Abstract
Fishes have diverse pigment patterns, yet mechanisms of pattern evolution remain poorly understood. In zebrafish, Danio rerio, pigment-cell autonomous interactions generate dark stripes of melanophores that alternate with light interstripes of xanthophores and iridophores. Here, we identify mechanisms underlying the evolution of a uniform pattern in D. albolineatus in which all three pigment cell classes are intermingled. We show that in this species xanthophores differentiate precociously over a wider area, and that cis regulatory evolution has increased expression of xanthogenic Colony Stimulating Factor-1 (Csf1). Expressing Csf1 similarly in D. rerio has cascading effects, driving the intermingling of all three pigment cell classes and resulting in the loss of stripes, as in D. albolineatus. Our results identify novel mechanisms of pattern development and illustrate how pattern diversity can be generated when a core network of pigment-cell autonomous interactions is coupled with changes in pigment cell differentiation.
Among vertebrates, teleost fishes have some of the most striking and diverse adult pigment patterns and these patterns can have important roles in behavior and speciation1, 2, 3, 4, 5, 6, 7, 8. Though mechanisms of pattern formation are starting to be elucidated, we still know very little about the genetic and cellular bases of pattern diversification. Fishes in the genus Danio can potentially shed light on pattern evolution because of their diversity of patterns9, 10, 11, 12, 13, and the phylogenetic proximity of these species to zebrafish D. rerio14, in which pattern development is being studied intensively. In zebrafish, dark stripes, comprising melanophores and a few iridescent iridophores, alternate with light interstripes of yellow-orange xanthophores and abundant iridophores, all of which are located in the hypodermis, between the skin and myotome15, 16, 17, 18. This striped pattern arises through interactions between pigment cells and their environment, as well as interactions within and between pigment cell classes19, 20, 21, 22, 23, 24, 25, 26; remarkably, the dynamics of some of these interactions resemble those predicted by Turing models of pattern formation27, 28, 29.
Within Danio a pattern that includes horizontal stripes and interstripes at some stage of post-embryonic development is likely to be ancestral10, 30, 31. These stripes and interstripes persist and can be reiterated in the adults of some species, and are most distinctive in D. rerio (Fig. 1a); similarly organized, albeit fewer, stripes and interstripes are found in the close zebrafish relative, spotted danio, D. nigrofasciatus. At the opposite end of a stripe continuum6 is the pearl danio, D. albolineatus, and very closely related species or subspecies (e.g., D. roseus), in which pigment cells of different classes are intermingled and nearly uniformly distributed, and only a residual interstripe remains (Fig. 1a). Although ambiguities in species relationships14, 32, 33, 34 preclude assessing the polarity of some evolutionary transformations in Danio, developmental and genetic analyses indicate that an ancestral pattern of stripes and interstripes has been elaborated upon in D. rerio and obscured in D. albolineatus9, 10, 31, 35, suggesting that further analyses of these divergent phenotypes could inform our understanding of pattern evolution more generally.
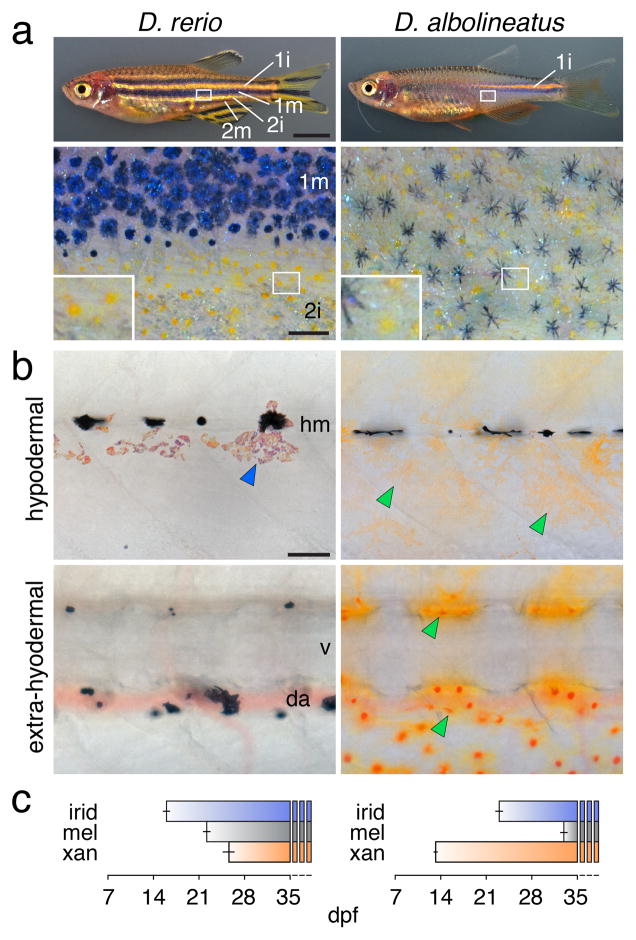
Figure 1. Different pigment patterns of D. rerio and D. albolineatus.
(a) D. rerio have interstripes with intervening stripes. The primary interstripe (1i) is first to develop, followed by primary stripes of melanophores (1m) and secondary interstripes (2i) and stripes (2m)42. Xanthophores (lower, inset) and melanophores are segregated spatially. D. al-bolineatus develop only a single incomplete interstripe, and melanophores and xanthophores are intermingled across the flank.
(b) Early in adult pigment pattern development in D. rerio (stage AR 42), xanthophores had not yet differentiated and only adult iridophores (blue arrowhead) and residual early larval melano-phores were observed hypodermally. At the same stage in D. albolineatus, numerous xantho-phores (green arrowheads) had differentiated hypodermally and extra-hypodermally. hm, horizontal myoseptum; v, vertebral column; da, dorsal aorta.
(c) In comparison to D. rerio, xanthophores of D. albolineatus developed precociously. Shown are mean±s.e.m. days post-fertilization when pigment cells of each class first appeared (species difference across all classes, F2,18=158, P<0.001; n=5 D. rerio; n=3 D. albolineatus).
Scale bars,1 mm (a upper); 200 μm (a lower); 60 μm (b).
