The incidence of esophageal adenocarcinoma (EAC) has been increasing steadily over the past few decades,1 despite widespread recognition of the problem and a vast body of research. This disease is believed to originate in the broadest sense from Barrett’s Esophagus (BE), the recognized precursor of EAC, but current methods for surveillance have not been found to be effective in defining patients at risk. While a number of molecular and cellular mechanisms that underlie the transformation of BE to EAC have been outlined, their utility in understanding and preventing the progression of this cancer or in managing the intervention-resistant clones has not been demonstrated in a number of proposed models. As the rate of progression of BE to EAC is very low, estimated at <0.5% per year,2–4 individual investigators rarely have access to a sufficient number of biospecimens or clinical data to rigorously validate their models in longitudinal studies. A multi-institutional approach that combines novel scientific resources is envisioned to be essential for elucidating early biological events that drive EAC formation, understanding the role of the precursor lesion BE including its cell of origin, and devising unique preventive methods.
The need for meaningful collaborations for conducting multi-disciplinary and multi-institutional studies was identified by the Barrett’s Esophagus Working Group in 2001, the Stomach/Esophageal Cancers Progress Review Group in 2002 and then further crystallized by the Barrett’s Esophagus Translational Research Working Group Meeting convened by the National Cancer Institute (NCI) in 2008. It became clear from these and other efforts that in order to address the challenge of EAC, understand the intricacies of the Barrett’s neoplasia, and facilitate the development of more translational strategies for early detection and prevention, it was imperative to create a trans-disciplinary team with key scientific expertise to “jump start” progress in this field. To that end, NCI formed the Barrett’s Esophagus Translational Research Network (BETRNet) in 2011, a program that is jointly funded through a cooperative agreement by the Division of Cancer Biology and the Division of Cancer Prevention (http://prevention.cancer.gov/programs-resources/programs/betrnet and https://dcb.nci.nih.gov/Pages/Programs.aspx).
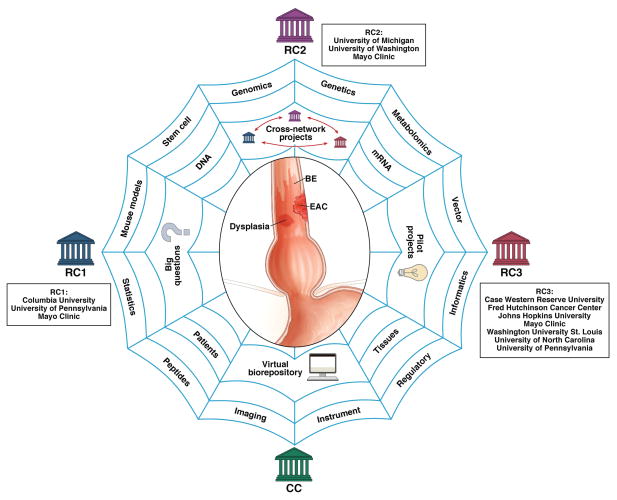
BETRNet encompasses three Research Centers, each representing a network of networks, and a Coordinating Center (Fig. 1). The translational Research Centers are headquartered at Columbia University, University of Michigan and Case Western Reserve University, and Coordinating Center is located at Vanderbilt University. The Research Centers are primarily focused on discovering biological pathways that trigger the development of BE and EAC, using state-of-the-art experimental models and human specimens for translation into early cancer detection, risk stratification and prevention strategies. The Coordinating Center functions to integrate cross-network activities with emphasis on a network-wide virtual biorepository. Through collective leveraging of novel animal and cellular models, genomics, proteomics and imaging as well as access to biospecimens, BETRNet is employing the concept of true team-based science to answer some of the “big” and most puzzling questions in EAC research.
Fig. 1. BETRNet.
This network of three Research Centers (RCs) and one Coordinating Center (CC) combines scientific expertise, patient access, and core facilities to accelerate the discovery of biological events underling EAC and to translate them into clinical practice for improved detection and prevention.
