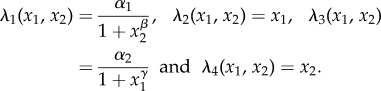Abstract
We consider the problem of estimating parameter sensitivity for Markovian models of reaction networks. Sensitivity values measure the responsiveness of an output with respect to the model parameters. They help in analysing the network, understanding its robustness properties and identifying the important reactions for a specific output. Sensitivity values are commonly estimated using methods that perform finite-difference computations along with Monte Carlo simulations of the reaction dynamics. These methods are computationally efficient and easy to implement, but they produce a biased estimate which can be unreliable for certain applications. Moreover, the size of the bias is generally unknown and hence the accuracy of these methods cannot be easily determined. There also exist unbiased schemes for sensitivity estimation but these schemes can be computationally infeasible, even for very simple networks. Our goal in this paper is to present a new method for sensitivity estimation, which combines the computational efficiency of finite-difference methods with the accuracy of unbiased schemes. Our method is easy to implement and it relies on an exact representation of parameter sensitivity that we recently proved elsewhere. Through examples, we demonstrate that the proposed method can outperform the existing methods, both biased and unbiased, in many situations.
Keywords: parameter sensitivity, stochastic reaction networks, Markov process, unbiased estimator
1. Introduction
Reaction networks are frequently encountered in several scientific disciplines, such as chemistry [1], systems biology [2], epidemiology [3], ecology [4] and pharmacology [5]. It is well known that when the population sizes of the reacting species are small, then a deterministic formulation of the reaction dynamics is inadequate for understanding many important properties of the system [6,7]. Hence, stochastic models are necessary and the most prominent class of such models is where the reaction dynamics is described by a continuous time Markov chain (CTMC). Our paper is concerned with sensitivity analysis of these Markovian models of reaction networks.
Generally, reaction networks involve many kinetic parameters that influence their dynamics. With sensitivity analysis, one can measure the receptiveness of an outcome to small changes in parameter values. These sensitivity values give insights about network design and its robustness properties [8]. They also help in identifying the important reactions for a given output, estimating the model parameters and in fine-tuning the system's behaviour [9]. The existing literature contains many methods for sensitivity analysis of CTMC models, but all these methods have certain drawbacks. They introduce a bias in the sensitivity estimate [10–13] or the associated estimator can have large variances [14] or the method becomes impractical for large networks [15]. Owing to these reasons, the search for better methods for sensitivity analysis of stochastic reaction networks is still an active research problem.
Consider a stochastic reaction network consisting of d species S1, … , Sd. Under the well-stirred assumption [16], the state of the system at any time is given by a vector in  , where
, where 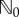 is the set of non-negative integers. When the state is x = (x1, … , xd), then the population size corresponding to species Si is xi. The state evolves as the species interact through K reaction channels. If the state is x, then the kth reaction fires at rate λk(x) and it displaces the state by the stoichiometric vector
is the set of non-negative integers. When the state is x = (x1, … , xd), then the population size corresponding to species Si is xi. The state evolves as the species interact through K reaction channels. If the state is x, then the kth reaction fires at rate λk(x) and it displaces the state by the stoichiometric vector
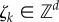 . Here, λk is called the propensity function for the kth reaction. We can represent the reaction dynamics by a CTMC and the distribution of this process evolves according to the Chemical Master Equation [16] which is quite difficult to solve except in some restrictive cases. Fortunately, the sample paths of this process can be easily simulated using Monte Carlo procedures such as Gillespie's stochastic simulation algorithm (SSA) and its variants [16,17].
. Here, λk is called the propensity function for the kth reaction. We can represent the reaction dynamics by a CTMC and the distribution of this process evolves according to the Chemical Master Equation [16] which is quite difficult to solve except in some restrictive cases. Fortunately, the sample paths of this process can be easily simulated using Monte Carlo procedures such as Gillespie's stochastic simulation algorithm (SSA) and its variants [16,17].
Suppose that the propensity functions of a reaction network depend on a scalar parameter θ. Hence, when the state is x, the kth reaction fires at rate λk(x, θ). Let 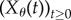 denote the CTMC representing the reaction dynamics with a known initial state x0. For a function
denote the CTMC representing the reaction dynamics with a known initial state x0. For a function 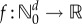 and an observation time T ≥ 0, our output of interest is f(Xθ(T)). We determine how much the expected value of this output changes with infinitesimal changes in the parameter θ. In other words, our aim is to compute
and an observation time T ≥ 0, our output of interest is f(Xθ(T)). We determine how much the expected value of this output changes with infinitesimal changes in the parameter θ. In other words, our aim is to compute
| 1.1 |
Since the mapping 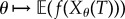 is generally unknown, it is difficult to evaluate Sθ(f, T) directly. Therefore, we need to estimate this quantity using simulations of the process Xθ. For this purpose, many methods use a finite-difference scheme such as
is generally unknown, it is difficult to evaluate Sθ(f, T) directly. Therefore, we need to estimate this quantity using simulations of the process Xθ. For this purpose, many methods use a finite-difference scheme such as
| 1.2 |
for a small h. These methods reduce the variance of the associated estimator by coupling the processes Xθ and Xθ+h in an intelligent way. Three such couplings are: common reaction numbers (CRN) [11], common reaction paths (CRP) [11] and coupled finite differences (CFD) [13]. The two best-performing finite-difference schemes are CRP and CFD, and we shall discuss them in §2.1. It is immediate that a finite-difference scheme produces a biased estimate of the true sensitivity value Sθ(f, T). To further compound this problem, the size and even the sign of the bias is unknown in most cases, thereby casting a doubt over the estimated sensitivity values. The magnitude of the bias must be proportional to h, but it can still be significant for small h (see §4). One can reduce the bias by decreasing h, but as h gets smaller, the variance of the finite-difference estimator gets larger, making it necessary to generate an extremely large sample to obtain a statistically accurate estimate. In many cases, for small values of h, the computational cost of generating a large sample is so high that finite-difference schemes become inefficient in comparison to the unbiased methods. Unfortunately, there is no way to select an optimum value of h, which is small enough to ensure that the bias is small and simultaneously large enough to ensure that the estimator variance is low. This is the main difficulty in using finite-difference schemes and we shall revisit this point in §4.
To obtain highly accurate estimates of Sθ(f, T), we need unbiased methods for sensitivity analysis. The first such scheme is the likelihood-ratio method which relies on the Girsanov measure transformation [14,18]. Hence, we refer to it as the Girsanov method in this paper. In this method, the sensitivity value is expressed as
| 1.3 |
where sθ(f, t) is a random variable whose realizations can be easily obtained by simulating the CTMC Xθ in the time interval [0, T]. The Girsanov method is easily implementable, and its unbiasedness guarantees convergence to the right value Sθ(f, T) as the sample size tends to infinity. However in many situations, the estimator associated with this method has a very high variance, making it extremely inefficient [11–13,15]. To remedy this problem, we proposed another unbiased scheme in [15] based on a different sensitivity formula of the form (1.3), which is derived using the random time-change representation of Kurtz (see ch. 7 in [19]). Unfortunately in this formula, the realizations of sθ(f, T) cannot be easily computed from the paths of the process Xθ as it involves several expectations of functions of the underlying CTMC at various times and various initial states. If all these expectations are estimated serially using independent paths, then the problem becomes computationally intractable. Hence, we devised a scheme in [15], called the Auxiliary Path Algorithm (APA), to estimate all these expectations in parallel using a fixed number of auxiliary paths. The implementation of APA is quite difficult and the memory requirements are high because one needs to store the paths and dynamically maintain a large table to estimate the relevant quantities. These reasons make APA impractical for large networks and also for large observation times T. However, in spite of these difficulties, we showed in [15] that APA can be far more efficient than the Girsanov method in certain situations.
The above discussion suggests that all the existing methods for sensitivity analysis, both biased and unbiased, have certain drawbacks. This motivated us to develop a new method for sensitivity estimation which is unbiased, easy to implement, has low memory requirements and is more versatile than the existing unbiased schemes. We use the main result in [15] to construct another random variable 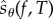 such that (1.3) holds. We then provide a simple scheme, called the Poisson path algorithm (PPA), to obtain realizations of
such that (1.3) holds. We then provide a simple scheme, called the Poisson path algorithm (PPA), to obtain realizations of 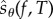 and this gives us a method to obtain unbiased estimates of parameter sensitivities for stochastic reaction networks. Similar to APA, PPA also requires estimation of certain quantities using auxiliary paths, but the number of these quantities can be controlled to be so low that they can be estimated serially using independent paths. Consequently, PPA does not require any storage of paths or the dynamic maintenance of a large table as in APA. Hence, the memory requirements are low, the implementation is easier and PPA works well for large networks and for large observation times T. In §4, we consider many examples to compare the performance of PPA with the Girsanov method, CFD and CRP. We find that PPA is usually far more efficient than the Girsanov method. Perhaps surprisingly, in many situations PPA can also outperform the finite-difference schemes (CRP and CRN) if we impose the restriction that the bias is small. This makes PPA an attractive method for sensitivity analysis because one can efficiently obtain sensitivity estimates, without having to worry about the (unknown) bias caused by finite-difference approximations.
and this gives us a method to obtain unbiased estimates of parameter sensitivities for stochastic reaction networks. Similar to APA, PPA also requires estimation of certain quantities using auxiliary paths, but the number of these quantities can be controlled to be so low that they can be estimated serially using independent paths. Consequently, PPA does not require any storage of paths or the dynamic maintenance of a large table as in APA. Hence, the memory requirements are low, the implementation is easier and PPA works well for large networks and for large observation times T. In §4, we consider many examples to compare the performance of PPA with the Girsanov method, CFD and CRP. We find that PPA is usually far more efficient than the Girsanov method. Perhaps surprisingly, in many situations PPA can also outperform the finite-difference schemes (CRP and CRN) if we impose the restriction that the bias is small. This makes PPA an attractive method for sensitivity analysis because one can efficiently obtain sensitivity estimates, without having to worry about the (unknown) bias caused by finite-difference approximations.
