Abstract
Extremely cold environments are a challenge for all organisms. They are mostly inhabited by psychrophilic and psychrotolerant bacteria, which employ various strategies to cope with the cold. Such harsh environments are often highly vulnerable to the influence of external factors and may undergo frequent dynamic changes. The rapid adjustment of bacteria to changing environmental conditions is crucial for their survival. Such “short-term” evolution is often enabled by plasmids—extrachromosomal replicons that represent major players in horizontal gene transfer. The genomic sequences of thousands of microorganisms, including those of many cold-active bacteria have been obtained over the last decade, but the collected data have yet to be thoroughly analyzed. This report describes the results of a meta-analysis of the NCBI sequence databases to identify and characterize plasmids of psychrophilic and psychrotolerant bacteria. We have performed in-depth analyses of 66 plasmids, almost half of which are cryptic replicons not exceeding 10 kb in size. Our analyses of the larger plasmids revealed the presence of numerous genes, which may increase the phenotypic flexibility of their host strains. These genes encode enzymes possibly involved in (i) protection against cold and ultraviolet radiation, (ii) scavenging of reactive oxygen species, (iii) metabolism of amino acids, carbohydrates, nucleotides and lipids, (iv) energy production and conversion, (v) utilization of toxic organic compounds (e.g., naphthalene), and (vi) resistance to heavy metals, metalloids and antibiotics. Some of the plasmids also contain type II restriction-modification systems, which are involved in both plasmid stabilization and protection against foreign DNA. Moreover, approx. 50% of the analyzed plasmids carry genetic modules responsible for conjugal transfer or mobilization for transfer, which may facilitate the spread of these replicons among various bacteria, including across species boundaries.
Keywords: plasmid, cold environment, psychrophilic and psychrotolerant bacteria, cold adaptation, horizontal gene transfer
Introduction
Cold environments represent a large fraction of the surface area of the Earth. They include polar regions as well as most of the oceans, which cover approximately 70% of the Earth's surface (Casanueva et al., 2010; Margesin and Miteva, 2011). Such habitats experience extreme conditions that are challenging to most life forms. However, even such harsh ecological niches are inhabited by various microorganisms, most of which are cold-adapted bacteria.
Cold-adapted bacteria can be classified into two groups based on their temperature tolerance: (i) psychrophiles, which can grow at temperatures not exceeding approx. 20°C, and (ii) psychrotrophs (or psychrotolerants), that tolerate a broader range of temperatures—between 0 and approx. 30°C (Morita, 1975). Both groups of microorganisms share basic molecular and physiological characteristics, which permit their survival in extremely cold environments: (i) increased fluidity of cellular membranes, (ii) the ability to accumulate compatible solutes (e.g., glycine, betaine and trehalose), (iii,iv) the expression of cold shock, antifreeze and ice-nucleating proteins, as well as (v) the production of cold-active enzymes (Casanueva et al., 2010).
The diversity, biology and ecology of psychrophilic or psychrotolerant bacteria have been extensively studied in recent years. Furthermore, numerous strains have been subjected to genomic investigations. This research has been mainly focused on their mechanisms of cold adaptation, which are interesting from both an evolutionary and a biotechnological point of view (Casanueva et al., 2010; Martinez-Rosales et al., 2012). Unfortunately, very little attention has been paid to the range and direction of horizontal gene transfer (HGT) among cold-active bacteria.
Extremely cold habitats are usually endemic and highly vulnerable to the influence of external factors, and therefore may undergo many dynamic changes. In such circumstances, rapid adaptation of bacterial cells to the changing environmental conditions is necessary for survival. Such “short-term” evolution is possible due to the presence of various mobile genetic elements (MGEs), including plasmids, which are considered major contributors to HGT.
Plasmids increase the plasticity of bacterial genomes, and consequently promote genetic diversity of their hosts (Siefert, 2009). Besides a set of basic structural genetic modules responsible for plasmid replication, maintenance and spread, these replicons very often carry an additional load of genetic information. These accessory genes serve as a reservoir of diverse genetic determinants whose presence may confer an evolutionary advantage to the host strains under appropriate environmental conditions (e.g., Wiedenbeck and Cohan, 2011; Heuer and Smalla, 2012).
Many plasmid-harboring bacterial strains have been isolated from permanently cold environments including the Arctic (e.g., Dziewit et al., 2013a; Moller et al., 2014), Antarctica (e.g., Kobori et al., 1984; Dziewit et al., 2013b) and Siberia (e.g., Petrova et al., 2014). Several plasmids were also identified during genomic projects of psychrophilic bacteria, e.g., five plasmids were recognized in the genome of Carnobacterium gilichinskyi WN1359T, a strain originating from Siberian permafrost (Leonard et al., 2013). Although plasmids appear to be common components of the genomes of psychrophilic and psychrotolerant bacteria, current knowledge about them is very fragmentary. A relatively small number have been fully sequenced and only a few analyzed in detail.
In this study we have performed a meta-analysis of the NCBI sequence databases, aimed at identifying plasmids of cold-active bacteria. The identified plasmids were thoroughly analyzed and their predicted adaptive role investigated.
Material and methods
Plasmid identification
The search for plasmids of cold-active bacteria was performed primarily in the NCBI databases (http://www.ncbi.nlm.nih.gov/), as well as the EMBL-EBI (http://www.ebi.ac.uk/) and Aclame databases (Leplae et al., 2010). The NCBI databases were searched using a set of words and phrases related to cold environments and cold adaptation. The genome browser in the Microbial Genomes Homepage (NCBI; http://www.ncbi.nlm.nih.gov/genome/browse/) was used to check for the presence of plasmids within the sequenced genomes of bacteria belonging to genera usually considered as psychrophilic (e.g., Psychrobacter, Pseudoalteromonas, Psychromonas etc). For further analyses only the plasmids of strains defined as psychrophilic or psychrotolerant according to the NCBI record or original research article were used.
Bioinformatic analyses
Plasmid nucleotide sequences were analyzed using Artemis software (Carver et al., 2008). Similarity searches were performed using the BLAST (Altschul et al., 1997) and Pfam 27.0 programs (Finn et al., 2014). The EC numbers for particular proteins were assigned using the PRIAM tool (Claudel-Renard et al., 2003). For the functional annotation of predicted proteins and their classification into appropriate Clusters of Orthologous Groups (COGs), the Batch Web CD-Search Tool (E value < 1e-05) (Marchler-Bauer et al., 2011) was used. The screening of metabolic pathways was performed using the KAAS—KEGG Automatic Annotation Server (Moriya et al., 2007) and then the KEGG ORTHOLOGY (KO) Database, linking genomes to pathways by ortholog annotation (Kanehisa and Goto, 2000; Kanehisa et al., 2014). The reference data set for the computational prediction of metabolic pathways was obtained from the MetaCyc database (Caspi et al., 2014). All predictions were verified manually.
The classification of the relaxases was performed using BALSTp analysis comparing the amino acid sequence of each relaxase with the manually curated databases of the relaxases representing particular families (Garcillan-Barcia et al., 2009). Additionally within the sequence of each analyzed relaxase the conserved amino acid motifs (specific for the particular relaxase family) were identified, confirming the classification.
The analysis of predicted restriction-modification systems was performed using REBASE (The Restriction Enzyme Database) (Roberts et al., 2010). Spacer sequences of CRISPR-Cas systems were identified using the CRISPRfinder program (Grissa et al., 2007).
Results and discussion
Diversity and general features of plasmids of cold-active bacteria
We performed in depth searches of available DNA databases in order to identify the sequences of plasmids of psychrophilic and psychrotolerant bacteria. This analysis revealed the presence of 66 such replicons (named pPSYCH plasmids) occurring in 39 bacterial strains (Table 1) isolated from various geographical locations: Antarctica (15 plasmids), Arctic (14), France (4), Japan (2), Norway (6), Poland (2), Puerto Rico (2), Russia (9), and the USA (5). For a few strains, information concerning the site of isolation was not available. It is important to note that the nucleotide sequences of 37 plasmids were obtained as a result of whole genome sequencing projects and have not previously been analyzed in detail.
Table 1.
Completely sequenced plasmids of psychrophilic and psychrotolerant bacteria (pPSYCH) analyzed in this study.