Research and Coordinating Centers
Perhaps the two most salient questions being asked by this network are 1) who is most likely to progress to cancer, and 2) what are the critical molecular and cellular mechanisms of pathogenesis of BE and EAC. The inclusion of multiple Research Centers will allow BETRNet to capture enough progression events to answer these questions in the setting of the low rate of progression from BE to EAC. Similarly, other uncommon, related conditions that hold great research interest because of their ability to yield novel insights into EAC pathogenesis, including the origin of buried BE, the source of stem cells for neosquamous epithelium (islands), and the risk for esophageal squamous cancers after BE ablation are being addressed through the network collaboration. With the established infrastructure, molecular analysis, biomarker exploration, and clinical research studies can be efficiently combined. In each of these studies, the network is more likely to accomplish robust molecular and clinical investigations in a timely manner than a single site.
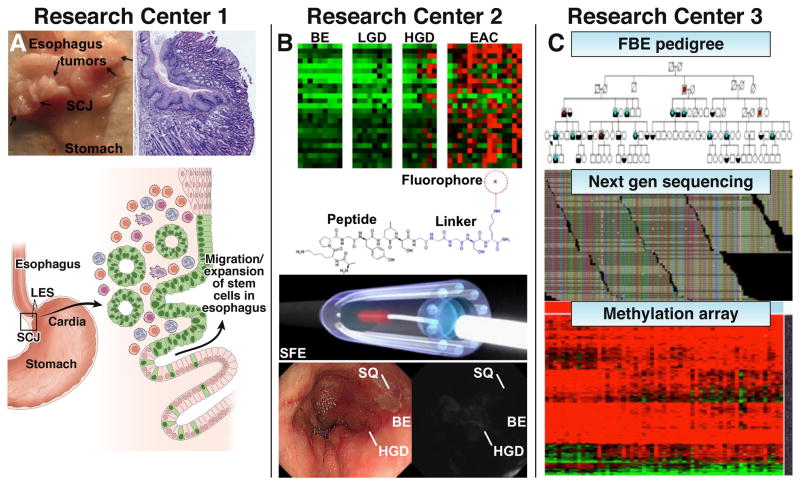
As for individual research centers, Research Center 1 (Columbia University, University of Pennsylvania, Mayo Clinic) aims to elucidate the cell of origin of BE in mouse and cell culture models by defining the role of active stem and progenitor cells normally present at the gastroesophageal junction, Fig. 2A–C. In particular, RC1 is exploring the role of stem cells in the gastric cardia, as well as squamous cells in the esophagus, as potential origins for BE and EAC. Both animal models and in vitro (3D or organotypic) culture systems are being applied to define the cell-of-origin and the biologic processes involved. Genetic mouse models, such as the L2-IL-1β transgenic mouse, provide evidence that the cell of origin for BE may reside in the proximal stomach, likely corresponding to the gastric cardia. This center also aims to determine whether early targeting of cardia progenitor cells may represent an effective chemoprevention strategy in human patients.
Fig. 2. Scientific focus of Research Centers.
A) Research Center 1 is studying the possible origins of BE and EAC using the pL2-IL-1β mouse model which develops multiple tumors at the squamocolumnar junction (SCJ) at 12 months. Photo and histology of pL2-IL-1β mouse with esophagitis and a tumor at the SJC at 15 months are shown. Schematic illustration of migrating stem cells (green, Lgr5+) at the cardia that expand into the inflamed esophagus. B) Research Center 2 is developing a molecular imaging approach by identifying targets that are gene amplified and/or highly overexpressed. Highly specific peptides are selected and fluorescently-labeled for in vivo imaging with a multi-spectral scanning fiber endoscope that can detect multiple targets simultaneously. This “red-flag” approach will help physicians to identify HGD in BE. C) Research Center 3 is investigating susceptibility genes in Familial Barrett’s Esophagus (FBE) and the role of methylated genes in BE and EAC. Pedigree shows affected subjects with BE (blue) and EAC (red). Alignment of reads from next generation sequencing run. Heat map from methylation arrays and non-coding and coding RNA signatures in BE and EAC.
Research Center 2 (University of Michigan, University of Washington, Mayo Clinic) is developing novel molecular imaging methodologies to visualize cell surface targets that are either gene amplified and/or highly overexpressed in EAC relative to BE. These targets are used to develop highly specific peptides that are fluorescently-labeled for real time endoscopic imaging to rapidly assess esophageal mucosa for neoplasia.5 This approach has the potential to detect pre-malignant lesions that are flat and would otherwise go undetected, Fig. 2D–H. Multiple targets are imaged at the same time to address disease heterogeneity over a broad patient population. A novel multi-spectral scanning fiber endoscope (SFE) that is small enough to pass through the instrument channel of a medical endoscope is used to collect the fluorescence images. This “red-flag” approach is provided to guide physicians by prioritizing tissue biopsy and thereby reducing overdiagnosis and distinguishing lethal cancers from non-lethal disease.