This paper is organized as follows. In §2, we provide the relevant mathematical background and discuss the existing schemes for sensitivity analysis in greater detail. We also present our main result which shows that for a suitably defined random variable 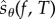 , relation (1.3) is satisfied. In §3, we describe PPA which is a simple method to obtain realizations of
, relation (1.3) is satisfied. In §3, we describe PPA which is a simple method to obtain realizations of 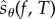 . In §4, we consider many examples to illustrate the efficiency of PPA and compare its performance with other methods. Finally, in §5, we conclude and discuss future research directions.
. In §4, we consider many examples to illustrate the efficiency of PPA and compare its performance with other methods. Finally, in §5, we conclude and discuss future research directions.
2. Preliminaries
Recall the description of the reaction network from the previous section and suppose that the propensity functions depend on a real-valued parameter θ. We can model the reaction dynamics by a CTMC whose generator1 is given by
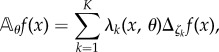 |
where 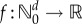 is any bounded function and
is any bounded function and 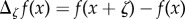 . Under some mild conditions on the propensity functions (see Condition 2.1 in [15]), a CTMC (Xθ(t))t≥0 with generator
. Under some mild conditions on the propensity functions (see Condition 2.1 in [15]), a CTMC (Xθ(t))t≥0 with generator 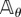 and any initial state x0 exists uniquely. The random time-change representation of Kurtz (see ch. 7 in [19]) shows that this CTMC can be expressed as
and any initial state x0 exists uniquely. The random time-change representation of Kurtz (see ch. 7 in [19]) shows that this CTMC can be expressed as
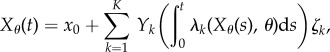 |
2.1 |
where 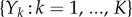 is a family of independent unit rate Poisson processes.
is a family of independent unit rate Poisson processes.
In this paper, we are interested in computing the sensitivity value Sθ(f, T) defined by (1.1). Assume that we can express the sensitivity value as (1.3) for some random variable sθ(f, T). If we are able to generate N independent samples  from the distribution of sθ(f, T), then Sθ(f, T) can be estimated as
from the distribution of sθ(f, T), then Sθ(f, T) can be estimated as
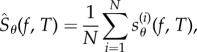 |
2.2 |
which is a random variable with mean μN and variance 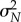 given by
given by
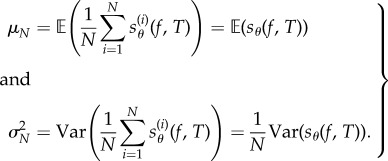 |
2.3 |
Owing to the central limit theorem, for large values of N, the distribution of 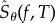 is approximately normal with mean μN and variance
is approximately normal with mean μN and variance 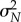 . Hence, for any interval
. Hence, for any interval 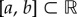 , the probability
, the probability 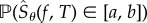 can be approximated as
can be approximated as
| 2.4 |
where 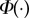 is the cumulative distribution function for the standard normal distribution and σN is the standard deviation. Generally, μN and σN are unknown but they can be estimated from the sample as
is the cumulative distribution function for the standard normal distribution and σN is the standard deviation. Generally, μN and σN are unknown but they can be estimated from the sample as
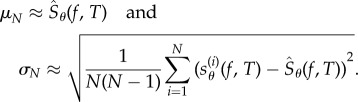 |
Note that the mean μN is just the one-point estimate for the sensitivity value, while the standard deviation σN measures the statistical spread around μN. The standard deviation σN can also be seen as the estimation error. From (2.3), it is immediate that σN is directly proportional to Var(sθ(f, T)) but inversely proportional to N. Hence, if Var(sθ(f, T)) is high then a larger number of samples is needed to achieve a certain statistical accuracy.
In finite-difference schemes, instead of Sθ(f, T) one estimates a finite difference Sθ,h(f, T) of the form (1.2) which can be written as 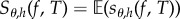 for
for
| 2.5 |
Here, the Markov processes Xθ and Xθ+h are defined on the same probability space and they have generators 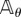 and
and 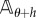 , respectively. In this case, the estimator mean μN and variance
, respectively. In this case, the estimator mean μN and variance 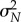 are given by (2.3) with sθ(f, T) replaced by sθ,h(f, T).
are given by (2.3) with sθ(f, T) replaced by sθ,h(f, T).
2.1. Finite-difference schemes
The main idea behind the various finite-difference schemes is that by coupling the processes Xθ and Xθ+h, one can increase the covariance between f(Xθ+h(T)) and f(Xθ(T)), thereby reducing the variance of sθ,h(f, T) and making the estimation procedure more efficient. As mentioned before, three such couplings suggested in the existing literature are: CRN [11], CRP [11] and CFD [13]. Among these, we only consider the two best-performing schemes, CRP and CFD, in this paper.
In CRP, the processes Xθ and Xθ+h are coupled by their random time-change representations according to
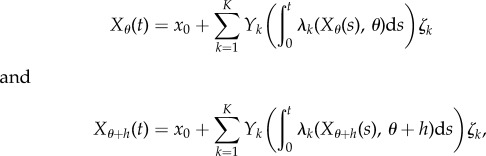 |
where 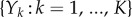 is a family of independent unit rate Poisson processes. Note that these Poisson processes are the same for both processes Xθ and Xθ+h, indicating that their reaction paths are the same.
is a family of independent unit rate Poisson processes. Note that these Poisson processes are the same for both processes Xθ and Xθ+h, indicating that their reaction paths are the same.
In CFD, the processes Xθ and Xθ+h are coupled by their random time-change representations as
 |
and
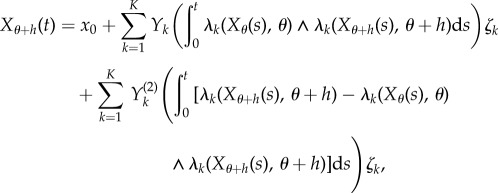 |
where 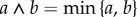 and
and 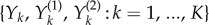 is again a family of independent unit rate Poisson processes. Under this coupling, the processes Xθ and Xθ+h have the same state until the first time the counting process corresponding to
is again a family of independent unit rate Poisson processes. Under this coupling, the processes Xθ and Xθ+h have the same state until the first time the counting process corresponding to 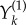 or
or 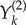 fires for some k. The probability that this separation time will come before T is proportional to h, which is usually small. This suggests that the processes Xθ and Xθ+h are strongly coupled.
fires for some k. The probability that this separation time will come before T is proportional to h, which is usually small. This suggests that the processes Xθ and Xθ+h are strongly coupled.
Note that both CRP and CFD are estimating the same quantity Sθ,h(f, T) (1.2) and hence they both suffer from the same bias 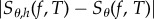 . The only difference between them is in the coupling of the processes Xθ and Xθ+h, which alters the variance of sθ,h(f, T). For a given example, the method with a lower variance is more efficient as it requires a smaller sample to achieve the desired statistical accuracy.
. The only difference between them is in the coupling of the processes Xθ and Xθ+h, which alters the variance of sθ,h(f, T). For a given example, the method with a lower variance is more efficient as it requires a smaller sample to achieve the desired statistical accuracy.
For a finite-difference scheme of the form (1.1), the bias 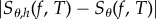 is of order h. However, replacing θ by (θ − h/2), we obtain a centred finite-difference scheme which has a bias of order h2. Such a centring does not cause any additional difficulties in simulating the coupled processes for CFD or CRN, but it may lead to a bias which is substantially smaller. Therefore, for the purpose of comparison, we work with the centred versions of CFD and CRN in this paper. In particular, we simulate processes X(θ−h/2) and X(θ+h/2) according to the appropriate coupling mechanism and estimate sensitivity as
is of order h. However, replacing θ by (θ − h/2), we obtain a centred finite-difference scheme which has a bias of order h2. Such a centring does not cause any additional difficulties in simulating the coupled processes for CFD or CRN, but it may lead to a bias which is substantially smaller. Therefore, for the purpose of comparison, we work with the centred versions of CFD and CRN in this paper. In particular, we simulate processes X(θ−h/2) and X(θ+h/2) according to the appropriate coupling mechanism and estimate sensitivity as 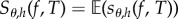 , where
, where
| 2.6 |
One can show that the variance Var(sθ,h(f, T)) of the random variable sθ,h(f, T) is proportional to 1/h. Hence, by making h smaller, we may reduce the bias but we will have to pay a computational cost by generating a large number of samples for the required estimation. This trade-off between bias and efficiency is the major drawback of finite-difference schemes as one generally does not know the right value of h for which Sθ,h(f, T) is close enough to Sθ(f, T), while the variance of sθ,h(f, T)) is not too large.
