Abstract
Bacteria produce unique volatile mixtures that could be used to identify infectious agents to the species, and possibly the strain level. However, due to the immense variety of human pathogens, and the close relatedness of some of these bacteria, the robust identification of the bacterium based on its volatile metabolome is likely to require a large number of volatile compounds for each species. We applied comprehensive two-dimensional gas chromatography–time-of-flight mass spectrometry (GC×GC-TOFMS) to the identification of the headspace volatiles of P. aeruginosa PA14 grown for 24 h in lysogeny broth. This is the first reported use of GC×GC-TOFMS for the characterization of bacterial headspace volatiles. The analytical purity that is afforded by this chromatographic method facilitated the identification of 28 new P. aeruginosa-derived volatiles, nearly doubling the list of volatiles for this species.
Keywords: GC×GC-TOFMS, SPME, P. aeruginosa, bacterial volatiles, metabolomics
1. Introduction
Bacteria produce suites of volatile metabolites, the whole of which can be used as unique chemical fingerprints of each species, and possibly the strain or serovar [1–3]. This bounty of information holds the promise for diagnosing infections in situ – for example, straight from a wound, a urine sample, or the breath – without recovering the microbes or their genetic material [2, 3], as is currently required for most routine clinical diagnostic procedures. In situ detection of infectious agents can reduce false-negative rates caused by the unsuccessful recovery of bacteria from infection sites, and provides the potential for faster diagnosis than biochemical methods that assay the presence of single metabolites (e.g., indole by Kovac’s reagent) of enriched cultures, particularly for slow-growing or difficult-to-isolate species. Volatiles-based analyses can also be applied to the phenotypic characterization of bacteria in situ, which has important implications on patient care. For instance, bacterial anaerobiosis in the infection site impacts antibiotic susceptibility [4], and cannot be characterized in vitro. Finally, volatile metabolomics can be used in non-invasive breath analyses of lung, sinus, or gastrointestinal infections [2, 3, 5]; for example, metabolomics are currently employed in the diagnosis of H. pylori stomach infections [6]. The applications for microbial volatile metabolomics extend beyond human health, and can be used for monitoring air quality and biofouling [7], detecting food contamination [3], or for tracking bioconversions in bioengineering processes [8].
Despite the potential uses of bacterial volatiles in medicine, science, and industry, less than three dozen volatiles have been identified for Pseudomonas aeruginosa [5, 9–18], a quantity that is typical for highly-studied bacterial species [3]. Lesser-known species are often characterized by one signature volatile compound [3]. The most commonly used technique for the discovery and identification of bacterial volatiles is one-dimensional gas chromatography-mass spectrometry (GC-MS). While this widely-available method has made it possible to identify the high-abundance volatiles produced by P. aeruginosa and other bacterial species, the method may not generate enough information for the robust identification of a bacterial species; certainly not to the serovar or strain level. The technical difficulty of using GC-MS to identify low-abundance compounds in complex mixtures is responsible, in part, for the paucity of information on the full suite of a bacterium’s volatile metabolome. High-abundance compounds can mask co-eluting low-intensity peaks, which is especially problematic for co-eluting isomers that can’t be deconvoluted because of similar mass fragmentation patterns. Late-eluting compounds may be difficult to identify by GC-MS, as their signal can be obscured by column bleed. Preconcentration of the volatiles may address the problems caused by column bleed, but it will not facilitate isomer deconvolution. The separation resolving power of comprehensive two-dimensional gas chromatography (GC×GC) can address both problems.
In GC×GC analysis, two GC columns of dissimilar stationary phase composition are connected in series through an on-column injector, called a modulator, that sequentially transfers effluent from the first separation dimension into the second dimension [19]. The modulator injection frequency is typically operated in the range between 0.1 and 0.5 Hz, and is chosen based on the desired secondary dimension retention period, which typically ranges between 2 and 10 s. The average width of second dimension peaks is approximately 200 ms, which requires the use of time-of-flight mass spectrometry (TOFMS) detection to provide comprehensive monitoring of mass-to-charge fragments over the small molecule range (0 to 1000 amu). TOFMS is the ideal detector for GC×GC because it can be operated to collect full-scan spectra at a rate of up to 500 Hz, which is fast enough to adequately sample the effluent coming out of the secondary column. TOFMS also provide consistent spectra profiles that are not concentration dependent (no spectral biasing), which allows for deconvolution algorithms to be used for peak “separation” in the MS domain [19]. Spectral deconvolution by TOFMS thus significantly enhances the overall resolving power of the GC×GC analysis.
The most frequently used GC×GC column set combinations separate sample compounds along two elution gradients; the first and second dimension separation gradients are related to the analyte’s relative volatility (boiling point) and polarity (polarizability), respectively. The additional resolving power of the second chromatographic dimension improves spectral purity, facilitating the detection and identification of low abundance compounds via matches to spectral databases (e.g., NIST Mass Spectral Libraries). GC×GC is routinely used for characterizing extremely complex samples, such as petroleum fractions [20], but has been applied to the analysis of microbial metabolites in only a few studies [21–24]. The work described herein is the first reported application of GC×GC to the identification of volatile bacterial metabolites. In this study, we identified 28 new volatile compounds in the headspace of Pseudomonas aeruginosa PA14, including alcohols, aldehydes, ketones, functionalized benzenes, and heteroaromatic molecules, nearly doubling the reported headspace compounds for P. aeruginosa.
