SUMMARY
DNA methylation in promoters is well known to silence genes and is the presumed therapeutic target of methylation inhibitors. Gene body methylation is positively correlated with expression yet its function is unknown. We show that 5-aza-2'-deoxycytidine treatment not only reactivates genes but decreases the over-expression of genes, many of which are involved in metabolic processes regulated by c-MYC. Down-regulation is caused by DNA demethylation of the gene bodies and restoration of high levels of expression requires remethylation by DNMT3B. Gene body methylation may therefore be an unexpected therapeutic target for DNA methylation inhibitors, resulting in the normalization of gene over-expression induced during carcinogenesis. Our results provide direct evidence for a causal relationship between gene body methylation and transcription.
Keywords: DNA methylation, DNA methylation inhibitor, gene body, demethylation, remethylation, DNMT3B
INTRODUCTION
Gene silencing mediated by aberrant promoter DNA hypermethylation is one of the key features of cancer (Baylin and Jones, 2011). Although somatically heritable, the reversibility of DNA methylation by pharmacological interventions makes it an attractive therapeutic target. Over the past few years, various DNA methyltransferase inhibitors have been developed with the goal of reactivating aberrantly silenced genes and many of them have shown encouraging results in both preclinical and clinical settings, highlighting the potential of epigenetic therapy(Azad et al., 2013; Balch and Nephew, 2013; Yamazaki and Issa, 2013). Two DNA methylation inhibitors, 5-Aza-2- deoxycytidine (5-Aza-CdR) and 5-Azacytidine (5-Aza-CR), were discovered to have hypomethylating activities at low concentrations (Jones and Taylor, 1980; Taylor and Jones, 1979). Both drugs are cytosine analogs that are incorporated into replicating DNA in place of cytosine and trap DNMTs, resulting in proteosomal degradation and heritable global demethylation as cells divide (Kelly et al., 2010a). They have been approved by the Food and Drug Administration (FDA) in the US for the treatment of myelodysplastic syndrome (MDS), a pre-leukemic disorder (Kantarjian et al., 2006; Silverman et al., 2002). More recently, clinical trials have been initiated to investigate the possibility of extending their utilization to solid tumors, such as ovarian and lung cancers, either alone or in combination with other (Azad et al., 2013; Juergens et al., 2011; Li et al., 2014; Matei et al., 2012; Wrangle et al., 2013). Transient exposure to low doses of DNA demethylating agents can exert durable antitumor effects in solid tumors and long-term stability of demethylation of promoter CpG islands (Kagey et al., 2010; Tsai et al., 2012).
Much effort has been invested to determine the molecular mechanisms underlining the clinical efficacies of 5-Aza-CR and 5-Aza-CdR (Heyn and Esteller, 2012; Kelly et al., 2010a). Until now, studies have been primarily confined to studying promoter demethylation followed by subsequent gene reactivation. Previously, we have shown that the transcribed regions of genes are heavily methylated and that the level of methylation is positively correlated with the level of expression (Bender et al., 1999; Salem et al., 2000). Recently more detailed genome wide studies also demonstrate that DNA methylation in transcribed regions is correlated with gene expression (Kulis et al., 2012; Maunakea et al., 2010; Varley et al., 2013). DNA methylation in transcribed regions could potentially silence alternative promoters, retrotransposon elements, and other functional elements to maintain the efficiency of transcription (Kulis et al., 2012; Maunakea et al., 2010; Varley et al., 2013; Wolff et al., 2010). However, the potential for demethylation of gene bodies induced by DNA methylation inhibitors to alter their expression does not seem to have been reported. In this study, we investigated whether demethylation of gene bodies induced by 5-Aza-CdR could alter gene expression and possibly be a therapeutic target in cancer.
RESULTS
Restoration of cell growth rate after 5-Aza-CdR treatment is associated with global remethylation
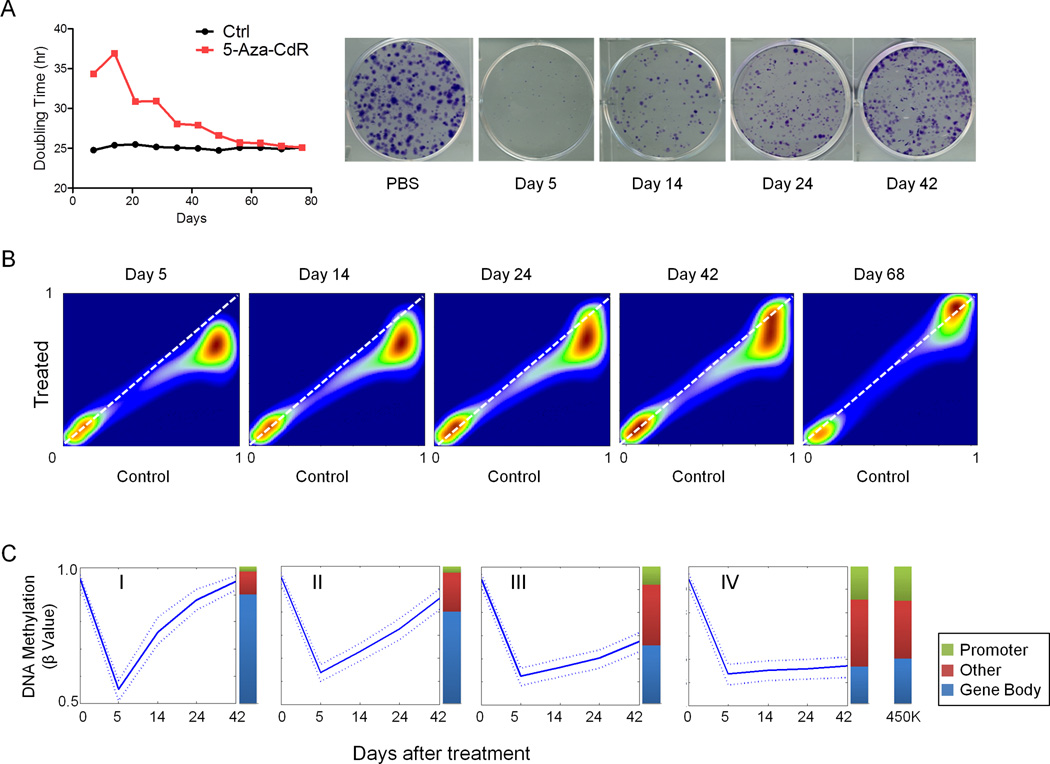
Transient exposure of dividing cells to low doses of 5-Aza-CdR or 5-Aza-CR leads to heritable increases in cell doubling time which are gradually reversed upon growth in the absence of further drug treatment (Bender et al., 1999; Tsai et al., 2012). To investigate potential mechanisms for the drug’s prolonged effects, we treated HCT116 cells transiently with low dose (0.3µM) 5-Aza-CdR for 24 hours and maintained the cells in drug-free medium until there were no further differences in the population doubling times (PDT) of the control and 5-Aza-CdR treated cells (Figure 1A). Drug treatment exerted an apparent biphasic effect on the PDT of HCT116 cells. Soon after exposure the PDT increased from 26 to 38 hr and then decreased quickly from 5 to 23 days before more gradually reverting to the untreated level around day 56 after drug treatment. The differences in growth rates between treated and controls completely disappeared by day 77. Low dose 5-Aza-CdR treatment also depressed the abilities of HCT116 cells to form colonies when plated at low density even at day 42 after drug treatment (Figure 1A).
Figure 1. Transient 5-Aza-CdR treatment shows prolonged effects on cell growth and DNA methylation.
A. Population doubling times (left panel) for HCT116 cells after 5-Aza-CdR treatment (red) and vehicle treatment (black). The y-axis denotes doubling time (hours); x-axis denotes time (days) after 5-Aza-CdR was withdrawn. Colony formation assay (right panel) for HCT116 cells at the indicated time points that the cells were seeded after the withdrawn of 5-Aza-CdR. Colonies were stained 10 days after seeding. B. Smooth Kernel scatter plots showing global DNA methylation patterns after 5-Aza-CdR treatment and at the indicated time points after treatment. The x-axis indicates beta values for untreated control, the y-axis indicates beta values for 5-Aza-CdR treated cells at indicated time points. C. Probes that were heavily methylated (beta value > 0.8) before 5-Aza-CdR treatment were classified, using consensus clustering, into four groups according to their rates of demethylation and remethylation after treatment. The solid line is the median value, while the dotted lines are the lower quartile (lower dotted line) and the upper quartile (upper dotted line). Distribution of probes: Locations of all probes on the 450K platform compared to probes in Group I to IV. See also Figure S1.