2.2. Unbiased schemes
Unbiased schemes are desirable because one does not have to worry about the bias, and the accuracy of the estimate can be improved by simply increasing the number of samples. The Girsanov method is the first such unbiased scheme [14,18] for sensitivity estimation. Recall the random time-change representation (2.1) of the process (Xθ(t))t≥0. In the Girsanov method, we express Sθ(f, T) as (1.3) where
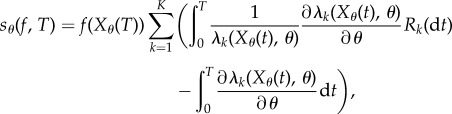 |
2.7 |
and (Rk(t))t≥0 is the counting process given by
In (2.7), Rk(dt) = 1 if and only if the kth reaction fires at time t. For any other t, Rk(dt) = 0. Hence, if the kth reaction fires at times  in the time interval [0, T], then
in the time interval [0, T], then
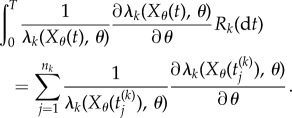 |
The Girsanov method is simple to implement because realizations of sθ(f, T) can be easily generated by simulating the paths of the process Xθ until time T. However, as mentioned before, the variance of sθ(f, T) can be very high [11–13], which is a serious problem as it implies that a large number of samples are needed to produce statistically accurate estimates. As simulations of the process Xθ can be quite cumbersome, generating a large sample can take an enormous amount of time.
Now consider the common situation where we have mass–action kinetics and θ is the rate constant of reaction k0. Then 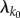 has the form
has the form 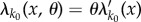 and for every k ≠ k0, λk does not depend on θ. In this case, the formula (2.7) for sθ(f, T) simplifies to
and for every k ≠ k0, λk does not depend on θ. In this case, the formula (2.7) for sθ(f, T) simplifies to
| 2.8 |
where 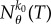 is the number of times reaction k0 fires in the time interval [0, T]. Clearly, the Girsanov method cannot estimate the sensitivity value at θ = 0 even though Sθ(f, T) (1.1) is well defined. This is a big disadvantage as the sensitivity value at θ = 0 is useful for understanding network design as it informs us whether the presence or absence of reaction k0 influences the output or not. Unfortunately, the problem with the Girsanov method is not just limited to θ = 0. Even for θ close to 0, the variance of sθ(f, T) can be very high, rendering this method practically infeasible [15]. This is again a serious drawback as reaction rate constants with small values are frequently encountered in systems biology and other areas.
is the number of times reaction k0 fires in the time interval [0, T]. Clearly, the Girsanov method cannot estimate the sensitivity value at θ = 0 even though Sθ(f, T) (1.1) is well defined. This is a big disadvantage as the sensitivity value at θ = 0 is useful for understanding network design as it informs us whether the presence or absence of reaction k0 influences the output or not. Unfortunately, the problem with the Girsanov method is not just limited to θ = 0. Even for θ close to 0, the variance of sθ(f, T) can be very high, rendering this method practically infeasible [15]. This is again a serious drawback as reaction rate constants with small values are frequently encountered in systems biology and other areas.
These issues with the Girsanov method severely restrict its use and also highlight the need for new unbiased schemes for sensitivity analysis. In our recent paper [15], we provide a new unbiased scheme based on another sensitivity formula of the form (1.3). To motivate this formula, we first discuss the problem of computing parameter sensitivity in the deterministic setting.
Consider the deterministic model for the parameter-dependent reaction network with reaction flux 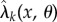 [20] and propensity vector ζk for reaction k. If
[20] and propensity vector ζk for reaction k. If 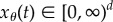 denotes the vector of species concentrations at time t, then (xθ(t))t≥0 is the solution of the following system of ordinary differential equations:
denotes the vector of species concentrations at time t, then (xθ(t))t≥0 is the solution of the following system of ordinary differential equations:
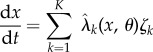 |
2.9 |
with initial condition x(0) = x0. Pick an observation time T > 0 and a differentiable output function 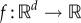 , and suppose we are interested in computing the sensitivity value
, and suppose we are interested in computing the sensitivity value
Using (2.9) we can write
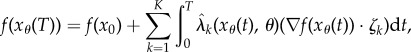 |
where 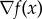 denotes the gradient of the map
denotes the gradient of the map 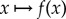 and · denotes the standard inner product on
and · denotes the standard inner product on 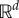 . Assuming differentiability of
. Assuming differentiability of 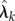 's we get
's we get
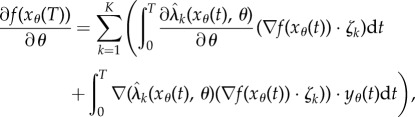 |
2.10 |
where 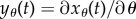 can be easily computed by solving the ordinary differential equation obtained by differentiating (2.9) with respect to θ. Hence, in the deterministic setting, computation of the parameter sensitivity is relatively straightforward.
can be easily computed by solving the ordinary differential equation obtained by differentiating (2.9) with respect to θ. Hence, in the deterministic setting, computation of the parameter sensitivity is relatively straightforward.
Relation (2.10) helps us in viewing parameter sensitivity as the sum of two parts: sensitivity of the reaction fluxes 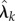 and the sensitivity of the states xθ(t). In [15], we provide such a decomposition for parameter sensitivity in the stochastic setting. However, unlike the deterministic scenario, measuring the second contribution is computationally very challenging. In order to present this result, we need to define certain quantities. Let (Xθ(t))t≥0 be a CTMC with generator
and the sensitivity of the states xθ(t). In [15], we provide such a decomposition for parameter sensitivity in the stochastic setting. However, unlike the deterministic scenario, measuring the second contribution is computationally very challenging. In order to present this result, we need to define certain quantities. Let (Xθ(t))t≥0 be a CTMC with generator 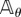 . For any
. For any 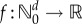 , t ≥ 0, k = 1, … , K and
, t ≥ 0, k = 1, … , K and  define
define
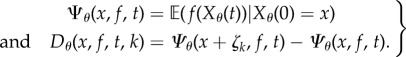 |
2.11 |
Define 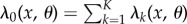 and let σ0, σ1, σ2 denote the successive jump times2 of the process Xθ. Theorem 2.3 in [15] shows that Sθ(f, T) satisfies (1.3) with
and let σ0, σ1, σ2 denote the successive jump times2 of the process Xθ. Theorem 2.3 in [15] shows that Sθ(f, T) satisfies (1.3) with
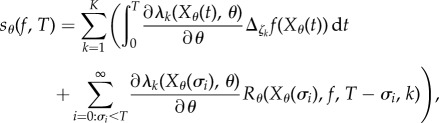 |
2.12 |
where
| 2.13 |
This result was proved by coupling the processes Xθ and Xθ+h as in CFD (see §2.1) and computing the limit of the finite difference Sθ,h(f, T) (see (1.2)) as h → 0. Similar to the deterministic case, this result shows that parameter sensitivity Sθ(f, T) can be seen as the sum of two parts: the sensitivity of the propensity functions (λk) and the sensitivity of the states Xθ(σi) of the Markov process at the jump times σi. Computing the latter contribution is difficult because it involves the function Rθ which generally does not have an explicit formula. To overcome this problem, one needs to estimate all the quantities of the form Rθ(x, f, t, k) that appear in (2.12). However, the number of these quantities is proportional to the number of jumps of the process before time T, which can be quite high even for small networks. If we estimate each of these quantities serially, as and when they appear, using a collection of independently generated paths of the process Xθ, then the problem becomes computationally intractable for most examples of interest. In [15], we devised a scheme, called the APA, to estimate all these quantities in parallel by generating a fixed number of auxiliary paths in addition to the main path. APA stores information about all the required quantities in a big Hashtable and tracks the states visited by the auxiliary paths to estimate those quantities. Owing to all the necessary book-keeping, APA is hard to implement and it also has high memory requirements. In fact, the space and time complexity of APA scales linearly with the number of jumps in the time interval [0, T] and this makes APA practically unusable for large networks or for large values of observation time T. Moreover, APA only works well if the stochastic dynamics visits the same states again and again, which may not be generally true.
The above discussion suggests that if we can modify the expression for sθ(f, T) in such a way that only a small fraction of unknown quantities (of the form Rθ(x, f, t, k)) require estimation, then it can lead to an efficient method for sensitivity estimation. We exploit this idea in this paper. By adding extra randomness to the random variable sθ(f, T), we construct another random variable 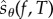 , which has the same expectation as sθ(f, T)
, which has the same expectation as sθ(f, T)
| 2.14 |
even though its distribution may be different. We then show that realizations of the random variable 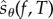 can be easily obtained through a simple procedure. This gives us a new method for unbiased parameter sensitivity estimation for stochastic reaction networks.
can be easily obtained through a simple procedure. This gives us a new method for unbiased parameter sensitivity estimation for stochastic reaction networks.
