Abstract
Airy beams maintain their intensity profiles over a large propagation distance without substantial diffraction and exhibit lateral bending during propagation1-5. This unique property has been exploited for micromanipulation of particles6, generation of plasma channels7 and guidance of plasmonic waves8, but has not been explored for high-resolution optical microscopy. Here, we introduce a self-bending point spread function (SB-PSF) based on Airy beams for three-dimensional (3D) super-resolution fluorescence imaging. We designed a side-lobe-free SB-PSF and implemented a two-channel detection scheme to enable unambiguous 3D localization of fluorescent molecules. The lack of diffraction and the propagation-dependent lateral bending make the SB-PSF well suited for precise 3D localization of molecules over a large imaging depth. Using this method, we obtained super-resolution imaging with isotropic 3D localization precision of 10-15 nm over a 3 μm imaging depth from ∼2000 photons per localization.
Super-resolution fluorescence imaging techniques overcome the diffraction limit and allow fluorescence imaging with near-molecular-scale resolution9, 10. These techniques include approaches that use spatially patterned illumination to control the emitting states of molecules in a spatially targeted manner or methods that are based on stochastic switching of individual molecules9, 10. The stochastic switching methods, such as stochastic optical reconstruction microscopy (STORM) and (fluorescence) photoactivation localization microscopy ((F)PALM), rely on sequential activation and precise localization of single molecules to reconstruct fluorescence images with sub-diffraction-limit resolution11-13. Various strategies have been used to localize fluorescent molecules in three dimensions for 3D super-resolution imaging, but these strategies suffer from either anisotropic image resolution14-17 or a limited imaging depth14, 15, 17-20. Hence, a localization method that provides isotropic 3D resolution and large imaging depth is highly desired.
Here we report an approach to localize individual fluorophores with both isotropic 3D localization precision and substantial improvement in the imaging depth based on a SB-PSF derived from Airy beams2, 3. Unlike standard Gaussian beams, Airy beams propagate over many Rayleigh lengths without appreciable diffraction2, 3 and are self-healing after being obscured in scattering media21. In addition, Airy beams undergo lateral displacement as they propagate, resulting in a bending light path. We reason that the propagation distance of an Airy beam along the axial direction, and hence the axial position of an emitter, can be determined from the lateral displacement of the beam.
Airy beams can be generated based on the consideration that a 2D exponentially truncated Airy function Ai(x/a0, y/a0) is the Fourier transform of a Gaussian beam modulated by a cubic spatial phase, where (x, y) and (kx, ky) are the spatial coordinates and corresponding wavevector components, respectively2, 3. Hence, fluorescence emissions from individual molecules can in principle be converted into Airy beams if we introduce the cubic spatial phase with a spatial light modulator (SLM) placed at the Fourier plane in the detection path of the microscope (Figure 1a and Supplementary Figures 1 and 2). Here, we used the cubic function ((kx + ky)/b0)3 + ((−kx + ky)/b0)3 for the spatial phase modulation such that light bending occurred in the x direction. In order to separate the desired first-order diffraction from the unmodulated, zeroth-order beam, an additional diffraction grating was added to the phase pattern on the SLM. To facilitate precise 3D localization of the emitters, we introduced two variations to the Airy beam. First, because the large side-lobes of Airy beams hinder accurate localization of individual emitters and prevent imaging of densely labeled samples (Supplementary Figure 3a), we eliminated these side-lobes by introducing an additional phase modulation on the SLM to divert all wavevectors with |ky|> kyc out of the detection path (Supplementary Figure 2). We optimized the parameters in the cubic phase and the cutoff value kyc to reduce the Airy beam side-lobes while still preserving 70-80% of photons (Supplementary Figures 2 and 3). Second, we split the unpolarized fluorescence emission into two orthogonally polarized beams and rotated one of the polarizations such that both beams were properly polarized for the SLM. This two-beam design not only reduced SLM-induced photon loss but also allowed us to separately direct the two beams so that they bent in opposite directions during propagation, which enables decoupling of the lateral position of the emitter from propagation-induced lateral displacement of the beam.