2. Experimental
2.1 Bacterial culturing and volatiles collection
P. aeruginosa PA14 was cultured aerobically at 37°C in 50 mL of lysogeny broth, Lennox (LB-Lennox; 10 g Tryptone, 5 g Yeast Extract, 5 g NaCl). After 24 h of growth, the bacterial culture was cooled on ice, and then centrifuged at 5000 rpm at 4°C for 20 min to pellet the cells. The supernatant was filter sterilized through a 0.22 μm PES membrane. Ten milliliters of the supernatant and a stir bar were sealed into a sterilized 20 mL GC headspace vial with a PTFE/silicone cap. Two PA14 cultures and one LB-Lennox blank were prepared. All samples were stored at −20°C until analyzed.
For all gas chromatography analyses (GC-MS and GC×GC-TOFMS), the samples were allowed to thaw at 4°C overnight, and then were heated and stirred for 10 min in a 50°C water bath. A solid-phase microextraction fiber (SPME; divinylbenzene/carboxen/polydimethylsiloxane, 50/30 μm; Supelco/Sigma-Aldrich, St. Louis, MO) was inserted into the vial and exposed for 10 min while the sample was stirred and maintained at 50°C.
2.2 Chromatographic and mass spectrometric methods
The GC×GC-TOFMS system (Pegasus 4D, Leco Corp., St. Joseph, MI) was fitted with a two-dimensional column set consisting of an Rxi-5MS (5% diphenyl/95% dimethyl polysiloxane; 30 m × 180 μm × 0.18 μm (length × internal diameter × film thickness); Restek, Bellefonte, PA) as the first column, and an Rxi-17 (50% diphenyl/50% dimethyl polysiloxane; 1 m × 100 μm × 0.1 μm; Restek) as the second column, joined by a press-fit connection. The columns were heated independently; the primary oven containing column 1 was initiated at 30°C (0.2 min hold), then heated at 10°C/min to 230°C (0.8 min hold); the secondary oven containing column 2 was initiated at 35°C (0.2 min hold), then heated at 10°C/min to 235°C (0.8 min hold). A quad-jet modulator was used with a 4 s modulation period (0.4 s hot pulses, 1.6 s cold pulses) and a 30°C temperature offset relative to the secondary oven. The carrier gas flow rate was 1 mL/min. A pulsed split injection was used, with a 10:1 split at 30 s. The inlet and transfer line temperatures were 250°C. Mass spectra were acquired over the range of 25 – 500 Da at the rate of 200 Hz. Data acquisition and analysis was performed using ChromaTOF software, Version 4.22 (LECO Corp.).
GC-MS was performed on an Rxi-5MS (5% diphenyl/95% dimethyl polysiloxane; 30 m × 250 μm × 0.25 μm; Restek) under conditions identical to the GC×GC parameters described above. Mass spectra were acquired over the range of 25 – 500 Da using mass selective detection.
Retention indices (RI) for the bacterial headspace volatiles were calculated using internal alkane standards for GC and GC×GC analyses. Specifically, after the SPME fiber had been exposed to the bacterial headspace volatiles for 10 min at 50°C, the fiber was exposed for one second at room temperature to the headspace of a mixture of alkanes (C5 – C20), then analyzed by the chromatographic methods described above.
2.3 Data Processing and Analysis for GC×GC-TOFMS
The GC×GC-TOFMS data for two samples and the blank were processed using the same ChromaTOF parameters. The baseline was drawn through the middle of the noise and the signal-to-noise (S/N) cutoff was set at 250. The resulting peaks were identified by a forward search of the NIST 08 Mass Spectral Library. Similarity scores were calculated by ChromaTOF and peaks with scores below 700 were designated as unknowns (named as “Unidentified” in the data Table). The number of unknown peaks was unrestricted during data processing. The identity of the peaks with similarity scores > 700 were further evaluated by their RIs, when available. Observed RIs that fall within the range of previously-reported values in the NIST database for an Rxi-5MS column (or equivalent) are recorded in bold in the Table. With the exception of acetic acid, all of the un-bolded RIs have insufficient data in the NIST database for comparison. The identities of the named compounds in the Table are tentative, based on MS library matching and RIs, and have not been confirmed by co-injection with authentic standards.
Table.
List of compounds identified in P. aeruginosa PA14 headspace.