2.3. Construction of 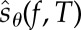
We now construct the random variable 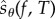 introduced in the previous section. Let (Xθ(t))t≥0 be the CTMC with generator
introduced in the previous section. Let (Xθ(t))t≥0 be the CTMC with generator 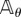 and initial state x0. Let σ0, σ1, … denote the successive jump times of this process. The total number of jumps until time T is given by
and initial state x0. Let σ0, σ1, … denote the successive jump times of this process. The total number of jumps until time T is given by
| 2.15 |
For any T > 0, we have η ≥ 1 because σ0 = 0. If the process Xθ reaches a state x for which λ0(x, θ) = 0, then x is an absorbing state and the process stays in x forever. From (2.15), it is immediate that for any non-negative integer i < η, Xθ(σi) cannot be an absorbing state. Let α indicate if the final state Xθ(ση) is absorbing
For each i = 0, … , (η − α), let γi be an independent exponentially distributed random variable with rate λ0(Xθ(σi), θ) and define
For each i = 0, … , (η − α) and k = 1, … , K, let βki be given by
where
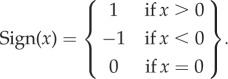 |
Fix a normalizing constant c > 0. The choice of c and its role will be explained later in the section. If βki≠ 0 and Γi = 1, then let 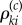 be an independent
be an independent 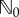 -valued random variable whose distribution is Poisson with parameter
-valued random variable whose distribution is Poisson with parameter
| 2.16 |
Here, the denominator λ0(Xθ(σi), θ) is non-zero because for i ≤ (η − α) the state Xθ(σi) cannot be absorbing. On the event 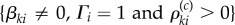 , we define another random variable as
, we define another random variable as
| 2.17 |
where (Z1(t))t≥0 and (Z2(t))t≥0 are two processes which are coupled by the following random time-change representations:
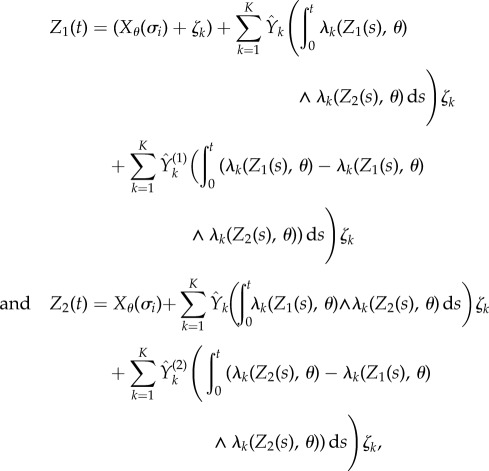 |
where 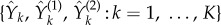 is an independent family of unit rate Poisson processes. This coupling is similar to the coupling used by CFD (see §2.1). Note that (Z1(t))t≥0 and (Z2(t))t≥0 are Markov processes with generator
is an independent family of unit rate Poisson processes. This coupling is similar to the coupling used by CFD (see §2.1). Note that (Z1(t))t≥0 and (Z2(t))t≥0 are Markov processes with generator 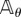 and initial states (Xθ(σi) + ζk) and Xθ(σi), respectively. Therefore,
and initial states (Xθ(σi) + ζk) and Xθ(σi), respectively. Therefore,
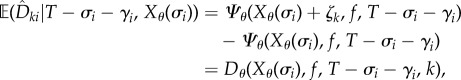 |
2.18 |
where Dθ is defined by (2.11). In other words, given T − σi − γi = t and Xθ(σi) = x, the mean of the random variable 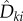 is just Dθ(x, f, t, k). The above coupling between (Z1(t))t≥0 and (Z2(t))t≥0 makes them strongly correlated, thereby lowering the variance of the difference
is just Dθ(x, f, t, k). The above coupling between (Z1(t))t≥0 and (Z2(t))t≥0 makes them strongly correlated, thereby lowering the variance of the difference 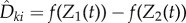 . This strong correlation is evident from the fact that if Z1(s) = Z2(s) for some s ≥ 0 then Z1(u) = Z2(u) for all u ≥ s. Finally, if the last state Xθ(ση) is absorbing (that is, α = 1) and ∂λk(Xθ(ση), θ)/∂θ is non-zero, then we define another random variable
. This strong correlation is evident from the fact that if Z1(s) = Z2(s) for some s ≥ 0 then Z1(u) = Z2(u) for all u ≥ s. Finally, if the last state Xθ(ση) is absorbing (that is, α = 1) and ∂λk(Xθ(ση), θ)/∂θ is non-zero, then we define another random variable 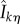 as
as
| 2.19 |
where (Z(t))t≥0 is an independent Markov process with generator 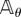 and initial state Xθ(ση) + ζk. Note that
and initial state Xθ(ση) + ζk. Note that
| 2.20 |
We are now ready to provide an expression for 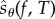 . Let
. Let
and define
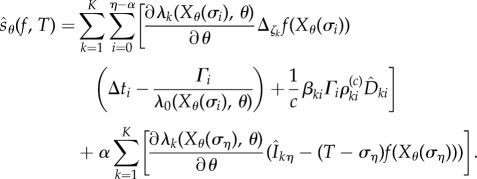 |
2.21 |
By a simple conditioning argument, we prove in the electronic supplementary material, §1, that relation (2.14) holds.
At first glance, it seems that obtaining realizations of 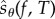 suffers from the same difficulties as obtaining realizations of sθ(f, T), because at each jump time σi < T, one needs to compute an unknown quantity
suffers from the same difficulties as obtaining realizations of sθ(f, T), because at each jump time σi < T, one needs to compute an unknown quantity 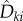 which requires simulation of new paths of the process Xθ. However, note that
which requires simulation of new paths of the process Xθ. However, note that 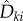 is only needed if the Poisson random variable
is only needed if the Poisson random variable 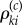 is strictly positive. Therefore, if we can effectively control the number of positive
is strictly positive. Therefore, if we can effectively control the number of positive 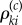 's, then we can efficiently generate realizations of
's, then we can efficiently generate realizations of 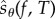 . This can be achieved using the positive parameter c (see (2.16)) as we later explain. The construction of
. This can be achieved using the positive parameter c (see (2.16)) as we later explain. The construction of 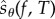 outlined above also provides a recipe for obtaining its realizations. Hence, we have a method for estimating the parameter sensitivity Sθ(f, T). We call this method the PPA because at each jump time σi, the crucial decision of whether new paths of the process Xθ are needed (for
outlined above also provides a recipe for obtaining its realizations. Hence, we have a method for estimating the parameter sensitivity Sθ(f, T). We call this method the PPA because at each jump time σi, the crucial decision of whether new paths of the process Xθ are needed (for 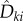 ) is based on the value of a Poisson random variable
) is based on the value of a Poisson random variable 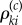 . We describe PPA in greater detail in §3.
. We describe PPA in greater detail in §3.
We now discuss how the positive parameter c can be selected. Let ηreq denote the total number of positive 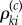 's that appear in (2.21). This is the number of
's that appear in (2.21). This is the number of 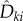 's that are required to obtain a realization of
's that are required to obtain a realization of 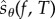 . It is immediate that ηreq is bounded above by
. It is immediate that ηreq is bounded above by 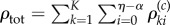 , which is a Poisson random variable with parameter cRtot, where
, which is a Poisson random variable with parameter cRtot, where
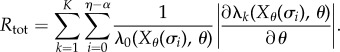 |
2.22 |
By picking a small c > 0, we can ensure that 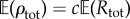 is small, which would also guarantee that ρtot and ηreq are small. Specifically, we choose a small positive integer M0 (like 10, for instance) and set
is small, which would also guarantee that ρtot and ηreq are small. Specifically, we choose a small positive integer M0 (like 10, for instance) and set
| 2.23 |
where 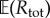 is estimated using N0 simulations of the process Xθ in the time interval [0, T]. The choice of N0 is not critical and typically a small value (like 100, for example) is sufficient to provide a decent estimate of
is estimated using N0 simulations of the process Xθ in the time interval [0, T]. The choice of N0 is not critical and typically a small value (like 100, for example) is sufficient to provide a decent estimate of 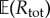 . The role of parameter M0 is also not very important in determining the efficiency of PPA. If M0 increases, then ηreq increases as well, and the computational cost of generating each realization of
. The role of parameter M0 is also not very important in determining the efficiency of PPA. If M0 increases, then ηreq increases as well, and the computational cost of generating each realization of 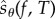 becomes higher. However, as ηreq increases, the variance of
becomes higher. However, as ηreq increases, the variance of 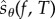 decreases and hence fewer realizations of
decreases and hence fewer realizations of 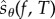 are required to achieve the desired statistical accuracy. These two effects usually offset each other and the overall efficiency of PPA remains relatively unaffected. Note that PPA provides unbiased estimates for the sensitivity values, regardless of the choice of N0 and M0.
are required to achieve the desired statistical accuracy. These two effects usually offset each other and the overall efficiency of PPA remains relatively unaffected. Note that PPA provides unbiased estimates for the sensitivity values, regardless of the choice of N0 and M0.
3. The Poisson path algorithm
We now describe PPA which produces realizations of the random variable 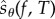 (defined by (2.21)) for some observation time T and some output function f. Since
(defined by (2.21)) for some observation time T and some output function f. Since 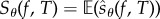 , we can estimate Sθ(f,T) by generating N realizations
, we can estimate Sθ(f,T) by generating N realizations  of the random variable
of the random variable 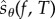 and then computing their empirical mean (2.2).
and then computing their empirical mean (2.2).
Let x0 denote the initial state of the process Xθ and assume that the function rand() returns independent samples from the uniform distribution on [0,1]. We simulate the paths of our CTMC using Gillespie's SSA [16]. When the state is x, the next time increment (Δt) and reaction index (k) is given by the function SSA(x) (see the electronic supplementary material, algorithm 1 in §2). We first fix a normalization parameter c according to (2.23), by estimating 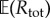 using N0 simulations of the CTMC (see the electronic supplementary material, algorithm 2 in §2).
using N0 simulations of the CTMC (see the electronic supplementary material, algorithm 2 in §2).
Once c is chosen, a single realization of 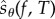 can be computed using GenerateSample(x0, T, c) (algorithm 1). This method simulates the process Xθ according to SSA and at each state x and jump time t, the following happens:
can be computed using GenerateSample(x0, T, c) (algorithm 1). This method simulates the process Xθ according to SSA and at each state x and jump time t, the following happens:
— If x is an absorbing state (λ0(x, θ) = 0), then t is the last jump time before T (t = ση). For each k = 1, … , K such that ∂λk(x, θ)/∂θ ≠ 0, the quantity
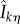 (see (2.19)) is evaluated using EvaluateIntergal(x + ζk, T − t) (see the electronic supplementary material, algorithm 3 in §2) and then used to update the sample value according to (2.21).