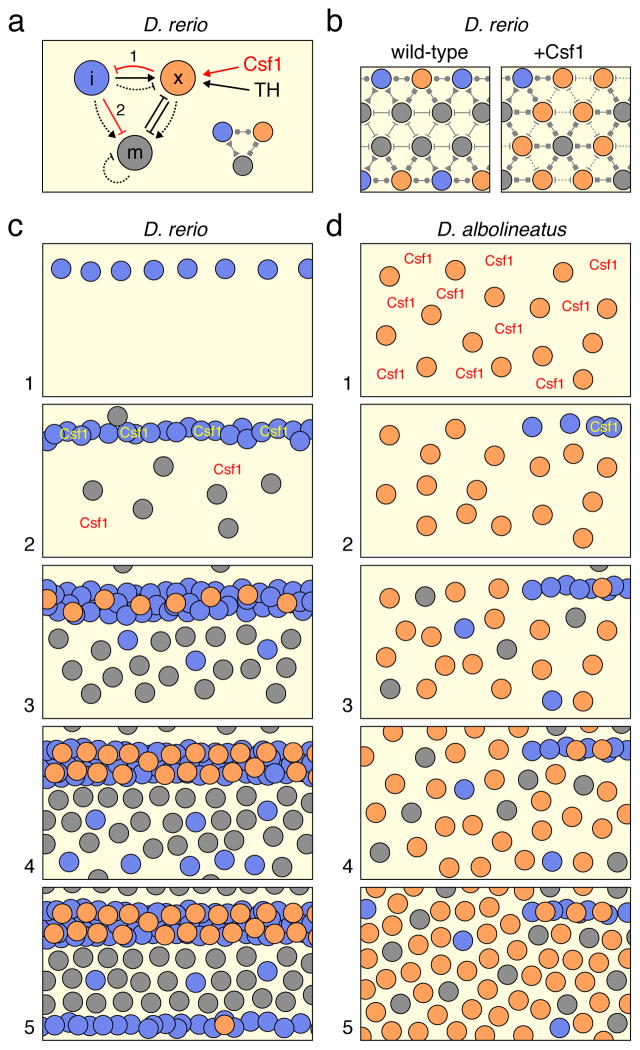
In this report, we investigated the mechanisms responsible for the nearly uniform pattern of D. albolineatus as compared to the reiterated stripes and interstripes of D. rerio. Besides overall pattern, these species differ strikingly in xanthophore abundance, with many more of these cells present in D. albolineatus hypodermally, and also “extra-hypodermally” in more medial locations (Fig. 1b). Although xanthophores of D. rerio require thyroid hormone (TH), many xanthophores of D. albolineatus develop independently of TH, suggesting the evolution of a compensatory factor in this species26. Here we show that a precocious, widespread differentiation of xanthopho-res in D. albolineatus is associated with increased expression of a xanthogenic factor, Colony stimulating factor 1 (Csf1), resulting in part from cis regulatory evolution at the csf1a locus. When Csf1 is expressed correspondingly in D. rerio, pigment cells are intermingled and a uniform pattern largely recapitulating that of D. albolineatus developed, owing to a xanthophore-dependent repression of iridophore organization and concomitant failure of stripe–interstripe boundary formation. Finally we show that interstripe iridophores in D. rerio not only specify melanophore stripe orientation and position22, 23, but also determine stripe width, and that development of these cells has been dramatically curtailed in D. albolineatus. Our results identify molecular and cellular changes contributing to a uniform pattern in D. albolineatus and illustrate how changes in the time and place of pigment cell differentiation have cascading effects on pattern formation.
Results
More xanthophores and increased Csf1 in D. albolineatus
Analyses of developing larvae revealed that xanthophores are the first adult pigment cell to differentiate in D. albolineatus, but the last to differentiate in D. rerio (Fig. 1b,c; Supplementary Fig. 1). We hypothesized that precocious development of especially abundant xanthophores in D. albolineatus evolved through changes in the expression of Colony Stimulating Factor 1 (Csf1). In D. rerio, Csf1 signaling through the Csf1 receptor (Csf1r)36, 37 is essential for xanthophore differentiation, proliferation and survival19, 20, 38. Csf1 is expressed by interstripe iridophores and promotes the development of interstripe xanthophores; this factor is also expressed in the skin and its ectopic expression drives ectopic xanthophore development22. Moreover, analyses of D. rerio x D. albolineatus hybrid phenotypes have suggested evolutionary changes in the Csf1 pathway9, 31. By RT-PCR of both species, we found that the two Csf1 loci, csf1a and csf1b, were expressed in skin and iridophores (Supplementary Fig. 2a). By quantitative RT-PCR of “internal” tissue denuded of skin, but including cells adjacent to hypodermis, csf1a and csf1b transcript abundances were elevated by as much 4–6 fold in D. albolineatus from early stages (Fig. 2a; Supplementary Fig. 2b). Thus, xanthogenic Csf1 is upregulated when xanthophores first develop in D. albolineatus, particularly internally where extra-hypodermal xanthophores arise, and adjacent to where hypodermal xanthophores develop.
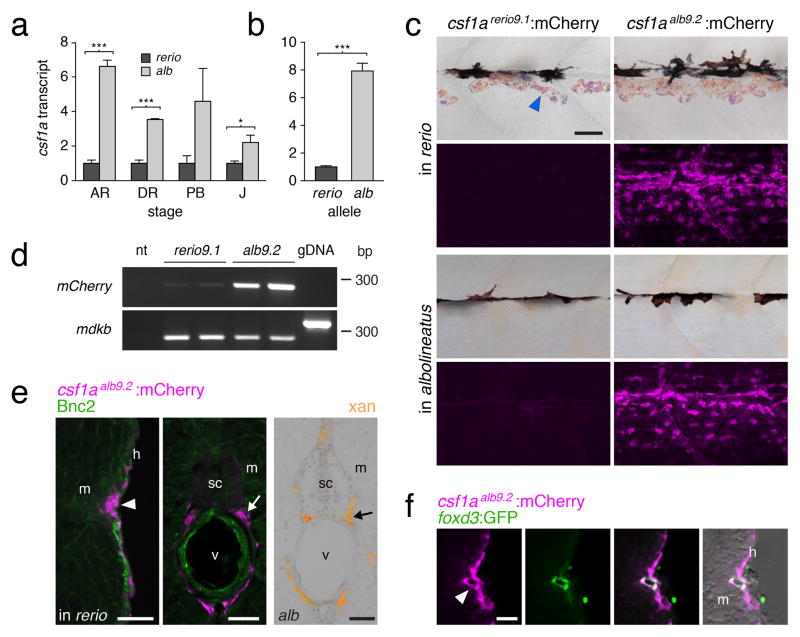
Figure 2. Enhanced Csf1 expression in D. albolineatus through cis regulatory evolution.
(a) csf1a transcript abundance during adult pigment pattern development in D. albolineatus relative to D. rerio for internal trunk tissue (mean±s.e.m.; n=3 biological replicates for all sample). ***P<0.0001; *P<0.05. (Stages: AR, anal fin ray appearance; DR, dorsal fin ray appearance; PB, pelvic bud appearance; J, juvenile.)
(b) Increased expression of D. albolineatus csf1a allele in D. rerio x D. albolineatus hybrids (mean±s.e.m.; paired t=12.8, d.f.=4, P<0.0005; n=5 biological replicates)
(c) mCherry in AR+ stage Tg(csf1arerio9.1:mCherry) (left) and Tg(csf1aalb9.2:mCherry) (right), in D. rerio (top) and D. albolineatus (bottom). Images for csf1arerio9.1:mCherry were exposed twice as long yet show only background fluorescence.
(d) csf1arerio9.1:mCherry was detectable at low levels. midkine b (mdkb), control target amplifying 259 bp from cDNA or 334 bp with intron from genomic DNA (gDNA); nt, no template.
(e) csf1aalb9.2:mCherry in D. rerio (magenta, arrowhead) in hypodermis (h), adjacent to the myo-tome (m) and also medially (arrow) in the vicinity of the spinal cord (sc) and vertebral column (v), corresponding to ventral motor nerves. Basonuclin-2 (Bnc2; green) promotes Csf1 expression in D. rerio22, but Bnc2+ cells did not co-express mCherry. Right, extra-hypodermal xantho-phores (arrow) in D. albolineatus.
(f) At the hypodermis, csf1aalb9.2:mCherry was expressed by Foxd3+ glia (green) of the lateral line nerve (arrow) and other cells.
Scale bars, 400 μm (a, for a–g), 400 μm (d); 60 μm (e for e–g); 60 μm (h).
cis regulatory evolution underlying csf1a expression
To understand the mechanism underlying evolutionary change in Csf1 expression, we focused on csf1a, for which the species difference lasts into late stages. To test for cis regulatory change at the csf1a locus, we crossed D. albolineatus to D. rerio and assayed species-specific transcript abundance in the averaged trans regulatory background of the hybrid. If differences are in cis, the D. albolineatus allele should be expressed higher than the D. rerio allele; if differences are in trans, alleles should be expressed similarly. These analyses revealed 8-fold higher expression of the D. albolineatus csf1a allele, strongly suggesting that cis regulatory changes contribute to the differential expression between species (Fig. 2b).
