Abstract
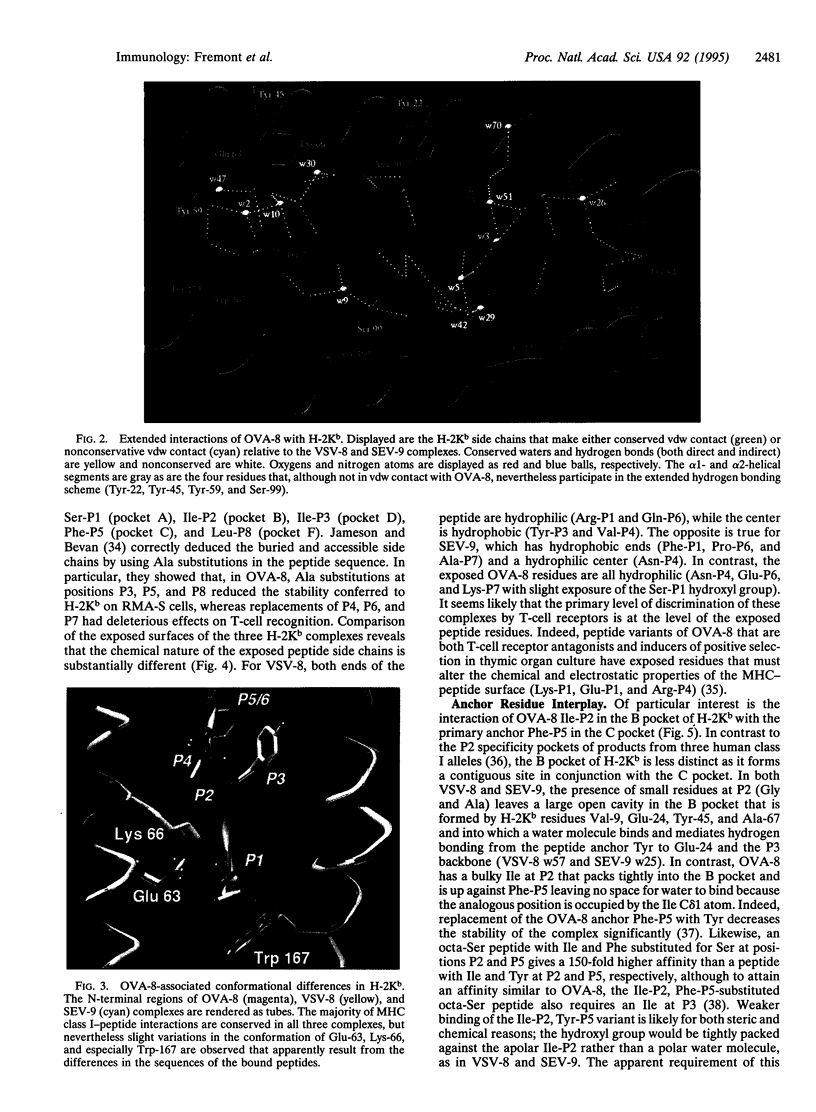
Sequence analysis of peptides naturally presented by major histocompatibility complex (MHC) class I molecules has revealed allele-specific motifs in which the peptide length and the residues observed at certain positions are restricted. Nevertheless, peptides containing the standard motif often fail to bind with high affinity or form physiologically stable complexes. Here we present the crystal structure of a well-characterized antigenic peptide from ovalbumin [OVA-8, ovalbumin-(257-264), SIINFEKL] in complex with the murine MHC class I H-2Kb molecule at 2.5-A resolution. Hydrophobic peptide residues Ile-P2 and Phe-P5 are packed closely together into binding pockets B and C, suggesting that the interplay of peptide anchor (P5) and secondary anchor (P2) residues can couple the preferred sequences at these positions. Comparison with the crystal structures of H-2Kb in complex with peptides VSV-8 (RGYVYQGL) and SEV-9 (FAPGNYPAL), where a Tyr residue is used as the C pocket anchor, reveals that the conserved water molecule that binds into the B pocket and mediates hydrogen bonding from the buried anchor hydroxyl group could not be likewise positioned if the P2 side chain were of significant size. Based on this structural evidence, H-2Kb has at least two submotifs: one with Tyr at P5 (or P6 for nonamer peptides) and a small residue at P2 (i.e., Ala or Gly) and another with Phe at P5 and a medium-sized hydrophobic residue at P2 (i.e., Ile). Deciphering of these secondary submotifs from both crystallographic and immunological studies of MHC peptide binding should increase the accuracy of T-cell epitope prediction.