We then monitored long term DNA methylation changes using the Illumina Infinium HumanMethylation450 (HM450) DNA methylation array. This platform contains probes for 482,421 CpG sites across the genome, covering 99% of RefSeq genes (more than 21,000 genes) with an average of 17 CpG sites per gene, and probes are distributed in promoters, gene bodies, enhancers, and intergenic regions (Sandoval et al., 2011). Global demethylation of all methylated probes (the mean decrease in beta value is 0.41) was observed at day 5 after drug treatment, and most of the demethylated probes regained their original methylation levels at day 68 in a time-dependent manner (Figure 1B). Using a beta value 0.2 as a threshold for differential DNA methylation between treated and mock-treated cells and examining heavily methylated probes in the controls (β >0.8), 188,631 probes were identified as becoming demethylated in HCT116 cells at day 5 (Figure 1B). At day 68, about 8% of the probes (14,762) remained demethylated under the same criteria. A positive correlation (r= 0.83) was apparent between PDT differences and median DNA demethylation changes in HCT116 cells after 5-Aza-CdR treatment (Figure S1A), which suggested that DNA methylation changes might determine cell growth recovery rate.
We next used consensus clustering to classify the behaviors of the top 10% of the most variable originally highly methylated probes in the controls, using k-means=4 to divide the probe sets into four groups according to their rates of remethylation after treatment (Figure 1C). More than 3,000 probes, which were demethylated at day 5 and regained DNA methylation quickly after drug withdrawal were classified as Group I probes. Their DNA methylation levels were almost completely restored to baseline at day 42. Alternatively, the 13,458 probes in Group IV showed the slowest rate of remethylation and remained demethylated at day 42. Rates of remethylation of the Group II probes (n=4,099) and Group III probes (n=5,674) had intermediate remethylation kinetics (Figure 1C) and were not included in subsequent analyses. Of the more than 21,000 total genes represented in the HM450 array, 34% (n=7,158) were included in these four groups. Group I-IV contain 1,507, 2,003, 2,657, and 3,990 genes respectively, some of which may overlap across the groups. We next studied the genomic locations of the probes specified in the HM450 annotation by defining 3 categories of probes; those located within 1,500 bp upstream and downstream of the transcription start site (TSS200 and TSS1500) were called “promoter regions”. Probes located in transcribed regions were called “gene bodies” and the remainder classified as “other”. About 80% of the probes that regained methylation in a rapid manner were located in gene bodies in Group I compared to about 30% in Group IV (Figure 1C) with intermediate kinetics visible in the Groups II and III, the detailed probe distribution in different groups were presented in Figure S1B.
Collectively, our data showed that 5-Aza-CdR induced immediate and prolonged cell growth inhibition as well as a genome-wide DNA demethylation immediately after treatment (see Day 5 in Figure S1C). The recovery rate of cell growth and the rate of global rebound DNA methylation were negatively correlated. Furthermore, the remethylation rates of individual probes were not uniform in that gene bodies were most rapidly remethylated and this might play a role in the prolonged effect of drug treatment.
The most rapid rebound methylation occurs in gene bodies and is positively correlated with gene expression levels
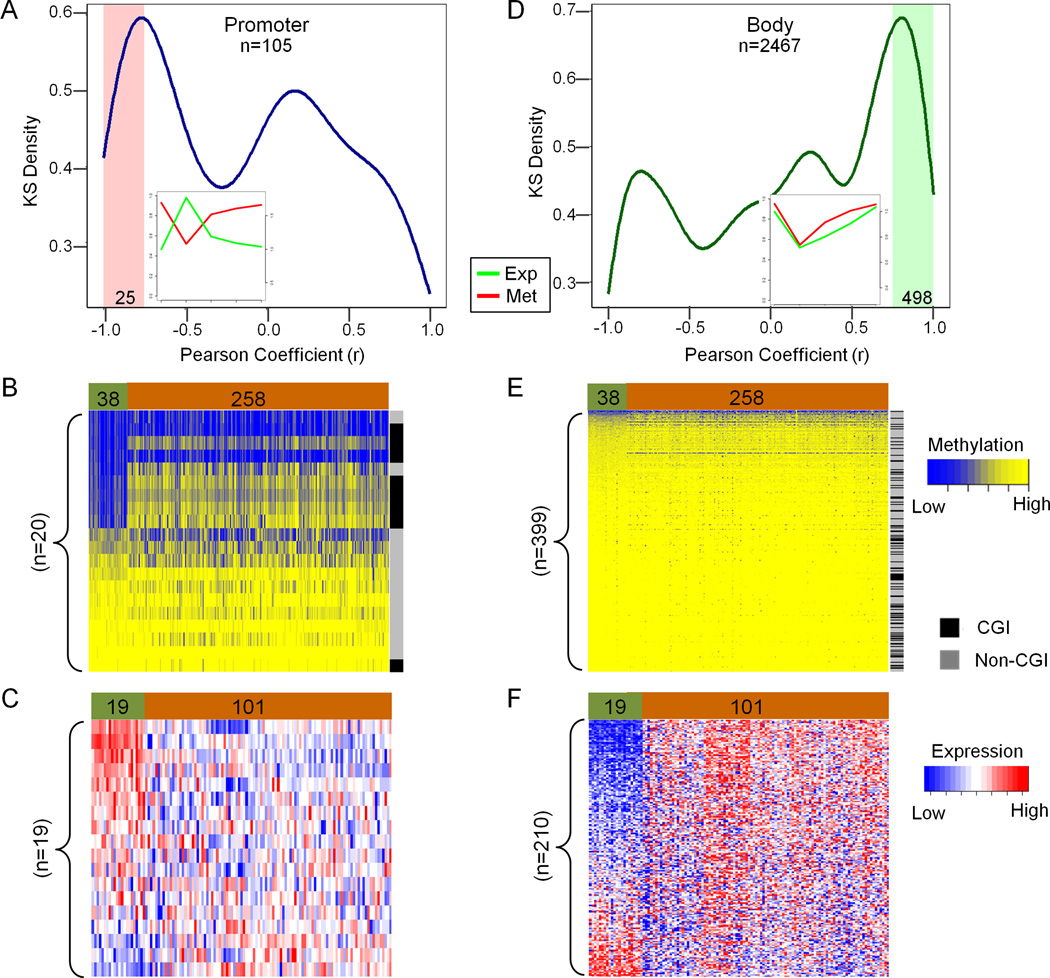
We next conducted genome-wide expression assays at the same time points as those analyzed by the DNA methylation assays. Separating Group I probes by genomic location, we first investigated the behaviors of the relatively small portion (less than 5%) within this group that were located in promoter regions. An enrichment peak of these probes (25/105, 24%) displayed the expected negative correlation between DNA methylation and gene expression (r < −0.75) in the kernel density plot (Figure 2A). Negatively correlated genes associated with these probes were reactivated at day 5 and re-silenced soon after treatment, and the original DNA methylation levels were reestablished at day 14 (Figure 2A). We then studied the methylation status of these negatively correlated probes in primary colorectal tumor samples and adjacent normal colonic tissues using The Cancer Genome Atlas (TCGA) colon database (http://cancergenome.nih.gov/). As shown in Figure 2B, approximately 50% of the probes were hypermethylated in uncultured colon carcinomas, while the other half of probes were methylated in both normal and tumor samples. Interestingly, nearly all the probes methylated in both normal and tumor samples were located outside of CpG islands (Figure 2B), suggestive of tissue-specific DNA methylation patterns. We also investigated the expression of the negatively correlated probes by comparing expression array data of 19 primary normal colonic tissues and 101 primary colon adenocarcinomas from the TCGA database (Figure 2C). Eight out of 20 annotated genes were significantly repressed in colon adenocarcinoma samples (p <0.001). Taken together, our data indicates that some of the fast rebounding CpGs located in promoter regions are likely silenced by tumor specific DNA hypermethylation in primary tumors.
Figure 2. DNA methylation and gene expression of fast rebound probes (Group I) in HCT116 cells and uncultured normal/tumor colon samples.
(A, D). Kernel density plots showing the correlation between DNA methylation at the promoter region (A) or gene body (D) and gene expression in HCT116 cells. The x-axis indicates the Pearson correlation (r) between DNA methylation and gene expression for a given probe at all the measured time-points. The y-axis indicates the density of probes. The 25 of 105 probes (r<−0.75) at the promoter region showing a significant inverse correlation between DNA methylation and gene expression are highlight in pink (A). Most of the probes in Group I (2467) are located in gene bodies (D). Among them, the 498 probes at gene bodies exhibit a positive correlation between DNA methylation and gene expression are highlighted in green (D). The small windows in A and D represent the average dynamic changes between DNA methylation (red, Met.) and expression (green, Exp.) from the shaded highlighted regions. The left y-axis represents percentage of DNA methylation and the right y-axis represents relative expression, while the x-axis represents times (from day 0 to day 42). (B , E). Supervised cluster analysis of DNA methylation in TCGA colon samples using the available probes identified in HCT116 (highlighted by shading in A and D). Twenty out of 25 are available for B, while 399 out of 498 are available for E. Green represents normal colon tissues (n=38); brown represents colon tumors (n=258). Blue indicates low beta values; yellow indicates high beta values. (C , F). Oncomine™ expression data analysis of 101 colon cancer samples (Brown) versus 19 normal colon samples (Green). The genes corresponding to the probes identified in B and E, 19 genes correspond to 25 probes in A, and 210 genes correspond to 498 probes in D. The expression data for genes was downloaded from Oncomine™ (TCGA Colon Adenocarcinoma vs. Normal set) and the heatmaps generated based on TCGA expression for the indicated groups. Blue denotes low expression and red denotes high expression. See also Figure S2.