To further test for cis regulatory evolution, we cloned ~9 kb regions between a conserved distal sequence and the csf1a translational start sites. We used these fragments, csf1arerio9.1 and csf1aalb9.2, to drive mCherry in multiple stable transgenic lines in each species. Each of 9 lines for csf1arerio9.1:mCherry failed to express mCherry at levels visualizable by native fluorescence or immunohistochemistry, though low levels of transcript were detectable by RT-PCR (Fig. 2c,d). By contrast, each of 6 lines for csf1aalb9.2:mCherry exhibited robust expression from early larval stages. Similar to native csf1a transcript in D. rerio22, csf1aalb9.2:mCherry was expressed in the hypodermis. csf1aalb9.2:mCherry also was expressed medially in peripheral nerves, adjacent to where extra-hypodermal xanthophores develop in D. albolineatus (Fig. 2e) and where xan-thophore precursors occur in D. rerio26; extra-hypodermal csf1a transcript has not been detectable by in situ hybridization in D. rerio22. Finally, csf1aalb9.2:mCherry was expressed by Foxd3+ glia of the lateral line (Fig. 2f) and in several other tissues as well (Supplementary Fig. 3).
Alignments of csf1arerio9.1 and csf1aalb9.2 revealed conserved regions missing or disrupted in D. albolineatus (Supplementary Fig. 3a), suggesting that increased expression may have resulted from the loss of one or more repressor elements. As an initial test of this possibility, we focused on an altered proximal region, where repressors often are found39, 40, and generated transgenic reporter lines for deletions of corresponding regions in D. rerio. In contrast to lines for csf1arerio9.1, in which mCherry was never detectable, deletion lines exhibited high mCherry expression from early stages that overlapped with sites of csf1aalb9.2 expression, yet specific domains varied markedly across constructs and replicates (Supplementary Fig. 3b), suggesting a derepression but also dysregulation subject to integration site effects. Though additional regulatory changes are presumed, these findings support a model in which loss of one or more rep-ressor elements contributed to earlier, higher and more widespread expression of csf1a in D. albolineatus, contributing to earlier, more numerous and more broadly distributed xanthophores.
Csf1 recruits extra xanthophores and drives stripe loss
We next asked whether changes in the time and place of xanthophore differentiation could account for the more uniform pattern of D. albolineatus. We reasoned that xanthophores, as the first adult pigment cells to appear in D. albolineatus (Fig. 1b,c), might have a critical role in specifying pattern, much as iridophores, the first adult pigment cell to appear in D. rerio, specify the location and orientation of the first stripes and interstripe22, 23. To test this possibility, we used the ubiquitous, heat-shock inducible promoter of hsp70l to overexpress Csf1a and thereby generated extra xanthophores in D. rerio, beginning when differences in Csf1 expression were first detectable between species. The resulting pattern resembled that of D. albolineatus, with (ectopic) extra-hypodermal xanthophores, supernumerary hypodermal xanthophores, and stripes of intermingled melanophores and xanthophores that extended to the ventral margin of the flank (Fig. 3a,c,d,l,m; Supplementary Fig. 4a). These fish also had extra melanophores, likely reflecting an indirect effect of Csf1 mediated through xanthophores, which provide trophic support to melanophores19, 28, 41 (Supplementary Fig. 4b).
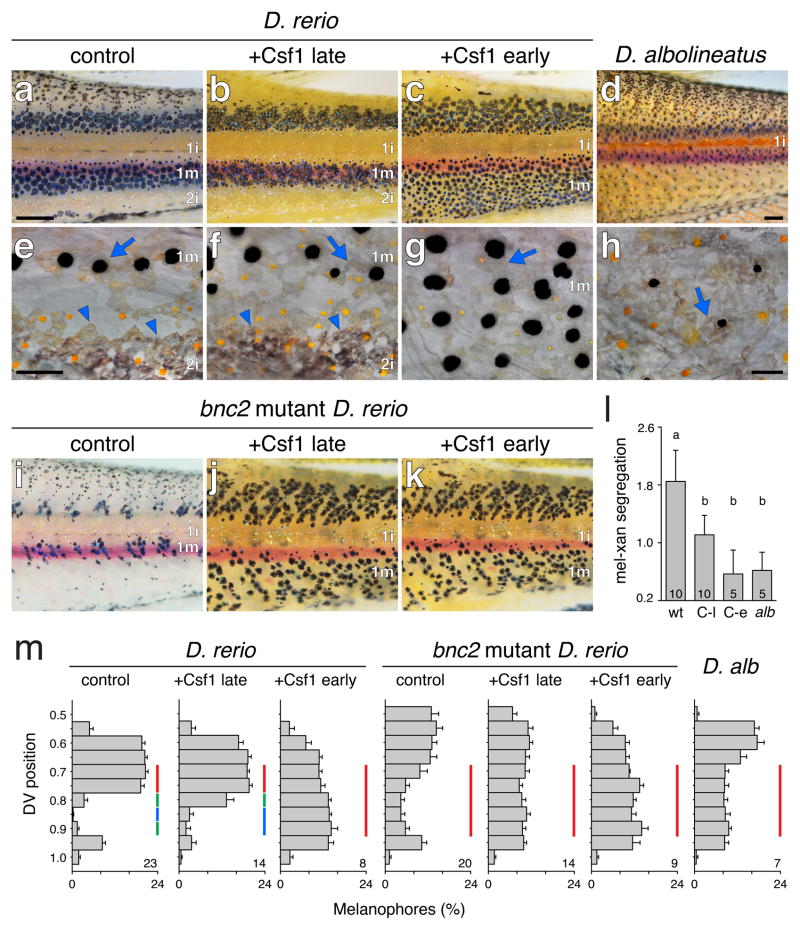
Figure 3. Time and pattern of xanthophore differentiation is critical for pattern outcome.
(a–d) When Csf1 was induced and extra xanthophores developed early in D. rerio (c; stage AR+ 42), stripes extended to the ventral margin of the flank, and secondary interstripes failed to develop similar to D. albolineatus (d). When induced late (b; SA+), stripes widths were similar to controls (a).
(e–h) Higher magnification images showing organized, interstripe iridophores (arrowheads in e, f) or their absence (g, h); scattered iridophores (arrows) are present in all panels. Yellow-orange xanthophores were segregated from melanophores in e, but intermingled with melanophores in f–h.
(i–k) bnc2 mutants fail to develop secondary interstripe iridophores and Csf1 overexpression either early or late resulted in a uniform pattern.
(l) Melanophore–xanthophore segregation was less in Csf1-overexpressing D. rerio and D. al-bolineatus (“wt,” non-transgenic siblings heat shock similarly) regardless of stripe presence (“C-l,” Csf1 late-overexpression) or absence (“C-e,” Csf1 early overexpression; alb, D. albolineatus). Common letters above bars indicate groups not significantly different (P>0.05) in Tukey Kramer post hoc comparisons; overall F3,27=9.2, P<0.0005). Shown are means±s.e.m.; sample sizes are indicated within each bar.
(m) Melanophore frequencies at dorsal–ventral positions on the ventral flank (0.5=horizontal myoseptum, 1=ventral margin of myotomes). Vertical bars of the same color indicate positions not significantly different in Tukey Kramer post hoc comparisons (P>0.05): a distinct discontinuity in melanophore distribution, representing the stripe–interstripe boundary, was evident in control and Csf1 late-overexpressing D. rerio but not other backgrounds. For clarity only comparisons across positions 0.7–0.9 are shown. Shown are means±s.e.m.; sample sizes are indicated at the lower right of each plot.
Scale bars, 400 μm (a, for a–g), 400 μm (d); 60 μm (a′ for a′–c′); 60 μm (d′).