| Name | Formula | MW | Sim. | Area % | Δ | R.T. (s) | R.I. | Classification |
|---|---|---|---|---|---|---|---|---|
| Hydrogen cyanide | CHN | 27 | 948 | 0.73 | + | 192, 1.150 | -- | Others |
| * Methyl alcohol | CH4O | 32 | 957 | 0.82 | + | 196, 1.180 | -- | Alcohols |
| Acetic acid | C2H4O2 | 60 | 948 | 0.12 | ++++ | 376, 1.375 | 755 | Acid |
| Dimethyl sulfide | C2H6S | 62 | 922 | 4.39 | ++++ | 224, 1.215 | 520 | Others |
| * Pyrrole | C4H5N | 67 | 943 | 0.04 | ++ | 376, 1.790 | 755 | Heteroaromatics |
| 2-Butanone | C4H8O | 72 | 899 | 3.23 | ++ | 256, 1.370 | 601 | Ketones |
| Thiocyanic acid methyl ester | C2H3NS | 73 | 926 | 0.02 | ++++ | 336, 1.735 | 710 | Others |
| * 2-Methyl-2-propanol | C4H10O | 74 | 929 | <0.01 | ++ | 224, 1.405 | 521 | Alcohols |
| * 2-Butanol | C4H10O | 74 | 840 | 0.44 | ++++ | 260, 1.380 | 606 | Alcohols |
| 1-Butanol | C4H10O | 74 | 868 | 0.30 | ++ | 300, 1.520 | 662 | Alcohols |
| * 2,3-Butanedione | C4H6O2 | 86 | 906 | 0.03 | + | 252, 1.115 | 590 | Ketones |
| * 3-Methyl-2-butanone | C5H10O | 86 | 913 | 0.01 | + | 296, 1.450 | 656 | Ketones |
| 2-Pentanone | C5H10O | 86 | 932 | 3.02 | ++ | 316, 1.490 | 684 | Ketones |
| * 2-Pentanol | C5H12O | 88 | 836 | 0.24 | ++++ | 324, 1.540 | 695 | Alcohols |
| 3-Methyl-1-butanol | C5H12O | 88 | 933 | 0.51 | ++ | 356, 1.615 | 732 | Alcohols |
| * Methyl thiolacetate | C3H6OS | 90 | 889 | 0.30 | ++++ | 328, 1.415 | 700 | Others |
| * Methyl pyrazine | C5H6N2 | 94 | 951 | 0.48 | + | 444, 1.600 | 828 | Heteroaromatics |
| * 4-Methyl-4-penten-2-one | C6H10O | 98 | 865 | 2.23 | ++ | 372, 1.545 | 750 | Ketones |
| * 3-Methyl-3-penten-2-one | C6H10O | 98 | 929 | 0.44 | ++++ | 456, 1.620 | 840 | Ketones |
| * 3-Methyl-2-pentanone | C6H12O | 100 | 935 | 2.42 | ++++ | 372, 1.530 | 750 | Ketones |
| * 2-Hexanone | C6H12O | 100 | 897 | 2.04 | + | 408, 1.565 | 791 | Ketones |
| * Benzonitrile | C7H5N | 103 | 963 | 0.08 | + | 608, 1.860 | 992 | Func. Benzenes |
| 2,5-Dimethyl-pyrazine | C6H8N2 | 108 | 944 | 5.07 | +++ | 532, 1.600 | 916 | Heteroaromatics |
| 2-Heptanone | C7H14O | 114 | 949 | 0.50 | + | 508, 1.600 | 892 | Ketones |
| * Benzoxazole | C7H5NO | 119 | 924 | 0.04 | ++ | 644, 1.705 | 1029 | Heteroaromatics |
| * Acetophenone | C8H8O | 120 | 974 | 1.22 | + | 688, 1.800 | 1075 | Func. Benzenes |
| * (R)- à-Methyl-benzenemethanol | C8H10O | 122 | 891 | 0.08 | ++++ | 684, 1.805 | 1071 | Func. Benzenes |
| * (E)-2-Octenal | C8H14O | 126 | 923 | 0.05 | ++ | 676, 1.695 | 1063 | Aldehydes |
| * Acetyl valeryl | C7H12O2 | 128 | 798 | 0.03 | + | 452, 1.555 | 836 | Ketones |
| * 2-Methyl-benzoxazole | C8H7NO | 133 | 854 | 0.01 | ++ | 736, 1.690 | 1128 | Heteroaromatics |
| 2-Aminoacetophenone | C8H9NO | 135 | 967 | 0.39 | ++ | 900, 1.955 | 1317 | Func. Benzenes |
| * 2-Methyl-3-isopropylpyrazine | C8H12N2 | 136 | 828 | 0.05 | + | 672, 1.585 | 1059 | Heteroaromatics |
| 2-Nonanone | C9H18O | 142 | 939 | 1.88 | ++ | 704, 1.630 | 1092 | Ketones |
| * 4-Methyl-quinazoline | C9H8N2 | 144 | 764 | 0.01 | + | 940, 1.850 | 1369 | Heteroaromatics |
| * 1-Phenyl-1-butanone | C10H12O | 148 | 803 | 0.01 | ++++ | 856, 1.760 | 1264 | Func. Benzenes |
| * 2-Butyl-3-methylpyrazine | C9H14N2 | 150 | 722 | 0.03 | + | 804, 1.610 | 1205 | Heteroaromatics |
| * 2-Ethyl-3-(methylthio)-pyrazine | C7H10N2S | 154 | 722 | 0.02 | ++++ | 856, 1.645 | 1264 | Heteroaromatics |
| * 3-Decanone | C10H20O | 156 | 807 | 0.02 | ++ | 792, 1.610 | 1191 | Ketones |
| 2-Undecanone | C11H22O | 170 | 954 | 0.80 | ++ | 884, 1.630 | 1296 | Ketones |
| * 2-Dodecanone | C12H24O | 184 | 867 | 0.02 | ++ | 964, 1.640 | 1400 | Ketones |
| * 2-Tridecanone | C13H26O | 198 | 932 | 0.11 | ++ | 1040, 1.645 | 1500 | Ketones |
| Unidentified 1 | 677 | 0.22 | ++++ | 324, 1.580 | 695 | Unknowns | ||
| Unidentified 2 | 562 | 3.25 | + | 532, 1.615 | 916 | Unknowns | ||
| Unidentified 3 | 739 | 0.03 | + | 572, 1.565 | 956 | Unknowns | ||
| Unidentified 4 | 597 | 0.01 | + | 620, 1.540 | 1004 | Unknowns | ||
| Unidentified 5 | C7H7N3 | 133 | 822 | 0.05 | ++++ | 760, 1.880 | 1155 | Heteroaromatics |