Full text
PDF

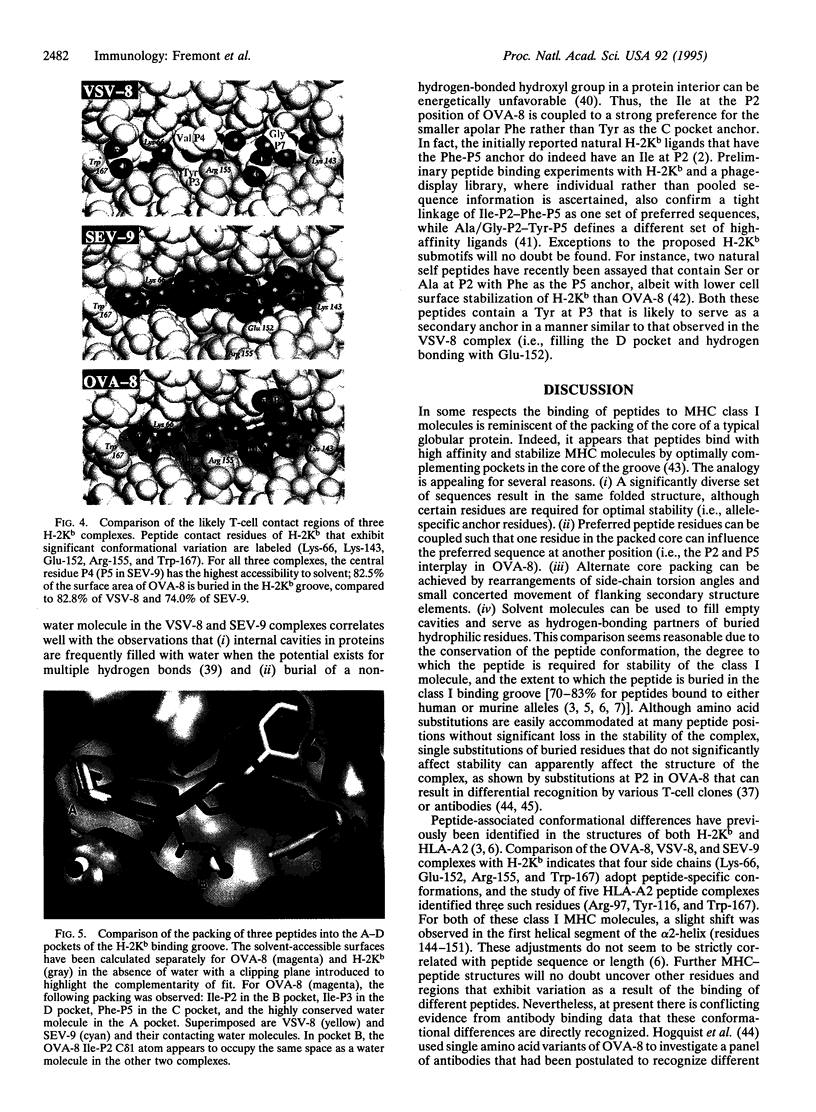

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blaber M., Lindstrom J. D., Gassner N., Xu J., Heinz D. W., Matthews B. W. Energetic cost and structural consequences of burying a hydroxyl group within the core of a protein determined from Ala-->Ser and Val-->Thr substitutions in T4 lysozyme. Biochemistry. 1993 Oct 26;32(42):11363–11373. doi: 10.1021/bi00093a013. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Carbone F. R., Bevan M. J. Induction of ovalbumin-specific cytotoxic T cells by in vivo peptide immunization. J Exp Med. 1989 Mar 1;169(3):603–612. doi: 10.1084/jem.169.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., McCluskey J., Rodda S., Carbone F. R. Changes at peptide residues buried in the major histocompatibility complex (MHC) class I binding cleft influence T cell recognition: a possible role for indirect conformational alterations in the MHC class I or bound peptide in determining T cell recognition. J Exp Med. 1993 Mar 1;177(3):869–873. doi: 10.1084/jem.177.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H. G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991 May 23;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- Fremont D. H., Matsumura M., Stura E. A., Peterson P. A., Wilson I. A. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 1992 Aug 14;257(5072):919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- Garrett T. P., Saper M. A., Bjorkman P. J., Strominger J. L., Wiley D. C. Specificity pockets for the side chains of peptide antigens in HLA-Aw68. Nature. 1989 Dec 7;342(6250):692–696. doi: 10.1038/342692a0. [DOI] [PubMed] [Google Scholar]
- Geluk A., van Meijgaarden K. E., Southwood S., Oseroff C., Drijfhout J. W., de Vries R. R., Ottenhoff T. H., Sette A. HLA-DR3 molecules can bind peptides carrying two alternative specific submotifs. J Immunol. 1994 Jun 15;152(12):5742–5748. [PubMed] [Google Scholar]
- Germain R. N., Margulies D. H. The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol. 1993;11:403–450. doi: 10.1146/annurev.iy.11.040193.002155. [DOI] [PubMed] [Google Scholar]
- Guo H. C., Jardetzky T. S., Garrett T. P., Lane W. S., Strominger J. L., Wiley D. C. Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature. 1992 Nov 26;360(6402):364–366. doi: 10.1038/360364a0. [DOI] [PubMed] [Google Scholar]
- Guo H. C., Madden D. R., Silver M. L., Jardetzky T. S., Gorga J. C., Strominger J. L., Wiley D. C. Comparison of the P2 specificity pocket in three human histocompatibility antigens: HLA-A*6801, HLA-A*0201, and HLA-B*2705. Proc Natl Acad Sci U S A. 1993 Sep 1;90(17):8053–8057. doi: 10.1073/pnas.90.17.8053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogquist K. A., Grandea A. G., 3rd, Bevan M. J. Peptide variants reveal how antibodies recognize major histocompatibility complex class I. Eur J Immunol. 1993 Nov;23(11):3028–3036. doi: 10.1002/eji.1830231145. [DOI] [PubMed] [Google Scholar]