Xanthophores can repress interstripe development
Besides alterations in melanophore and xanthophore distributions, D. rerio overexpressing Csf1 from early stages had changes to iridophore patterning. D. rerio normally develop a single “primary” interstripe of iridophores followed by two primary stripes of melanophores above and below. Later, “secondary” interstripes are added further dorsally and ventrally, followed by secondary stripes42. In Csf1-overexpressing D. rerio, however, iridophores were fewer and aggregations of secondary interstripe iridophores failed to develop, similar to D. albolineatus (Fig. 3e,g,h). Because changes in iridophore pattern could have resulted from Csf1-dependent increases in either xanthophores or melanophores, we used a transgenic line22 for melanogenic Kit ligand-a43, 44 to generate extra melanophores, but not xanthophores, at the same stages: in these fish iridophores were distributed similarly to the wild type (Supplementary Fig. 5). Together these findings indicate that overexpression of Csf1 and xanthophore population expansion can repress iridophore organization, despite the normal role of iridophores in promoting xanthophore development22. This suggests that changes in differentiation timing and location can have cascading effects, dramatically altering the pattern even without changes in the network architecture of pigment-cell autonomous interactions19, 20, 22, 23, 27, 28.
To assess the limits of such pattern plasticity, we overexpressed Csf1 in D. rerio at a later stage, when secondary interstripe iridophores had started to develop already. In these fish, supernumerary xanthophores were intermingled with melanophores, yet the secondary interstripe persisted and melanophores were confined to the normal domain of the ventral primary stripe (Fig. 3b,f,l,m), suggesting that once a secondary interstripe has been established it is resistant to Csf1-dependent xanthophore repression. To further test that persistence of a stripe–secondary interstripe boundary in late Csf1-overexpressing fish depended on iridophores, we repeated these experiments in bnc2 mutant D. rerio, which have severe deficiencies in all three pigment cell classes and fail to form secondary interstripes22, 45. In this background, Csf1 over-expression resulted in intermingled melanophores and xanthophores that extended to the margin of the flank, regardless of stage (Fig. 3i–k,m). Together these experiments suggest a “priority effect” in which the first pigment cell class to differentiate in a given region can have a critical influence in specifying the pattern.
Iridophores delimit stripe width
To place these findings in an evolutionary context, we further examined iridophore development in both species, focusing on the ventral region of the flank (Fig. 4a). In D. rerio, iridophores initially populated the primary interstripe but were later found sparsely within the ventral primary stripe, and, subsequently, in aggregations further ventrally where they established a secondary interstripe. As iridophores increased in number, melanophores that had started to differentiate in the prospective secondary interstripe died or migrated away (Fig. 4b; Supplementary Movie 1). These observations suggested that iridophores not only initiate melanophore stripe patterning22, 23 but also terminate stripes once patterning has started. We tested this by ablating secondary interstripe iridophores using a transgene, pnp4a:nVenus-2a-nfnB, in which bacterial nitroreduc-tase is expressed by the iridophore-specific promoter of purine nucleoside phosphorylase 4a, allowing targeted killing of iridophores by treatment with metronidazole22, 46. In the absence of iridophores, melanophores persisted and the ventral primary melanophore stripe extended to the margin of the flank (Fig. 4c; Supplementary Fig. 6a–c).
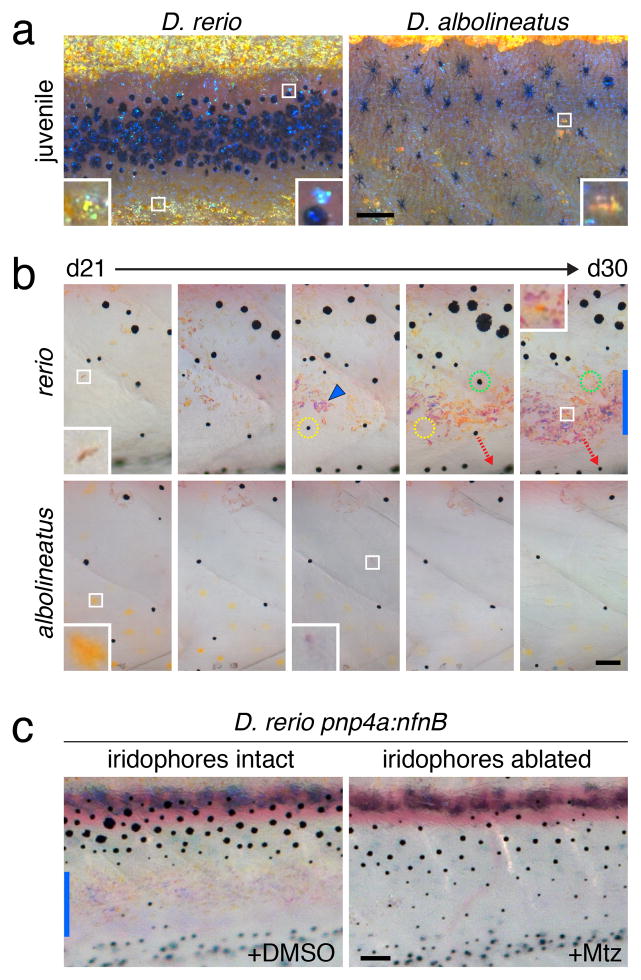
Figure 4. Iridophore distributions and role in stripe termination.
(a) Juvenile D. rerio and D. albolineatus imaged to highlight iridophores. For D. rerio, insets show iridophores at the edge of the ventral secondary interstripe (left) and an iridophore in the stripe (right). For D. albolineatus, inset shows one of only a few small aggregates of iridophores
(b) Development of ventral iridophores between 21 and 30 dpf (stages PB+ to SA). In D. rerio, iridophores (e.g., inset, d21) were initially scattered among melanophores in the developing ventral primary melanophore stripe but subsequently formed aggregates (e.g., arrowhead) at the site of the secondary ventral interstripe. Some melanophores initially in this region died; yellow and green dashed circles show initial positions for each of two melanophores and their absence on subsequent days. Other melanophores in this region translocated further dorsally or ventrally; dashed red arrow indicates one melanophore that moved ventrally). At d30, inset shows first xanthophore to differentiate in the secondary interstripe and vertical blue bar indicates overall interstripe width. In D. albolineatus at d21, xanthophores are abundant (inset) but iridophores are scarce (e.g., inset, middle panel) as are melanophores.
(c) In D. rerio, ablation of ventral secondary interstripe iridophores by treatment with Mtz results in a failure of stripe termination. Blue bar, secondary interstripe in DMSO control. Fish in b and c were treated with epinephrine to contract pigment granules towards cell centers.
Scale bars, 60 μm (a, b); 100 μm (c).
In D. albolineatus, iridophores were fewer initially and never formed ventral aggregations (Fig. 4a,b; Supplementary Movie 2) though in the adult these cells were scattered widely over the flank (Supplementary Fig. 6d). Thus, interstripe iridophores delimit the width of melanophore stripes in D. rerio whereas development of these cells has been curtailed in D. albolineatus, a change that may itself be xanthophore-dependent (compare Figures 3e,g,h).
Discussion
Our results identify new interactions among pigment cell classes in D. rerio, elaborating upon a model for stripe development, and also suggest a new model for how evolutionary changes in pigment cell differentiation have contributed to the strikingly different pattern of D. albolineatus, and perhaps other species.