| Unidentified 6 | C10H20O | 156 | 808 | 0.02 | ++ | 764, 1.630 | 1159 | Ketones |
| Unidentified 7 | 668 | 0.02 | + | 836, 1.620 | 1241 | Unknowns | ||
| Unidentified 8 | 627 | 0.01 | ++ | 860, 1.645 | 1268 | Unknowns | ||
| Unidentified 9 | 797 | 0.05 | ++ | 892, 1.640 | 1306 | Unknowns | ||
| Unidentified 10 | C9H8N2 | 144 | 879 | 0.02 | ++ | 904, 1.765 | 1322 | Heteroaromatics |
| Unidentified 11 | 716 | 0.03 | + | 932, 1.745 | 1358 | Unknowns | ||
| Unidentified 12 | 565 | 0.01 | + | 976, 1.460 | 1416 | Unknowns | ||
| Unidentified 13 | C15H22 | 202 | 840 | 0.02 | ++++ | 1004, 1.525 | 1453 | Unknowns |
| Unidentified 14 | C12H22O2 | 198 | 817 | 0.13 | ++++ | 1024, 1.665 | 1479 | Unknowns |
| Unidentified 15 | 890 | 0.05 | + | 1044, 1.480 | 1506 | Unknowns |
Notes:
Indicates volatiles that have not been previously reported for P. aeruginosa.
MW = Molecular weight
Sim. = Similarity to reference spectrum on a scale of 0 – 999, with higher scores indicating greater similarity
Area % = Area percent of the Total Ion Chromatogram (TIC) of the peak with respect to the total area of the 181 peaks in the sample
Δ = Fold-increase over blank: (+) 2–10; (++) 11–100; (+++) 101–1000; (++++) Present in the sample, but not the blank
RTs = Retention times: first dimension, second dimension
RI = Retention index; Bolded values indicate a match to previously reported RIs. RIs could not be calculated for hydrogen cyanide or methanol based on the internal standards used.
All peaks were assigned to one of the following compound classifications: aliphatic hydrocarbons (CxHy), including alkanes, alkenes, and alkynes), aromatic hydrocarbons (CxHy), heteroaromatic hydrocarbons (containing a N, O, or S heteroatom in the aromatic ring), functionalized benzenes (benzene rings with various heteroatomic functional groups), alcohols, aldehydes, ketones, acids, esters, or other (any other compound class, or compounds containing multiple functional groups). When no compound classification could be assigned for an unidentified compound, the peak was designated as class “Unknown”.
Peaks that were assigned as P. aeruginosa PA14 headspace volatiles were present in both samples at concentrations at least two-fold greater than the blank. The fold-increase over the blank (Δ in the Table) was calculated by unique masses for each compound, and the reported value is the least of the two sample replicates, if different. The area percent (Area % in the Table) was calculated using the TICs of the sample peaks (excluding artifacts).
3. Results and Discussion
Solid-phase microextraction (SPME) was used to sample the headspace of P. aeruginosa strain PA14 grown for 24 h in LB-Lennox. The volatile and semi-volatile PA14 metabolites, which are dissolved in the growth medium, are predicted to achieve equilibrium between the liquid medium, headspace, and SPME stationary phase within minutes at 50°C with stirring [25]. As a brief validation of this prediction, we measured the amount of 2-aminoacetophenone (2-AA) and indole that were adsorbed onto the triphase SPME fiber as a function of exposure time, temperature, and concentration in water. 2-AA is a low-abundance, but highly specific semi-volatile biomarker responsible for the iconic grape-like fragrance of P. aeruginosa, and indole is a high-abundance semi-volatile compound that gives E. coli its mothball-like scent [12, 26, 27]. We observed that 2-AA and indole reach equilibrium concentrations on the SPME fiber within 10 min at 40°C with stirring (data not shown). Furthermore, the adsorbed concentrations of both compounds are linear over several orders of magnitude when sampled at equilibrium: 0.5 – 500 μM 2-AA in water (R2 = 0.999), and 5 – 500 μM indole in water (R2 = 0.999). While this chromatographic analysis of PA14 headspace compounds is designed for qualitative volatile discovery, based on the range of Henry’s (KH) and octanol-water partition (KOW) coefficients of the compounds observed in this study, we estimate that the majority of the PA14 headspace volatiles were sampled at or near equilibrium concentrations[25].