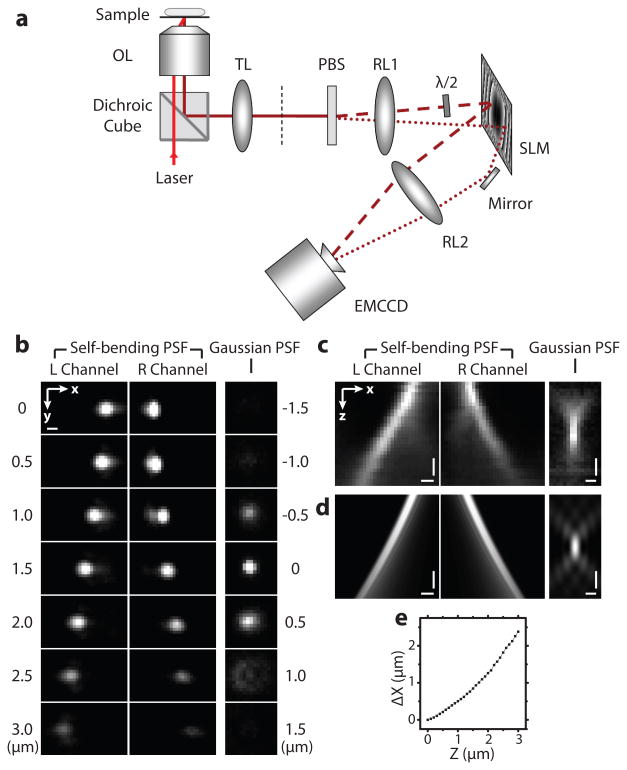
Figure 1. Self-bending point spread function (SB-PSF).
(a) Schematic of the experimental setup. The objective lens (OL) and tube lens (TL) form an image of the sample at an intermediate plane (black dashed line), which is relayed to the EMCCD camera by the relay lenses (RL1 and RL2). The emission is split into two polarizations by a polarizing beam splitter (PBS), one of which is rotated by a half-wave plate (λ/2), such that the polarizations of both beams are aligned along the active polarization direction of the spatial light modulator (SLM). The SLM situated at the focal plane of RL1 and RL2 imparts a phase modulation that converts the emission into the desired SB-PSF. Detailed setup is shown in Supplementary Figure 1. (b) The x-y cross-sections of the SB-PSF and the standard Gaussian PSF recorded as the images of fluorescence microspheres at several axial positions over a 3 μm z range. (c) x-z views of the SB-PSF and the standard Gaussian PSF. (d) The corresponding x-z views of the PSFs obtained from numerical simulation. (e) Calibration curve of the lateral bending distance, Δx = (xR −xL)/2, as a function of the z positions of the beads. Scale bars, 500 nm.
To test the performance of the SB-PSF, we first used 100 nm fluorescent beads as point emitters and recorded their images in the two polarization channels, denoted as left (L) and right (R) channels. It is worth noting that the PSF tends to bend in the same direction above and below the focal plane, it is thus necessary to select only one side of the focal plane for imaging to avoid ambiguity. In addition, the refractive index mismatch between the sample and the oil-immersion lens causes a spherical aberration, which decreases the localization precision22. Considering that the spherical aberration for emitters below the focal plane is smaller than that above the focal plane22, we only imaged below the focal plane. The bead sample was thus placed at the focal plane initially and then scanned towards the objective along the axial (z) direction. As the sample was translated in z, the images of individual beads in the two channels shifted laterally in opposite directions in x (Figure 1b). The SB-PSF maintained a compact profile over a ∼3 μm range, expanding only 2.4 times in width (Figure 1b). In contrast, the bead images recorded without using the SLM, i.e. in the form of a standard Gaussian PSF (or Airy-disk to be exact) expanded by more than 15 times over the same z range, and became barely detectable when placed >500 nm from the focal plane (Figure 1b). Measurement of the lateral bending of the SB-PSF as a function of axial displacement (Figure 1c) was in good agreement with numerical simulation results (Figure 1d, see Supplementary Note 1 for simulation details).
We next generated a calibration curve that relates the known z positions of the beads to the observed lateral bending Δx = (xR − xL)/2 of the bead images, where xR and xL represent the peak positions of the bead images along the x direction in the R and L channels, respectively (Figure 1e). Notably, in spite of the removal of the side-lobes and the use of incoherent fluorescence light, the observed lateral bending agreed well with the prediction based on a coherent Airy beam , where k is the wavenumber, x0 describes the transverse size of the beam (Supplementary Note 2). For any emitter, we can then determine its transverse coordinates (x, y) from the average peak positions of the images in the L and R channels, i.e. (x, y) = ((xL + xR)/2,(yL + yR), and its z coordinate from Δx = (xR − xL)/2 using the calibration curve.