Previous studies elucidated a complex network of interactions involving all three pigment cell classes in D. rerio (Fig. 5a)19, 20, 21, 22, 23, 24, 25, 26, 27, 28. Iridophores promote the differentiation and localization of xanthophores at short range and repress their differentiation at long range. By producing extra pigment cells transgenically, we identified a reciprocal interaction in which xanthophores repress the development and organization of iridophores (interaction 1 in Fig. 5a). A contribution of xanthophores to patterning iridophores during normal development is likewise suggested by defects in iridophore organization in xanthophore-deficient csf1r mutants22, 23. Our analyses of phenotypes resulting from iridophore ablation further support the idea that iridopho-res influence melanophore distributions either directly or indirectly (interaction 2 in Fig. 5a), presumably through a combination of short range inhibition and longer range attraction22, 23. These experiments that either increased or decreased the numbers of pigment cells illustrate the critical role of cellular context in pattern formation: even with the core network of interactions unchanged, differences in the abundance and distribution of particular pigment cell classes can limit the sort of interactions in which cells participate, with consequences for the pattern that develops (Fig. 5b).
Figure 5. Models for pattern formation and evolution.
(a) Network of interactions amongst iridophores (i), xanthophores (x) and melanophores (m).Interactions are positive (→) or negative (⊣); long-range are indicated by dashed lines. Csf1 and TH both promote xanthophore development; Csf1 is also supplied by iridophores to xantho-phores (i→x)22. When xanthophores are highly abundant, these cells can repress iridophore development (1). Iridophores attract melanophores (i→m), but also repress melanophore survival and localization, and terminate stripe development (2). Simplified interaction diagram at lower right, corresponding to (b).
(b) When Csf1 is overexpressed In D. rerio, melanophores encounter more xanthophores and fewer iridophores, participate in only a subset of potential interactions, and fail to receive positional information necessary to terminate primary stripes or initiate secondary stripes. Interactions between xanthophores are hypothetical.
(c) Pattern development in D. rerio. (1) Iridophores differentiate at the horizontal myoseptum. (2) Melanophores arise dispersed over the flank while iridophores express Csf1, which is also expressed in the skin. (3) Locally high levels of Csf1 promote xanthophore differentiation in the interstripe as interactions involving all three classes of pigment cells refine the pattern of stripes and interstripes and additional iridophores infiltrate the stripe. (4) Iridophores begin to emerge beyond the stripe and initiate a secondary interstripe. (5) Iridophores of the secondary inter-stripe terminate the primary stripe, promote differentiation of secondary interstripe xanthopho-res, and specify the location of secondary stripe development. Adult xanthophore precursors, and, later, incompletely differentiated xanthophores are distributed sparsely in stripes as well26,50 (not shown).
(d) Pattern development in D. albolineatus. (1) Initially high expression of iridophore-independent Csf1 promotes early, widespread xanthophore development. (2) A few iridophores develop near the horizontal myoseptum posteriorly. (3) Melanophores begin to appear scattered over the flank as do additional iridophores. (4–5) Residual stripes begin to form posteriorly around the residual interstripe35 but this pattern is obscured by widespread melanophores and xanthophores that fail to resolve into stripes and interstripes.
In D. rerio, these several interactions, combined with tissue-derived positional information and other factors, generate the body stripes and interstripes (Fig. 5c). Iridophores are the first adult pigment cells to differentiate22, 23, 42. Their precursors migrate from extra-hypodermal locations to the hypodermis where they differentiate, proliferate and organize in the vicinity of the horizontal myoseptum47, 48, thereby establishing the primary interstripe (Fig. 5c1). Melanophores are the second adult pigment cell to appear, and arise from highly motile precursors that also travel from extra-hypodermally, via peripheral nerves and other tissues, to reach the hypoder-mis44, 49. Once in the hypodermis, these cells initiate their differentiation relatively widely over the flank49 (Fig. 5c2). Some newly differentiating melanophores—as well as persisting early larval melanophores—occur in the interstripe and migrate short distances to join the stripes; others initially in the interstripe die or are covered by iridophores17, 21, 22, 31, 38. Iridophores help to organize melanophores, and thereby specify the location and orientation of the primary stripes, and later colonize the primary stripe in smaller numbers (this study; 22, 23). Interstripe xanthophores are the last to appear (Fig. 5c3), and develop from cells already in the hypodermis including embryonic xanthophores that transiently lose their pigment only to reacquire it later26. Xan-thophore differentiation depends on TH as well as Csf1, which is supplied locally by iridophores and more globally by other cells in the skin22; interactions between xanthophores and melano-phores promote the segregation of these cell types into stripes and interstripes19,20,26,28 (Fig. 5c4). During later development interstripes and stripes are added. Our analyses of iridophore ablation phenotypes demonstrate an important role for iridophores in this process of pattern reiteration, specifically in terminating the primary melanophore stripe and specifying the position of the next, secondary melanophore stripe (Fig. 5c6). These inferences and our observations of iridophore development (e.g., Supplementary Movie 1) are consistent with a recent description of irido-phore locations and clonal expansion47.
Our study reveals very different events in D. albolineatus. In this species, pattern formation begins with an abundance of iridophore-independent Csf1, which promotes early, wide-spread xanthophore differentiation (Fig. 5d1). Unlike primary interstripe iridophores in D. rerio, which impart positional information to melanophores22, 23, the scattered population of xanthophores in D. albolineatus lacks positional information for melanophores. In addition, the primary interstripe is itself reduced and secondary interstripes do not form, possibly owing to repressive effects of xanthophores, though a definitive assessment of this idea requires additional analyses now underway. Thus, although D. albolineatus initiate interstripe and stripe development, especially posteriorly (Fig. 5d2, 5d3), and exhibit latent-stripe forming potential31, 35, the combination of widespread xanthophores and an absence of secondary interstripe iridophores promotes a nearly uniform pattern of melanophores.
We found that D. albolineatus express more Csf1 than D. rerio owing in part to cis regulatory changes at csf1a, and that enhanced Csf1 expression can drive the development of supernumerary xanthophores in D. rerio, resulting in the loss of stripes, similar to D. albolineatus. These findings also suggest excess Csf1 as a candidate mechanism allowing some xanthophores to differentiate in D. albolineatus even without TH26. Genetic analyses previously identified Csf1r or its pathway as potentially contributing to stripe loss in D. albolineatus and pattern alterations in other danios9, 31: hybrids between D. albolineatus and wild-type D. rerio developed stripes, yet hybrids between D. albolineatus and xanthophore-deficient csf1r mutant D. rerio had disrupted stripes and xanthophores in a pattern resembling D. albolineatus. Our observations are consistent with a model in which excess Csf1 derived from D. albolineatus alleles, in conjunction with evolutionary changes affecting receptor–ligand interactions31, explain the disruption of stripes in hybrids between D. albolineatus and D. rerio mutant for csf1r.
We have identified roles for Csf1 and differential xanthophore recruitment in the evolution of pattern differences between D. albolineatus and D. rerio. Yet, our findings do not exclude roles for additional factors, including changes to pigment cell-autonomous interactions themselves, or other modifications to the tissue environment or hormonal milieu. Indeed, experiments with D. rerio did not recapitulate the smaller population of melanophores in D. albolineatus: in Csf1-overexpressing D. rerio, extra xanthophores were accompanied by extra melanophores, whereas in D. albolineatus, melanophores are fewer and more likely to die than in D. rerio31. This disparity may reflect species differences in the relative strengths of supportive and repressive interactions between melanophores and xanthophores19, 28, 41, as has also been suggested by genetic analyses31. Such possibilities are currently being investigated.
In conclusion, our analyses demonstrate that changing the initial time and place at which pigment cells differentiate can result in “priority effects” that alter subsequent pattern development. If pigment-cell autonomous interactions are an engine for pattern formation, our study illustrates that very different pattern outcomes can occur depending on the context in which this engine operates. Such considerations may help to explain the extraordinary diversification of pigment pattern in teleosts8, 11, 51, 52 and other ectothermic vertebrates53, 54, 55.