Comprehensive two-dimensional gas chromatography (GC×GC) of PA14 culture headspace reveals a complexity that has not been previously described for bacterial volatiles (Fig. 1). Two headspace samples of PA14 were analyzed by GC×GC, yielding an average of 500 peaks per sample with a total ion chromatograph (TIC) signal-to-noise (S/N) greater than 250. By GC-MS, an order-of-magnitude fewer peaks are distinguishable at this conservative S/N cutoff (Fig. 1, top). After the removal of artifacts (e.g., siloxanes, phthalates, and plasticizers), there were 181 peaks (S/N > 250) arising from a combination of the metabolic activities of PA14 and the LB-Lennox medium, represented by aliphatic and aromatic hydrocarbons, alcohols, aldehydes, ketones, acids, esters, and heteroaromatics (Fig 1, Bottom). The separation of the volatiles mixture by two chemical properties, boiling point on the first column, and polarity on the second, creates chromatographic bands of chemically and structurally related compounds in GC×GC [19], which can be visualized most easily in Figure 1 (Bottom) for the aliphatic hydrocarbons (red spheres), the ketones (blue), and the aromatic hydrocarbons (cyan). This chromatographic information complements mass fragmentation data in the identification of chemical isomers of unknowns [19].
Figure 1.
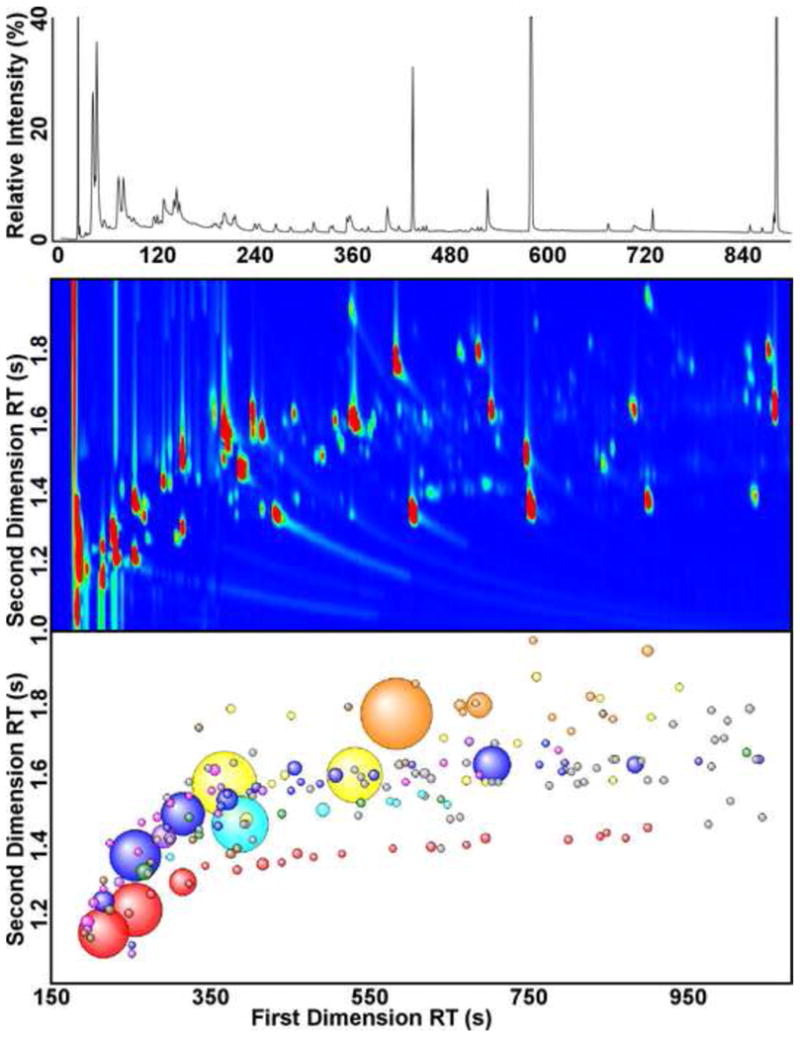
Gas chromatography of the headspace volatiles of P. aeruginosa PA14 grown aerobically in LB-Lennox. (Top) One-dimensional gas chromatogram (GC) of the headspace volatiles. (Middle) Two-dimensional gas chromatogram (GC×GC) of the headspace volatiles, excluding regions containing only column bleed (2nd RT < 1.0). The GC and GC×GC chromatograms were aligned using the measured retention indices of the headspace volatiles. (Bottom) Bubble plot of the GC×GC chromatogram, after chromatographic artifacts were removed. The colors reflect the compound classes detected for the sample and blank: aliphatic hydrocarbons (red), alcohols (pink), aldehydes (purple), ketones (blue), esters (green), aromatic hydrocarbons (cyan), heteroaromatics (yellow), functionalized benzenes (orange), and all other classes (brown). Unidentified compounds are shaded in gray. The radii of the bubbles correspond to the relative TIC peak intensities.