- Hogquist K. A., Jameson S. C., Bevan M. J. The ligand for positive selection of T lymphocytes in the thymus. Curr Opin Immunol. 1994 Apr;6(2):273–278. doi: 10.1016/0952-7915(94)90101-5. [DOI] [PubMed] [Google Scholar]
- Hogquist K. A., Jameson S. C., Heath W. R., Howard J. L., Bevan M. J., Carbone F. R. T cell receptor antagonist peptides induce positive selection. Cell. 1994 Jan 14;76(1):17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Huczko E. L., Bodnar W. M., Benjamin D., Sakaguchi K., Zhu N. Z., Shabanowitz J., Henderson R. A., Appella E., Hunt D. F., Engelhard V. H. Characteristics of endogenous peptides eluted from the class I MHC molecule HLA-B7 determined by mass spectrometry and computer modeling. J Immunol. 1993 Sep 1;151(5):2572–2587. [PubMed] [Google Scholar]
- Jackson M. R., Song E. S., Yang Y., Peterson P. A. Empty and peptide-containing conformers of class I major histocompatibility complex molecules expressed in Drosophila melanogaster cells. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12117–12121. doi: 10.1073/pnas.89.24.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson S. C., Bevan M. J. Dissection of major histocompatibility complex (MHC) and T cell receptor contact residues in a Kb-restricted ovalbumin peptide and an assessment of the predictive power of MHC-binding motifs. Eur J Immunol. 1992 Oct;22(10):2663–2667. doi: 10.1002/eji.1830221028. [DOI] [PubMed] [Google Scholar]
- Kast W. M., Brandt R. M., Sidney J., Drijfhout J. W., Kubo R. T., Grey H. M., Melief C. J., Sette A. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J Immunol. 1994 Apr 15;152(8):3904–3912. [PubMed] [Google Scholar]
- Madden D. R., Garboczi D. N., Wiley D. C. The antigenic identity of peptide-MHC complexes: a comparison of the conformations of five viral peptides presented by HLA-A2. Cell. 1993 Nov 19;75(4):693–708. doi: 10.1016/0092-8674(93)90490-h. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Fremont D. H., Peterson P. A., Wilson I. A. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 1992 Aug 14;257(5072):927–934. doi: 10.1126/science.1323878. [DOI] [PubMed] [Google Scholar]