| Plasmid name | Host strain | Strain origin | Size (kb) | GC (%) | No. of genes | References/Accession No. |
|---|---|---|---|---|---|---|
| pVSAL43 | Aliivibrio salmonicida LFI1238 | Norway, Atlantic cod | 4.3 | 35.6 | 3 | (Hjerde et al., 2008)/FM178384 |
| pVSAL54 | A. salmonicida LFI1238 | Norway, Atlantic cod | 5.4 | 38.1 | 3 | (Hjerde et al., 2008)/FM178383 |
| pVSAL320 | A. salmonicida LFI1238 | Norway, Atlantic cod | 30.8 | 37.3 | 32 | (Hjerde et al., 2008)/FM178382 |
| pVSAL840 | A. salmonicida LFI1238 | Norway, Atlantic cod | 83.5 | 40.1 | 72 | (Hjerde et al., 2008)/FM178381 |
| pVSAL111 | A. salmonicida TEO83.001 | Norway, Salmon salar | 11.1 | 37.4 | 8 | –/HG983279 |
| pVSAL68 | A. salmonicida Vs289 | Norway, Salmon salar | 6.8 | 38.5 | 5 | –/HG983280 |
| pGIAK1 | Bacillaceae bacterium JMAK1 | Antarctica, stairway of Concordia station | 38.4 | 37.5 | 49 | (Guo and Mahillon, 2013)/JX406743 |
| pBWB401 | Bacillus weihenstephanensis KBAB4 | France, forest soil near Versailles | 417.1 | 33.7 | 332 | –/CP000904 |
| pBWB402 | B. weihenstephanensis KBAB4 | France, forest soil near Versailles | 75.1 | 33.4 | 75 | –/CP000905 |
| pBWB403 | B. weihenstephanensis KBAB4 | France, forest soil near Versailles | 65.0 | 43.4 | 76 | –/CP000906 |
| pBWB404 | B. weihenstephanensis KBAB4 | France, forest soil near Versailles | 52.8 | 35.4 | 71 | –/CP000907 |
| pWNCR9 | Carnobacterium gilichinskyi WN1359T | Siberia, permafrost | 9.6 | 29.7 | 14 | (Leonard et al., 2013)/CP006817 |
| pWNCR12 | C. gilichinskyi WN1359T | Siberia, permafrost | 12.7 | 37.0 | 18 | (Leonard et al., 2013)/CP006813 |
| pWNCR15 | C. gilichinskyi WN1359T | Siberia, permafrost | 15.5 | 34.6 | 18 | (Leonard et al., 2013)/CP006814 |
| pWNCR47 | C. gilichinskyi WN1359T | Siberia, permafrost | 47.1 | 32.3 | 46 | (Leonard et al., 2013)/CP006815 |
| pWNCR64 | C. gilichinskyi WN1359T | Siberia, permafrost | 64.5 | 34.6 | 62 | (Leonard et al., 2013)/CP006816 |
| plasmid small | Desulfotalea psychrophila LSv54 | Arctic, sediments | 14.7 | 28.6 | 17 | (Rabus et al., 2004)/CR522872 |
| plasmid large | D. psychrophila LSv54 | Arctic, sediments | 121.6 | 43.6 | 101 | (Rabus et al., 2004)/CR522871 |
| pEXIG01 | Exiguobacterium sibiricum 255-15 | Siberia, permafrost | 4.9 | 37.1 | 5 | (Rodrigues et al., 2006)/CP001023 |
| pEXIG02 | E. sibiricum 255-15 | Siberia, permafrost | 1.8 | 41.4 | 3 | (Rodrigues et al., 2006)/CP001024 |
| pCP1 | Flavobacterium psychrophilum D12 | salmonid fish | 3.4 | 27.4 | 4 | (Alvarez et al., 2004)/AY277637 |
| pFL1 | Flavobacterium sp. KP1 | unknown | 2.3 | 32.7 | 2 | (Ashiuchi et al., 1999)/AB007196 |
| pGLAAG01 | Glaciecola sp. 4H-3-7+YE-5 | Japan, sediments at Suruga Bay | 341.3 | 42.0 | 332 | (Klippel et al., 2011)/CP002527 |
| pTA144 Dw | Moraxella sp. TA144 | Antarctica | 1.3 | 46.2 | 0 | (Tutino et al., 2000)/AJ224744 |
| pTA144 Up | Moraxella sp. TA144 | Antarctica | 1.9 | 40.6 | 1 | (Tutino et al., 2000)/AJ224743 |
| pOA307_63 | Octadecabacter antarcticus 307 | Antarctica, McMurdo Sound | 62.9 | 52.8 | 60 | (Vollmers et al., 2013)/CP003741 |
| pOA238_160 | Octadecabacter arcticus 238 | USA, 150 km offshore, Alaska | 159.7 | 54.4 | 157 | (Vollmers et al., 2013)/CP003744 |
| pOA238_118 | O. arcticus 238 | USA, 150 km offshore, Alaska | 118.3 | 51.5 | 126 | (Vollmers et al., 2013)/CP003743 |
| pMWHK1 | Pedobacter cryoconitis BG5 | Antarctica | 6.2 | 34.8 | 8 | (Wong et al., 2013)/FJ613505 |
| pPBPR1 | Photobacterium profundum SS9 | deap sea | 80.0 | 44.0 | 67 | (Vezzi et al., 2005)/CR377818 |
| pMtBL | Pseudoalteromonas haloplanktis TAC 125 | Antarctica | 4.1 | 39.3 | 2* | (Tutino et al., 2001)/AJ224742 |
| pKW1 | Pseudoalteromonas sp. 643A | Antarctica, krill Euphasia superba | 4.6 | 43.2 | 7 | (Cieslinski et al., 2008)/EU636993 |
| pSM327 | Pseudoalteromonas sp. BSi20327 | Arctic, sea ice | 6.1 | 37.5 | 4 | –/GU198194 |
| pSM429 | Pseudoalteromonas sp. Bsi429 | Arctic, sea ice | 3.9 | 28.4 | 4 | (Zhao et al., 2011)/EU627679 |
| pPS1M3 | Pseudoalteromonas sp. PS1M3 | deap see | 3.1 | 37.1 | 1 | (Kurusu et al., 2001)/AB022096 |
| pGLE121P1 | Pseudomonas sp. GLE121 | Antarctica, glacier ice | 6.9 | 49.7 | 9 | (Dziewit et al., 2013b)/KC542381 |
| pGLE121P2 | Pseudomonas sp. GLE121 | Antarctica, glacier ice | 8.3 | 53.7 | 11 | (Dziewit et al., 2013b)/KC542382 |
| pGLE121P3 | Pseudomonas sp. GLE121 | Antarctica, glacier ice | 39.6 | 52.2 | 44 | (Dziewit et al., 2013b)/KC542383 |
| plasmid KOPRI126573 | Pseudomonas sp. MC1 | Antarctica, waste water treatment plant | 81.8 | 55.8 | 83 | –/JN248563 |
| plasmid 1 | Psychrobacter cryohalolentis K5 | Siberia, permafrost | 41.2 | 38.3 | 44 | –/CP000324 |
| pKLH80 | Psychrobacter maritimus MR29-12 | Siberia, permafrost | 14.8 | 40.3 | 19 | (Petrova et al., 2014)/HF953351 |
| pP109bwP1 | Psychrobacter sp. DAB_AL109bw | Arctic, little auks guano | 4.4 | 42.9 | 6 | (Dziewit et al., 2013a)/JQ245702 |
| pP12P1 | Psychrobacter sp. DAB_AL12 | Arctic, little auks guano | 2.9 | 35.7 | 2 | (Dziewit et al., 2013a)/JQ231228 |
| pP32BP1 | Psychrobacter sp. DAB_AL32B | Arctic, little auks guano | 4.6 | 42.7 | 6 | (Dziewit et al., 2013a)/JQ245699 |
| pP43BP1 | Psychrobacter sp. DAB_AL43B | Arctic, little auks guano | 4.4 | 37.2 | 6 | (Dziewit et al., 2013a)/JQ245700 |
| pP43BP2 | Psychrobacter sp. DAB_AL43B | Arctic, little auks guano | 5.4 | 37.3 | 6 | (Dziewit et al., 2013a)/JQ245701 |
| pP43BP3 | Psychrobacter sp. DAB_AL43B | Arctic, little auks guano | 5.0 | 39.2 | 7 | (Dziewit et al., 2013a)/JQ348845 |
| pP43BP4 | Psychrobacter sp. DAB_AL43B | Arctic, little auks guano | 6.5 | 41.3 | 9 | (Dziewit et al., 2013a)/JQ348844 |
| pP60P1 | Psychrobacter sp. DAB_AL60 | Arctic, little auks guano | 5.4 | 41.7 | 8 | (Dziewit et al., 2013a)/JQ245703 |
| pP60P2 | Psychrobacter sp. DAB_AL60 | Arctic, little auks guano | 14.9 | 37.8 | 13 | (Dziewit et al., 2013a)/JQ245704 |
| pP62BP1 | Psychrobacter sp. DAB_AL62B | Arctic, little auks guano | 34.5 | 36.5 | 33 | (Lasek et al., 2012)/JQ065022 |
| plasmid PsyG_3 | Psychrobacter sp. G | Antarctica, King George Island | 4.0 | 38.5 | 5 | (Che et al., 2013)/CP006268 |
| plasmid PsyG_4 | Psychrobacter sp. G | Antarctica, King George Island | 4.5 | 36.5 | 5 | (Che et al., 2013)/CP006267 |
| plasmid PsyG_26 | Psychrobacter sp. G | Antarctica, King George Island | 26.1 | 41.2 | 26 | (Che et al., 2013)/CP006266 |
| pRWF101 | Psychrobacter sp. PRwf-1 | Puerto Rico, fish Lutjanus vivanus | 14 | 38.3 | 15 | –/CP000714 |
| pRWF102 | Psychrobacter sp. PRwf-1 | Puerto Rico, fish Lutjanus vivanus | 2.1 | 40.4 | 1 | –/CP000715 |
| pRUNSL01 | Runella slithyformis DSM 19594 | USA, University Lake near Baton Rouge | 107.0 | 46.3 | 88 | (Copeland et al., 2012)/CP002860 |
| pRUNSL02 | R. slithyformis DSM 19594 | USA, University Lake near Baton Rouge | 93.5 | 41.4 | 91 | (Copeland et al., 2012)/CP002861 |
| pRUNSL03 | R. slithyformis DSM 19594 | USA, University Lake near Baton Rouge | 66.9 | 40.8 | 70 | (Copeland et al., 2012)/CP002862 |
| pRUNSL04 | R. slithyformis DSM 19594 | USA, University Lake near Baton Rouge | 44.8 | 43.2 | 46 | (Copeland et al., 2012)/CP002863 |
| pRUNSL05 | R. slithyformis DSM 19594 | USA, University Lake near Baton Rouge | 38.8 | 44.2 | 32 | (Copeland et al., 2012)/CP002864 |
| pSFKW33 | Shewanella sp. 33B | Poland, Baltic Sea | 8.0 | 38.3 | 8 | (Werbowy et al., 2009)/FJ626843 |
| pSinA | Sinorhizobium sp. M14 | Poland, Zloty Stok gold mine | 108.9 | 59.5 | 102 | (Drewniak et al., 2013)/JF809815 |
| plasmid F | Sphingopyxis alaskensis RB2256 | USA, sea water near Alaska | 28.5 | 60.4 | 30 | (Lauro et al., 2009)/CP000357 |
| pSP01 | Streptomyces sp. PAMC26508 | Antarctica, lichen Cladonia borealis | 104.0 | 68.5 | 104 | –/CP003991 |
| pSCD | Sulfuricella denitrificans skB26 | Japan, freshwater lake | 86.6 | 56.7 | 91 | (Watanabe et al., 2014)/AP013067 |
The data were obtained from the GenBank databases (NCBI).
*all ORFs were distinguished in the course of this study (plasmid sequence deposited at the NCBI database has not been annotated).