More interestingly, 80% of Group I probes were located in gene body regions and about 20% of these exhibited a positive correlation between DNA methylation and expression (498/2,467 probes (20%), r > 0.75) (Figure 1C, Figure S1B and Figure 2D). Their expression was reduced 5 days after 5-Aza-CdR treatment, increased at day 14 and almost reverted to original levels of both expression and DNA methylation at day 42 (Figure 2D). In addition, the positively correlated gene body CpGs were methylated in both primary normal and tumor colon tissues in the TCGA colon data set (Figure 2E), and the majority of these are located in non-CpG island regions (Figure 2E). Surprisingly, a large proportion of the positively-correlated genes were over expressed in primary tumor samples compared to normal colon tissues (Figure 2F). Most of the promoter regions corresponding to these body probes were not differentially methylated in normal and uncultured tumor samples (Figure S2). The overexpression of the genes in cancer was therefore not being driven by DNA methylation changes in their promoters.
Slow-rebounding genomic regions are de novo methylated in primary colon tumors
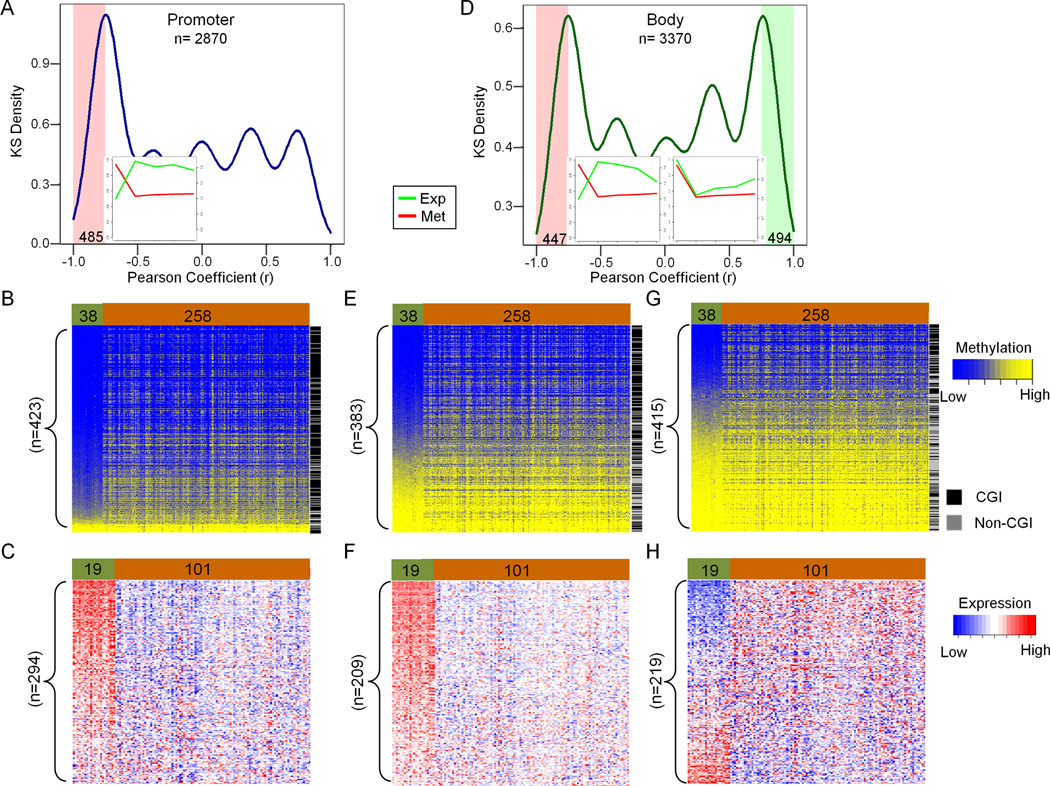
Next, we investigated the Group IV probes shown in Figure 1C which displayed the slowest rates of rebound methylation compared to the other three groups. Only 27% of probes in the slowest remethylation group were located in gene body regions, whereas 24% were located in promoter regions. A set of 485 probes displayed an inverse correlation between DNA methylation and gene expression after 5-Aza-CdR treatment, and these remained expressed for over 42 days, while their promoters had sustained DNA demethylation (Figure 3A). In contrast to the fast rebounding promoter CpGs (Figure 2B), the majority of the slow rebounding CpGs were de novo methylated targets in primary colon cancers (319/423, 75%), p < 0.001, Figure 3B), and were also located within CpG islands (348/423 probes, 82%) in the TCGA data set (Figure 3B). Additionally, at least 57% of these genes had been silenced or down-regulated in TCGA colon tumor samples according to the Oncomine™ database (Figure 3C).
Figure 3. DNA methylation and gene expression of slow rebound probes (Group IV) in HCT116 cells and normal/tumor colon samples.
(A, D). Kernel density plots show the correlation between DNA methylation (promoter region A, and gene body D) and gene expression in HCT116 cells. The x-axis indicates the Pearson correlation (r) between DNA methylation and gene expression for a given probe at all the measured time-points. The y-axis indicates the density of probes. Four hundred eighty-five of 2870 probes (r<−0.75) show a significant inverse correlation between DNA methylation and gene expression (A). A similar number of probes in Group IV (3370) are located in gene bodies (D). Among them, 447 probes (left highlighted region in D) exhibit a negative correlation between DNA methylation and gene expression, while 494 probes (right highlighted region in D) along with a positive correlation. The small windows in A and D represent the average dynamic changes between DNA methylation (red, Met.) and expression (green, Exp.) from the shaded highlighted regions. The left y-axis represents percentage DNA methylation and the right y-axis represents relative expression, while the x-axis represents times (from day 0 to day 42). (B, E, G). Supervised cluster analysis of DNA methylation in TCGA colon samples using the available probes identified in HCT116 cells (highlighted by shading in A and D). Four hundred twenty-three of 485 probes are available for B, while 383 out of 447 probes (negative correlation, E) and 415 out of 494 (positive correlation, G) are available. Green represents normal colon tissues (n=38); Brown represents colon tumors (n=258). Blue indicates low beta values; yellow indicated high beta values. (C, F, H). Oncomine™ expression data analysis of 101 colon cancer samples (Brown) versus 19 normal colon (Green); The genes corresponding to the probes identified in B, E, and G, 294 genes correspond to 485 probes in A, and 209 genes correspond to 447 probes in D, while 219 genes correspond to 494 probes in D. The expression data for genes was downloaded from Oncomine™ (TCGA Colon Adenocarcinoma vs. Normal set); the heatmaps were generated based on TCGA expression data for the indicated groups. Blue denotes low expression and red denotes high expression. See also Figure S3
On the other hand, 3,370 probes located in gene bodies displayed a multimodal distribution of correlation coefficients between DNA methylation and gene expression (Figure 3D). A set of 494 probes showed a positive correlation with expression (Figure 3D, right side), as had been observed in the fast rebounding group (Figure 2D). Furthermore, less than half of these probes had become de novo methylated in primary colon cancers (Figure 3G) and some corresponding genes were also over expressed in primary colon tumors (Figure 3H). In addition, 447 probes located in gene bodies were negatively correlated with gene expression despite their genomic location. Almost half of these were methylated in primary colon tumors (Figure 3E) and 62% were located in CpG islands (Figure 3E). Consistent with the DNA methylation data, the expression of more than 90% of negatively-correlated genes and probes were also down regulated in colon cancers (Figure 3F). To understand why gene body DNA methylation and gene expression showed a negative correlation, we found that about 40% of these inversely correlated gene body probes were associated with H3K4m1 and H2A.Z enrichment peaks in HMEC cells (Encyclopedia of DNA Elements, ENCODE, ChIP-seq data). These findings suggest that some of the probes located in gene bodies may acutely represent functional elements, such as enhancers or alternative promoters under normal circumstances (Figure S3).