- Matsumura M., Saito Y., Jackson M. R., Song E. S., Peterson P. A. In vitro peptide binding to soluble empty class I major histocompatibility complex molecules isolated from transfected Drosophila melanogaster cells. J Biol Chem. 1992 Nov 25;267(33):23589–23595. [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. J Mol Biol. 1968 Apr 28;33(2):491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- McDonald I. K., Thornton J. M. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994 May 20;238(5):777–793. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- Rammensee H. G., Falk K., Rötzschke O. Peptides naturally presented by MHC class I molecules. Annu Rev Immunol. 1993;11:213–244. doi: 10.1146/annurev.iy.11.040193.001241. [DOI] [PubMed] [Google Scholar]
- Rashin A. A., Iofin M., Honig B. Internal cavities and buried waters in globular proteins. Biochemistry. 1986 Jun 17;25(12):3619–3625. doi: 10.1021/bi00360a021. [DOI] [PubMed] [Google Scholar]
- Rohren E. M., McCormick D. J., Pease L. R. Peptide-induced conformational changes in class I molecules. Direct detection by flow cytometry. J Immunol. 1994 Jun 1;152(11):5337–5343. [PubMed] [Google Scholar]
- Ruppert J., Sidney J., Celis E., Kubo R. T., Grey H. M., Sette A. Prominent role of secondary anchor residues in peptide binding to HLA-A2.1 molecules. Cell. 1993 Sep 10;74(5):929–937. doi: 10.1016/0092-8674(93)90472-3. [DOI] [PubMed] [Google Scholar]
- Saito Y., Peterson P. A., Matsumura M. Quantitation of peptide anchor residue contributions to class I major histocompatibility complex molecule binding. J Biol Chem. 1993 Oct 5;268(28):21309–21317. [PubMed] [Google Scholar]
- Sheriff S., Hendrickson W. A., Smith J. L. Structure of myohemerythrin in the azidomet state at 1.7/1.3 A resolution. J Mol Biol. 1987 Sep 20;197(2):273–296. doi: 10.1016/0022-2836(87)90124-0. [DOI] [PubMed] [Google Scholar]
- Silver M. L., Guo H. C., Strominger J. L., Wiley D. C. Atomic structure of a human MHC molecule presenting an influenza virus peptide. Nature. 1992 Nov 26;360(6402):367–369. doi: 10.1038/360367a0. [DOI] [PubMed] [Google Scholar]
- Stura E. A., Matsumura M., Fremont D. H., Saito Y., Peterson P. A., Wilson I. A. Crystallization of murine major histocompatibility complex class I H-2Kb with single peptides. J Mol Biol. 1992 Dec 5;228(3):975–982. doi: 10.1016/0022-2836(92)90881-j. [DOI] [PubMed] [Google Scholar]
- Young A. C., Zhang W., Sacchettini J. C., Nathenson S. G. The three-dimensional structure of H-2Db at 2.4 A resolution: implications for antigen-determinant selection. Cell. 1994 Jan 14;76(1):39–50. doi: 10.1016/0092-8674(94)90171-6. [DOI] [PubMed] [Google Scholar]
- Zhang W., Young A. C., Imarai M., Nathenson S. G., Sacchettini J. C. Crystal structure of the major histocompatibility complex class I H-2Kb molecule containing a single viral peptide: implications for peptide binding and T-cell receptor recognition. Proc Natl Acad Sci U S A. 1992 Sep 1;89(17):8403–8407. doi: 10.1073/pnas.89.17.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]