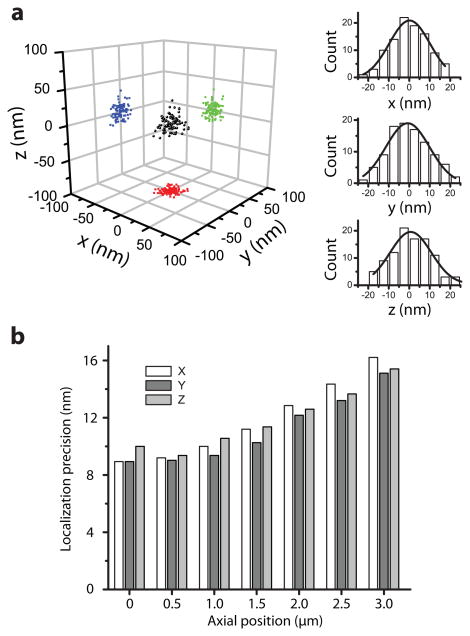
To characterize the 3D localization precisions using this SB-PSF, we imaged individual molecules of Alexa 647, a photoswitchable dye23, 24, immobilized on a glass surface. The dye molecules were switched on and off for multiple cycles, and the localization precisions were determined from the standard deviation (SD) of repetitive localizations of each molecule (Figure 2a)14. At the focal plane (z = 0), the localization precisions were 9.2, 8.9 and 10.0 nm in x, y and z, respectively (Figure 2a). The corresponding full width at half maximum (FWHM) values, which describes the minimum separation required for two molecules to be resolved, were 21.6 nm, 20.9 nm and 23.5 nm. Notably, the localization precisions were isotropic and did not change rapidly with the axial position of the molecules over a 3 μm range (Figure 2b).
Figure 2. Three-dimensional localization precision of single fluorescent molecules using the SB-PSF.
(a) Repetitive activation gives a cluster of localizations for each individual molecule. Multiple clusters were aligned by their centers of mass to generate the overall 3D localization distribution (left). The right panels show the distributions in x (top), y (middle), and z (bottom). The distributions were fit to a Gaussian function (black line), yielding standard deviations of 9.2 nm, 8.9 nm and 10.0 nm along x, y and z, respectively, for molecules at the focal plane (z near zero). (b) The localization precision values determined at various axial (z) positions over a 3 μm range. The localization precision values were obtained with an average of ∼2000 photons per switching localization.
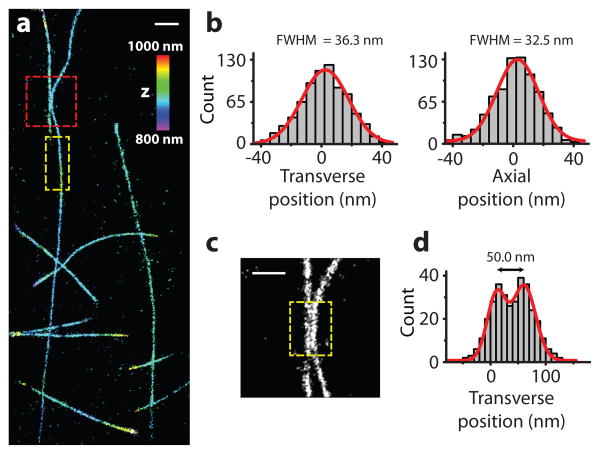
Next, we demonstrated the use of the SB-PSF for super-resolution STORM imaging of biological samples. We first imaged in vitro polymerized microtubules directly labeled with the photoswitchable HyLite 647 dye (a structural analog of Alexa 647). During imaging, we activated only a sparse subset of dye labels at a time and localized the activated dye molecules using the SB-PSF. Continuous activation and localization allowed numerous dye labels to be localized over time and a 3D super-resolution image to be constructed from the localizations (Figure 3a). Both lateral and axial cross-sectional profiles of the microtubules gave consistent, isotropic 3D widths of ∼35 nm (Figure 3b), which is in agreement with the convolution of the known ∼25 nm diameter of microtubules and the image resolution. Microtubules separated by ∼50 nm were well resolved (Figure 3c,d).