Rapid rebound DNA methylation requires DNMT3B
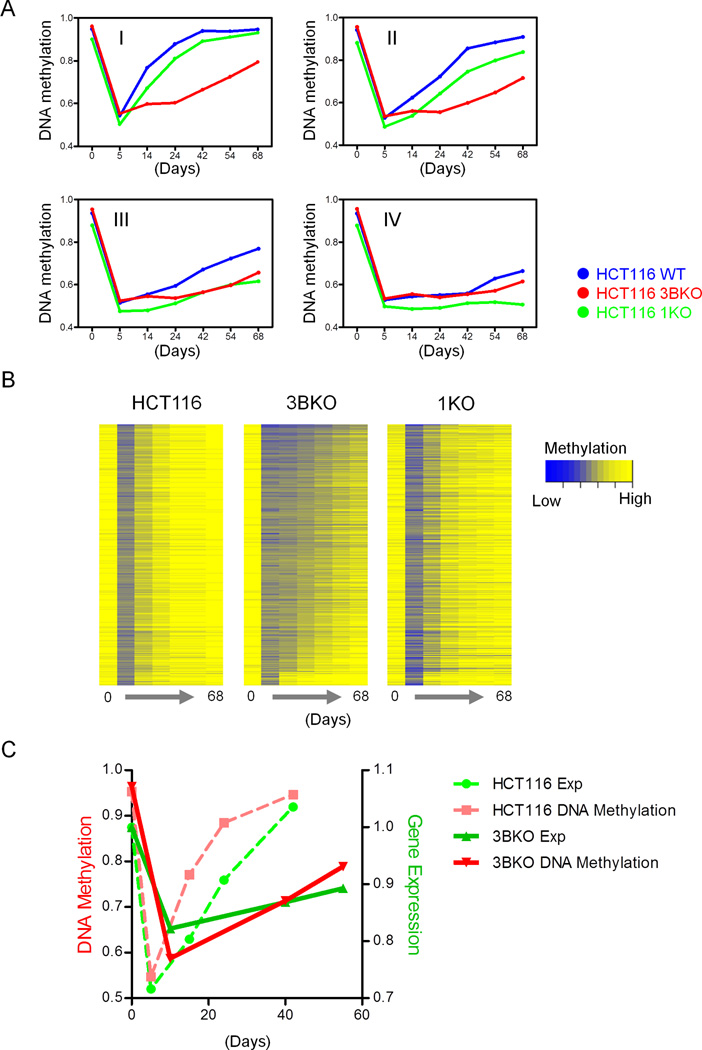
We next monitored global DNA methylation and gene expression profiles in two HCT116 derivative cell lines with genetic knockouts for DNMT3B (3BKO) or for DNMT1 (1KO) (Rhee et al., 2002; Rhee et al., 2000) to determine the specific DNA methyltransferases responsible for restoring the demethylation after 5-Aza-CdR treatment. All probes examined were the same in all three cell lines and the rates of remethylation were strongly retarded in 3BKO cells, especially in the Group I gene set (Figure 4A). Compared to the wild type cells, 5-Aza-CdR treatment strongly inhibited the PDT of 3BKO cells at early time points, consistent with our hypothesis that the rapid rebound probes seen in Group I might be responsible for the early effects of 5-Aza-CdR (Figure S4). A heatmap generated to visualize the detailed DNA methylation changes of individual probes demonstrated that DNMT3B knockout reduced the rates of remethylation for the majority of the rapid rebound genes in Group I (Figure 4B). In addition, the probes in Group I dependent on DNMT3B for rapid remethylation were already methylated in the 3BKO cells suggesting that their methylation status was maintained by DNMT1and/or DNMT3A in the steady state. De novo remethylation of the regions in Group I after drug treatment was accelerated by the presence of the DNMT3B but not by DNMT3A or DNMT1. Interestingly, the rate of remethylation was not strongly influenced by the absence of DNMT1, consistent with DNMT1 functioning mainly, although not exclusively, as a maintenance enzyme (Jones and Liang, 2009).
Figure 4. Comparison of DNA methylation and gene expression prior to and after 5-Aza-CdR treatment of HCT116 and derivative HCT116 1KO and HCT116 3BKO cells.
A. Line graphs show the median DNA methylation changes in HCT116, HCT116 1KO and HCT116 3BKO cells for each group of probes previously identified in Figure 1. B. Heatmaps show the remethylation behaviors of Group I probes that were originally heavily methylated in all 3 cell lines. Blue means unmethylated; yellow means fully methylated. The x-axis denotes the days after 5-Aza-CdR treatment. C. Median gene expression and DNA methylation changes of Group I positively correlated probes and the associated genes in HCT116 and HCT116 3BKO cells. See also Figure S4.
We then focused our study on the genes whose body methylation was positively correlated with gene expression to elucidate how DNMT3B affected the expression of Group I genes. After 5-Aza-CdR treatment, 210 genes regained body DNA methylation as well as gene expression in a rapid manner in HCT116 wild type cells (Figure 2D). However, DNA methylation and gene expression levels remained low in 3BKO cells (Figure 4C), suggesting that DNA methylation changes in the gene bodies caused gene expression changes. A critical additional point is that the failure of the DNA methylation to rebound shows that we were observing de novo methylation rather than the outgrowth of cells that were never demethylated in the first place. Together, these data imply a casual role for 5-Aza-CdR induced gene body demethylation in the down-regulation of gene expression. Fast rebounding DNA methylation required DNMT3B and, while it is possible that it is simply the presence of the protein that is responsible for the increased expression, this seems unlikely because the down-regulation of gene expression was initially seen after the methylation had been removed by 5-Aza-CdR treatment. Therefore, while neither observation by itself establishes causality, the two together strongly suggest that DNA methylation can lead to increased expression rather than this being due to the presence of DNMT3B only.
Decreased DNA methylation in gene bodies is associated with increased chromatin accessibility, occupancy by H2A.Z and alterations of histone modifications
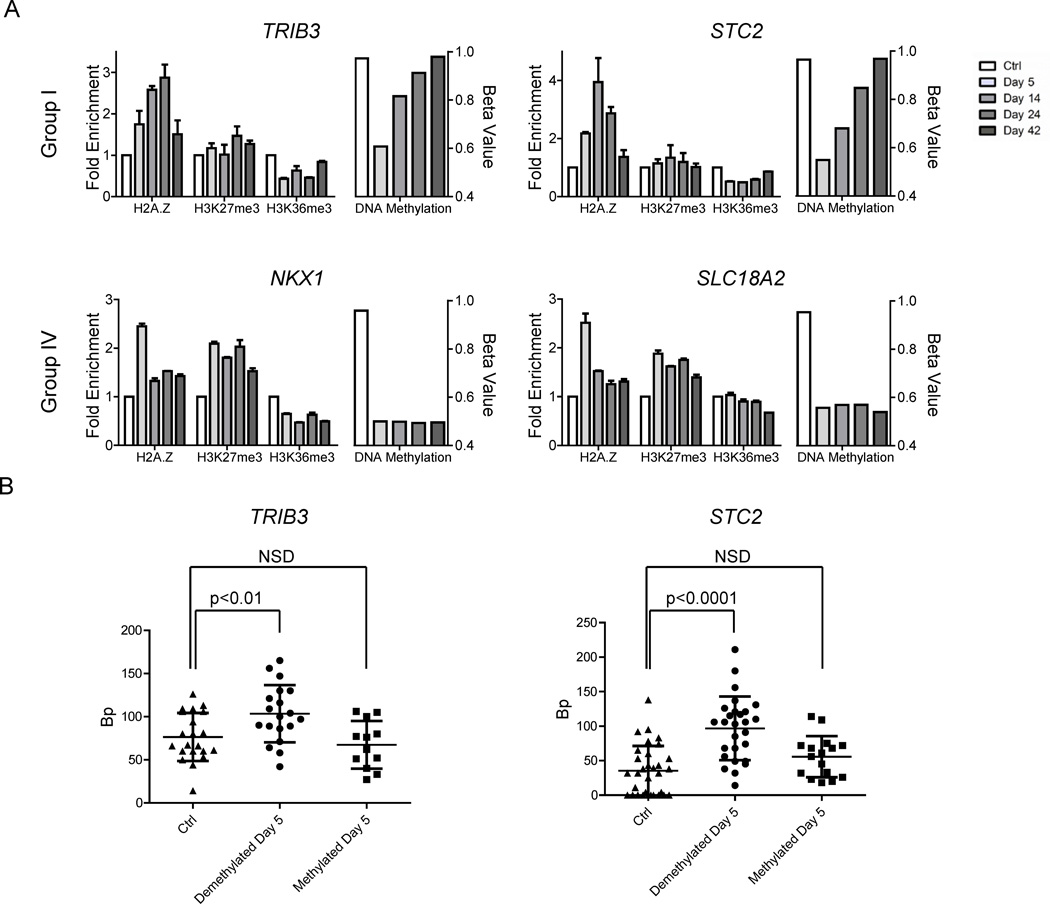
We selected two genes, TRIB3 and STC2, from the fast rebounding Group I for detailed analysis for expression, histone modifications, histone variants and chromatin accessibility in gene body regions after 5-Aza-CdR treatment. The expression of both genes decreased upon 5-Aza-CdR treatment (Figure S5A) and this was accompanied by gene body DNA demethylation at day 5, with increasing gene body DNA methylation thereafter (Figure 5). Enrichment of H2A.Z, H3K27m3, and H3K36m3 has been reported in gene bodies (Coleman-Derr and Zilberman, 2012; Nag et al., 2013; Venkatesh et al., 2012; Wagner and Carpenter, 2012). Therefore, we investigated the changes in their focal enrichments (see Figure S5B for detailed maps on these genes). An increase in H2A.Z occupancy was observed starting at day 5, peaking at day 24, which then declined to near basal levels at day 42 (Figure 5A). Enrichment of H3K27m3 generally remained constant throughout the experiment, whereas H3K36m3 levels decreased modestly at day 5 and gradually reverted back to the original level by day42, echoing the changes in gene expression (Figure 5A). To potentially explain the different kinetics of remethylation between Groups I and IV, we also performed ChIP assays on H3K27m3, H3K36m3, and H2A.Z distribution on two selected genes, NKX1 and SLC18A2, from the slow remethylation group (Group IV) (Figure 5A). We observed similar changes in distribution of H3K36m3 and H2A.Z in the bodies of these Group IV genes but dramatically increased levels of H3K27m3 were present in the Group IV relative to the Group I examples (Figure 5A). This result suggested that the kinetics of remethylation may be related to the presence or absence of the H3K27m3 mark following demethylation.
