacterium with an extremely high magnesium requirement. Int. J. Syst. Bacteriol. 33, 381–386. ( 10.1099/00207713-33-2-381) [DOI] [Google Scholar]
- 32.Eimhjellen KE, Jensen SL. 1964. Biosynthesis of carotenoids in Phodopseudomonas gelatinosa. Biochim. Biophys. Acta 82, 21–40. ( 10.1016/0304-4165(64)90004-2) [DOI] [Google Scholar]
- 33.de Oliveira VE, Castro HV, Edwards HGM, de Oliveira LFC. 2010. Carotenes and carotenoids in natural biological samples: a Raman spectroscopic analysis. J. Raman Spectrosc. 41, 642–650. ( 10.1002/jrs.2493) [DOI] [Google Scholar]
- 34.Rosch P, et al. 2005. Chemotaxonomic identification of single bacteria by micro-Raman spectroscopy: application to clean-room-relevant biological contaminations. Appl. Environ. Microbiol. 71, 1626–1637. ( 10.1128/aem.71.3.1626-1637.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulte F, Maeder J, Kroh LW, Panne U, Kneipp J. 2009. Characterization of pollen carotenoids with in situ and high-performance thin-layer chromatography supported resonant Raman spectroscopy. Anal. Chem. 81, 8426–8433. ( 10.1021/ac901389p) [DOI] [PubMed] [Google Scholar]
- 36.Jehlička J, Edwards HGM, Oren A. 2013. Bacterioruberin and salinixanthin carotenoids of extremely halophilic Archaea and Bacteria: a Raman spectroscopic study. Spectrochim. Acta Part A 106, 99–103. ( 10.1016/j.saa.2012.12.081) [DOI] [PubMed] [Google Scholar]
- 37.Vítek P, Camara-Gallego B, Edwards HGM, Jehlička J, Ascaso C, Wierzchos J. 2013. Phototrophic community in gypsum crust from the Atacama Desert studied by Raman spectroscopy and microscopic imaging. Geomicrobiol. J. 30, 399–410. ( 10.1080/01490451.2012.697976) [DOI] [Google Scholar]
- 38.Edwards HGM, Vandenabeele P, Jorge-Villar SE, Carter EA, Perez FR, Hargreaves MD. 2007. The Rio Tinto Mars analogue site: an extremophilic Raman spectroscopic study. Spectrochim. Acta Part A 68, 1133–1137. ( 10.1016/j.saa.2006.12.080) [DOI] [PubMed] [Google Scholar]
- 39.Edwards HGM. 2014. Will-o′-the-Wisp: an ancient mystery with extremophile origins?. Phil. Trans. R. Soc. A 372, 20140206 ( 10.1098/rsta.2014.0206) [DOI] [PubMed] [Google Scholar]
- 40.Edwards HGM, Hutchinson IB, Ingley R, Jehlička J. 2014. Biomarkers and their Raman spectroscopic signatures: a spectral challenge for analytical astrobiology. Phil. Trans. R. Soc. A 372, 20140193 ( 10.1098/rsta.2014.0193) [DOI] [PubMed] [Google Scholar]
- 41.Edwards HGM, Edwards KAE, Farwell DW, Lewis IR, Seaward MRD. 1994. An approach to stone and fresco lichen biodeterioration through Fourier-transform Raman microscopic investigation of Thallus substratum encrustations. J. Raman Spectrosc. 25, 99–103. ( 10.1002/jrs.1250250114) [DOI] [Google Scholar]
- 42.Edwards HGM, Farwell DW, Seaward MRD. 1997. FT-Raman spectroscopy of Dirina massiliensis f sorediata encrustations growing on diverse substrata. Lichenologist 29, 83–90. [Google Scholar]
- 43.Russell NC, Edwards HGM, Wynn-Williams DD. 1998. FT-Raman spectroscopic analysis of endolithic microbial communities from beacon sandstone in Victoria Land, Antarctica. Antarct. Sci. 10, 63–74. ( 10.1017/S0954102098000091) [DOI] [Google Scholar]
- 44.Vítek P, Edwards HGM, scaso Jehlička CA, De Les Rios A, Valea S, Jorge-villar SE, Davila AF, Wierzchos J. 2010. Microbial colonization of halite from the hyper-arid Atacama Desert studied by Raman spectroscopy. Phil. Trans. R. Soc. A 368, 3205–3221. ( 10.1098/rsta.2010.0059) [DOI] [PubMed] [Google Scholar]
- 45.Jehlička J, Oren A. 2013. Use of a handheld Raman spectrometer for fast screening of microbial pigments in cultures of halophilic microorganisms and in microbial communities in hypersaline environments in nature. J. Raman Spectrosc. 44, 1285–1291. ( 10.1002/Jrs.4362) [DOI] [Google Scholar]
- 46.Varnali T, Edwards HGM, Hargreaves MD. 2009. Scytonemin: molecular structural studies of a key extremophilic biomarker for astrobiology. Int. J. Astrobiol. 8, 133–140. ( 10.1017/s1473550409004455) [DOI] [Google Scholar]
- 47.Brambilla L, Tommasini M, Zerbi G, Stradi R. 2012. Raman spectroscopy of polyconjugated molecules with electronic and mechanical confinement: the spectrum of Corallium rubrum. J. Raman Spectrosc. 43, 1449–1458. ( 10.1002/jrs.4057) [DOI] [Google Scholar]
- 48.Withnall R, Chowdhry BZ, Silver J, Edwards HGM, de Oliveira LFC. 2003. Raman spectra of carotenoids in natural products. Spectrochim. Acta Part A 59, 2207–2212. ( 10.1016/s1386-1425(03)00064-7) [DOI] [PubMed] [Google Scholar]
- 49.Karampelas S, Fritsch E, Mevellec J-Y, Sklavounos S, Soldatos T. 2009. Role of polyenes in the coloration of cultured freshwater pearls. Eur. J. Mineral. 21, 85–97. ( 10.1127/0935-1221/2009/0021-1897) [DOI] [Google Scholar]
- 50.Bergamonti L, Bersani D, Csermely D, Lottici PP. 2011. The nature of the pigments in corals and pearls: a contribution from Raman spectroscopy. Spectrosc. Lett. 44, 453–458. ( 10.1080/00387010.2011.610399) [DOI] [Google Scholar]
- 51.Jehlička J, Osterrothová K, Oren A, Edwards HGM. 2013. Raman spectrometric discrimination of flexirubin pigments from two genera of Bacteroidetes. FEMS Microbiol. Lett. 348, 97–102. ( 10.1111/1574-6968.12243) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.