The majority of the identified plasmids (53; 80.3%) occur in Gram-negative bacteria and the most numerous group of the replicons (17; 25.8%) was found in strains of the genus Psychrobacter (Gammaproteobacteria) (Table 1). The plasmids range in size from 1.3 kb (pTA144 Dw of Moraxella sp. TA144) to 417 kb (pBWB401 of Bacillus weihenstephanensis KBAB4) (Table 1), and they contain between 0 (pTA144 Dw of Moraxella sp. TA144) and 332 (pGLAAG01 of Glaciecola sp. 4H-3-7+YE-5) open reading frames (ORFs). Many of the plasmids (30; 45.5%) are small, cryptic replicons not exceeding 10 kb in size (Figure 1A). The average GC content of the pPSYCH nucleotide sequences ranges from 27.4 to 68.5%. This GC value is <45% in 53 (80.3%) plasmids, and >55% in only 5 (7.6%) replicons (Figure 1B).
Figure 1.
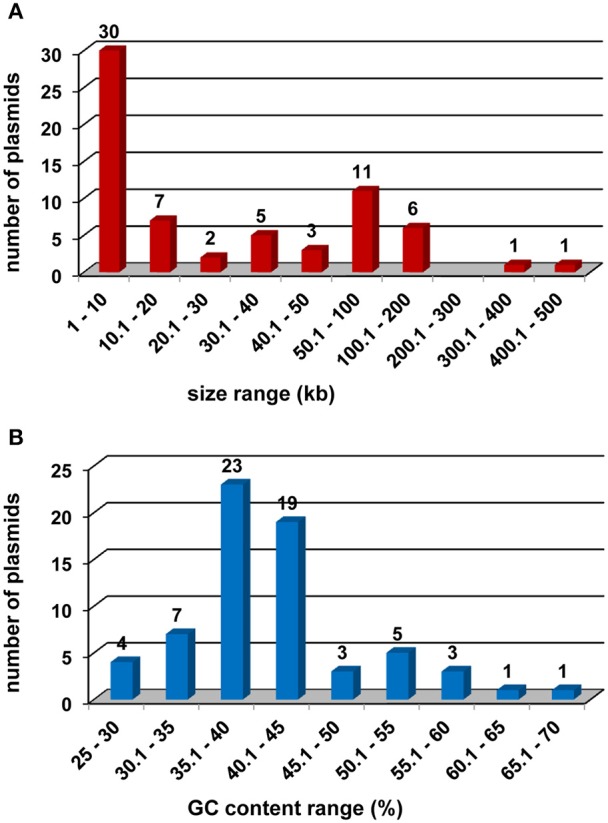
Size range (A) and GC content range (B) of plasmids of cold-active bacteria.
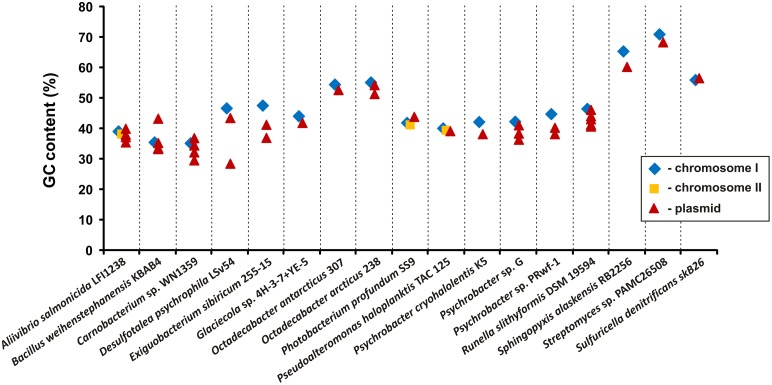
A more complex analysis was performed to compare the relative GC contents of the plasmids and the chromosomes of their hosts. For this analysis we used the completely sequenced genomic DNA of 17 bacterial strains, containing 37 pPSYCH plasmids. The analysis revealed that 32 (86.5%) of the plasmids have a lower GC content than their host's chromosome. The greatest difference (18.2%) was observed between “plasmid small” and the chromosomal DNA of Desulfotalea psychrophila LSv54 (Figure 2). This finding is in good agreement with the observation that bacteria usually fail to stably maintain horizontally-acquired DNAs whose GC content is considerably higher than that of the chromosome (Nishida, 2012).
Figure 2.
Comparison of the GC content of the nucleotide sequences of plasmids of cold-active bacteria and their hosts chromosomes.
In the next step of the pPSYCH plasmids analysis we performed the characterization of their conserved backbones, composed of replication and conjugal transfer modules. The replication machinery is a crucial component of each plasmid, since it is responsible for its maintenance in bacterial host cells (Thomas, 2000). Therefore, for the initial classification of pPSYCH plasmids we analyzed their replication modules.
The analysis revealed that 56 plasmids carry a single replication modules, while plasmid 1 of Psychrobacter cryohalolentis K5 and pBWB402 of B. weihenstephanensis KBAB4 are composite replicons and carry two replication systems. We were not able to distinguish the replication modules of 8 pPSYCH plasmids.
We found that 29 (43.9%) plasmids encode the RepB replication protein of the Rep3 superfamily, and the structure of their replication systems is typical for many theta-replicating plasmids (Chattoraj, 2000). The replication modules of the second most numerous group of plasmids (8; 12.1%) contain a single gene encoding a putative protein with three conserved regions: the replicase domain, an alpha helical domain of primases (PriCT-1) and a HTH motif. Moreover, their predicted oriV regions have the structure typical for the ColE2-type plasmids (Dziewit et al., 2013a).
Analysis of the remaining plasmids revealed that they encode replication proteins which were classified into the following families: (i) RepL (2 plasmids), (ii) RepZ (ColIB-P9) (3), (iii) RepC of repABC system (3), (iv) replication initiation protein containing predicted helix-turn-helix (HTH_36) domain (2), (v) Rep1 (rolling circle replication initiator protein) (1), (vi) RstA (phage-related replication proteins) (1), and (vii) II/X (phage/plasmid replication protein) (1). Moreover, 10 plasmids encode unclassified replication initiation proteins.
Many plasmids are capable of horizontal transfer by conjugation. According to their transfer machineries, they may be grouped into two categories comprising self-transmissible (conjugative) and mobilizable replicons (Garcillan-Barcia et al., 2009). However, it is worth mentioning, that the conjugal transfer phenotype of a particular plasmid depends not only on the genes it carries, but also the genomic background (i.e., other co-residing replicons) (Torres Tejerizo et al., 2014).
The conjugative plasmids encode a large set of proteins responsible for the processing of conjugative DNA (MOB) and mating pair formation (MPF), while the mobilizable plasmids carry only MOB regions, composed of an origin of transfer oriT and a gene encoding relaxase, which nicks DNA at the oriT sites. The transfer of mobilizable plasmids requires a membrane-associated mating pair formation complex, which may be provided by self-transmissible plasmids or integrative and conjugative elements (ICE) (Garcillan-Barcia et al., 2009; Smillie et al., 2010). ICE primarily reside in a host chromosome but they can excise and form plasmid-like circular molecules that are intermediates for conjugal transfer. In contrast to plasmids, ICEs do not contain their own replication systems (Wozniak and Waldor, 2010).
Our analysis revealed that 32 of the pPSYCH plasmids (48.5%) carry conjugal transfer modules. Of these, 12 (18.2%) are predicted to be conjugative plasmids (size range 38.4–121.6 kb) and 20 (30.3%), mobilizable replicons (sizes not exceeding 28.5 kb).
Based on a detailed comparative analysis of the relaxase amino acid sequences, we found that these proteins (and the MOB modules encoding them) can be classified within five different families: MOBP, MOBQ, MOBV, MOBC and MOBF. As shown in Figure 3, members of the MOBPfamily are the most numerous. It is also important to note that the relaxases of the MOBF family are exclusively present in large self-transmissible plasmids. Both observations are in a good agreement with the results of a complex survey of the sequences of plasmid-encoded MOB modules (Smillie et al., 2010).
Figure 3.
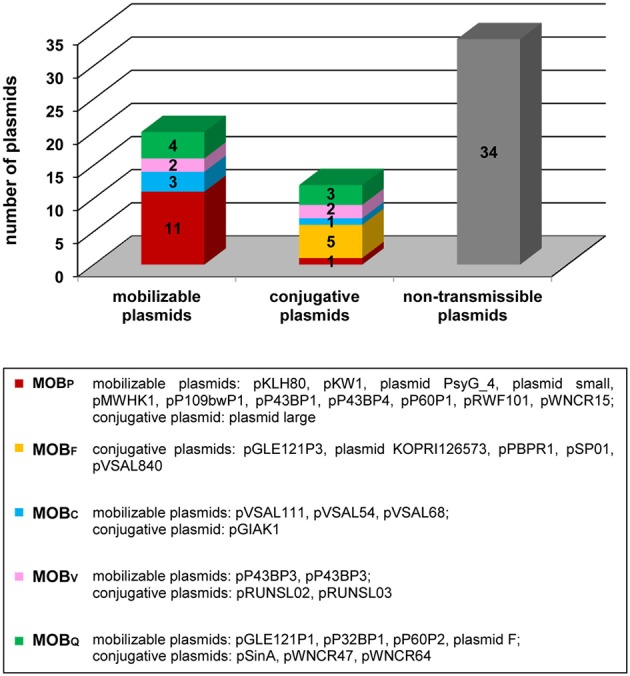
Distribution and classification of relaxases encoded within the pPSYCH plasmids.
The presence of TRA and/or MOB modules within plasmid genomes (including those of cold-active bacteria) may facilitate dissemination of these replicons among evolutionarily-distinct bacterial species. Our analyses have revealed that the conjugative and mobilizable pPSYCH plasmids carry numerous genes of adaptive value, and therefore their transfer may strongly influence adaptation of the host bacteria to the harsh environmental conditions they encounter.
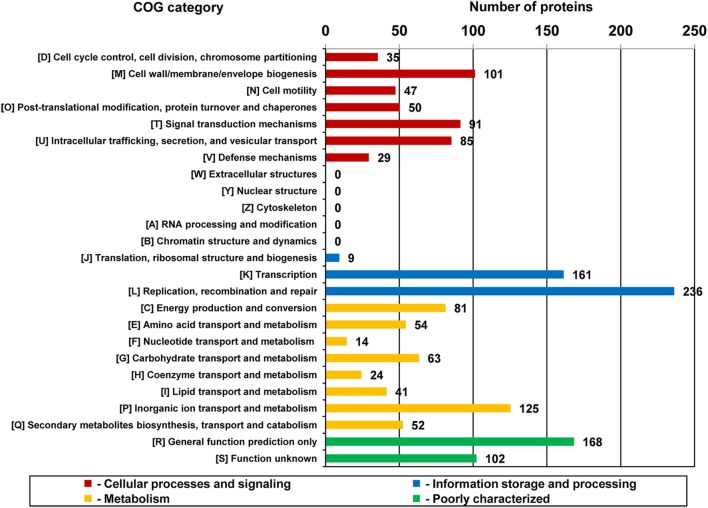
Plasmids are natural genetic vectors, which strongly influence bacterial evolution by disseminating genetic information determining various phenotypes (e.g., Harrison and Brockhurst, 2012). To investigate the adaptive role of pPSYCH plasmids, functional categorization of the proteins encoded by these replicons was performed. We were able to assign COG numbers to approximately 50% of the predicted proteins and calculate the proportion of the proteins in each COG category (Figure 4, Table S1).