Figure 5. Chromatin architecture is disrupted in gene body regions after 5-Aza-CdR treatment.
A. ChIP results for H2A.Z, H3K27m3, and H3K36m3 before and after 5-Aza-CdR treatment at two representative gene body regions from Group I (top panel) and two representative gene body regions from Group IV (bottom panel). Each of the four panels shows histone modifications on the left and DNA methylation changes on the right. Fold enrichment of ChIP data was normalized to input and compared to control. Error bars represent standard deviation of the mean from three independent biological experiments. Regarding DNA methylation changes, before and after 5-Aza-CdR treatment, shown on the right side for each gene: the y-axis represents the beta value of DNA methylation, based on data from the 450K Illumina DNA methylation array. For both data sets: ChIP and DNA methylation, the x-axis represents the days after 5-Aza-CdR treatment as indicated in the legend. B. Column scatter plots indicate the NOMe-seq results for changes in chromatin accessibility separating the data, the parent methylated and daughter unmethylated DNA strands before and after 5-Aza-CdR treatment at the two representative gene body regions. The y-axis represents the average sizes of accessible regions where each dot represents the size of accessible regions in individual DNA strand as detected by bisulfite sequencing. Error bars represent standard deviation of the mean of the accessible regions from the independent sequenced DNA strands. NSD represents no significant difference of chromatin accessibility. See also Figure S5.
We also used the highly sensitive single-molecule nucleosome occupancy and methylome sequencing (NOMe-seq) assay (Kelly et al., 2012) to analyze potential chromatin accessibility changes in selected regions of TRIB3 and STC2 gene body regions (Figure S5C). We calculated the lengths of accessible regions on the methylated (parent) and demethylated (daughter) strands before and after treatment for each sequenced strand and showed accessibility using scatter box plots (Figure 5B). Although both regions were heavily methylated before treatment, the chromatin architecture was not completely closed in gene body regions compared to promoter regions. This may suggest a specific architecture for highly transcribed gene bodies. Transient exposure to 5-Aza-CdR significantly increased the accessibility on the demethylated (daughter) DNA strands in both regions at day 5. Consistent with our previous study (Yang et al., 2012), the DNA strands that remained methylated after treatment did not show alterations in chromatin accessibility in both promoters and gene body regions.
Collectively, our results showed that the 5-Aza-CdR treatment induced gene body demethylation, deposition of H2A.Z, and increased accessibility in demethylated DNA regions, potentially leading to a disruption of chromatin structure. In addition, the presence or absence of the polycomb mark, H3K27m3, after treatment might alter remethylation kinetics.
Differential chromatin signatures associated with Group I and IV probes
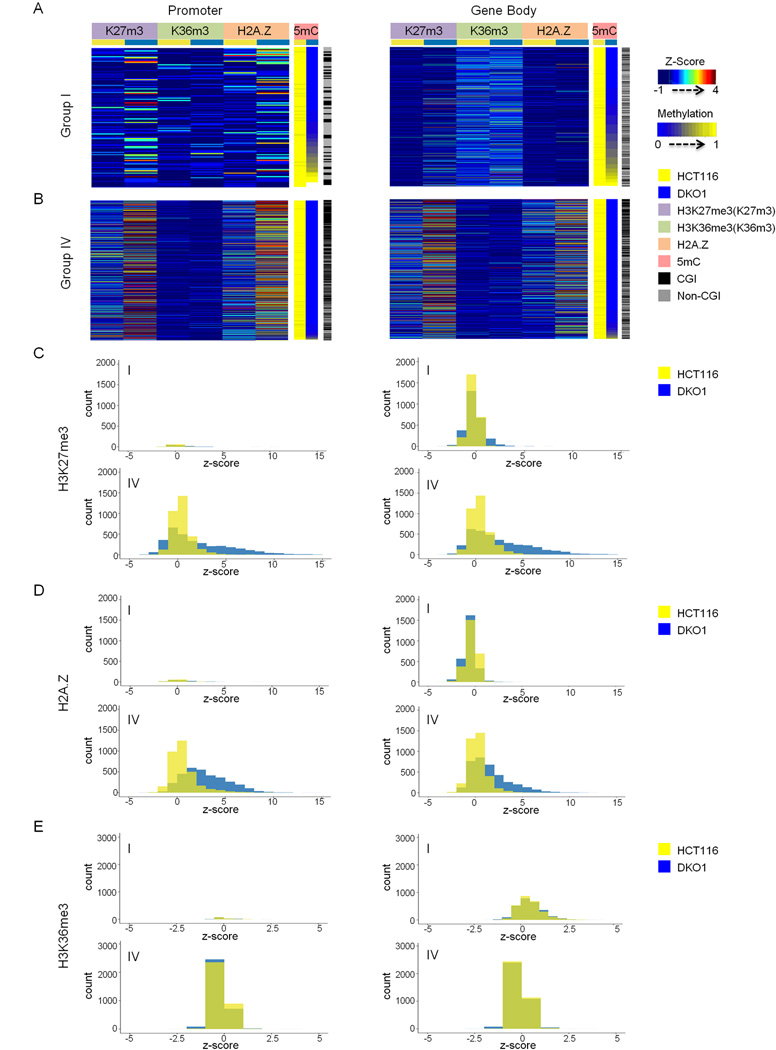
To probe potential mechanisms leading to different DNA remethylation kinetics in more detail, we also tested whether there is a relationship between pre-existing gene expression levels and remethylation kinetics among the four groups. The mean level of pre-existing expression of Group I genes was the highest, while the Group IV set displayed the lowest mean expression (Figure S6). These data, together with those shown in Figure 5, show that chromatin signatures may play a role to maintain expression levels in addition to DNA methylation. Next we asked whether the differences in remethylation kinetics between Group I and IV probes might be explained by altered chromatin signatures. For this we used HCT116 and HCT116-derived DKO1 cells with markedly decreased (~95%) DNA methylation engineered by genetic knockdowns of DNMT3B and DNMT1 (Egger et al., 2006; Rhee et al., 2002). Stable knockdowns were used to increase the clarity of the analysis. The promoters of both Group I and Group IV probes generally showed increases in H3K27m3 and H2.A.Z in the absence of DNA methylation (Figure 6A and B), although this was enhanced in the Group IV set (Figure 6C and D). On the other hand, the bodies of Group IV genes showed a strong enrichment of both H3K27m3 and H2.A.Z compared to Group I in the absence of DNA methylation (Figure 6A, B, C, and D). Both of these marks are considered antagonistic to DNA methylation, which might explain why they were refractory to rebound methylation (Gal-Yam et al., 2008; Kondo et al., 2008; Zilberman et al., 2008). In contrast, the bodies of Group I genes were marked with H3K36m3 independently of the DNA methylation level (Figure 6A and E) suggesting that this well-known mark of actively transcribed regions may make them receptive to re-methylation.
Figure 6. Differential chromatin signatures are associated with Group I and IV probes.
(A , B). Heatmaps showing the enrichment of H3K27m3, H3K36m3 and H2A.Z for each category of probes (promoter and gene body) in A for Group I and B for Group IV. Wiggler was used to normalize the data into a single value for each genomic position and mean wiggler value was calculated in a 10bp-bin. Z-score was then calculated by transforming the wiggler value in each bin as (Xi-Xmean)/Xstdev where Xi is experimental wiggler value subtracted by the input in the same bin and Xmean and Xstdev is an average and standard deviation of Xi in all of the bins in the whole genome. Each methylation probes was then correlated with a z-score (Bernstein et al., 2012; Xie et al., 2013). C-E. Bar charts showing the distribution of probes across a range of z-scores for H3K27m3 (C), H2A.Z (D), and H3K36m3 (E). The x-axes represent the range of z-scores for each modification and y-axes show the number of probes falling in a particular range of z-scores. Z-score of =< 0 indicates no enrichment. See also Figure S6.