Research Center 3 (Case Western Reserve University, Fred Hutchinson Cancer Center, Johns Hopkins University, Mayo Clinic, Washington University at St. Louis, University of North Carolina, University of Pennsylvania) aims to improve the understanding of genetic susceptibility, genomic alterations, and epigenetic changes in BE and EAC (Fig. 3I–K). This Center is identifying susceptibility genes using whole exome sequencing of genomic DNA from affected distant relative pairs and cancer DNA from familial BE pedigrees. The data are used to develop models for predicting the presence of BE in family members. Genome wide epigenetic changes associated with the development of BE and its progression to EAC are also being defined. Aberrantly methylated CpG sites are being translated into noninvasive DNA based methods for detecting BE, dysplasia, and early EAC. Furthermore, coding and non-coding RNA signatures associated with an increased risk of progression are being identified by deep sequencing the transcriptome and microRNA fraction isolated from tissues.
The Coordinating Center (Vanderbilt-Ingram Cancer Center) has developed a web-based infrastructure for a virtual biorepository with patient registry to integrate and standardize clinical data from the Research Centers, including information on the availability of high quality and ethically acquired specimens to enable the rapid sharing of samples and data across the network for use in the spectrum of projects. This Center also facilitates interactions among the network by providing logistical and administrative support for cross-network communications and establishing special focus working groups.
Virtual Biorepository
The virtual biorepository (https://betrnet.mc.vanderbilt.edu) with patient registry catalogs and tracks esophageal biospecimens that can be made available for sharing across the network. Key annotations, including demographics, clinical outcomes, and pathology, are standardized to accelerate searches for relevant cases. Currently, data on ~2000 specimens have been included in this repository. A sophisticated query interface allows users to access the virtual biorepository and identify disease cohorts for pilot or cross-network projects. After a cohort of sufficient sample size is identified, investigators can request specimens by submitting an application along with proposed case inclusion for review by the utilization committee. With project approval, investigators then use the on-line interface to request cohorts for fulfillment of sample distribution requests. Thus, the virtual biorepository provides full functionality to support specimen sharing across BETRNet to accelerate the validations of biological discoveries. Access is currently available through collaborations with the BETRNet research centers.
Pilot and Cross-Network Projects
BETRNet provides support for pilot and cross-network projects to capitalize on network resources. These projects allow for exploring new ideas, developing emerging opportunities, and recruiting outside expertise to contribute to the main projects. The short-term studies could develop into larger research projects as they mature and achieve independent funding. The questions asked and resources dedicated to these programs have led to a number of innovative studies, including the contribution of genetics and ethnic factors to the risk for BE and EAC, the effects of biological aging on the risk for EAC and the factors that mediate recurrence of BE after radio-frequency ablation. These studies arose out of interactive network-wide face-to-face meetings and teleconferences that demonstrate the potential of BETRNet to ask the “big” questions.
Opportunity to Answer “Big” Questions
The BETRNet environment provides a unique opportunity to ask and answer some of the “big” questions about the pathogenesis of EAC. As examples:
Do factors such as the microenvironment, obesity, acid and bile reflux explain the rising incidence of BE and EAC and how can this knowledge be translated to improve the approach to this disease?
Can the marked gender and ethnicity differences in BE and EAC be addressed and used with other demographic (or environmental) risk factors to develop novel screening strategies that identify the majority of patients with metaplasia who are at risk for progressing to EAC?
What are the genetic, epigenetic, and proteomic differences between Barrett’s metaplasia, dysplasia, and adenocarcinoma and can these differences be translated into prognostic biomarkers and/or in vivo imaging modalities that effectively detect BE patients who will progress to high grade dysplasia and early cancer as well as detect BE patients who will not progress?
Can the molecular understanding of EAC carcinogenesis be exploited to develop novel chemopreventive agents such as Notch inhibitors and demonstrate the efficacy of potential drugs such as statins and non-steroidals in clinical trials?
Can the biology of ablative therapy be outlined and translated into effective and permanent reduction of cancer risk in BE utilizing uniformly rigorous definitions of ablation-refractory high-grade dysplasia and post-ablation recurrent BE?