Figure 4.
Number of proteins encoded within the plasmids of cold-active bacteria associated with the general COG functional categories.
The largest fraction of classified proteins (15.1%) was assigned to the L category by COG annotation (Figure 4). This group gathers proteins involved in replication, recombination and repair that may be directly linked with both plasmid maintenance and the mobility of transposable elements (TEs) carried by plasmids. Our finding is in agreement with the results of previous global plasmid relationships analyses, performed with the application of gene-sharing networks, which revealed that the majority of genes shared by plasmids encode proteins related to the process of HGT itself (DNA replication and transposition) (Tamminen et al., 2012).
A significant proportion of the analyzed proteins were assigned to the K COG category, gathering proteins associated with transcription (Figure 4). Proteins of this type are often involved in the stable maintenance of bacterial plasmids (e.g., components of toxin-antitoxin systems) and overall transcriptional regulation.
A total of 454 (29%) of the pPSYCH-encoded proteins (with assigned COG numbers) appear to be involved in the overall cellular metabolism. The most numerous of these are members of COG category P, gathering proteins involved in inorganic ion transport and metabolism, while others were assigned to functional categories C, G, E, F, H, I, and Q (Figure 4).
A similar number of proteins (438; 27.9%) were assigned to categories D, M, N, O, T, U, V, whose members are involved in various cellular processes and signaling, including cell wall/membrane/envelope biogenesis (category M—the most highly represented), cellular defense (V), but also cell motility (N) and secretion (U) (Figure 4).
These data strongly suggest that plasmids of psychrophilic and psychrotolerant bacteria (as plasmid mobilome of any particular ecological niche) may determine or modulate different metabolic properties of their hosts and influence the overall functioning of bacterial cells.
Cold protection
One of the most prominent responses of bacteria to low temperatures is the induction of cold shock protein (CSP) synthesis, which counteracts the harmful effects of the temperature downshift. The proteins comprising this diverse group include helicases, nucleases and ribosome-associated components that directly or indirectly interact with DNA and RNA. Several of these proteins are essential for survival in the cold, but none participates exclusively in cold adaptation (Weber and Marahiel, 2003).
A comparative BLASTp analysis of two manually curated databases was performed to identify cold shock proteins. One database represented a summarized proteome of all the plasmids analyzed in this study and the second included the sequences of 31 known cold shock proteins (Gualerzi et al., 2003; Barria et al., 2013). This comparative analysis revealed the presence of three types of predicted plasmid-encoded cold shock protein: (i) putative pyruvate dehydrogenases, (ii) global DNA-binding transcriptional regulators (H-NS), and (iii) nucleoid-associated proteins (HUβ).
The role of pyruvate dehydrogenase in cold adaptation is not well understood. It was proposed that this enzyme is involved in the intensification of glycolysis and suppression of the tricarboxylic acid cycle, which may lead to the retardation of cell growth, and adaptation to extreme conditions (Qiu et al., 2006). Pyruvate dehydrogenase (E1), together with two other enzymes, dihydrolipoamide acetyltransferase (E2) and dihydrolipoyl dehydrogenase (E3), forms the pyruvate dehydrogenase complex (PDC), which converts pyruvate into acetyl-CoA in a process called pyruvate decarboxylation (de Kok et al., 1998). The PDC components E1 and E2 (AceEF, DPPB38-40) were found to be encoded by “plasmid large” of D. psychrophila LSv54. Moreover, our analysis revealed that plasmid pOA238_160 of Octadecabacter arcticus 238 encodes a predicted protein, designated OA238_160p1370, with 71% amino-acid sequence identity to an E1 component-like protein of the Hyphomonas jannaschiana VP2 pyruvate dehydrogenase (AceE protein, EC 1.2.4.1). Interestingly, the gene encoding OA238_160p1370 (predicted AceE protein) is flanked within the pOA238_160 genome by insertion sequences, which suggests that it has been horizontally acquired.
Two other types of plasmid-encoded cold shock protein (H-NS and HUβ) most probably participate in the modulation of DNA supercoiling. It is well known that negative DNA supercoiling is increased when cells are exposed to cold shock. The reorganization of DNA structure may influence the pattern of gene expression and lead to remodeling of the metabolic state of the cell, which is crucial under stress conditions (Mizushima et al., 1997; Giangrossi et al., 2002). Detailed inspection of the plasmids' proposed proteomes revealed the presence of one H-NS family histone-like protein (OA238_118p0910 of pOA238_118) and two HUβ nucleoid-associated proteins (Glaag_4392 of pGLAAG01 and OA238_160p1030 of pOA238_160).
Ultraviolet (UV) radiation protection
Bacteria inhabiting polar regions have to cope not only with cold, but also with increased solar UV radiation due to stratospheric ozone depletion (Madronich, 1994). UV-B exposure may cause DNA damage and increased production of reactive oxygen species (ROS), which can further induce mutagenesis and ultimately lead to genomic instability (Santos et al., 2012). We have investigated whether plasmids of cold-active bacteria have the potential to provide UV protection via mechanisms such as ROS scavenging.
Two highly homologous genetic modules are involved in conferring tolerance to UV radiation—the rulAB and umuDC DNA repair operons. Both are components of the global SOS system and therefore their expression is regulated by the transcriptional repressor LexA (Sundin and Murillo, 1999; Kim and Sundin, 2000). In response to DNA damage, proteins encoded by the operons regulate DNA synthesis and function as a DNA polymerase that facilitates replication over any DNA lesions. Therefore, both modules play an important role in maintaining of genome stability and therefore they increase the evolutionary fitness of their hosts (Tark et al., 2005). It was also shown that elevated expression of the the umuDC gene products confer a cold-sensitive growth phenotype that correlates with a rapid inhibition of DNA synthesis (Marsh and Walker, 1985). A crucial role seems to play UmuC, since alterations in the protein sequence resulted in loss of the cold-sensitive phenotype (Beuning et al., 2006). Further studies are necessary to understand the molecular basis and the importance of the observed phenotypes.
The aforementioned operons were identified within three of the analyzed plasmids: (i) pGLE121P3 of Arctic Pseudomonas sp. strain GLE121 (rulAB), (ii) plasmid KOPRI126573 of Antarctic Pseudomonas sp. strain MC1 (pYIC1_09- pYIC1_10) and (iii) pSCD of Sulfuricella denitrificans skB26 (SCD_n03037-SCD_n03038), originating from Japan (Table 1).
Bacteria respond to increased ROS production by the expression of alkyl hydroperoxide reductase complexes, AhpCD or AhpCF, which serve as important antioxidants. The AhpC component, considered to be the major scavenger of ROS produced by aerobic metabolism (Steele et al., 2010), is encoded by plasmid pP60P2 of Arctic Psychrobacter sp. strain DAB_AL60. Acquisition of the ahpC-encoding plasmid may have increased the adaptation abilities of the host strain to the polar environment with enhanced UV radiation (Dziewit et al., 2013a).
Basic cellular metabolism
The COG categorization of the pPSYCH-encoded proteins revealed that nearly one third of those with assigned COG numbers are predicted to be involved in cellular metabolism, including amino acid (COG functional category E), nucleotide (F), carbohydrate (G) and lipid (I) transport and metabolism, as well as energy production and conversion (C) (Figure 5). The proteins (in number from 1 to 50, depending on the replicon) classified into these COG categories were encoded within 32 plasmids (Figure 5, Table S1).
Figure 5.
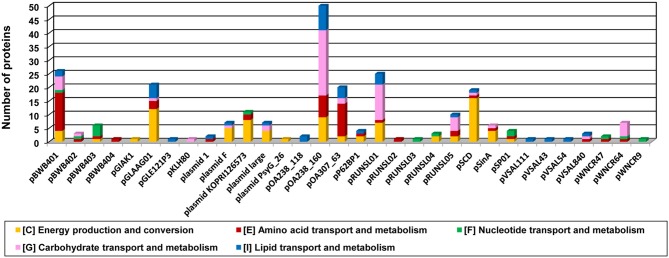
Distribution of plasmid-encoded proteins predicted to be involved in amino acid (COG functional category E), nucleotide (F), carbohydrate (G) and lipid (I) transport and metabolism, as well as energy production and conversion (C).
One of the pPSYCH plasmids, pOA238_160 of O. arcticus 238 (Table 1), is of particular interest because it encodes as many as 50 proteins, which may be directly linked with cellular metabolism (COG functional categories E, F, G, I or C). Almost 50% of these proteins appear to be involved in carbohydrate transport and metabolism. Analysis of the genetic content of this plasmid also revealed the presence of genes encoding (i) DctPQM periplasmic solute transporters of the C4-dicarboxylates malate, succinate, and fumarate (4 copies of each gene—OA238_160p0130, OA238_160p0140, OA238_160p0150; OA238_160p0940, OA238_160p0950, OA238_160p0960; OA238_160p1290, OA238_160p1300, OA238_160p1310; OA238_160p1460, OA238_160p1470, OA238_160p1480) (Forward et al., 1997); (ii) two enzymes of the citrate (TCA) cycle—aconitate hydratase (OA238_160p0470; EC 4.2.1.3), responsible for the bidirectional conversion of citrate into isocitrate, and succinate dehydrogenase (OA238_160p0460; EC 1.3.5.1), catalyzing the bidirectional transformation of succinate into fumarate; (iii) alpha-ketoglutarate-dependent taurine dioxygenase TauD (OA238_160p0180; EC 1.14.11.17), responsible for taurine utilization under sulfur starvation conditions, resulting in the release of sulfites (Eichhorn et al., 1997), and (iv) histidinol dehydrogenase HisD (OA238_160p0290; EC 1.1.1.23), catalyzing the final reaction in histidine biosynthesis (Thoma et al., 1998).