Rapidly remethylating genes are enriched for metabolic processes and are c-MYC regulated
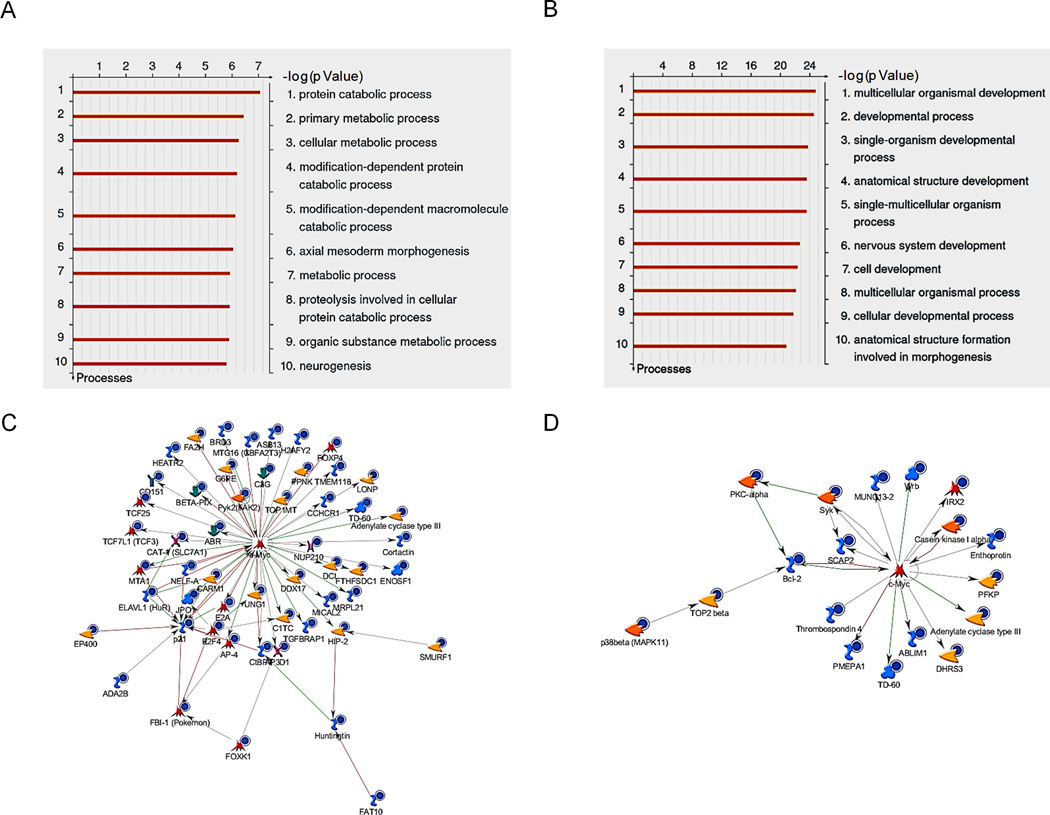
We next investigated the functions of the Group I and Group IV genes using the “Metacore™” commercial program. First, we analyzed the down-regulated genes that displayed gene body demethylation after 5-Aza-CdR treatment using Gene Ontology (GO) analysis (Table S1). This showed that metabolic processes and protein catabolic processes were overrepresented in the Group I gene set (Figure 7A). Remarkably, for the slow rebounding genes (Group IV positively-correlated genes), seven out of the 10 most significant GO processes were development related (Figure 7B), and included well-known tumor suppressor genes, such as SFRP1, TXNIP, KL, CHFR, TCF4, PRICKLE1, LTF, CYGB, GALR1, PHOX2A. In addition, we incorporated these two groups of genes in the “Analyze network (transcription factors)” program of Metacore™ to create networks centered on transcription factors (Table S2). These analyses revealed that 56 Group I genes either regulated or were regulated by c-MYC (p = 1.5 × 10−144) (Figure 7C). The Group IV genes did not show this enrichment pattern (Figure 7D). Therefore, by altering gene body DNA methylation, 5-Aza-CdR strongly affected several well-known tumor-related oncogenic pathways, especially metabolic pathways and those regulated by c-MYC.
Figure 7. GO analyses of gene body probes which exhibit a positive correlation between DNA methylation and gene expression.
A and B. Top ten significantly enriched cellular processes in Groups I and IV probes. The lengths of the orange bars denote significance. C and D. Network diagrams indicate the relationship between c-MYC and genes in Group I (C) and in Group IV (D). Ontology analysis was performed using built-in functions of MetaCore™. The networks are constructed from the basic algorithm "analyze networks (Transcription Factors)" of MetaCore™ (Thomson Reuters Inc.). The genes down-regulated by 5-Aza-CdR treatment are marked by blue solid circle. The other symbols used are as seen on the Metacore™ website. See also Table S1 and S2.
DISCUSSION
The role of DNA methylation in controlling the activities of gene promoters, whether CpG islands or non-CpG islands, has been extensively investigated over the last three decades (Jones, 2012). These studies have conclusively demonstrated that the establishment of a closed chromatin state associated with the presence of nucleosomes at the transcription start site that effectively blocks transcription initiation by RNA polymerase II (Jones and Liang, 2009). The function of gene body DNA methylation is poorly understood, although several recent studies have begun to investigate the potential for how gene body DNA methylation impacts gene expression (Kulis et al., 2012; Lister et al., 2009; Maunakea et al., 2010; Varley et al., 2013). While it is well-known that the bodies of actively expressed genes, particularly those located on the active X chromosome, show increased DNA methylation relative to their inactive counterparts, the issue of whether this causally affects transcription rates has been very difficult to discern (Hellman and Chess, 2007). Our experiments show that gene body DNA methylation increases gene expression and that the rapid establishment of this is dependent on the presence of the DNMT3B.
Several reports have suggested that gene body DNA methylation may increase transcriptional activity by blocking the initiation of intragenic promoters or by affecting the activities of repetitive DNAs within the transcriptional unit (Maunakea et al., 2010). An alternative explanation may be that the formation of an ordered structure within the transcribed unit increases the rate of transcription either by elongation or splicing. It is known that the nucleosomes in transcribed region are positioned non-randomly, are enriched at intron-exon junctions and the enrichment of nucleosomes at intron-exon junctions may be involved in regulating splicing events. Therefore, nucleosome destabilization in the exon-intron junctions may alter mRNA levels (Andersson et al., 2009; Luco et al., 2011; Tilgner et al., 2009). We observed an increase in H2A.Z occupancy and chromatin accessibility especially in the slow rebounding Group IV probes. The histone variant H2A.Z is mutually exclusive with DNA methylation (Edwards et al., 2010; Xiao et al., 2012; Zemach et al., 2010; Zilberman et al., 2008), and destabilizes nucleosomes and mediates nucleosome depletion in contrast to DNA methylation (Jones and Liang, 2009; Li et al., 2012; Sharma et al., 2011). In addition, the mark for active transcription elongation H3K36m3, which we found to be enriched only in the fast remethylation group (Group I), has been shown to suppress histone exchange on transcribed genes thus maintaining the accuracy and efficiency of transcription (Venkatesh et al., 2012; Wagner and Carpenter, 2012). This may also explain why the pretreatment expression levels are strongly associated with remethylation kinetics. Based on our current results and those of others, gene body DNA demethylation may lead to nucleosome destabilization in transcribed regions and reduced efficiencies of transcription elongation or splicing.
Interestingly, not all of the gene body probes were positively correlated with expression as has been reported in other studies (Kulis et al., 2012; Maunakea et al., 2010; Varley et al., 2013). A potential explanation is that transcribed regions often include functional elements, such as alternative promoters, enhancers, transcription factor binding sites, repetitive elements and enrichment of nucleosomes at intron-exon junctions. Most of these genomic elements are suppressed or stabilized by DNA methylation and hypomethylation may therefore result in their activation and interference with expression of the host gene. However, the demethylation of these elements is only a necessary but not a sufficient condition for them to regain their activities, because the presence of transcription factors specific for these elements (such as highly tissue specific enhancers) is a requirement for their reactivation. In addition, we recently reported that only small percentage of DNA hypomethylated regions gain functionality and chromatin accessibility after 5-Aza-CdR treatment (Pandiyan et al., 2013). Taken together, gene repression due to gene body DNA demethylation may be dependent on several key factors including: 1) whether embedded demethylated functional elements regain activity, 2) whether demethylated regions are located at intron-exon junctions together with destabilized nucleosomes, 3) possible effects on the rates of transcript elongation and splicing.