Methods
Fish stocks, staging and rearing conditions
D. rerio wild-type stock fish, WT(WA), were produced by crosses between the inbred genetic strains ABwp and wik or the progeny of these crosses. Iridophore-deficient bonaparte mutants are presumptive null alleles bnc2utr16e1 (45). D. aff. albolineatus31 stocks were obtained from tropical fish suppliers and have been inbred and maintained in the lab for more than 8 generations. Transgenic lines were: Tg(hsp70l:csf1a-IRES-nlsCFP)wp.r.t4; Tg(hsp70l:kitlga)wp.r.t2, Tg(csf1arerio:mCherry) and Tg(csf1aalb:mCherry) in D. rerio; Tg(csf1arerio:mCherry) and Tg(csf1aalb:mCherry) in D. albolineatus; and TgBAC(foxd3:EGFP)n15 D. rerio (56), kindly provided by Alex Nechiporuk (Oregon Health Sciences University, Portland OR). Post-embryonic staging followed (42). D. albolineatus reached each developmental stage at a larger size than D. rerio so criteria such as fin ray development were used as an indicator of stage rather than standardized standard length (SSL). All fish stocks were reared in standard conditions of 14L:10D at 28.5°C, and fed Rotimac-fortified marine rotifers followed by brine shrimp and flake food. For transgene inductions of Tg(hsp70l:csf1a-IRES-nlsCFP)wp.r.t4 and Tg(hsp70l:kitlga)wp.r.t2, fish were heat-shocked at 38°C twice daily for 1 h through adult pigment pattern formation. All experiments were conducted with approval of the University of Washington Animal Care and Use Committee and complied with United States federal guidelines for ethical use of animals in research.
Transgene construction and microinjection
To generate csf1a reporter lines, we cloned ~9 kb proximal to the csf1a start site from both species into a 5′ Gateway vector. Transgenes were then assembled by Gateway cloning of entry plasmids into pDestTol2CG2 containing Tol2 repeats for efficient genomic integration and cmlc2:EGFP as a transgenesis marker57, 58. Microinjection of plasmids and Tol2 mRNA followed standard methods. Progeny of F0 injected fish were screened for cmlc2:EGFP and used to establish independent, stable transgenic lines in D. rerio [Tg(csf1arerio9.1:mCherry), n=8; Tg(csf1aalb9.2:mCherry), n=3); Tg(csf1arerio9.1_338:mCherry), n=3; Tg(csf1arerio9.1_1039:mCherry), n=6) and D. albolineatus [Tg(csf1arerio9.1:mCherry), n=1; Tg(csf1aalb9.2:mCherry), n=3)]. Conservation and divergence of csf1a regulatory regions was evaluated using the UCSC Genome Browser59. Ablation of secondary interstripe iridophores in larvae mosaic for pnp4a:nVenus-2a-nfnB followed (22), with Mtz treatment beginning at stage PR.
RT-PCR and quantitative RT-PCR
For quantitative RT-PCR in D. rerio and D. albolineatus, skins and underlying tissue were collected from AR+, DR+, PB+ and J stage larvae. To generate interspecific hybrids for quantitative RT-PCR, a D. rerio female was crossed to a D. albolineatus male by in vitro fertilization and resulting larvae were collected at AR+. All tissues were placed directly into either Trizol Reagent (Invitrogen) or RNAlater (Ambion). RNA was isolated using either RNaqueous Microkit (Ambion) or Trizol, followed by LiCl precipitation. cDNA was synthesized with iScript cDNA Synthesis Kit (Bio-Rad). Quantitative RT-PCRs were performed and analyzed on a StepOnePlus System (Life Technologies) using Custom Taqman Gene Expression Assays designed to bind either conserved sites (csf1a, csf1b, rpl13a) or species specific sites (csf1a hybrid analysis) and Taqman Gene Expression Master Mix (Life Technologies). For assays of expression from species-specific alleles in hybrids, abundances estimated from probes specific to each allele were normalized to a common probe recognizing both alleles. All assays were replicated across ≥3 biological samples each having three technical replicates.
To detect mCherry transcripts in D. rerio Tg(csf1arerio9.1:mCherry) and Tg(csf1aalb9.2:mCherry) transgenic lines, individual larvae were collected at stage PR. Heads and tails were removed and trunks were placed in RNAlater (Ambion). RNA was isolated using the Direct-zol RNA Kit (Zymo Research, R2050) and cDNA synthesized using the iScript cDNA Synthesis Kit (Bio-Rad).
For RT-PCR of isolated iridophores, late metamorphic/early juvenile larvae were euthanized and skins were collected in PBS. Skins were vortexed briefly to remove scales, then centrifuged gently and washed again in PBS. Tissue was incubated 10 min at 37°C in 0.25% trypsin-EDTA (Invitrogen). Trypsin was removed and tissue incubated 10 min at 37°C in tyrpsin-inhibitor (Sigma T6414) with 3 mg/ml collagenase, and 2 μl Dnase I, RNase-Free (Thermo Scientific), followed by gentle pipetting until skins were completely dissociated. Cells were then washed in PBS and filtered through a 40 μm cell strainer. Cell were placed on a glass bottom dish and examined on a Zeiss Observer inverted compound microscope. A minimum of 50 iridophores were picked using a Narishige 1M 9B microinjector, expelled into PBS, then re-picked and expelled directly into Resuspension Buffer. cDNA was synthesized using Superscript III Cells Direct cDNA Synthesis Kit (Invitrogen). RT-PCR was performed with the following primer sets (forward, reverse): actb1, ACTGGGATGACATGGAGAAGAT, GTGTTGAAGGTCTCGAACATGA; csf1a, CAACAACTGAGCCAACACATAAATA, GGGATCTGTGGTCTTTGCTGAT; csf1b, AACACCCCTGTTAACTGGACCT, GAGGCAGTAGGCAGTGAGAAGA; mdkb, AGTGAATGG-CAGTATGGGAAAT, TGGACACTTTAATGGTGGTCTG; mCherry, CCAGCTTGATGTTGAC-GTTG, AGGACGGCGAGTTCATCTAC; pnp4a, GAAAAGTTTGGTCCACGATTTC, TACTCATTCCAACTGCATCCAC.
Immunohistochemistry
For immunohistochemistry, fish were fixed in 4% paraformaldehyde for 30 minutes at room temperature, washed with PBS, transferred to 15% sucrose, followed by 30% sucrose, and then embedded and frozen in OCT media. Cryosections of 20_m thickness were collected on Mi-crofrost Plus slides (Fisher) and allowed to dry. Slides were washed in PBS and then fixed in 4% paraformaldehyde for 10 minutes at room temperature followed by additional PBS washes. Sections were blocked in PBS containing 5% heat inactivated goat serum and 0.1% Triton-X, then incubated overnight at 4°C with primary antibody. Antibodies were monoclonal rat anti-mCherry (1:300; Life Technologies, M11217), and polyclonal rabbit anti-Human BNC2 antibody (1:350). After washes, slides were incubated with secondary antibodies (Alexa Fluor 488, 568), washed in PBS and imaged on a Zeiss Observer inverted microscope equipped with Yokogawa CSU-X1M5000 laser spinning disk.
Imaging and Quantitative Analysis
Fish were imaged on an Olympus SZX-12 stereomicroscope, Zeiss Axioplan 2i compound microscope or Zeiss Observer inverted compound microscope using Zeiss Axiocam HR or MRc cameras, or a Photometrics Evolve EMCCD camera and Axiovision software. Images were color balanced and in some instances processed to facilitate visualization of xanthophores in Adobe Photoshop; comparable sets of images across genetic backgrounds or treatments were processed identically.
For repeated imaging of individuals, fish were reared separately in glass beakers, then immediately prior to imaging were treated with 10 mM epinephrine to contract pigment granules towards cell centers, then anesthetized briefly, imaged, and allowed to recover. Resulting images were re-scaled and aligned in Photoshop then exported as animated movies.