(see (2.19)) is evaluated using EvaluateIntergal(x + ζk, T − t) (see the electronic supplementary material, algorithm 3 in §2) and then used to update the sample value according to (2.21).— If x is not an absorbing state, then t = σi for some jump time σi with i < η. An exponential random variable γ (where γ = γi in (2.21)) is generated, and for each k = 1, … , K such that ∂λk(x, θ)/∂θ ≠ 0, the Poisson random variable n (where
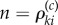 in (2.21)) is also generated. If γ < (T − t) and n > 0, then the quantity
in (2.21)) is also generated. If γ < (T − t) and n > 0, then the quantity 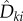 (see (2.17)) is evaluated using EvaluateCoupledDifference(x, x + ζk, T − t − γ) (see the electronic supplementary material, algorithm 5 in §2) and then used to update the sample value according to (2.21). A Poisson random variable with parameter r is generated using the function GeneratePoisson(r) (see the electronic supplementary material, algorithm 4 in §2).
(see (2.17)) is evaluated using EvaluateCoupledDifference(x, x + ζk, T − t − γ) (see the electronic supplementary material, algorithm 5 in §2) and then used to update the sample value according to (2.21). A Poisson random variable with parameter r is generated using the function GeneratePoisson(r) (see the electronic supplementary material, algorithm 4 in §2).
4. Numerical examples
In this section, we present many examples to compare the performance of PPA with the Girsanov method which is unbiased, and the two best-performing finite-difference schemes, CFD and CRP, which are of course biased. To compare the performance of different methods, we compare the time they need to produce a statistically accurate estimate of the true sensitivity value Sθ(f, T) given by (1.1). Our first task is to find a way to judge that an estimate of Sθ(f, T) is statistically accurate.
A common way to assess statistical accuracy of an estimator (biased or unbiased) is by computing its mean-squared error (m.s.e.), which is the sum of its variance and its bias (squared). However m.s.e. is not well-suited for our situation. To see this, consider two sensitivity estimators whose means are μ1 = 2 and μ2 = 1.1, and standard deviations are σ1 = 0.1 and σ2 = 1. Assuming that the true sensitivity value is s0 = 1 and the distribution of the estimators is approximately normal, the two estimators have the same m.s.e., which is equal to 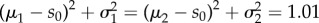 . However, the second estimator is clearly more accurate than the first one, because it is far more likely to yield a sensitivity estimate which is ‘close’ to the actual value s0. This explains how m.s.e. can be misleading and therefore we now describe another method to compare the statistical accuracy of estimators.
. However, the second estimator is clearly more accurate than the first one, because it is far more likely to yield a sensitivity estimate which is ‘close’ to the actual value s0. This explains how m.s.e. can be misleading and therefore we now describe another method to compare the statistical accuracy of estimators.
Algorithm 1. Generates one realization of 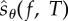 according to (2.21). according to (2.21). | |
| 1: | function GenerateSample(x0, T, c) |
| 2: | Set x = x0, t = 0 and s = 0 |
| 3: | while t < T do |
| 4: | Calculate (Δt, k0) = SSA(x) |
| 5: | Update Δt ← min{Δt, T − t} |
| 6: | if λ0(x, θ) > 0 then |
| 7: | Set 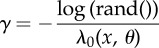
|
| 8: | end if |
| 9: | for k = 1 to K do |
| 10: | Set 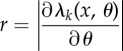 and and 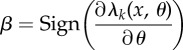
|
| 11: | if r > 0 then |
| 12: | if λ0(x, θ) = 0 then |
| 13: | Update 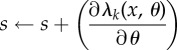 (EvaluateIntegral (EvaluateIntegral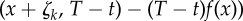
|
| 14: | else |
| 15: | Set n = GeneratePoisson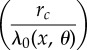
|
| 16: | if γ < (T − t) then |
| 17: | Update 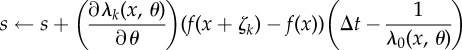
|
| 18: | if n > 0 then |
| 19: | Update 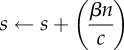 EvaluateCoupledDifference EvaluateCoupledDifference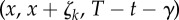
|
| 20: | end if |
| 21: | else |
| 22: | 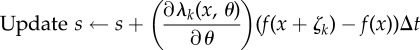
|
| 23: | end if |
| 24: | end if |
| 25: | end if |
| 26: | end for |
| 27: | Update t ← t + Δt and x ← x + ζk0 |
| 28: | end while |
| 29: | return s |
| 30: | end function |
Suppose that a method estimates parameter sensitivity using N samples whose mean is μN and standard deviation is σN (see §2). Assume that the true value of Sθ(f, T) is s0. Let 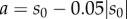 and
and 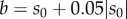 , where
, where  denotes the absolute value function. Then [a, b] is the 5% interval around the true value s0. Under the central limit approximation, the estimator
denotes the absolute value function. Then [a, b] is the 5% interval around the true value s0. Under the central limit approximation, the estimator 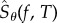 can be seen as a normally distributed random variable with mean μN and variance
can be seen as a normally distributed random variable with mean μN and variance 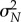 . Hence, using (2.4) we can calculate the probability
. Hence, using (2.4) we can calculate the probability
that the estimator lies within the 5% interval around s0. Henceforth, we refer to p as the confidence level for the estimator. The statistical accuracy of an estimate can be judged from the value of p: the higher the value of p, the more accurate is the estimate. As we calculate p using the 5% interval around s0, we ensure that the statistical precision is high or low depending on whether the sensitivity value is small or large.
The standard deviation σN is inversely proportional to the number of samples N (see §2), which shows that σN
→ 0 as N → ∞. For an unbiased scheme (Girsanov and PPA), we have μN → s0 as N → ∞, due to the law of large numbers and therefore (2.4) implies that the confidence level p can be made as close to 1 as desired by picking a large sample size N. However, the same is not true for finite-difference schemes such as CFD and CRP. In such schemes, μN → Sθ,h(f, T) as N → ∞, where Sθ,h(f, T) is generally different from Sθ(f, T) = s0 because of the bias. If this bias is large, then Sθ,h(f, T) may lie outside the 5% interval [a, b] around s0, and hence p is close to 0 for large N. Therefore, if high confidence levels are desired with finite-difference schemes, then a small h must be chosen to ensure that the bias 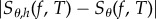 is low. However, the variance
is low. However, the variance 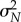 of the associated estimator scales as 1/h (see §2.1), implying that if h is too small then an extremely large sample size is required to achieve high confidence levels, and this can impose a heavy computational burden.
of the associated estimator scales as 1/h (see §2.1), implying that if h is too small then an extremely large sample size is required to achieve high confidence levels, and this can impose a heavy computational burden.
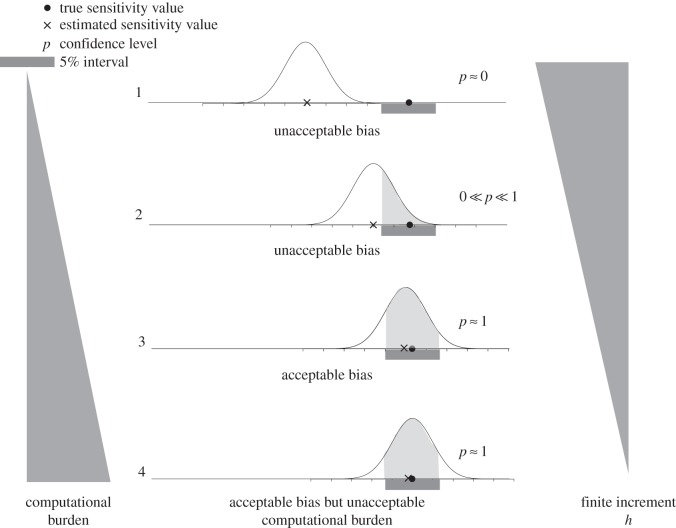
In this paper, we compare the efficiency of different methods by measuring the CPU time3 that is needed to produce a sample with least possible size, such that the corresponding estimate has a certain threshold confidence level. This threshold confidence level is close to 1 (either 0.95 or 0.99) and hence the produced estimate is statistically quite accurate. Previous discussion suggests that for finite-difference schemes, there is a trade-off between the statistical accuracy and the computational burden. This trade-off is illustrated in figure 1 which depicts four cases that correspond to four values of h arranged in decreasing order. We describe these cases below.
Case 1. Here, h is large and so the computational burden is low. However, due to the large bias, the estimator distribution (normal curve in figure 1) puts very little mass on the 5% interval around the true sensitivity value. Therefore, the confidence level p is low and the estimate is unacceptable.
Case 2. Now h is smaller, the computational burden is higher and the bias is lower. However, the estimator distribution still does not put enough mass on the 5% interval and so the value of p is not high enough for the estimate to be acceptable.
Case 3. Now h is even smaller and so the computational burden is higher, but fortunately the bias is small enough to ensure that the estimator distribution puts nearly all its mass on the 5% interval. Therefore, the confidence level p is close to 1 and the estimate is acceptable.
Case 4. Here, h is smallest and so the computational burden is the highest among all the cases. However in comparison to Case 3, the additional computational burden is unnecessary because there is only a slight improvement in the bias and the confidence level.
Figure 1.