The genes with rapid remethylation in their gene bodies are associated with increased cellular growth. In addition, these genes are enriched with metabolic pathway functions or are up-regulated by c-MYC in uncultured human cancers. These findings are strongly suggestive of an active selection process within the cultures that allows for the selection for genes that are rapidly remethylated in their gene bodies regions. DNA methyltransferase inhibitors such as 5-aza-CR and 5-aza-CdR have long been considered to act as non-specific inhibitors of DNA methylation and act primarily by reactivating genes silenced by promoter methylation. Indeed, we found that transient treatment with low doses of 5-aza-CdR results in promoter DNA demethylation and gene re-expression. Moreover, the effects of this DNA demethylation are generally more durable than what we observed in the gene body regions. Thus, the DNA demethylating agents do not show specificity with respect to their demethylating activities, however, the remethylation kinetics are very different depending to a large extent on the location of CpGs within the transcriptional unit. The focus on promoter demethylation has provided insufficient knowledge of the genome-wide effects of DNA methylation inhibitors (Oki et al., 2007). Our finding suggests a potential role for DNA methylation inhibitors in blunting the effects of c-MYC which plays a key role in the pathogenesis of many human cancers (Dang, 2012). Metabolic reprogramming is now considered to be one of cancer’s hallmarks because the metabolites themselves can be oncogenic by altering cell signaling and blocking cellular differentiation (Hanahan and Weinberg, 2011; Ward and Thompson, 2012). The c-MYC and metabolic reprogramming pathways can mutually regulate each other in producing synergistic oncogenic effects, and inhibition of these activities by bromodomain protein inhibitors has become an attractive cancer therapeutic target (Butler et al., 2013; Delmore et al., 2011). We found that the most dramatic inhibition of cell growth occurs within the first two weeks after treatment, during the fast rebound period, suggesting that down-regulated genes become up-regulated by the ensuing DNA remethylation, which seems to be dependent on DNMT3B and the local chromatin structure.
We demonstrate that the DNA remethylation of these transcribed regions is not simply a consequence of gene expression changes but can increase gene expression levels. Also, selection of small subpopulations of cells that escaped demethylation after drug treatment seems unlikely because the Group I genes failed to remethylate quickly in the 3BKO cells. These findings do not negate the possibility that the increased DNA methylation establishes a feedback loop by which DNA methylation encourages increased gene expression, which in turn facilitates further DNA methylation. Murine DNMT3B promotes tumorigenesis in vivo (Linhart et al., 2007) so that DNMT3B could also be a therapeutic target to prevent rebound methylation and prolong antitumor effects.
Another surprising result from our study is that most de novo methylated promoter and gene body regions (Group IV) show sustained DNA demethylation and gene reactivation or down-regulation of expression after drug withdrawal. This may explain the observed long-term antitumor effects in cancer cells after treatment with DNA methylation inhibitors (Tsai et al., 2012). In general, those regions seem to be more resistant to remethylation, possibly due to a chromatin signature in which the loss of DNA methylation is followed by an enrichment of H3K27m3 marks, which in turns might block access to DNMT3B. This may also be due to transcription factor occupancy of reactivated genes and epigenetic switching for silenced genes, since most of de novo methylated promoter regions are targets of the polycomb repressive complex(Gal-Yam et al., 2008; Widschwendter et al., 2007).
Taken together, our detailed genome-wide analyses demonstrate that DNA methylation inhibitors induce DNA demethylation across all genomic features. In addition to the well-known and well-studied direct activation of tumor suppressor genes, gene body DNA methylation might be an intriguing additional target for therapy, since it could lead to the down-regulation of oncogenes and metabolic genes. Therefore, these findings will have direct implications in cancer treatment, not only for the understanding of the overall role of DNA methylation in the epigenome, but also in understanding patient’s response to FDA-approved drugs.
EXPERIMENTAL PROCEDURES
Cell lines and drug treatment
HCT116 WT, HCT116 3BKO and HCT116 1KO cells were treated with 0.3µM of 5-Aza-CdR (Sigma-Aldrich, St. Louis, MO). The medium was changed 24 hours later. All three cell lines were maintained in McCoy’s 5A medium, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. DNA and RNA purifications were performed as previously described (Egger et al., 2006).
Colony formation
Cells were seeded into 6-well plates at 1,000 cells per well either with or without indicated treatment at indicated days. The culture medium was changed every three days. After 10 days of incubation at 37°C, cells were washed with PBS, fixed with methanol and stained with 0.5% crystal violet. The colony formation results are scanned images of 6-well plates with indicated treatment.
Real-Time RT-PCR
Total RNA was isolated from cells using Trizol reagent (Invitrogen) at indicated time points. One µg of RNA was reverse transcribed using M-MLV and random hexamers (Invitrogen). PCR reactions were performed using KAPA SYBR® FAST University 2X qPCR Master Mix. The sequences of gene-specific primers are available upon request.
Illumina Infinium HM450 DNA methylation assay
The Infinium DNA methylation assay was performed at the USC Epigenome Center according to the manufacturer’s specifications (Illumina, San Diego, CA). The Illumina Infinium HumanMethylation450 (HM450) BeadChip examines DNA methylation status of 482,421 CpG sites, covering 99% of RefSeq Genes and intergenic regions. The DNA methylation level for each interrogated CpG site is reported as a beta value, ranging from 0 (not methylated) to 1 (fully methylated). Downstream processing and beta value calculations were performed as previously described (De Carvalho et al., 2012). Processed genomic DNA methylation datasets from primary tissues were downloaded from TCGA public data portal (http://cancergenome.nih.gov/dataportal/). A description of TCGA data types, platforms and analyses are as previously described (De Carvalho et al., 2012).
NOMe-seq assay
Nuclei preparation and GpC Methyltransferase treatment were performed as described previously (Kelly et al., 2012). Briefly, freshly extracted nuclei were treated with 200 U of GpC methyltransferase in the presence of SAM for 15 min at 37°C. An equal volume of stop solution (20 nM Tris–HCl, 600 mM NaCl, 1% SDS, 10 mM EDTA, 400 mg/ml proteinase K) was added to stop the reaction. The entire reaction mixture was incubated at 55°C overnight. DNA was purified by phenol/chloroform extraction and ethanol precipitation. PCRs were performed using bisulfite converted DNA and the product were cloned using TOPO TA cloning kit (Invitrogen). Individual colony was sequenced to investigate accessibility and DNA methylation status of each unique strand. Primer sequences are available upon request. NOMe-seq results were analyzed using student t tests on GraphPad Prism version 5 (GraphPad Software).
Chromatin immunoprecipitation (ChIP) and ChIP-Seq library generation and sequencing
ChIP assays were performed as described previously using commercially available antibodies (Liang et al., 2004). H3 and H2A.Z antibodies were purchased from Abcam (Cambridge, MA). H3K27m3 and IgG antibodies were purchased from Millipore (Billerica, MA). H3K36m3 antibodies were purchased from Active Motif (Carlsbad, CA).
Genome-wide libraries were generated using 20ng of purified ChIP and input DNA using previously described protocol (Kelly et al., 2010b). Samples were barcoded and sequenced on HiSeq2000 (Illumina) to generate 50 single-end reads. Sequencing reads were aligned to hg19 using bwa and we filtered out non-unique reads and PCR duplicates. We normalized each experimental data into a wiggler value (Bernstein et al., 2012) which was further transformed into z-score to normalize variations between experiments based on previously described method (Xie et al., 2013). In addition, processed HMEC ChIP-seq datasets can be downloaded from the Encyclopedia of DNA Elements (ENCODE) public data portal (https://genome.ucsc.edu/ENCODE/).
Gene expression assay and statistical analysis
Expression analysis was performed at Sanford-Burnham Medical Institute(La Jolla, CA) using the Illumina genome-wide expression BeadChip (HumanHT-12_V4_0_R1) (Illumina). Gene expression data were processed using the lumi package in R. The data were log2-transformed and normalized using Robust Spline Normalization (RSN). Comparisons between the control and treated samples for all three cell lines were performed using the R package limma. Oncomine™ (Compendia Bioscience, Ann Arbor, MI) was used for analysis of publicly available gene expression data. Metacore™ (Thomson Reuters Inc. Version, 6.14.61508) was used to identify and visualize biological processes and pathways that were enriched due to 5- Aza-CdR treatment. Relevant Gene Ontology (GO) lists are ranked according to their p value as determined by MetaCore™. We performed this analysis using the default setting which calculated the p value based on hypergeometric intersection in comparison to Illumina expression array background. Please note that Metacore GO analysis algorithm only showed the top ten most enriched cellular processes of the total GO cellular processes.
Statistical tests were done using R software (R version 2.15.2 , R Development Core Team, 2012). "ConsensusClusterPlus" package from Bioconductor was used to cluster DNA methylation probes into different groups with variant remethylation patterns (Wilkerson and Hayes, 2010). Consensus clustering was performed using a maximum evaluated cluster counts (k) of 10 (therefore cluster counts of 2, 3, 4, 5, 6, 7, 8, 9, 10 were evaluated) and 10,000 resamplings with k-means. The R package “polycor” was used to calculate a matrix consisting of Pearson correlations between DNA methylation and gene expression. The R package “Gviz” was used to generate genes’ schematic maps. The following CRAN packages were used to generate plots: "ggplot2" and "gplots".
Supplementary Material
Highlights.