Of the 181 sample peaks, we detected 56 compounds that are present in the 24 h culture headspace of P. aeruginosa PA14 in at least two-fold greater abundance than in the headspace of the LB-Lennox medium blank (Table). The named compounds in the Table were identified by matching the observed mass spectra to their respective library reference spectra with a similarity score of 700 or greater (70% match), and many were further confirmed by their retention indices (RI), indicated in bold in the Table. The compounds in the Table that are reported as “Unidentified” fall into three categories: 1) Those with molecular formulae and assigned classifications, 2) those with molecular formulae, but classified as “Unknown”, and 3) those without molecular formulae. The peaks in the first and second categories had good matches to more than one isomer with the given molecular formula, and in the case of “Unknown” classification, those isomers belong to two or more different classifications of molecules (e.g., Unidentified 13). The peaks in the third category – without molecular formulae – either had good matches to two or more compounds with dissimilar molecular formulae (e.g., Unidentified 15), or did not have a suitable match (Similarity > 700) to a NIST library spectrum (e.g., Unidentified 2). The majority of the peaks in the third category had very low headspace concentrations, and may be present in the liquid phase in greater quantities. Though the compounds that are reported here require confirmation by co-injections with authentic standards, the high-confidence mass spectral data generated by GC×GC-TOFMS, in conjunction with RI data, provide relatively confident identifications for bacterial headspace volatiles.
Half of the 56 PA14 headspace compounds that we observed are reported here for the first time for P. aeruginosa, nearly doubling the list of volatiles available in the literature for this species. Previous studies have established that P. aeruginosa produces a variety of ketones and heteroaromatic molecules in high abundance [5, 12, 14, 15, 17], and the newly-described compounds here uphold that pattern (Fig. 2). The odd-chain-length 2-ketones (C5 –C11) were identified as P. aeruginosa volatiles more than thirty years ago [12], but with the additional chromatographic dimension provided by GC×GC, we were also able to observe the presence of 2-dodecanone and 2-tridecanone – late-eluting compounds with relative abundances at a fraction of a percent (Table).
Figure 2.
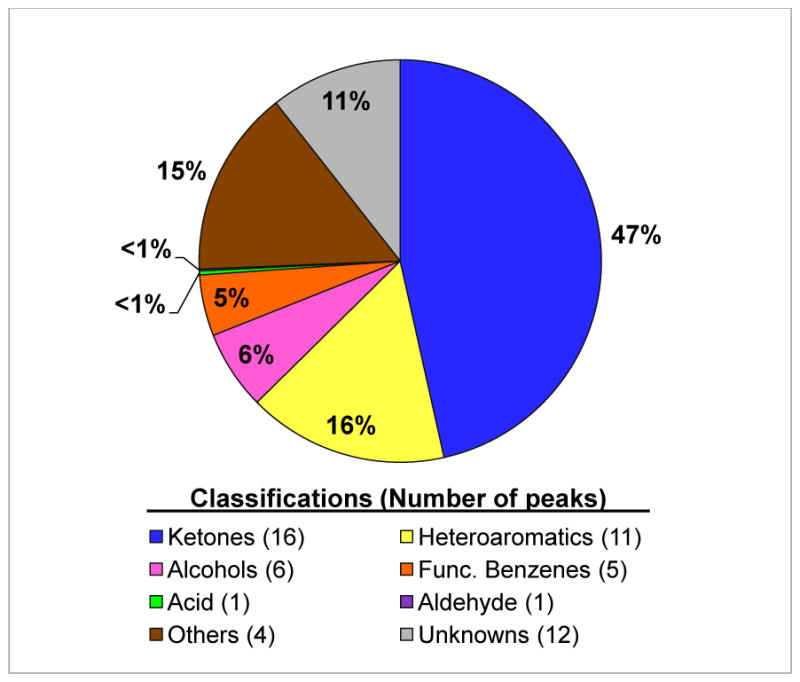
Classifications of P. aeruginosa PA14 headspace compounds, represented by their relative abundance in area percent.
The superior resolving power of GC×GC provides data with improved signal-to-noise (S/N) and analytical purity relative to GC [28], revealing low-abundance compounds in densely-populated regions of one-dimensional chromatograms. For example, the peak for 2-aminoacetophenone (2-AA) is obscured by column bleed in GC (Fig. 1, Top; RT = 723 s, RI = 1317) [5], but is well-resolved by GC×GC, with a S/N = 23158 (Fig. 3, RTs = 900 s, 1.955 s). GC×GC-TOFMS also reveals that there are eight compounds with a S/N>100 co-eluting with 2-AA in the first dimension, which are undetectable by GC-MS (Fig. 3). Separating the 2-AA from these co-eluting peaks and the column bleed enables the acquisition of a 2-AA mass spectrum that is free of foreign peaks (high mass spectral purity), assisting detection using chromatographic processing software, and high-confidence identification by spectral library match (Fig 4, Table). The high proportion and abundance of artifact peaks in the bacterial headspace mixture that arise from column bleed, SPME fibers, and the headspace vial caps are well separated from compounds of interest by GC×GC, facilitating the discovery of new bacterial volatiles.
Figure 3.