Figure 3. STORM imaging of in vitro polymerized microtubules using the SB-PSF.
(a) 3D STORM image of in vitro assembled microtubules, with z-positions color-coded according to the color scale bar. (b) The left and right panels show the transverse (x) and axial (z) cross-sectional profiles of a 2 μm segment outlined in the yellow box, respectively. The profiles exhibit isotropic FWHM of 36.3 nm and 32.5 nm, respectively. (c) Zoom-in image of the red-boxed region in (a). (d) Transverse cross-sectional profile of the two nearby microtubules shown in the yellow-boxed region in (c). Scale bars, (a) 1 μm, (c) 500 nm.
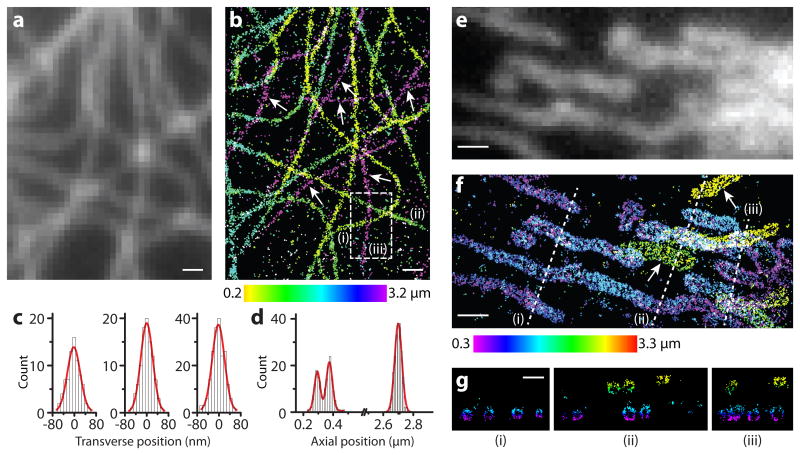
Finally, we recorded STORM images of immunolabeled microtubules and mitochondria in mammalian cells using the SB-PSF and compared with conventional images taken using the Gaussian PSF without any modulation at the SLM (Figure 4 and Supplementary Figure 4). Remarkably, the STORM images not only exhibited substantially higher resolution than the conventional images, but also captured microtubules and mitochondria that were completely undetectable in the conventional images due to diffraction of the Gaussian PSF (Figure 4b, f and Supplementary Figure 4b, h). STORM images taken with the SB-PSF maintained a high, isotropic 3D resolution over a ∼3 μm range without the need of any sample or focal-plane scanning (Figure 4b,f). The transverse and axial profiles of three microtubules spanning a ∼2.5 μm axial range showed nearly isotropic widths of 50-60 nm (Figure 4c, d), which is expected for microtubules that are broadened by immunolabeling with primary and secondary antibodies. The hollow structures of the immunolabeled microtubules were also clearly observed for densely labeled microtubule samples25 (Supplementary Figure 4c-f, i, j). Similarly, STORM images over ∼3 μm z range were acquired for mitochondria using the SB-PSF. The hollow outer-membrane structure of mitochondria was clearly observable throughout this range (Figure 4g). Analysis of the repetitive localizations of isolated antibodies spread across the 3 μm imaging depth gave an average localization precision of 13 nm, 13 nm and 15 nm in the x, y and z directions, respectively, consistent with the average localization precisions over the same z range derived from surface immobilized molecules (Figure 2b).
Figure 4. STORM imaging of microtubules and mitochondria in cells using the SB-PSF.
(a,e) Conventional immunofluorescence images of microtubules (a) and mitochondria (e) in different BS-C-1 cells taken with the standard Gaussian PSF. (b,f) The 3D STORM images of the same areas taken with the SB-PSF, respectively. The z-position information is color-coded according to the color scale bar. White arrows indicate microtubules and mitochondria that are undetectable in the conventional images but are captured in the STORM images. (c) From left to right, transverse cross-sectional profiles of the three microtubule filaments (i), (ii) and (iii) in the boxed region in (b), respectively. The FWHM of the three peaks are 52.1 nm, 55.9 nm and 53.5 nm, respectively. (d) Axial cross-sectional profiles of the same microtubules. The FWHM of the three peaks are 52.4 nm, 50.8 nm and 58.0 nm, respectively. (g) The cross-sections along the dashed lines (i), (ii) and (iii) in (f) show hollow outer-membrane structures of mitochondria. Scale bars, (a,b) 500 nm, (e-g) 1 μm.