The most numerous proteins encoded by the pPSYCH plasmids are carbohydrate (COG category G) and amino-acid (COG category E) transporters and enzymes involved in the metabolism of these compounds. The G category is mainly represented by carbohydrate transporters, while the E category proteins include a broad spectrum of enzymes involved in the transport, biosynthesis and degradation of amino acids. COG functional category E representatives include, among others, four enzymes encoded within a single plasmid—pBWB401 of Bacillus weihenstephanensis KBAB4: (i) asparagine synthase (glutamine-hydrolysing) (BcerKBAB4_5684; EC 6.3.5.4), (ii) alanine dehydrogenase (BcerKBAB4_5593; EC 1.4.1.1), (iii) L-serine/L-threonine ammonia-lyase (BcerKBAB4_5570; EC 4.3.1.17) and (iv) homocysteine S-methyltransferase (BcerKBAB4_5466; EC 2.1.1.10) (Table S1).
These findings suggest that an efficient amino acid metabolism plays a very important role in cold-active bacteria. In nutrient-limiting cold environments, amino acids can be used as carbon and nitrogen sources, and therefore enhanced proteolytic capabilities may be beneficial to bacteria. This would explain the presence of plasmid-encoded enzymes involved in the degradation of amino acids.
Increased intracellular protein degradation has been observed in bacterial cells exposed to cold-shock (Jozefczuk et al., 2010). It seems obvious that the synthesis of induced cold shock proteins is more efficient when the availability of amino acids is increased. Therefore, the presence of additional, plasmid-encoded enzymes involved in amino acid synthesis may also be a beneficial feature.
A significant fraction of the pPSYCH-encoded proteins were identified as enzymes responsible for energy production and conversion (COG functional category C). When analyzing the distribution of genes encoding proteins of this group, we identified three plasmids carrying gene clusters encoding Ni, Fe-hydrogenases: (i) pGLAAG01 of Glaciecola sp. 4H-3-7+YE-5, (ii) pRUNSL01 of Runella slithyformis DSM 19594 and (iii) plasmid F of Sphingopyxis alaskensis RB2256 (Table 1). Such oxidative enzymes catalyze both the production and the consumption of hydrogen, and provide a redox mechanism by which hydrogen-metabolizing microorganisms can store and utilize energy (Fritsch et al., 2013). Since H2 oxidation may contribute significantly to the bacterial energy budget (Anantharaman et al., 2013), it is probable that the plasmid-encoded hydrogenases play an important role by providing an alternative and additional mechanism of energy production for bacteria inhabiting extreme environments.
Utilization of toxic organic compounds
Increasing anthropogenic activity in cold environments results in the growing contamination of these regions with various toxic organic compounds, including long-chain and polycyclic aromatic hydrocarbons (PAH) (e.g., Aislabie et al., 1999; Macdonald et al., 2005). However, some specialized bacteria are able to utilize such compounds as carbon, nitrogen and energy sources. Therefore, the acquisition of genetic information enabling the degradation of toxic organic compounds is highly beneficial, especially under nutrient limitation, which often occurs in extremely cold environments.
Detailed analysis of the genetic content of the pPSYCH plasmids identified one replicon, KOPRI126573 of Antarctic Pseudomonas sp. strain MC1, containing a complete set of genes responsible for the utilization of naphthalene. This self-transferable plasmid of the IncP-9 incompatibility group is almost identical (99% nucleotide sequence identity over its entire length) to the well characterized catabolic plasmid NAH7 of Pseudomonas putida G7 (Dunn and Gunsalus, 1973; Sota et al., 2006). A 27-kb region of plasmid KOPRI126573 (33% of the plasmid genome) is composed of two clusters of genes, encoding (i) the upper-pathway enzymes, involved in the conversion of naphthalene to salicylate and (ii) the lower-pathway enzymes, responsible for the conversion of salicylate to acetyl-CoA via meta-ring cleavage (Figure S1). Similar nah-like gene clusters have previously been identified in several other strains of Pseudomonas (Habe and Omori, 2003).
Resistance to heavy metals, metalloids and antibiotics
Environmental contamination with heavy metals and metalloids is a very common phenomenon. This is a consequence of both natural processes (e.g., occurrence of metal-containing minerals and their geobiochemical transformations, volcanic emissions, forest fires, deep-sea vents and geysers) and anthropogenic activities (e.g., large-scale burning of fossil fuels, mining, metal manufacturing, paint production and tanneries) (Adriano, 2001). Another emerging problem is the increase in antibiotics and other antimicrobial agents, originating from the medical (including veterinary) and agricultural sectors, contaminating the natural environment (Hektoen et al., 1995; Macdonald et al., 2005; Cantas et al., 2013). To combat the toxicity of these compounds bacteria develop or, more often, acquire (via HGT) various genetic modules conferring resistance. This phenomenon is also observed in extremely cold environments (e.g., Moller et al., 2014).
We investigated the potential of pPSYCH replicons for the determination of resistance phenotypes and identified 10 (15.2%) plasmids (Table 2) containing modules conferring heavy metal/metalloid resistance (7 replicons) or antibiotic resistance (2 replicons). One plasmid contained both types of module (Table 2). Interestingly, 8 of the plasmids carried two or more different resistance modules and may be defined as multi-resistance plasmids.
Table 2.
The pPSYCH plasmids containing heavy metal, metalloid and antibiotic resistance determinants.
| Plasmid name | Heavy metal or metalloid resistance [locus_tag or gene name] | Antibiotic resistance [locus_tag or gene name] |
|---|---|---|
| pGIAK1 | ArsDRKRBC—arsenite and arsenate resistance [pGIAK1_24-29] | N/A |
| CadDCA—cadmium resistance [pGIAK1_32-34; pGIAK1_39] | ||
| pGLAAG01 | MerRTPCA—mercury resistance [Glaag_4409-4413] | N/A |
| CnrA—nickel and cobalt resistance [Glaag_4419; Glaag_4433; Glaag_4453; Glaag_4522] | ||
| CzcD—cobalt, zinc and cadmium resistance [Glaag_4423; Glaag_4425; Glaag_4457; Glaag_4459] | ||
| CopABCD—copper resistance [Glaag_4445-4448; Glaag_4478-4479; Glaag_4518-4519] | ||
| Copper exporting ATPase (ZntA family)—copper resistance [Glaag_4490] | ||
| pKLH80 | N/A | Carbenicillin-hydrolysing betta-lactamase-class A penicillinase—beta-lactams resistance [blaRTG-6] |
| Streptomycin phosphotransferase—streptomycin resistance [strAB] | ||
| Tetracycline efflux protein of class H—tetracycline resistance [tetH] | ||
| pRUNSL02 | Copper exporting ATPase (ZntA family)—copper resistance [Runsl_5918; Runsl_5924] | N/A |
| Copper-(or silver)-translocating P-type ATPase—copper (or silver) resistance [Runsl_5929] | ||
| pSCD | CzcD—cobalt, zinc and cadmium resistance [SCD_n03083] | N/A |
| Arsenite efflux pump (ACR3 family)—arsenite resistance [SCD_n03106] | ||
| pSinA | ArsRCBH—arsenite and arsenate resistance [arsRCBH] | N/A |
| CzcD—cobalt, zinc and cadmium resistance [czcD] | ||
| MerRTPA—mercury resistance [merRTPA] | ||
| pSP01 | N/A | Virginiamycin B hydrolase—virginiamycin resistance [F750_7148] |
| pWNCR12 | CadD—cadmium resistance [Q783_11480] | N/A |
| pWNCR15 | Heavy metal-(Cd/Co/Hg/Pb/Zn)-translocating P-type | Chloramphenicol acetyltransferase—chloramphenicol resistance [Q783_11585] |
| ATPase—cadmium, cobalt, mercury, lead and zinc rsistance [Q783_11505] | ||
| CzcD—cobalt, zinc and cadmium resistance [Q783_11515] | ||
| pWNCR64 | Copper exporting ATPase (ZntA family)—copper resistance [Q783_11905] | N/A |
| Copper-(or silver)-translocating P-type ATPase—copper (or silver) resistance [Q783_11920] |
Only four of the identified resistance plasmids have been thoroughly analyzed previously. The ability to confer resistance to various heavy metals or metalloids was demonstrated for pGIAK1, originating from the obligate alkaliphilic and halotolerant Bacillaceae strain JMAK1, pSinA of arsenite oxidizing Sinorhizobium sp. M14 and pSCD of sulfur oxidizer S. denitrificans skB26 (Drewniak et al., 2013; Guo and Mahillon, 2013; Watanabe et al., 2014); while pKLH80 of Psychrobacter maritimus MR29-12, a strain isolated from ancient permafrost, is a novel multidrug-resistant plasmid, carrying genes determining streptomycin, tetracycline and β-lactam resistance (Petrova et al., 2014) (Table 2).
The nucleotide sequences of the remaining pPSYCH resistance plasmids (pGLAAG01, pRUNSL02, pSP01, pWNCR12, pWNCR15, and pWNCR64) were determined in the course of several whole genome sequencing projects and these plasmids have not previously been described. We found that plasmid pGLAAGO1 of Glaciecola sp. 4H-3-7+YE-5 carries as many as 13 predicted heavy metal resistance modules, conferring resistance to cadmium, cobalt, copper, mercury and zinc. The only replicon that appears to determine resistance to both heavy metals and an antibiotic (chloramphenicol) is pWNCR15 of C. gilichinskyi WN1359T (Table 2). Interestingly, the strain WN1359T also carries two other plasmids, pWNCR12 and pWNCR64, containing heavy metal resistance modules (Table 2). The remaining two pPSYCH plasmids, pRUNSL02 (R. slithyformis DSM 19594) and pSP01 (Streptomyces sp. PAMC26508), may confer resistance to copper and virginiamycin, respectively (Table 2).
We investigated whether the identified resistance modules are associated with transposable elements and we found that the merRTPCA gene cluster of pGLAAG01 is most probably a component of a Tn3-family transposon. Moreover, it was previously shown that insertion sequence ISPpy1 of pKLH80 can mobilize for transposition streptomycin (strAB), tetracycline (tetH) and β-lactam (blaRTG−6) resistance modules (Petrova et al., 2009, 2014). The presence of the resistance modules within transposable elements is of a great importance, since these elements contribute highly in dissemination of the resistance determinants among diverse plasmids co-residing in a single bacterial cell.