Because the burden of BE extends beyond U.S. boundaries, BETRNet has initiated dialog with international investigators to provide a global perspective. By pursuing worldwide collaborations, we can learn much from nationalized healthcare systems, such as in Europe, on how to track patient outcomes over time and about geographical and sociodemographic differences that influence the natural history of this disease. Ultimately, the success of BETRNet will be measured by its ability to meet the challenge of advancing the knowledge of EAC biology and reducing its mortality. With a focus on BE, it’s cell of origins, and risk for progression into cancer, the collective knowledge gained from this network effort must be translated into more effective methods for early detection and prevention of EAC. Genetic and epigenetic profiling may enable improved risk stratification as well as identification of patient-specific therapeutic targets to reduce risk. Molecular imaging technologies that visualize overexpressed cell surface targets that herald the onset of EAC, independent of histology, can improve surveillance and hence early detection. For those patients requiring endoscopic therapy, the identification of prognostic markers as well as the development of adjunctive therapies could enhance the durability of treatment response. Using the multi-pronged approach that simultaneously encompasses biological models, imaging, biomarkers, and clinical outcomes, BETRNet can facilitate new discoveries and approaches to fully understand some of the persistent questions regarding BE and its progression to EAC. Furthermore, by leveraging the resources of the newly established biorepository, network investigators can quickly carry out large scale validation studies based on initial findings. While long-term success will require continued coordinated efforts, BETRNet represents a distinct paradigm in the research enterprise that is now properly equipped to face highly complex and challenging clinical problems such as EAC.
Acknowledgments
Funding: U54 CA163004 (TCW), U54 CA163059 (TDW), U54 CA163060 (AC), U01 CA163056 (YS), U01CA152756 (WMG), U01CA182940 (WMG)
We thank M Quante (Technical University of Munich) for his thoughtful review and images, and JJ Bergman (Academic Medical Centre, Amsterdam), NA Berger, K Guda, JS Barnholtz-Sloan, JE Willis (Case Western Reserve University), HF Andrews, CJ Lightdale, R Friedman, TA Gonda, Z Jin, JM Poneros, A Sepulveda (Columbia University), AM Kaz, G Luebeck (Fred Hutchinson Cancer Research Center), MI Canto (John Hopkins University), NSButtar, PGIyer (Mayo Clinic), JM Taylor, RDKuick (University of Michigan), and JS Wang (Washington University at St. Louis) for technical support.
Abbreviations
- BE
Barrett’s Esophagus
- BETRNet
Barrett’s Esophagus Translational Research Network
- CC
Coordinating Center
- EAC
esophageal adenocarcinoma
- FBE
Familial Barrett’s Esophagus
- HGD
high grade dysplasia
- LGD
low grade dysplasia
- NCI
National Cancer Institute
- RC
Research Center
- SCJ
squamocolumnar junction
- SFE
scanning fiber endoscope
Footnotes
Conflicts of Interest: The authors report no conflicts of interest
References
- 1.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–6. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 2.Desai TK, Krishnan K, Samala N, Singh J, Cluley J, Perla S, Howden CW. The incidence of oesophageal adenocarcinoma in non-dysplastic Barrett’s oesophagus: a meta-analysis. Gut. 2012;61:970–6. doi: 10.1136/gutjnl-2011-300730. [DOI] [PubMed] [Google Scholar]
- 3.Bhat S, Coleman HG, Yousef F, Johnston BT, McManus DT, Gavin AT, Murray LJ. Risk of malignant progression in Barrett’s esophagus patients: results from a large population-based study. J Natl Cancer Inst. 2011;103:1049–57. doi: 10.1093/jnci/djr203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hvid-Jensen F, Pedersen L, Drewes AM, Sørensen HT, Funch-Jensen P. Incidence of adenocarcinoma among patients with Barrett’s esophagus. N Engl J Med. 2011;365:1375–83. doi: 10.1056/NEJMoa1103042. [DOI] [PubMed] [Google Scholar]
- 5.Sturm MB, Joshi BP, Lu S, Piraka C, Khondee S, Elmunzer BJ, Kwon RS, Beer DG, Appelman HD, Turgeon DK, Wang TD. Targeted endoscopic imaging of Barrett’s neoplasia with specific fluorescent-labeled peptide: first in-human results. Science Translational Medicine. 2013;5:184ra61. doi: 10.1126/scitranslmed.3004733. [DOI] [PMC free article] [PubMed] [Google Scholar]