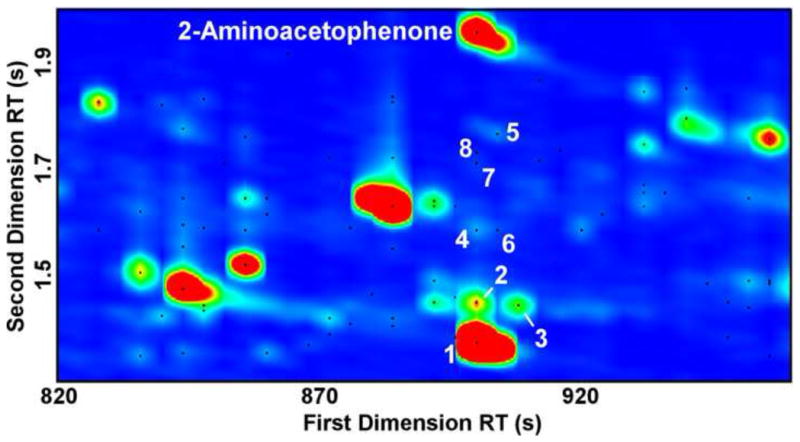
GC×GC chromatographic region containing the 2-aminoacetophenone (2-AA) peak. Each black dot marks a peak with a TIC S/N > 100. Eight other peaks co-elute with 2-AA in the first chromatographic dimension, but are separated from 2-AA in the second dimension. The co-eluting compounds are numbered in order of peak area, with dodecamethyl cyclohexasiloxane (peak 1, Sim.=842, S/N=79182) as the largest peak. The other compounds are unsaturated hydrocarbons (peaks 2 and 3, S/N=754 and 537, respectively), a heteroaromatic compound (peak 5, S/N=1010), and Unknowns (peaks 4, 6, 7, and 8, S/N = 585, 226, 185, and 131, respectively).
Figure 4.
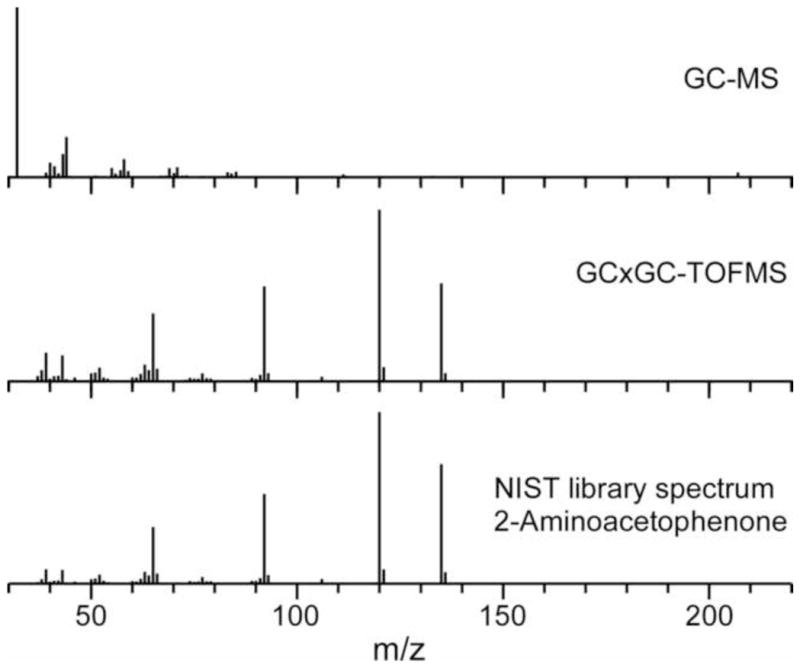
Mass spectra of the chromatographic regions containing 2-aminoacetophenone (2-AA; RI =1317) as acquired by GC-MS (Top) and GC×GC-TOFMS (Middle). Column bleed and co-eluting compounds in GC chromatography obscure the mass spectrum of the low-abundance compound 2-AA. GC×GC affords higher chromatographic purity, making it possible to identify low-abundance compounds by library matching. The NIST library spectrum of 2-AA is provided for reference (Bottom).
4. Conclusions
The key to bacterial identification via volatile metabolomics lies in assigning unique patterns of volatile biomarkers to individual bacterial species. Due to the sheer abundance of microbial species and infectious agents to identify, and the close relatedness of some species within their genus, selective and specific fingerprinting methods will require a large selection of volatiles to be used in combination for correct identifications. Therefore, as we establish bacterial volatile profiles, we must go beyond the obvious (e.g., easy to smell), high-abundance, and common bacterial volatiles, and begin identifying the peaks in the chromatographic grass. Through the enhanced analytical purity that can be achieved by GC×GC-TOFMS, the discovery of these hence-unknown compounds is readily attainable.
Highlights.
First application of SPME-GC×GC-TOFMS to the identification of bacterial headspace volatiles
Twenty-eight new P. aeruginosa headspace volatiles
The list of P. aeruginosa headspace volatiles is nearly doubled
Acknowledgments
This project was supported by grants from the National Center for Research Resources (5P20RR021905-07; JEH) and the National Institute of General Medical Sciences (8 P20 GM103496-07; JEH) from the National Institutes of Health, the Cystic Fibrosis Foundation Research Development Program (STANTO07R0; JEH), NASA (NNH09ZNE002C; JEH), and the Major Research Instrumentation Program from the National Science Foundation (0923295; JMD).