In summary, we developed a SB-PSF based on a modified Airy beam for 3D super-resolution imaging and obtained an isotropically high resolution in all three dimensions over an imaging depth of several microns. The SB-PSF provides 2-3 fold higher z localization precision than previous 3D localization approaches using PSF engineering, such as astigmatism14, bi-plane15 and double-helix PSF16, 26, 27 imaging. The imaging depth of the SB-PSF approach is ∼50% larger than that of the double-helix PSF method and 4-5 fold larger than the astigmatism and bi-plane methods. The z localization precision of the interferometric methods18, 19 is currently higher than that of the SB-PSF approach, but the imaging depth of SB-PSF is 5-10 fold larger than that of the interferometric approaches. Although it is possible to increase the imaging depth of all methods by scanning the focal plane, photobleaching of the out of focus fluorophores prior to their imaging substantially reduces the localization density and compromises the effective image resolution. Selective-plane microscopy can help alleviate this photobleaching problem, but the complexity of the imaging setup is much increased, especially for cell imaging28. The SB-PSF approach would thus be particularly useful for high-resolution imaging of relatively thick samples. It is, however, worth noting that the relatively large area of the SB-PSF, as compared to the simple PSFs (e.g. the size of the SB-PSF is 1.3 times that of the astigmatism PSF) reduces the number of localizable fluorophores per imaging frame and hence may reduce the imaging speed moderately. Furthermore, because imaging a larger depth simultaneously requires a lower density of activated fluorophores, the SB-PSF approach also imposes a stronger requirement on the duty cycle (on-time to off-time ratio) of the photoswitchable probes25.
The image resolution afforded by the SB-PSF, like other single-molecule approaches, depends on the number of photons detected from individual fluorophores. In this work, due to SLM-imposed photon loss (detailed in Supplementary Note 3), only ∼2000 photons were detected per switching cycle of Alexa 647, whereas 5000-6000 photons were detected when the SLM was not used. However, we note that the SLM-imposed photon loss can be largely mitigated by using a continuous phase mask fabricated using gray-scale photolithography29. Moreover, the SB-PSF approach can also be combined with the recently reported dual-objective detection scheme17 or ultra-bright photoactivatable fluorophores30, which should further increase the number of photons detected and allow for even higher image resolutions.
Methods
Optical setup
All measurements were performed on a home-built inverted microscope (Supplementary Figure 1) configured for either total internal reflection fluorescence (TIRF) or oblique incidence excitation. The microscope utilized a 100×, 1.4 NA oil-immersion objective lens (Olympus UPlanSApo 100×, 1.4 NA) mounted beneath a three-axis nanopositioning system (Nano-LPS100, Mad City Labs), which controls the position of the sample. Activation of the Alexa 647 or HyLite 647 dye was provided by a 405 nm solid-state laser (CUBE, Coherent) and excitation of the activated dye molecules was provided by a 647 nm krypton gas laser (Innova 70C, Coherent). A 660 nm longpass dichroic mirror (Z660DCXRU, Chroma) was used to reflect the 405 nm and 647 nm lasers, and the transmitted fluorescence light was passed through a 700/75 emission filter (ET700/75m, Chroma). A 200 mm achromatic doublet lens (Thorlabs) functioned as a tube lens and formed an intermediate image plane situated at the input of the SB-PSF module. The SB-PSF module consisted of a two-channel 4-f imaging system with a programmable spatial light modulator (SLM, Custom XY Nematic Series, Boulder Nonlinear Systems) located at the Fourier-plane. The emission light was split into two orthogonal polarizations, which were then directed with mirrors onto different regions of the 256 × 256 pixel display of the SLM. Since the SLM can only modulate light with a defined polarization, the polarization of one beam was rotated 90° prior to impinging upon the SLM. Finally, the two beams reflected off the SLM, denoted as the left (L) and right (R) channels, were recorded on the left and right halves of an electron-multiplying CCD camera (iXon897, Andor), respectively.