Motility
Many bacteria exhibit motility and most of them move by the use of flagella. Motility is defined as the ability to move actively, employing ion motive forces to power flagellar rotation. This is an adaptive trait that is advantageous to bacteria. Motile bacteria have the capacity to move toward favorable niches and avoid detrimental conditions, which allows them to succeed in competition with other groups of microorganisms (Duan et al., 2013; Guttenplan and Kearns, 2013).
Upon analyzing the distribution of genes encoding proteins of different COG categories within the pPSYCH genomes we found that 33 (70.2%) of the proteins classified in the N (cell motility) category are encoded by plasmid pOA238_118 of O. arcticus 238. This plasmid carries three clusters of genes that are possibly involved in the formation of flagella. However, the host strain of pOA238_118, as well as other strains of the genus Octadecabacter, are non-motile, so the role of the identified gene clusters remains unknown (Vollmers et al., 2013).
A second mechanism enabling “motility” of bacterial cells may be determined by another plasmid (pOA238_160) of O. arcticus 238. This replicon carries a gvp gene cluster encoding proteins that mediate the formation of gas vesicles (Vollmers et al., 2013). The presence of gas vesicles in polar heterotrophic bacteria may constitute an important selective advantage, since they provide buoyancy which regulates bacterial position in vertically stratified water columns and therefore functions as a dispersal mechanism (Gosink and Staley, 1995).
Protection against exogenous DNA (restriction-modification and CRISPR-Cas systems)
Bacteria have developed several mechanisms to protect their cells against invading DNA of bacteriophages. The most common are restriction-modification (R-M) and CRISPR-Cas (clustered regularly interspaced short palindromic repeats) systems. In general, the R-M systems are composed of two sequence-specific DNA recognition enzymes—a restriction endonuclease and a methyltransferase. The presence of R-M modules allows horizontally-acquired foreign DNA (non-methylated and so not protected against restriction) to be discriminated from the fully methylated DNA of the host. CRISPR-Cas systems recognize and inactivate exogenous genetic elements by crRNA-guided Cas proteins (Tock and Dryden, 2005; van Der Oost et al., 2014).
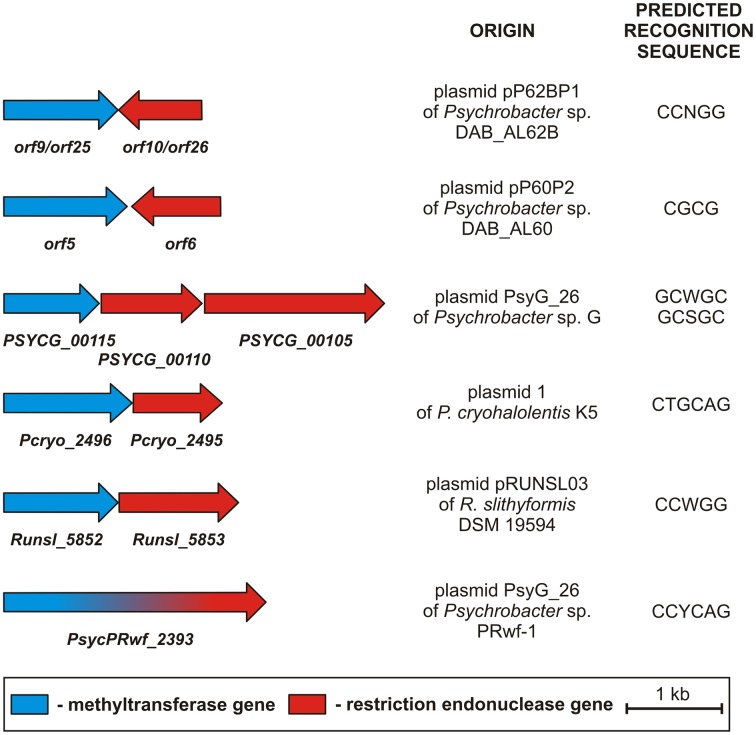
Both systems may play a dual role in bacteria inhabiting extreme environments. Not only do they protect their host strains against infecting bacteriophages, they could also help to eliminate any other invading selfish DNAs (including plasmids), which may consume valuable cellular energy and substrates for their own maintenance. Within the analyzed pPSYCH plasmids we found 7 putative type II R-M modules. The majority of these (6) were present within five plasmids occurring in bacteria of the genus Psychrobacter: (i) pP62BP1 (carries two, nearly identical R-M modules), (ii) pP60P2, (iii) plasmid 1, (iv) plasmid PsyG_26 and (v) pRWF101 (Table 1). The remaining type II R-M system was identified within plasmid pRUNSL03 of R. slithyformis DSM 19594 (Figure 6).
Figure 6.
Genetic organization and the predicted recognition sequences of type II R-M systems carried by pPSYCH plasmids.
Six of the identified R-M systems encode DNA m5C methyltransferases and have been classified into subtype IIP, while one R-M module (of plasmid pRWF101), encoding only one polypeptide with cleavage and modification domains, belongs to the subtype IIG (Pingoud et al., 2005). The likely recognition sequences of the restriction endonucleases and methyltransferases, predicted on the basis of detailed comparative sequence analyses performed at the REBASE database, are shown in Figure 6.
It is highly probable that the identified R-M systems, besides protecting the cell against foreign DNA, may also function as a type of post-segregational cell killing system that increases the stability of the plasmids in the bacterial population. Such activity has been previously demonstrated for several plasmid-encoded R-M modules (e.g., Dziewit et al., 2011).
The CRISPR-Cas systems mediating immunity to foreign DNA sequences that are integrated as spacers between repeats in the CRISPR locus, are widely distributed among bacterial genomes. Our analysis revealed that only one pPSYCH plasmid (pSP01 of Streptomyces sp. PAMC26508; Table 1) carries such a system, classified as type I-E (Makarova et al., 2011). The identified locus is composed of 8 genes encoding Cas/Cse proteins and the upstream region containing 22 short spacer sequences (21 of 32 bp in length and 1 of 33 bp) (Figure 7). BLASTn comparative analysis revealed that two of the spacers show sequence similarity to genomic sequences of Caulobacter and Mycobacterium phages and one spacer shows similarity to plasmid 1 of Gemmatimonadetes bacterium KBS708 (Figure 7).
Figure 7.
Genetic organization (A) and nucleotide sequences of the spacers (B) of the CRISPR-Cas locus of Streptomyces sp. plasmid pSP01.
Conclusions
This study represents the first comprehensive survey of plasmids (pPSYCH) maintained by psychrophilic and psychrotolerant bacteria. Our analysis revealed the presence of 66 such replicons occurring in 39 cold-active bacteria isolated from different geographical regions. The pPSYCH plasmids differ greatly in their genetic content. A thorough comparative analysis and the classification of the plasmid-encoded proteins into appropriate COG functional categories has identified several adaptive mechanisms that may benefit bacteria inhabiting cold environments.
We found that the plasmids of cold-active bacteria encode different types of predicted cold shock proteins as well as DNA-repair proteins enabling the cells to cope with enhanced UV radiation. Moreover, the plasmids encode numerous proteins possibly involved in energy production (including hydrogenases), transport of carbohydrates and amino acids, and amino acid synthesis and degradation, which may represent a form of adaptation to nutrient deprivation. They also contain genes conferring resistance to phage infection, heavy metals and metalloids, antibiotics, and (in one case) genes encoding catabolic enzymes involved in naphthalene utilization.
The vast majority of the aforementioned phenotypic traits are encoded within self-transmissible or mobilizable plasmids, which enable their dissemination by HGT. Thus, the pPSYCH plasmids may play an important role in the adaptation of psychrophilic and psychrotolerant bacteria to low temperatures and to rapid environmental changes.
Author contributions
Lukasz Dziewit conceived, designed and performed the analyses, and wrote the first draft of the manuscript. Dariusz Bartosik contributed to the critical revision of the draft manuscript. Both authors read and approved the final manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the National Science Centre, Poland (grant 2013/09/D/NZ8/03046).