Trade-off between accuracy and computational burden for finite-difference schemes. The four cases correspond to four values of h arranged in decreasing order. For Case 2, 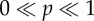 should be interpreted as ‘p is far away from 0 and 1’.
should be interpreted as ‘p is far away from 0 and 1’.
To use a finite-difference scheme efficiently, we need to know the largest value of h for which an estimate with a high confidence level can be produced (Case 3 in figure 1). However, in general, it is impossible to determine this value of h. A blind choice of h is likely to correspond to Cases 1 or 4 in figure 1. To make matters worse, since the true sensitivity value Sθ(f, T) is usually unknown, one cannot measure the statistical accuracy without re-estimating Sθ(f, T) using an unbiased method. Most practitioners who use finite-difference schemes assume that the statistical accuracy is high if h is small. However, this is not true in general (see example 4.1). Therefore, if high accuracy is desired, then an unbiased method should be preferred over finite-difference schemes.
As it is difficult to determine the optimum value of h, we adopt the following strategy to gauge the efficiency of a finite-difference scheme. We find the largest value of h in the sequence 0.1, 0.01, 0.001 … , for which the threshold confidence level p is achievable for a large enough sample size. For performance comparison, we only use the CPU time corresponding to this value of h and we disregard all the CPU time wasted in testing other values of h. We have intentionally made our comparison strategy more lenient towards the finite-difference schemes, in order to compensate for the arbitrariness in choosing the ‘test’ values of h.
To compute the confidence level p, we require the exact sensitivity value Sθ(f, T). In some examples, Sθ(f, T) can be analytically evaluated and the computation of p is straightforward. In other examples, where Sθ(f, T) cannot be explicitly computed, we use PPA to produce an estimate with a very low sample variance and since PPA is unbiased, this estimate would be close to the true sensitivity value Sθ(f, T) and hence it can be used in the calculation of p.
We now discuss the examples. In all the examples, the propensity functions λk follow mass–action kinetics unless stated otherwise. Our method PPA depends on two parameters N0 and M0 (see §2.3) and in this paper we always use N0 = 100 and M0 = 10. In each example, we provide a bar chart with the CPU times required by different methods to produce an estimate with the desired confidence level p = 0.95 or p = 0.99. This facilitates an easy performance comparison between various methods. The exact values of the CPU times, sample size N, estimator mean μN, estimator standard derivation σN and h (for finite-difference schemes) are provided in the electronic supplementary material, §3.
Example 4.1. (Single-species birth–death model) —
Our first example is a simple birth–death model in which a single species
is created and destroyed as
Let θ1 = θ2 = 0.1 and assume that the sensitive parameter is θ = θ2. Suppose that X(0) = 0 and let (X(t))t≥0 be the CTMC representing the reaction dynamics. Hence the population of 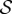 at time t is
at time t is 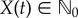 . For f(x) = x, we wish to estimate
. For f(x) = x, we wish to estimate
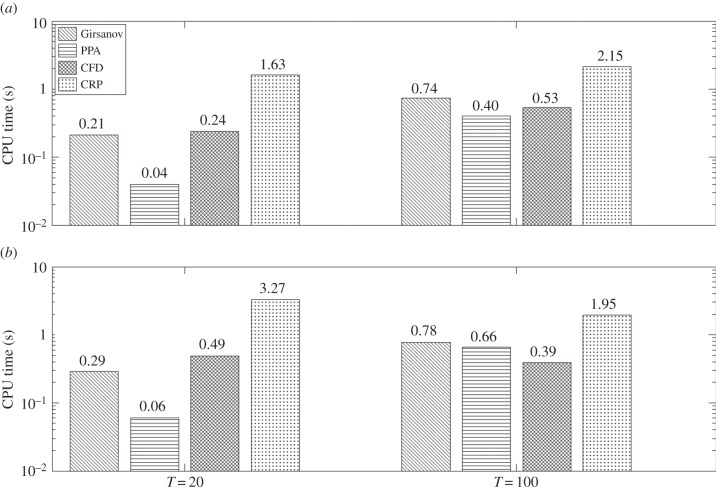
for T = 20 and 100. As the propensity functions of this network are affine, we can compute Sθ(f, T) exactly (see the electronic supplementary material, §3) and use this value in computing the confidence level p of an estimate. We estimate Sθ(f, T) with all the methods with two threshold confidence levels (p = 0.95 and p = 0.99) and plot the corresponding CPU times in figure 2. For CFD and CRP, the desired confidence level was achieved in all the cases for h = 0.01.
Figure 2.
Efficiency comparison for birth–death model with two threshold confidence levels: p = 0.95 (a) and p = 0.99 (b) and two observation times: T = 20 and T = 100. The exact CPU time is shown at the top of each bar. Note that efficiency is inversely related to the CPU time. For finite-difference schemes, h = 0.01 was used in all the cases.
From figure 2, it is immediate that PPA generally performs better than the other methods. To measure this gain in performance more quantitatively, we compute the average speed-up factor of PPA with respect to other methods. For each threshold confidence level, this factor is calculated by simply taking the ratio of the aggregate CPU times required by a certain method and PPA to perform all the sensitivity estimations. In this example, this aggregate involves the CPU times for T = 20 and T = 100. These average speed-up factors are presented in table 1. Note that in this example, PPA is significantly faster than CRP but only slightly faster than CFD and the Girsanov method.
Table 1.
Birth–death model: average speed-up factors of PPA with respect to other methods.
| p | Girsanov | CFD | CRP |
|---|---|---|---|
| 0.95 | 2.16 | 1.75 | 8.59 |
| 0.99 | 1.5 | 1.22 | 7.25 |
Now we use this example to demonstrate the pitfalls of choosing a ‘wrong’ value of h for finite-difference schemes. We know from figure 2 that for T = 100, the desired confidence level of 0.99 was achieved with h = 0.01. This ‘right’ value of h corresponds to Case 3 in figure 1. If we select a higher value of h, such as h = 0.1 or h = 0.05, then with 10 000 samples we see that the achieved confidence level p is much lower than the desired value 0.99 (table 2), indicating that the estimate is not sufficiently accurate. Note that h = 0.1 and h = 0.05 correspond to Cases 1 and 2 in figure 1. Observe that for h = 0.1, the estimate produced by finite-difference schemes (μN) is more than 30% off from the exact sensitivity value of −9.995, which shows that the bias can be large for h that appears small. On the other hand, if we select h which is really small, such as h = 0.001, then we do produce an estimate with the desired confidence level of 0.99 (see table 2), but the required sample size is very large and hence the computational burden is much higher than the ‘right’ value of h, which is h = 0.01. The value h = 0.001 corresponds to Case 4 in figure 1.
Table 2.
Birth–death model: pitfalls of choosing a ‘wrong’ h.
| h | method | N | mean (μN) | s.d. (σN) | CPU time (s) | p |
|---|---|---|---|---|---|---|
| 0.1 | CFD | −13.286 | 0.0998 | 10000 | 0.15 | 0 |
| CRP | −13.144 | 0.1206 | 10000 | 0.62 | 0 | |
| 0.05 | CFD | −10.634 | 0.1168 | 10000 | 0.13 | 0.1165 |
| CRP | −10.558 | 0.1504 | 10000 | 0.64 | 0.3370 | |
| 0.001 | CFD | −9.9424 | 0.1869 | 281723 | 3.65 | 0.99 |
| CRP | −10.0625 | 0.1829 | 300721 | 19.8 | 0.99 |
Example 4.2. (Gene expression network) —
Our second example considers the model for gene transcription given in [2]. It has three species, Gene (G), mRNA (M) and protein (P), and there are four reactions given by
The rate of translation of a gene into mRNA is θ1, while the rate of transcription of mRNA into protein is θ2. The degradation of mRNA and protein molecules occurs at rates θ3 and θ4, respectively. Typically, 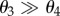 implying that a protein molecule lives much longer than a mRNA molecule. In accordance with the values given in [2] for lacA gene in bacterium E. coli, we set θ1 = 0.6 min−1, θ2 = 1.7329 min−1 and θ3 = 0.3466 min−1. Our sensitive parameter is θ = θ4.
implying that a protein molecule lives much longer than a mRNA molecule. In accordance with the values given in [2] for lacA gene in bacterium E. coli, we set θ1 = 0.6 min−1, θ2 = 1.7329 min−1 and θ3 = 0.3466 min−1. Our sensitive parameter is θ = θ4.
Let (X(t))t≥0 be the CTMC representing the dynamics with initial state (X1(0), X2(0)) = (0, 0). For any time t, X(t) = (X1(t), X2(t)), where X1(t) and X2(t) are the number of mRNA and protein molecules, respectively. Define 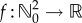 by f(x1, x2) = x2. We would like to estimate the logarithmic sensitivity
by f(x1, x2) = x2. We would like to estimate the logarithmic sensitivity
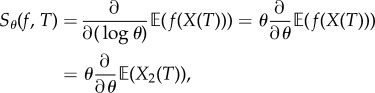 |
4.27 |
which measures the sensitivity of the mean of the protein population at time T with respect to perturbations in the protein degradation rate at the logarithmic scale. For finite-difference schemes, this corresponds to replacing h by θh in (2.6).
For sensitivity estimation, we consider two values of T, 20 and 100 min, and two values of θ, 0.0693 and 0.0023 min−1. These values of θ correspond to the protein half-life of 10 min and 5 h. Like example 4.1, this network also has affine propensity functions and hence we can compute Sθ(f, T) exactly (see the electronic supplementary material, §3) and use these values to compute the confidence levels.