Abbreviations
- 2-AA
2-aminoacetophenone
- GC×GC-TOFMS
comprehensive two-dimensional gas chromatography – time of flight mass spectrometry
- LB
lysogeny broth
- RI
retention index
- RT
retention time
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhu J, Bean HD, Kuo YM, Hill JE. J Clin Microbiol. 2010;48:4426–4431. doi: 10.1128/JCM.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casalinuovo IA, Di Pierro D, Coletta M, Di Francesco P. Sensors. 2006;6:1428–1439. doi: 10.1111/j.1472-765X.2005.01792.x. [DOI] [PubMed] [Google Scholar]
- 3.Wilson AD, Baietto M. Sensors. 2011;11:1105–1176. doi: 10.3390/s110101105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borriello G, Werner E, Roe F, Kim AM, Ehrlich GD, Stewart PS. Antimicrob Agents Chemother. 2004;48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Preti G, Thaler E, Hanson CW, Troy M, Eades J, Gelperin A. J Chromatogr B. 2009;877:2011–2018. doi: 10.1016/j.jchromb.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 6.Schuman R, Rigas B, Prada A, Minoli G. Gastroenterology. 1995;108:A215–A215. doi: 10.1097/00004836-199806000-00009. [DOI] [PubMed] [Google Scholar]
- 7.Moularat S, Robine E, Ramalho O, Oturan MA. Chemosphere. 2008;72:224–232. doi: 10.1016/j.chemosphere.2008.01.057. [DOI] [PubMed] [Google Scholar]
- 8.Chippendale TWE, Spanel P, Smith D. Rapid Commun Mass Spectrom. 2011;25:2163–2172. doi: 10.1002/rcm.5099. [DOI] [PubMed] [Google Scholar]
- 9.Allardyce RA, Hill AL, Murdoch DR. Diagn Microbiol Infect Dis. 2006;55:255–261. doi: 10.1016/j.diagmicrobio.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 10.Allardyce RA, Langford VS, Hill AL, Murdoch DR. J Microbiol Methods. 2006;65:361–365. doi: 10.1016/j.mimet.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Carroll W, Lenney W, Wang TS, Spanel P, Alcock A, Smith D. Pediatr Pulmonol. 2005;39:452–456. doi: 10.1002/ppul.20170. [DOI] [PubMed] [Google Scholar]
- 12.Labows JN, McGinley KJ, Webster GF, Leyden JJ. J Clin Microbiol. 1980;12:521–526. doi: 10.1128/jcm.12.4.521-526.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scholler C, Molin S, Wilkins K. Chemosphere. 1997;35:1487–1495. doi: 10.1016/s0045-6535(97)00209-9. [DOI] [PubMed] [Google Scholar]
- 14.Zechman JM, Labows JN. Can J Microbiol. 1985;31:232–237. doi: 10.1139/m85-045. [DOI] [PubMed] [Google Scholar]
- 15.Rudzinski CM, Herzig-Marx R, Lin J, Szpiro A, Johnson B. Proceedings of the Scientific Conference on Chemical and Biological Defense Research; 2004. [Google Scholar]
- 16.Julak J, Stranska E, Rosova V, Geppert H, Spanel P, Smith D. J Microbiol Methods. 2006;65:76–86. doi: 10.1016/j.mimet.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Senecal AG, Magnone J, Yeomans W, Powers EM. Proceedings of SPIE. 2002;4575:121–131. [Google Scholar]
- 18.Shestivska V, Nemec A, Dřevínek P, Sovová K, Dryahina K, Španěl P. Rapid Commun Mass Spectrom. 2011;25:2459–2467. doi: 10.1002/rcm.5146. [DOI] [PubMed] [Google Scholar]
- 19.Dimandja JMD. Anal Chem. 2004;76:167A–174A. [Google Scholar]
- 20.Blomberg J, Schoenmakers PJ, Beens J, Tijssen R. J High Res Chrom. 1997;20:539–544. [Google Scholar]
- 21.Humston EM, Dombek KM, Tu BP, Young ET, Synovec RE. Anal Bioanal Chem. 2011;401:2387–2402. doi: 10.1007/s00216-011-4800-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gardner JY, Brillhart DE, Benjamin MM, Dixon LG, Mitchell LM, Dimandja JMD. J Sep Sci. 2011;34:176–185. doi: 10.1002/jssc.201000612. [DOI] [PubMed] [Google Scholar]
- 23.Purcaro G, Tranchida PQ, Dugo P, La Camera E, Bisignano G, Conte L, Mondello L. J Sep Sci. 2010;33:2334–2340. doi: 10.1002/jssc.201000160. [DOI] [PubMed] [Google Scholar]
- 24.Yang S, Sadilek M, Synovec RE, Lidstrorn ME. J Chromatogr A. 2009;1216:3280–3289. doi: 10.1016/j.chroma.2009.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Pawliszyn J. Anal Chem. 1993;65:1843–1852. [Google Scholar]
- 26.Cox CD, Parker J. J Clin Microbiol. 1979;9:479–484. doi: 10.1128/jcm.9.4.479-484.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falkow S, Mekalanos J. The enteric bacilli and vibrios. In: Davis BD, Dulbecco R, Eisen HN, Ginsberg HS, editors. Microbiology. Lippincott Williams and Wilkins; Philadelphia: 1990. p. 1233. [Google Scholar]
- 28.Welthagen W, Shellie RA, Spranger J, Ristow M, Zimmermann R, Fiehn O. Metabolomics. 2005;1:65–73. doi: 10.1016/j.chroma.2005.05.088. [DOI] [PubMed] [Google Scholar]