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00596/abstract
References
- Adriano D. C. (2001). Trace Elements in Terrestrial Environments: Biogeochemistry, Bioavailability, and Risks of Metals. New York, NY: Springer. [Google Scholar]
- Aislabie J., Balks M., Astori N., Stevenson G., Symons R. (1999). Polycyclic aromatic hydrocarbons in fuel-oil contaminated soils, Antarctica. Chemosphere 39, 2201–2207. 10.1016/S0045-6535(99)00144-7 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez B., Secades P., Mcbride M. J., Guijarro J. A. (2004). Development of genetic techniques for the psychrotrophic fish pathogen Flavobacterium psychrophilum. Appl. Environ. Microbiol. 70, 581–587. 10.1128/AEM.70.1.581-587.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman K., Breier J. A., Sheik C. S., Dick G. J. (2013). Evidence for hydrogen oxidation and metabolic plasticity in widespread deep-sea sulfur-oxidizing bacteria. Proc. Natl. Acad. Sci. U.S.A. 110, 330–335. 10.1073/pnas.1215340110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiuchi M., Zakaria M. M., Sakaguchi Y., Yagi T. (1999). Sequence analysis of a cryptic plasmid from Flavobacterium sp. KP1, a psychrophilic bacterium. FEMS Microbiol. Lett. 170, 243–249. 10.1111/j.1574-6968.1999.tb13380.x [DOI] [PubMed] [Google Scholar]
- Barria C., Malecki M., Arraiano C. M. (2013). Bacterial adaptation to cold. Microbiology 159, 2437–2443. 10.1099/mic.0.052209-0 [DOI] [PubMed] [Google Scholar]
- Beuning P. J., Sawicka D., Barsky D., Walker G. C. (2006). Two processivity clamp interactions differentially alter the dual activities of UmuC. Mol. Microbiol. 59, 460–474. 10.1111/j.1365-2958.2005.04959.x [DOI] [PubMed] [Google Scholar]
- Cantas L., Shah S. Q., Cavaco L. M., Manaia C. M., Walsh F., Popowska M., et al. (2013). A brief multi-disciplinary review on antimicrobial resistance in medicine and its linkage to the global environmental microbiota. Front. Microbiol. 4:96. 10.3389/fmicb.2013.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T., Berriman M., Tivey A., Patel C., Bohme U., Barrell B. G., et al. (2008). Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics 24, 2672–2676. 10.1093/bioinformatics/btn529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanueva A., Tuffin M., Cary C., Cowan D. A. (2010). Molecular adaptations to psychrophily: the impact of ‘omic’ technologies. Trends Microbiol. 18, 374–381. 10.1016/j.tim.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Caspi R., Altman T., Billington R., Dreher K., Foerster H., Fulcher C. A., et al. (2014). The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 42, D459–D471. 10.1093/nar/gkt1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj D. K. (2000). Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol. 37, 467–476. 10.1046/j.1365-2958.2000.01986.x [DOI] [PubMed] [Google Scholar]
- Che S., Song L., Song W., Yang M., Liu G., Lin X. (2013). Complete genome sequence of Antarctic bacterium Psychrobacter sp. strain G. Genome Announc. 1, e00725–e00713. 10.1128/genomeA.00725-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieslinski H., Werbowy K., Kur J., Turkiewicz M. (2008). Molecular characterization of a cryptic plasmid from the psychrotrophic antarctic bacterium Pseudoalteromonas sp. 643A. Plasmid 60, 154–158. 10.1016/j.plasmid.2008.06.002 [DOI] [PubMed] [Google Scholar]
- Claudel-Renard C., Chevalet C., Faraut T., Kahn D. (2003). Enzyme-specific profiles for genome annotation: PRIAM. Nucleic Acids Res. 31, 6633–6639. 10.1093/nar/gkg847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland A., Zhang X., Misra M., Lapidus A., Nolan M., Lucas S., et al. (2012). Complete genome sequence of the aquatic bacterium Runella slithyformis type strain (LSU 4(T)). Stand. Genomic Sci. 6, 145–154. 10.4056/sigs.2475579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok A., Hengeveld A. F., Martin A., Westphal A. H. (1998). The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim. Biophys. Acta 1385, 353–366. 10.1016/S0167-4838(98)00079-X [DOI] [PubMed] [Google Scholar]
- Drewniak L., Dziewit L., Ciezkowska M., Gawor J., Gromadka R., Sklodowska A. (2013). Structural and functional genomics of plasmid pSinA of Sinorhizobium sp. M14 encoding genes for the arsenite oxidation and arsenic resistance. J. Biotechnol. 164, 479–488. 10.1016/j.jbiotec.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Duan Q., Zhou M., Zhu L., Zhu G. (2013). Flagella and bacterial pathogenicity. J. Basic Microbiol. 53, 1–8. 10.1002/jobm.201100335 [DOI] [PubMed] [Google Scholar]
- Dunn N. W., Gunsalus I. C. (1973). Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J. Bacteriol. 114, 974–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziewit L., Cegielski A., Romaniuk K., Uhrynowski W., Szych A., Niesiobedzki P., et al. (2013a). Plasmid diversity in arctic strains of Psychrobacter spp. Extremophiles 17, 433–444. 10.1007/s00792-013-0521-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziewit L., Grzesiak J., Ciok A., Nieckarz M., Zdanowski M. K., Bartosik D. (2013b). Sequence determination and analysis of three plasmids of Pseudomonas sp. GLE121, a psychrophile isolated from surface ice of Ecology Glacier (Antarctica). Plasmid 70, 254–262. 10.1016/j.plasmid.2013.05.007 [DOI] [PubMed] [Google Scholar]
- Dziewit L., Kuczkowska K., Adamczuk M., Radlinska M., Bartosik D. (2011). Functional characterization of the type II PamI restriction-modification system derived from plasmid pAMI7 of Paracoccus aminophilus JCM 7686. FEMS Microbiol. Lett. 324, 56–63. 10.1111/j.1574-6968.2011.02388.x [DOI] [PubMed] [Google Scholar]
- Eichhorn E., Van Der Ploeg J. R., Kertesz M. A., Leisinger T. (1997). Characterization of alpha-ketoglutarate-dependent taurine dioxygenase from Escherichia coli. J. Biol. Chem. 272, 23031–23036. 10.1074/jbc.272.37.23031 [DOI] [PubMed] [Google Scholar]
- Finn R. D., Bateman A., Clements J., Coggill P., Eberhardt R. Y., Eddy S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. 10.1093/nar/gkt1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forward J. A., Behrendt M. C., Wyborn N. R., Cross R., Kelly D. J. (1997). TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J. Bacteriol. 179, 5482–5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch J., Lenz O., Friedrich B. (2013). Structure, function and biosynthesis of O(2)-tolerant hydrogenases. Nat. Rev. Microbiol. 11, 106–114. 10.1038/nrmicro2940 [DOI] [PubMed] [Google Scholar]
- Garcillan-Barcia M. P., Francia M. V., De La Cruz F. (2009). The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33, 657–687. 10.1111/j.1574-6976.2009.00168.x [DOI] [PubMed] [Google Scholar]
- Giangrossi M., Giuliodori A. M., Gualerzi C. O., Pon C. L. (2002). Selective expression of the beta-subunit of nucleoid-associated protein HU during cold shock in Escherichia coli. Mol. Microbiol. 44, 205–216. 10.1046/j.1365-2958.2002.02868.x [DOI] [PubMed] [Google Scholar]
- Gosink J. J., Staley J. T. (1995). Biodiversity of gas vacuolate bacteria from Antarctic sea ice and water. Appl. Environ. Microbiol. 61, 3486–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I., Vergnaud G., Pourcel C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. 10.1093/nar/gkm360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualerzi C. O., Giuliodori A. M., Pon C. L. (2003). Transcriptional and post-transcriptional control of cold-shock genes. J. Mol. Biol. 331, 527–539. 10.1016/S0022-2836(03)00732-0 [DOI] [PubMed] [Google Scholar]
- Guo S., Mahillon J. (2013). pGIAK1, a heavy metal resistant plasmid from an obligate alkaliphilic and halotolerant bacterium isolated from the Antarctic Concordia station confined environment. PLoS ONE 8:e72461. 10.1371/journal.pone.0072461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenplan S. B., Kearns D. B. (2013). Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 37, 849–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habe H., Omori T. (2003). Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 67, 225–243. 10.1271/bbb.67.225 [DOI] [PubMed] [Google Scholar]
- Harrison E., Brockhurst M. A. (2012). Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 20, 262–267. 10.1016/j.tim.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Hektoen H., Berge J. A., Hormazabal V., Yndestad M. (1995). Persistence of antibacterial agents in marine sediments. Aquaculture 133, 175–184 10.1016/0044-8486(94)00310-K [DOI] [Google Scholar]
- Heuer H., Smalla K. (2012). Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol. Rev. 36, 1083–1104. 10.1111/j.1574-6976.2012.00337.x [DOI] [PubMed] [Google Scholar]
- Hjerde E., Lorentzen M. S., Holden M. T., Seeger K., Paulsen S., Bason N., et al. (2008). The genome sequence of the fish pathogen Aliivibrio salmonicida strain LFI1238 shows extensive evidence of gene decay. BMC Genomics 9:616. 10.1186/1471-2164-9-616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefczuk S., Klie S., Catchpole G., Szymanski J., Cuadros-Inostroza A., Steinhauser D., et al. (2010). Metabolomic and transcriptomic stress response of Escherichia coli. Mol. Syst. Biol. 6, 364. 10.1038/msb.2010.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. (2000). KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Kawashima M., Furumichi M., Tanabe M. (2014). Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205. 10.1093/nar/gkt1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J., Sundin G. W. (2000). Regulation of the rulAB mutagenic DNA repair operon of Pseudomonas syringae by UV-B (290 to 320 nanometers) radiation and analysis of rulAB-mediated mutability in vitro and in planta. J. Bacteriol. 182, 6137–6144. 10.1128/JB.182.21.6137-6144.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel B., Lochner A., Bruce D. C., Davenport K. W., Detter C., Goodwin L. A., et al. (2011). Complete genome sequence of the marine cellulose- and xylan-degrading bacterium Glaciecola sp. strain 4H-3-7+YE-5. J. Bacteriol. 193, 4547–4548. 10.1128/JB.05468-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobori H., Sullivan C. W., Shizuya H. (1984). Bacterial plasmids in antarctic natural microbial assemblages. Appl. Environ. Microbiol. 48, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurusu Y., Yoshimura S., Tanaka M., Nakamura T., Maruyama A., Higashihara T. (2001). Genetic transformation system for a psychrotrophic deep-sea bacterium: isolation and characterization of a psychrotrophic plasmid. Mar. Biotechnol. 3, 96–99. 10.1007/s101260000067 [DOI] [PubMed] [Google Scholar]