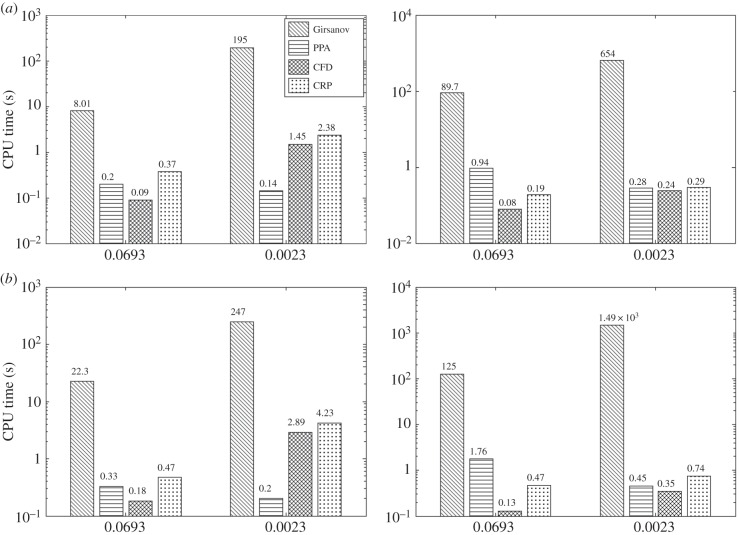
The CPU times required by all the methods for the two threshold confidence levels (p = 0.95 and p = 0.99) are presented in figure 3. For CFD and CRP, the desired confidence level was achieved in all cases with h = 0.1. We compute the average speed-up factor for PPA with respect to other methods, by aggregating CPU times for the two values of θ and T. These average speed-up factors are presented in table 3 and they show that PPA can be far more efficient than the Girsanov method, but its performance is comparable to CFD and CRP. Furthermore, the results in figure 3 indicate that unlike PPA, the performance of the Girsanov method deteriorates drastically as θ gets smaller: on average PPA is 76 times faster for θ = 0.0693 min−1 and 2413 times faster for θ = 0.0023 min−1. This deterioration in the performance of the Girsanov method is consistent with the results in [15].
Figure 3.
Efficiency comparison for gene expression network with two threshold confidence levels: p = 0.95 (a) and p = 0.99 (b) and two observation times: T = 20 min (left-hand panels) and T = 100 min (right-hand panels). For finite-difference schemes, h = 0.1 was used in all the cases.
Table 3.
Gene expression network: average speed-up factors of PPA with respect to other methods.
| p | Girsanov | CFD | CRP |
|---|---|---|---|
| 0.95 | 606.74 | 1.19 | 2.07 |
| 0.99 | 686.35 | 1.30 | 2.16 |
Even though PPA is only slightly faster than CFD and CRP in this example, its unbiasedness guarantees that one does not require any additional checks to verify its accuracy. This gives PPA a distinct advantage over finite-difference schemes.
Example 4.3. (Circadian clock network) —
We now consider the network of a circadian clock studied by Vilar et al. [21]. It is a large network with nine species S1, …, S9 and 16 reactions. These reactions along with the rate constants are given in table 4. Let (X(t))t≥0 be the CTMC corresponding to the dynamics with initial state X(0) = (1, 0, 0, 0, 1, 0, 0, 0, 0). For T = 5 and f(x) = x4, we wish to estimate
for θ = θi and i = 5, 6, 8, 12 and 14.
Table 4.
Circadian clock network: list of reactions.
| no. | reaction | rate constant | no. | reaction | rate constant |
|---|---|---|---|---|---|
| 1 | S6 + S2 → S7 | θ1 = 1 | 9 | 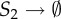 |
θ9 = 100 |
| 2 | S7 → S6 + S2 | θ2 = 50 | 10 | S9 → S9 + S3 | θ10 = 0.01 |
| 3 | S8 + S2 → S9 | θ3 = 50 | 11 | S8 → S8 + S3 | θ11 = 50 |
| 4 | S9 → S8 + S2 | θ4 = 500 | 12 | 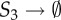 |
θ12 = 0.5 |
| 5 | S7 → S7 + S1 | θ5 = 10 | 13 | S3 → S3 + S4 | θ13 = 5 |
| 6 | S6 → S6 + S1 | θ6 = 50 | 14 | 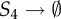 |
θ14 = 0.2 |
| 7 | 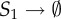 |
θ7 = 1 | 15 | S2 + S4 → S5 | θ15 = 20 |
| 8 | S1 → S1 + S2 | θ8 = 1 | 16 | S5 → S4 | θ16 = 1 |
Owing to the presence of many bimolecular interactions, exact values of Sθ(f, T) are difficult to compute analytically, but they are required in computing the confidence levels of estimates. To solve this problem, we use PPA to produce sensitivity estimates with very low variance, and then use these approximate values in computing confidence levels. These approximate values are given in the electronic supplementary material, §3.
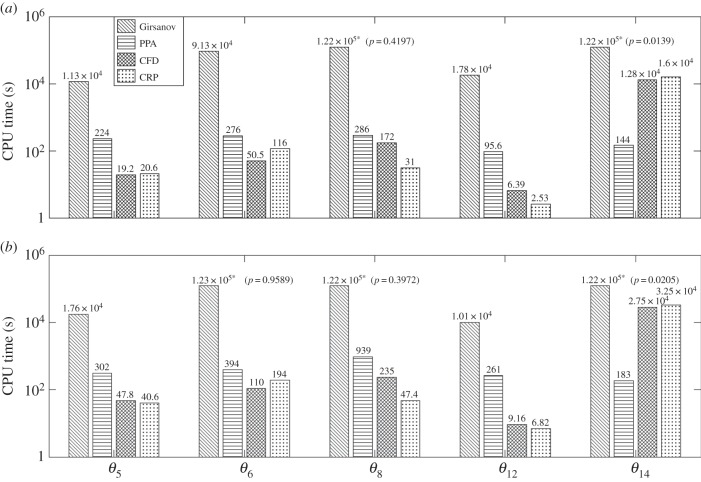
The CPU times needed by all the methods for two threshold confidence levels (p = 0.95 and p = 0.99) are shown in figure 4. As in the previous example, for CFD and CRP, the desired confidence level was achieved in all the cases with maximum allowed h value (h = 0.1). Results in figure 4 indicate that PPA has much better performance than the Girsanov method. In fact in many cases, the Girsanov method could not produce an estimate with the desired confidence level even with the maximum sample size of 107. These values are marked with asterisks in figure 4 and the confidence level they correspond to is shown in parentheses.
Figure 4.
Efficiency comparison for circadian clock network with two threshold confidence levels: p = 0.95 (a) and p = 0.99 (b) and observation time T = 5. The values marked with asterisks correspond to cases where the desired confidence level was not reached even with the maximum sample size of 107. The final confidence level reached is shown in parentheses. For finite-difference schemes, h = 0.1 was used in all the cases.
If we compare the performance of PPA with finite-difference schemes, then we find that PPA is slower for all parameters except θ = θ14, in which case PPA is far more efficient than either CFD or CRP. The overall efficiency of PPA can be judged from its average speed-up factor with respect to other methods. These average speed-up factors are displayed in table 5 and they are calculated by aggregating CPU times for the five values of θ. As in many cases the Girsanov method did not yield an estimate with the desired confidence level even with the maximum sample size, the actual speed-up factor of PPA with respect to this method may be much higher than what is shown in table 5.
Example 4.4. (Genetic toggle switch) —
As our last example, we look at a simple network with nonlinear propensity functions. Consider the network of a genetic toggle switch proposed by Gardner et al. [22]. This network has two species
and
that interact through the following four reactions:
where the propensity functions λi are given by
Table 5.
Circadian clock network: average speed-up factors of PPA with respect to other methods.
| p | Girsanov | CFD | CRP |
|---|---|---|---|
| 0.95 | 355.08 | 12.76 | 15.73 |
| 0.99 | 189.82 | 13.44 | 15.78 |
In the above expressions, x1 and x2 denote the number of molecules of 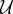 and
and  , respectively. We set α1 = 50, α2 = 16, β = 2.5 and γ = 1. Let (X(t))t≥0 be the CTMC representing the dynamics with initial state (X1(0), X2(0)) = (0, 0). For T = 10 and f(x) = x1, our goal is to estimate
, respectively. We set α1 = 50, α2 = 16, β = 2.5 and γ = 1. Let (X(t))t≥0 be the CTMC representing the dynamics with initial state (X1(0), X2(0)) = (0, 0). For T = 10 and f(x) = x1, our goal is to estimate
for θ = α1, α2, β and γ. In other words, we would like to measure the sensitivity of the mean of the number of 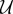 molecules at time T = 10, with respect to all the model parameters.
molecules at time T = 10, with respect to all the model parameters.