To quantify melanophore dorsal–ventral positions, we measured the distance of each mel-anophore to the dorsal and ventral margins of the myotome and divided dorsal length by the total distance. For pnp4a:nVenus-2a-nfnB mosaic iridophore ablations and controls, positions were determined for all melanophores ventral to the horizontal myoseptum, in the area bordered by the anterior and posterior ends of the anal fin. For D. albolineatus, wild-type D. rerio, bnc2 mutant D. rerio, and Tg(hsp70l:csf1a-IRES-nlsCFP)wp.r.t4, melanophore positions were assessed for all melanophores ventral to the horizontal myoseptum, in the anterior third of the area between the anterior and posterior ends of the anal fin.
To assess segregation vs. intermingling of melanophores and xanthophore k-nearest-neighbor (kNN) classifications were conducted using a MATLAB routine written in-house (MAT-LAB R2011a, Mathworks, Natick, MA, USA). For each fish, an average of the five nearest neighbor distances were calculated for each cell from images taken at the border of the ventral melanophore stripe and the secondary ventral iridophore stripe or this region in individuals that did develop iridophores. Melanophore–xanthophore structure indices shown are the ratios of mean melanophore–xanthophore nearest neighbor distances relative to melanophore–melanophore nearest neighbor distance. Other metrics for describing spatial segregation from nearest neighbor values yielded equivelent results.
Analyses of total melanophore numbers and melanophore–xanthophore segregation used ln-transformed dependent values to correct for heteroscedasticity of residuals. All statistical analyses were performed using JMP 8.0.2 (SAS Institute, Cary NC).
Supplementary Material
Acknowledgments
Thanks to Michael Nishizaki for pigment cell k-nearest-neighbor (kNN) analysis in MATLAB, Jessica Spiewak for performing AC stage quantitative RT-PCR and assistance with daily image series, Anna McCann for staging and embedding fish for cyrosectioning, and Amandine Van-houtteghem and Philippe Djian for BNC2 antibody. Supported by NIH R01 GM096906 and NIH R01 GM062182 to DMP.
Footnotes
Author Contributions
Conceived and designed the experiments: LBP DMP. Performed the experiments: LBP EJB. Analyzed the data: LBP DMP. Wrote the paper: LBP DMP.
The authors have no competing financial interests.
References
- 1.Endler JA. Natural-Selection on Color Patterns in Poecilia-Reticulata. Evolution. 1980;34:76–91. doi: 10.1111/j.1558-5646.1980.tb04790.x. [DOI] [PubMed] [Google Scholar]
- 2.Houde AE. Sex, Color, and Mate Choice in Guppies. Princeton University Press; 1997. [Google Scholar]
- 3.Price AC, Weadick CJ, Shim J, Rodd FH. Pigments, patterns, and fish behavior. Zebraf-ish. 2008;5:297–307. doi: 10.1089/zeb.2008.0551. [DOI] [PubMed] [Google Scholar]
- 4.Seehausen O, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455:620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- 5.Theis A, Salzburger W, Egger B. The function of anal fin egg-spots in the cichlid fish As-tatotilapia burtoni. PLoS One. 2012;7:e29878. doi: 10.1371/journal.pone.0029878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engeszer RE, Wang G, Ryan MJ, Parichy DM. Sex-specific perceptual spaces for a vertebrate basal social aggregative behavior. Proc Natl Acad Sci U S A. 2008;105:929–933. doi: 10.1073/pnas.0708778105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mills MG, Patterson LB. Not just black and white: Pigment pattern development and evolution in vertebrates. Semin Cell Dev Biol. 2008;20:72–81. doi: 10.1016/j.semcdb.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts RB, Ser JR, Kocher TD. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science. 2009;326:998–1001. doi: 10.1126/science.1174705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parichy DM, Johnson SL. Zebrafish hybrids suggest genetic mechanisms for pigment pattern diversification in Danio. Dev Genes Evol. 2001;211:319–328. doi: 10.1007/s004270100155. [DOI] [PubMed] [Google Scholar]
- 10.Quigley IK, et al. Pigment pattern evolution by differential deployment of neural crest and post-embryonic melanophore lineages in Danio fishes. Development. 2004;131:6053–6069. doi: 10.1242/dev.01526. [DOI] [PubMed] [Google Scholar]
- 11.Parichy DM. Evolution of danio pigment pattern development. Heredity. 2006;97:200–210. doi: 10.1038/sj.hdy.6800867. [DOI] [PubMed] [Google Scholar]
- 12.Fang F. Barred Danio species from the Irrawaddy River drainage (Teleostei, Cyprini-dae) Ichthyol Res. 2000;47:13–26. [Google Scholar]
- 13.Fang F, Kottelat M. Ichthyol Explor Freshwaters. 1999. Danio species from northern Laos, with descriptions of three new species (Teleostei: Cyprinidae) [Google Scholar]
- 14.Tang KL, et al. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cy-prinidae) Mol Phylogenet Evol. 2010;57:189–214. doi: 10.1016/j.ympev.2010.05.021. [DOI] [PubMed] [Google Scholar]
- 15.Kirschbaum F. Untersuchungen über das Farbmuster der Zebrabarbe Brachydanio rerio (Cyprinidae, Teleostei) Wilhelm Roux’s Arch. 1975;177:129–152. doi: 10.1007/BF00848526. [DOI] [PubMed] [Google Scholar]
- 16.Johnson SL, Africa D, Walker C, Weston JA. Genetic control of adult pigment stripe development in zebrafish. Dev Biol. 1995;167:27–33. doi: 10.1006/dbio.1995.1004. [DOI] [PubMed] [Google Scholar]
- 17.Parichy DM, Turner JM. Zebrafish puma mutant decouples pigment pattern and somatic metamorphosis. Developmental Biology. 2003;256:242–257. doi: 10.1016/s0012-1606(03)00015-0. [DOI] [PubMed] [Google Scholar]
- 18.Hirata M, Nakamura K, Kanemaru T, Shibata Y, Kondo S. Pigment cell organization in the hypodermis of zebrafish. Dev Dyn. 2003;227:497–503. doi: 10.1002/dvdy.10334. [DOI] [PubMed] [Google Scholar]
- 19.Parichy DM, Turner JM. Temporal and cellular requirements for Fms signaling during zebrafish adult pigment pattern development. Development. 2003;130:817–833. doi: 10.1242/dev.00307. [DOI] [PubMed] [Google Scholar]
- 20.Maderspacher F, Nusslein-Volhard C. Formation of the adult pigment pattern in zebraf-ish requires leopard and obelix dependent cell interactions. Development. 2003;130:3447–3457. doi: 10.1242/dev.00519. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi G, Kondo S. Melanophores in the stripes of adult zebrafish do not have the nature to gather, but disperse when they have the space to move. Pigment Cell Mela-noma Res. 2008;21:677–686. doi: 10.1111/j.1755-148X.2008.00504.x. [DOI] [PubMed] [Google Scholar]
- 22.Patterson LB, Parichy DM. Interactions with iridophores and the tissue environment required for patterning melanophores and xanthophores during zebrafish adult pigment stripe formation. PLoS Genet. 2013;9:e1003561. doi: 10.1371/journal.pgen.1003561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frohnhofer HG, Krauss J, Maischein HM, Nusslein-Volhard C. Iridophores and their interactions with other chromatophores are required for stripe formation in zebrafish. Development. 2013;140:2997–3007. doi: 10.1242/dev.096719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamanaka H, Kondo S. In vitro analysis suggests that difference in cell movement during direct interaction can generate various pigment patterns in vivo. Proc Natl Acad Sci U S A. 2014;111:1867–1872. doi: 10.1073/pnas.1315416111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe M, Kondo S. Changing clothes easily: connexin41.8 regulates skin pattern variation. Pigment Cell Melanoma Res. 2012;25:326–330. doi: 10.1111/j.1755-148X.2012.00984.x. [DOI] [PubMed] [Google Scholar]