- Lasek R., Dziewit L., Bartosik D. (2012). Plasmid pP62BP1 isolated from an Arctic Psychrobacter sp. strain carries two highly homologous type II restriction-modification systems and a putative organic sulfate metabolism operon. Extremophiles 16, 363–376. 10.1007/s00792-012-0435-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauro F. M., Mcdougald D., Thomas T., Williams T. J., Egan S., Rice S., et al. (2009). The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U.S.A. 106, 15527–15533. 10.1073/pnas.0903507106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard M. T., Panayotova N., Farmerie W. G., Triplett E. W., Nicholson W. L. (2013). Complete genome sequence of Carnobacterium gilichinskyi strain WN1359T (DSM 27470T). Genome Announc. 1, e00985–e00913. 10.1128/genomeA.00985-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leplae R., Lima-Mendez G., Toussaint A. (2010). ACLAME: a CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 38, D57–D61. 10.1093/nar/gkp938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald R. W., Harner T., Fyfe J. (2005). Recent climate change in the Arctic and its impact on contaminant pathways and interpretation of temporal trend data. Sci. Total Environ. 342, 5–86. 10.1016/j.scitotenv.2004.12.059 [DOI] [PubMed] [Google Scholar]
- Madronich S. (1994). Increases in biologically damaging UV-B radiation due to stratospheric ozone depletion: a brief review. Arch. Hydrobiol. Beih. 43, 17–30. [Google Scholar]
- Makarova K. S., Haft D. H., Barrangou R., Brouns S. J., Charpentier E., Horvath P., et al. (2011). Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 9, 467–477. 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., Deweese-Scott C., et al. (2011). CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margesin R., Miteva V. (2011). Diversity and ecology of psychrophilic microorganisms. Res. Microbiol. 162, 346–361. 10.1016/j.resmic.2010.12.004 [DOI] [PubMed] [Google Scholar]
- Marsh L., Walker G. C. (1985). Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J. Bacteriol. 162, 155–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Rosales C., Fullana N., Musto H., Castro-Sowinski S. (2012). Antarctic DNA moving forward: genomic plasticity and biotechnological potential. FEMS Microbiol. Lett. 331, 1–9. 10.1111/j.1574-6968.2012.02531.x [DOI] [PubMed] [Google Scholar]
- Mizushima T., Kataoka K., Ogata Y., Inoue R., Sekimizu K. (1997). Increase in negative supercoiling of plasmid DNA in Escherichia coli exposed to cold shock. Mol. Microbiol. 23, 381–386. 10.1046/j.1365-2958.1997.2181582.x [DOI] [PubMed] [Google Scholar]
- Moller A. K., Barkay T., Hansen M. A., Norman A., Hansen L. H., Sorensen S. J., et al. (2014). Mercuric reductase genes (merA) and mercury resistance plasmids in High Arctic snow, freshwater and sea-ice brine. FEMS Microbiol. Ecol. 87, 52–63. 10.1111/1574-6941.12189 [DOI] [PubMed] [Google Scholar]
- Morita R. Y. (1975). Psychrophilic bacteria. Bacteriol. Rev. 39, 144–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A. C., Kanehisa M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, W182–W185. 10.1093/nar/gkm321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida H. (2012). Genome DNA sequence variation, evolution, and function in Bacteria and Archaea. Curr. Issues Mol. Biol. 15, 19–24. [PubMed] [Google Scholar]
- Petrova M., Gorlenko Z., Mindlin S. (2009). Molecular structure and translocation of a multiple antibiotic resistance region of a Psychrobacter psychrophilus permafrost strain. FEMS Microbiol. Lett. 296, 190–197. 10.1111/j.1574-6968.2009.01635.x [DOI] [PubMed] [Google Scholar]
- Petrova M., Kurakov A., Shcherbatova N., Mindlin S. (2014). Genetic structure and biological properties of the first ancient multiresistance plasmid pKLH80 isolated from a permafrost bacterium. Microbiology 160, 2253–2263. 10.1099/mic.0.079335-0 [DOI] [PubMed] [Google Scholar]
- Pingoud A., Fuxreiter M., Pingoud V., Wende W. (2005). Type II restriction endonucleases: structure and mechanism. Cell. Mol. Life Sci. 62, 685–707. 10.1007/s00018-004-4513-1 [DOI] [PubMed] [Google Scholar]
- Qiu Y., Kathariou S., Lubman D. M. (2006). Proteomic analysis of cold adaptation in a Siberian permafrost bacterium - Exiguobacterium sibiricum 255-15 by two-dimensional liquid separation coupled with mass spectrometry. Proteomics 6, 5221–5233. 10.1002/pmic.200600071 [DOI] [PubMed] [Google Scholar]
- Rabus R., Ruepp A., Frickey T., Rattei T., Fartmann B., Stark M., et al. (2004). The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ. Microbiol. 6, 887–902. 10.1111/j.1462-2920.2004.00665.x [DOI] [PubMed] [Google Scholar]
- Roberts R. J., Vincze T., Posfai J., Macelis D. (2010). REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 38, D234–D236. 10.1093/nar/gkp874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues D. F., Goris J., Vishnivetskaya T., Gilichinsky D., Thomashow M. F., Tiedje J. M. (2006). Characterization of Exiguobacterium isolates from the Siberian permafrost. Description of Exiguobacterium sibiricum sp. nov. Extremophiles 10, 285–294. 10.1007/s00792-005-0497-5 [DOI] [PubMed] [Google Scholar]
- Santos A. L., Gomes N. C., Henriques I., Almeida A., Correia A., Cunha A. (2012). Contribution of reactive oxygen species to UV-B-induced damage in bacteria. J. Photochem. Photobiol. B 117, 40–46. 10.1016/j.jphotobiol.2012.08.016 [DOI] [PubMed] [Google Scholar]
- Siefert J. L. (2009). Defining the mobilome. Methods Mol. Biol. 532, 13–27. 10.1007/978-1-60327-853-9_2 [DOI] [PubMed] [Google Scholar]
- Smillie C., Garcillan-Barcia M. P., Francia M. V., Rocha E. P., De La Cruz F. (2010). Mobility of plasmids. Microbiol. Mol. Biol. Rev. 74, 434–452. 10.1128/MMBR.00020-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sota M., Yano H., Ono A., Miyazaki R., Ishii H., Genka H., et al. (2006). Genomic and functional analysis of the IncP-9 naphthalene-catabolic plasmid NAH7 and its transposon Tn4655 suggests catabolic gene spread by a tyrosine recombinase. J. Bacteriol. 188, 4057–4067. 10.1128/JB.00185-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele K. H., Baumgartner J. E., Valderas M. W., Roop R. M., 2nd. (2010). Comparative study of the roles of AhpC and KatE as respiratory antioxidants in Brucella abortus 2308. J. Bacteriol. 192, 4912–4922. 10.1128/JB.00231-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin G. W., Murillo J. (1999). Functional analysis of the Pseudomonas syringae rulAB determinant in tolerance to ultraviolet B (290-320 nm) radiation and distribution of rulAB among P. syringae pathovars. Environ. Microbiol. 1, 75–87. 10.1046/j.1462-2920.1999.00008.x [DOI] [PubMed] [Google Scholar]
- Tamminen M., Virta M., Fani R., Fondi M. (2012). Large-scale analysis of plasmid relationships through gene-sharing networks. Mol. Biol. Evol. 29, 1225–1240. 10.1093/molbev/msr292 [DOI] [PubMed] [Google Scholar]
- Tark M., Tover A., Tarassova K., Tegova R., Kivi G., Horak R., et al. (2005). A DNA polymerase V homologue encoded by TOL plasmid pWW0 confers evolutionary fitness on Pseudomonas putida under conditions of environmental stress. J. Bacteriol. 187, 5203–5213. 10.1128/JB.187.15.5203-5213.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma R., Schwander M., Liebl W., Kirschner K., Sterner R. (1998). A histidine gene cluster of the hyperthermophile Thermotoga maritima: sequence analysis and evolutionary significance. Extremophiles 2, 379–389. 10.1007/s007920050082 [DOI] [PubMed] [Google Scholar]
- Thomas C. M. (2000). Paradigms of plasmid organization. Mol. Microbiol. 37, 485–491. 10.1046/j.1365-2958.2000.02006.x [DOI] [PubMed] [Google Scholar]
- Tock M. R., Dryden D. T. (2005). The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 8, 466–472. 10.1016/j.mib.2005.06.003 [DOI] [PubMed] [Google Scholar]
- Torres Tejerizo G., Pistorio M., Althabegoiti M. J., Cervantes L., Wibberg D., Schluter A., et al. (2014). Rhizobial plasmid pLPU83a is able to switch between different transfer machineries depending on its genomic background. FEMS Microbiol. Ecol. 88, 565–578. 10.1111/1574-6941.12325 [DOI] [PubMed] [Google Scholar]
- Tutino M. L., Duilio A., Moretti M. A., Sannia G., Marino G. (2000). A rolling-circle plasmid from Psychrobacter sp. TA144: evidence for a novel rep subfamily. Biochem. Biophys. Res. Commun. 274, 488–495. 10.1006/bbrc.2000.3148 [DOI] [PubMed] [Google Scholar]
- Tutino M. L., Duilio A., Parrilli R., Remaut E., Sannia G., Marino G. (2001). A novel replication element from an Antarctic plasmid as a tool for the expression of proteins at low temperature. Extremophiles 5, 257–264. 10.1007/s007920100203 [DOI] [PubMed] [Google Scholar]
- van Der Oost J., Westra E. R., Jackson R. N., Wiedenheft B. (2014). Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat. Rev. Microbiol. 12, 479–492. 10.1038/nrmicro3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vezzi A., Campanaro S., D'angelo M., Simonato F., Vitulo N., Lauro F. M., et al. (2005). Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307, 1459–1461. 10.1126/science.1103341 [DOI] [PubMed] [Google Scholar]
- Vollmers J., Voget S., Dietrich S., Gollnow K., Smits M., Meyer K., et al. (2013). Poles apart: Arctic and Antarctic Octadecabacter strains share high genome plasticity and a new type of xanthorhodopsin. PLoS ONE 8:e63422. 10.1371/journal.pone.0063422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Kojima H., Fukui M. (2014). Complete genomes of freshwater sulfur oxidizers Sulfuricella denitrificans skB26 and Sulfuritalea hydrogenivorans sk43H: genetic insights into the sulfur oxidation pathway of betaproteobacteria. Syst. Appl. Microbiol. 37, 387–395. 10.1016/j.syapm.2014.05.010 [DOI] [PubMed] [Google Scholar]
- Weber M. H., Marahiel M. A. (2003). Bacterial cold shock responses. Sci. Prog. 86, 9–75. 10.3184/003685003783238707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werbowy K., Cieslinski H., Kur J. (2009). Characterization of a cryptic plasmid pSFKW33 from Shewanella sp. 33B. Plasmid 62, 44–49. 10.1016/j.plasmid.2009.03.003 [DOI] [PubMed] [Google Scholar]
- Wiedenbeck J., Cohan F. M. (2011). Origins of bacterial diversity through horizontal genetic transfer and adaptation to new ecological niches. FEMS Microbiol. Rev. 35, 957–976. 10.1111/j.1574-6976.2011.00292.x [DOI] [PubMed] [Google Scholar]
- Wong C. M., Tam H. K., Ng W. M., Boo S. Y., Gonzalez M. (2013). Characterisation of a cryptic plasmid from an Antarctic bacterium Pedobacter cryoconitis strain BG5. Plasmid 69, 186–193. 10.1016/j.plasmid.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Wozniak R. A., Waldor M. K. (2010). Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8, 552–563. 10.1038/nrmicro2382 [DOI] [PubMed] [Google Scholar]
- Zhao D. L., Yu Z. C., Li P. Y., Wu Z. Y., Chen X. L., Shi M., et al. (2011). Characterization of a cryptic plasmid pSM429 and its application for heterologous expression in psychrophilic Pseudoalteromonas. Microb. Cell. Fact. 10, 30. 10.1186/1475-2859-10-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.