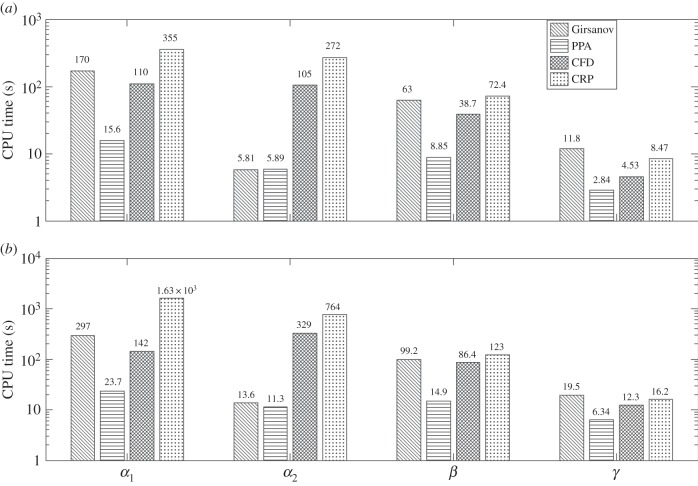
As the propensity functions λ1 and λ3 are nonlinear, it is difficult to compute Sθ(f, T) exactly. As before, we obtain close approximations of Sθ(f, T) using PPA (values given in the electronic supplementary material, §3), and then use these values in computing confidence levels of estimates. The CPU times needed by all the methods for two threshold confidence levels (p = 0.95 and p = 0.99) are presented in figure 5. As in the previous two examples, both CFD and CRP yield an estimate with the desired confidence level with h = 0.1 in all the cases.
Figure 5.
Efficiency comparison for genetic toggle switch with two threshold confidence levels: p = 0.95 (a) and p = 0.99 (b) and observation time T = 10. For finite-difference schemes, h = 0.1 was used in all the cases.
From figure 5, it can be easily seen that PPA is the best-performing method. The average speed-up factors for PPA with respect to other methods are given in table 6. They are calculated by aggregating CPU times for the four values of θ. Note that in this example, the unbiased schemes perform much better than the finite-difference schemes, possibly due to nonlinear parameter-dependence of the propensity functions.
Table 6.
Genetic toggle switch: average speed-up factors of PPA with respect to other methods.
| p | Girsanov | CFD | CRP |
|---|---|---|---|
| 0.95 | 7.57 | 7.81 | 21.34 |
| 0.99 | 7.62 | 10.11 | 45.02 |
The above examples illustrate that in producing an estimate with a specified statistical accuracy, PPA can be much more efficient than other methods, both biased and unbiased. The finite-difference schemes may outperform PPA in some cases but they can also be much slower in other cases. As it is difficult to independently verify the statistical accuracy of finite-difference schemes, PPA is an attractive alternative to these schemes, as its unbiasedness presents a theoretical guarantee for statistical accuracy.
5. Conclusion and future work
We provide a new unbiased method for estimating parameter sensitivity of stochastic reaction networks. Using a result from our recent paper [15], we construct a random variable whose expectation is the required sensitivity value. We then present a simple procedure, called the PPA, to compute the realizations of this random variable. This gives us a way to generate samples for estimating the parameter sensitivity. Our method can be viewed as an improved version of the APA, that we presented in [15]. Unlike APA, the proposed method is easy to implement, has low memory requirements and it works well for large networks and large observation times.
Through examples, we compare the performance of PPA with other methods for sensitivity estimation, both biased and unbiased. Our results indicate that PPA easily outperforms the unbiased Girsanov method. Moreover in many cases, it can even be faster than the best-performing finite-difference schemes (CRP and CFD) in producing a statistically accurate estimate. This makes PPA an appealing method for sensitivity estimation because it is computationally efficient and one does not have to tolerate a bias of an unknown size that is introduced by finite-difference approximations.
In our method, we simulate the paths of the underlying Markov process using Gillespie's SSA [16]. However, SSA is very meticulous in the sense that it simulates the occurrence of each and every reaction in the dynamics. This can be very cumbersome for large networks with many reactions. To resolve this problem, a class of τ-leaping methods have been developed [23,24]. These methods are approximate in nature, but they significantly reduce the computational effort that is required to simulate the reaction dynamics. In future, we would like to develop a version of PPA that uses a τ-leaping method instead of SSA and still produces an accurate estimate for the sensitivity values. Such a method would greatly simplify the sensitivity analysis of large networks. Recently, hybrid approaches that combine several existing sensitivity estimation methods in a multi-level setting have been introduced in [25]. Integrating PPA into this hybrid framework may further enhance its efficiency. This will be investigated in a future work.
Endnotes
The generator of a Markov process is an operator which specifies the rate of change of the distribution of the process. See ch. 4 in [19] for more details.
We define σ0 = 0 for convenience.
All the computations in this paper were performed using C++ programs on a Linux machine with a 2.4 GHz Intel Xeon processor.
Funding statement
We acknowledge funding from Human Frontier Science Programme (RGP0061/2011).
References
- 1.Érdi P, Tóth J. 1989. Mathematical models of chemical reactions. Nonlinear science: theory and applications. Theory and applications of deterministic and stochastic models. Princeton, NJ: Princeton University Press. [Google Scholar]
- 2.Thattai M, van Oudenaarden A. 2001. Intrinsic noise in gene regulatory networks. Proc. Natl Acad. Sci. USA 98, 8614–8619. ( 10.1073/pnas.151588598) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hethcote H. 2000. The mathematics of infectious diseases. SIAM Rev. 42, 599–653. ( 10.1137/S0036144500371907) [DOI] [Google Scholar]
- 4.Bascompte J. 2010. Structure and dynamics of ecological networks. Science 329, 765–766. ( 10.1126/science.1194255) [DOI] [PubMed] [Google Scholar]
- 5.Berger SI, Iyengar R. 2009. Network analyses in systems pharmacology. Bioinformatics 25, 2466–2472. ( 10.1093/bioinformatics/btp465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elowitz MB, Levine AJ, Siggia ED, Swain PS. 2002. Stochastic gene expression in a single cell. Science 297, 1183–1186. ( 10.1126/science.1070919) [DOI] [PubMed] [Google Scholar]
- 7.McAdams H, Arkin A. 1997. Stochastic mechanisms in gene expression. Proc. Natl Acad. Sci. USA 94, 814–819. ( 10.1073/pnas.94.3.814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stelling J, Gilles ED, Doyle FJ. 2004. Robustness properties of circadian clock architectures. Proc. Natl Acad. Sci. USA 101, 13 210–13 215. ( 10.1073/pnas.0401463101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feng XJ, Hooshangi S, Chen D, Li R, Genyuan W, Rabitz H. 2004. Optimizing genetic circuits by global sensitivity analysis. Biophys. J. 87, 2195–2202. ( 10.1529/biophysj.104.044131) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunawan R, Cao Y, Doyle FJ. 2005. Sensitivity analysis of discrete stochastic systems. Biophys. J. 88, 2530–2540. ( 10.1529/biophysj.104.053405) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rathinam M, Sheppard PW, Khammash M. 2010. Efficient computation of parameter sensitivities of discrete stochastic chemical reaction networks. J. Chem. Phys. 132, 034103 ( 10.1063/1.3280166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sheppard PW, Rathinam M, Khammash M. 2012. A pathwise derivative approach to the computation of parameter sensitivities in discrete stochastic chemical systems. J. Chem. Phys. 136, 034115 ( 10.1063/1.3677230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson D. 2012. An efficient finite difference method for parameter sensitivities of continuous time Markov chains. SIAM J. Numer. Anal. 50, 2237–2258. ( 10.1137/110849079) [DOI] [Google Scholar]
- 14.Plyasunov S, Arkin AP. 2007. Efficient stochastic sensitivity analysis of discrete event systems. J. Comput. Phys. 221, 724–738. ( 10.1016/j.jcp.2006.06.047) [DOI] [Google Scholar]
- 15.Gupta A, Khammash M. 2013. Unbiased estimation of parameter sensitivities for stochastic chemical reaction networks. SIAM J. Sci. Comput. 35, A2598–A2620. ( 10.1137/120898747) [DOI] [Google Scholar]
- 16.Gillespie DT. 1977. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81, 2340–2361. ( 10.1021/j100540a008) [DOI] [Google Scholar]
- 17.Gibson MA, Bruck J. 2000. Efficient exact stochastic simulation of chemical systems with many species and many channels. J. Phys. Chem. A 104, 1876–1889. ( 10.1021/jp993732q) [DOI] [Google Scholar]
- 18.Glynn PW. 1990. Likelihood ratio gradient estimation for stochastic systems. Commun. ACM 33, 75–84. ( 10.1145/84537.84552) [DOI] [Google Scholar]
- 19.Ethier SN, Kurtz TG. 1986. Markov processes: characterization and convergence. Wiley Series in Probability and Mathematical Statistics, vol. 44. New York, NY: John Wiley. [Google Scholar]
- 20.Goutsias J. 2007. Classical versus stochastic kinetics modeling of biochemical reaction systems. Biophys. J. 92, 2350–2365. ( 10.1529/biophysj.106.093781) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vilar JMG, Kueh HY, Barkai N, Leibler S. 2002. Mechanisms of noise-resistance in genetic oscillator. Proc. Natl Acad. Sci. USA 99, 5988–5992. ( 10.1073/pnas.092133899) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner TS, Cantor CR, Collins JJ. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342. ( 10.1038/35002131) [DOI] [PubMed] [Google Scholar]
- 23.Gillespie DT. 2001. Approximate accelerated stochastic simulation of chemically reacting systems. J. Chem. Phys. 115, 1716–1733. ( 10.1063/1.1378322) [DOI] [Google Scholar]
- 24.Cao Y, Gillespie DT, Petzold LR. 2006. Efficient step size selection for the tau-leaping simulation method. J. Chem. Phys. 124, 044109 ( 10.1063/1.2159468) [DOI] [PubMed] [Google Scholar]
- 25.Wolf E, Anderson D. 2014. Hybrid pathwise sensitivity methods for discrete stochastic models of chemical reaction systems. (http://arxiv.org/abs/14083655) [DOI] [PubMed]