- 26.McMenamin SK, et al. Thyroid hormone-dependent adult pigment cell lineage and pattern in zebrafish. Science. 2014;345:1358–1361. doi: 10.1126/science.1256251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi M, Yoshimoto E, Kondo S. Pattern regulation in the stripe of zebrafish suggests an underlying dynamic and autonomous mechanism. Proc Natl Acad Sci U S A. 2007;104:4790–4793. doi: 10.1073/pnas.0607790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamasu A, Takahashi G, Kanbe A, Kondo S. Interactions between zebrafish pigment cells responsible for the generation of Turing patterns. Proc Natl Acad Sci U S A. 2009;106:8429–8434. doi: 10.1073/pnas.0808622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kondo S, Miura T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science. 2010;329:1616–1620. doi: 10.1126/science.1179047. [DOI] [PubMed] [Google Scholar]
- 30.Parichy DM. Evolutionary genetics of Danio pigment pattern development. Oxford Univ Press Inc; 2005. [Google Scholar]
- 31.Quigley IK, et al. Evolutionary diversification of pigment pattern in Danio fishes: differential fms dependence and stripe loss in D. albolineatus. Development. 2005;132:89–104. doi: 10.1242/dev.01547. [DOI] [PubMed] [Google Scholar]
- 32.Fang F. Phylogenetic analysis of the Asian cyprinid genus Danio (Teleostei, Cyprinidae) Copeia. 2003;2003:714–728. [Google Scholar]
- 33.Fang F, Norén M, Liao TY, Källersjö M, Kullander SO. Molecular phylogenetic interrelationships of the south Asian cyprinid genera Danio, Devario and Microrasbora (Teleostei, Cyprinidae, Danioninae) Zoologica Scripta. 2009;38:237–256. [Google Scholar]
- 34.Mayden RL, et al. Phylogenetic relationships of Danio within the order Cypriniformes: a framework for comparative and evolutionary studies of a model species. J Exp Zoolog B Mol Dev Evol. 2007;308:642–654. doi: 10.1002/jez.b.21175. [DOI] [PubMed] [Google Scholar]
- 35.Mills MG, Nuckels RJ, Parichy DM. Deconstructing evolution of adult phenotypes: genetic analyses of kit reveal homology and evolutionary novelty during adult pigment pattern development of Danio fishes. Development. 2007;134:1081–1090. doi: 10.1242/dev.02799. [DOI] [PubMed] [Google Scholar]
- 36.Braasch I. Asymmetric Evolution in Two Fish-Specifically Duplicated Receptor Tyrosine Kinase Paralogons Involved in Teleost Coloration. Molecular Biology and Evolution. 2006;23:1192–1202. doi: 10.1093/molbev/msk003. [DOI] [PubMed] [Google Scholar]
- 37.Stanley ER, et al. Biology and action of colony stimulating factor-1. Mol Reprod Dev. 1997;46:4–10. doi: 10.1002/(SICI)1098-2795(199701)46:1<4::AID-MRD2>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 38.Parichy DM, Ransom DG, Paw B, Zon LI, Johnson SL. An orthologue of the kit-related gene fms is required for development of neural crest-derived xanthophores and a sub-population of adult melanocytes in the zebrafish, Danio rerio. Development. 2000;127:3031–3044. doi: 10.1242/dev.127.14.3031. [DOI] [PubMed] [Google Scholar]
- 39.Heisig J, et al. Target gene analysis by microarrays and chromatin immunoprecipitation identifies HEY proteins as highly redundant bHLH repressors. PLoS Genet. 2012;8:e1002728. doi: 10.1371/journal.pgen.1002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. Comprehensive analysis of transcriptional promoter structure and function in 1% of the human genome. Genome Res. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamada H, et al. Involvement of Delta/Notch signaling in zebrafish adult pigment stripe patterning. Development. 2014;141:318–324. doi: 10.1242/dev.099804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. Normal table of postem-bryonic zebrafish development: staging by externally visible anatomy of the living fish. Developmental Dynamics. 2009;238:2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hultman KA, Bahary N, Zon LI, Johnson SL. Gene Duplication of the zebrafish kit ligand and partitioning of melanocyte development functions to kit ligand a. PLoS Genet. 2007;3:e17. doi: 10.1371/journal.pgen.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dooley CM, Mongera A, Walderich B, Nusslein-Volhard C. On the embryonic origin of adult melanophores: the role of ErbB and Kit signalling in establishing melanophore stem cells in zebrafish. Development. 2013;140:1003–1013. doi: 10.1242/dev.087007. [DOI] [PubMed] [Google Scholar]
- 45.Lang MR, Patterson LB, Gordon TN, Johnson SL, Parichy DM. Basonuclin-2 requirements for zebrafish adult pigment pattern development and female fertility. PLoS Genet. 2009;5:e1000744. doi: 10.1371/journal.pgen.1000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Curado S, Stainier DY, Anderson RM. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc. 2008;3:948–954. doi: 10.1038/nprot.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh AP, Schach U, Nusslein-Volhard C. Proliferation, dispersal and patterned aggregation of iridophores in the skin prefigure striped colouration of zebrafish. Nat Cell Biol. 2014;16:607–614. doi: 10.1038/ncb2955. [DOI] [PubMed] [Google Scholar]
- 48.Budi EH, Patterson LB, Parichy DM. Embryonic requirements for ErbB signaling in neural crest development and adult pigment pattern formation. Development. 2008;135:2603–2614. doi: 10.1242/dev.019299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budi EH, Patterson LB, Parichy DM. Post-embryonic nerve-associated precursors to adult pigment cells: genetic requirements and dynamics of morphogenesis and differentiation. PLoS Genet. 2011;7:e1002044. doi: 10.1371/journal.pgen.1002044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mahalwar P, Walderich B, Singh AP, Nüsslein-Volhard C. Local reorganization of xan-thophores fine-tunes and colors the striped pattern of zebrafish. Science. 2014;345:1362–1364. doi: 10.1126/science.1254837. [DOI] [PubMed] [Google Scholar]
- 51.Baldwin CC. The phylogenetic significance of colour patterns in marine teleost larvae. Zool J Linn Soc. 2013;168:496–563. doi: 10.1111/zoj.12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maan ME, Sefc KM. Colour variation in cichlid fish: developmental mechanisms, selective pressures and evolutionary consequences. Semin Cell Dev Biol. 2013;24:516–528. doi: 10.1016/j.semcdb.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parichy DM. Pigment patterns of ectothermic vertebrates: heterochronic vs. non-heterochronic models for pigment pattern evolution. In: Zellditch ML, editor. Beyond Heterochrony. Ch 7. John Wiley & Son, Inc; 2001. [Google Scholar]
- 54.Olsson M, Stuart-Fox D, Ballen C. Genetics and evolution of colour patterns in reptiles. Semin Cell Dev Biol. 2013;24:529–541. doi: 10.1016/j.semcdb.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Rudh A, Qvarnstrom A. Adaptive colouration in amphibians. Semin Cell Dev Biol. 2013;24:553–561. doi: 10.1016/j.semcdb.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Drerup CM, Nechiporuk AV. JNK-interacting protein 3 mediates the retrograde transport of activated c-Jun N-terminal kinase and lysosomes. PLoS Genet. 2013;9:e1003303. doi: 10.1371/journal.pgen.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kwan KM, et al. The Tol2kit: a multisite gateway-based construction kit for Tol2 transpo-son transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
- 58.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Research. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.