Fast remethylation is often seen in the gene bodies of oncogenic regulated genes
Slowly remethylating loci are frequently located in promoters methylated in tumors
Rapid remethylation and restoration of expression levels require DNMT3B
There is a causal relationship between gene body DNA methylation and gene expression
SIGNIFICANCE.
Although promoter methylation is known to silences genes it has not yet been demonstrated whether there is a causal relationship between gene body methylation and expression. We show that the DNA methylation inhibitor 5-aza-2'deoxycytidine induces the demethylation of gene bodies and alters expression of the associated genes in cancer. The rapid remethylation after drug treatment requires the DNMT3B showing a causal relationship between gene body methylation and altered expression. Therefore, gene body methylation could be a potential therapeutic target since its inhibition could lead to the down regulation of oncogenes and metabolic genes which are commonly overexpressed in various cancers. It has direct implications in understanding the responses of patients to FDA approved DNA methylation inhibitor drugs.
ACKNOWLEDGMENTS
Funding for this work was provided by NIH R37 CA-082422 (P.J.) and NIH RO1 CA-124518 (G.L.). The project described was supported in part by award number P30CA014089 from the National Cancer Institute. D.C is supported by funding from the Princess Margaret Cancer Foundation. We thank Drs. Bert Vogelstein and Stephen B. Baylin for kindly providing HCT116, HCT116 1KO and HCT116 3BKO cells. The authors thank Dr. Daniel J. Weisenberge for discussing, reading, and revising the manuscript. We also thank Dr. Peggy J. Farnham to share the ChiP-seq data for this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
All genome-wide DNA methylation and gene expression data utilized in the study has been deposited in GEO under the accession number GSE51815. And all ChIP-Seq data has been deposited in GEO under the accession number GSE58638.
AUTHOR CONTRIBUTIONS
Conceived and designed the experiments: X.Y., H.H., D.C., P.A.J. and G.L. Performed the experiments: X.Y. and H.H. Analyzed the data: X.Y., H.H., D.C. and F.D.L. Wrote the paper: X.Y., H.H., P.A.J. and G.L.
REFERENCES
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours--lessons from the past. Nat Rev Clin Oncol. 2013;10:256–266. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch C, Nephew KP. Epigenetic targeting therapies to overcome chemotherapy resistance. Adv Exp Med Biol. 2013;754:285–311. doi: 10.1007/978-1-4419-9967-2_14. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender CM, Gonzalgo ML, Gonzales FA, Nguyen CT, Robertson KD, Jones PA. Roles of cell division and gene transcription in the methylation of CpG islands. Mol Cell Biol. 1999;19:6690–6698. doi: 10.1128/mcb.19.10.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler EB, Zhao Y, Munoz-Pinedo C, Lu J, Tan M. Stalling the engine of resistance: targeting cancer metabolism to overcome therapeutic resistance. Cancer Res. 2013;73:2709–2717. doi: 10.1158/0008-5472.CAN-12-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, Yang X, Liang G, Jones PA. DNA methylation screening identifies driver epigenetic events of cancer cell survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JR, O'Donnell AH, Rollins RA, Peckham HE, Lee C, Milekic MH, Chanrion B, Fu Y, Su T, Hibshoosh H, et al. Chromatin and sequence features that define the fine and gross structure of genomic methylation patterns. Genome Res. 2010;20:972–980. doi: 10.1101/gr.101535.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger G, Jeong S, Escobar SG, Cortez CC, Li TW, Saito Y, Yoo CB, Jones PA, Liang G. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci U S A. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, Lin JC, Liang G, Jones PA, Tanay A. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1:598–607. doi: 10.1158/2159-8290.CD-11-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey JD, Kapoor-Vazirani P, McCabe MT, Powell DR, Vertino PM. Long-term stability of demethylation after transient exposure to 5-aza-2'-deoxycytidine correlates with sustained RNA polymerase II occupancy. Mol Cancer Res. 2010;8:1048–1059. doi: 10.1158/1541-7786.MCR-10-0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantarjian H, Issa JP, Rosenfeld CS, Bennett JM, Albitar M, DiPersio J, Klimek V, Slack J, de Castro C, Ravandi F, et al. Decitabine improves patient outcomes in myelodysplastic syndromes: results of a phase III randomized study. Cancer. 2006;106:1794–1803. doi: 10.1002/cncr.21792. [DOI] [PubMed] [Google Scholar]
- Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010a;28:1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Liu Y, Lay FD, Liang G, Berman BP, Jones PA. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res. 2012 doi: 10.1101/gr.143008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly TK, Miranda TB, Liang G, Berman BP, Lin JC, Tanay A, Jones PA. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell. 2010b;39:901–911. doi: 10.1016/j.molcel.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- Kulis M, Heath S, Bibikova M, Queiros AC, Navarro A, Clot G, Martinez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012 doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RW, Vatapalli R, Topper MJ, Luo J, Connolly RM, Azad NS, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014 doi: 10.18632/oncotarget.1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gadue P, Chen K, Jiao Y, Tuteja G, Schug J, Li W, Kaestner KH. Foxa2 and H2A.Z mediate nucleosome depletion during embryonic stem cell differentiation. Cell. 2012;151:1608–1616. doi: 10.1016/j.cell.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, Weisenberger DJ, Egger G, Takai D, Gonzales FA, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhart HG, Lin H, Yamada Y, Moran E, Steine EJ, Gokhale S, Lo G, Cantu E, Ehrich M, He T, et al. Dnmt3b promotes tumorigenesis in vivo by gene-specific de novo methylation and transcriptional silencing. Genes Dev. 2007;21:3110–3122. doi: 10.1101/gad.1594007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei D, Fang F, Shen C, Schilder J, Arnold A, Zeng Y, Berry WA, Huang T, Nephew KP. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012;72:2197–2205. doi: 10.1158/0008-5472.CAN-11-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Savova V, Fung HL, Miron A, Yuan GC, Zhang K, Gimelbrant AA. Chromatin signature of widespread monoallelic expression. Elife. 2013;2:e01256. doi: 10.7554/eLife.01256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki Y, Aoki E, Issa JP. Decitabine--bedside to bench. Crit Rev Oncol Hematol. 2007;61:140–152. doi: 10.1016/j.critrevonc.2006.07.010. [DOI] [PubMed] [Google Scholar]
- Pandiyan K, You JS, Yang X, Dai C, Zhou XJ, Baylin SB, Jones PA, Liang G. Functional DNA demethylation is accompanied by chromatin accessibility. Nucleic Acids Res. 2013;41:3973–3985. doi: 10.1093/nar/gkt077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- Salem CE, Markl ID, Bender CM, Gonzales FA, Jones PA, Liang G. PAX6 methylation and ectopic expression in human tumor cells. Int J Cancer. 2000;87:179–185. doi: 10.1002/1097-0215(20000715)87:2<179::aid-ijc4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, Esteller M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet. 2011;7:e1001286. doi: 10.1371/journal.pgen.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman LR, Demakos EP, Peterson BL, Kornblith AB, Holland JC, Odchimar-Reissig R, Stone RM, Nelson D, Powell BL, DeCastro CM, et al. Randomized controlled trial of azacitidine in patients with the myelodysplastic syndrome: a study of the cancer and leukemia group B. J Clin Oncol. 2002;20:2429–2440. doi: 10.1200/JCO.2002.04.117. [DOI] [PubMed] [Google Scholar]
- Taylor SM, Jones PA. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979;17:771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, et al. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–446. doi: 10.1016/j.ccr.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh S, Smolle M, Li H, Gogol MM, Saint M, Kumar S, Natarajan K, Workman JL. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489:452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–1573. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff EM, Byun HM, Han HF, Sharma S, Nichols PW, Siegmund KD, Yang AS, Jones PA, Liang G. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6:e1000917. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrangle J, Wang W, Koch A, Easwaran H, Mohammad HP, Vendetti F, Vancriekinge W, Demeyer T, Du Z, Parsana P, et al. Alterations of immune response of non-small cell lung cancer with Azacytidine. Oncotarget. 2013 doi: 10.18632/oncotarget.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Xie D, Cao X, Yu P, Xing X, Chen CC, Musselman M, Xie M, West FD, Lewin HA, et al. Comparative epigenomic annotation of regulatory DNA. Cell. 2012;149:1381–1392. doi: 10.1016/j.cell.2012.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki J, Issa JP. Epigenetic aspects of MDS and its molecular targeted therapy. Int J Hematol. 2013;97:175–182. doi: 10.1007/s12185-012-1197-4. [DOI] [PubMed] [Google Scholar]
- Yang X, Noushmehr H, Han H, Andreu-Vieyra C, Liang G, Jones PA. Gene reactivation by 5-aza-2'-deoxycytidine-induced demethylation requires SRCAP-mediated H2A.Z insertion to establish nucleosome depleted regions. PLoS Genet. 2012;8:e1002604. doi: 10.1371/journal.pgen.1002604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–919. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.