Image alignment and channel registration
Prior to imaging, an alignment between the two channels (L and R) was performed. In brief, 100 nm fluorescent microspheres (TetraSpeck, Invitrogen) were immobilized on the surface of a glass coverslip at a density of ∼0.2 microspheres/μm2. Each imaging field of view contains more than 100 beads. Starting from the focal plane, the sample was sequentially displaced in 100 nm increments over a range of slightly more than 3 μm while images of the beads in both channels were recorded for each z position of the sample. A new region was then chosen and images of 10 regions are super-imposed to create an image with a high density of fiducial markers. Multiple steps of third-order polynomial transformations were used to correct for aberration in each of the L and R channels such that the bead images in the two channels were identical to each other at any z position of the bead sample, but with exactly anti-symmetric z-dependent lateral bending (Supplementary Figure 5). After channel alignment, the x and y positions of the beads do not move appreciably as the sample was translated along the axial direction, indicating a high accuracy for the alignments. The residual alignment error is <8 nm (Supplementary Figure 6). For each channel, the multiple steps of third-order polynomial transformations were combined into a single transformation matrix (the channel alignment matrix) for analysis of individual STORM frames. The calibration curve for the lateral bending as a function of the axial position (shown in Figure 1e) was also generated in this process.
Single-molecule and STORM imaging
A 647 nm laser at an intensity of 2 kW/cm2 was used for excitation of the dyes. Under this condition, the dye molecules are in the fluorescent state initially but rapidly switch to a dark state. All STORM movies were recorded at a frame rate of 50 Hz using home-written Python-based data acquisition software. The movies typically consisted of 30,000 – 100,000 frames. During each movie, a 405 nm laser light (ramped between 0.1 and 2 W/cm2) was used to activate fluorophores and to maintain a roughly constant density of activated molecules. In STORM imaging of in vitro microtubules, a weak 561 nm laser (∼20 W/cm2) was used to illuminate fiducial markers.
STORM image analysis
Single-molecule and STORM movies from the L and R channels (recorded on the left and right halves of the same camera, respectively) were first split and individually analyzed using similar methods as previously described14. Fiducial markers and image correlations14 were used for drift corrections for single-molecule and STORM imaging, respectively. Channel alignment matrices were applied to drift-corrected molecule localizations, resulting in molecule lists in each channel (xLmol, yLmol) and (xRmol, yRmol), respectively. Localizations in the two channels were linked as arising from the same molecule if they fulfilled the following four criteria: 1) their separation along the x-dimension is less than the maximum bending distance (0 < xRmol − xLmol < 5 μm); 2) their separation along y-dimension was less than the size of a single pixel (140 nm); 3) both localizations appeared and disappeared in the same frame; 4) localizations that appeared to have more than one pairing candidates in the other channel were rejected to avoid ambiguity. After linking, the lateral position (x, y) of the molecule was determined using x = (xLmol + xRmol)/2 and y = (yLmol + yRmol)/2, while the axial position z was determined from Δx = (xRmol − xLmol)/2 using the calibration curve shown in Figure 1e, followed by a correction for refractive index mismatch as previously described14, 31. Because of the sparsity of the activated molecules in each imaging frame, only a small fraction (<20%) of the localizations were rejected by the above-described criteria. Among the retained localizations, very few (< 1%) appeared to be erroneous according to our manual examination.
Detailed sample preparation procedures for single-molecule characterization, in vitro assembled microtubule, and immunofluorescence staining of cellular structures are described in the Supplementary Information.
Supplementary Material
Acknowledgments
This project is in part supported by the National Institutes of Health. X.Z. is a Howard Hughes Medical Institute investigator. J.C.V. is supported in part by a Burroughs Wellcome Fund Career Award at the Scientific Interface.
Footnotes
Author contributions: S.J. and X.Z. conceived and designed the project. S.J. performed the experiments, simulations and analysis. J.C.V. helped with the experimental set-up. S.J., J.C.V., and X.Z. wrote the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Berry MV, Balazs NL. Non-Spreading Wave Packets. Am J Phys. 1979;47:264–267. [Google Scholar]
- 2.Siviloglou GA, Broky J, Dogariu A, Christodoulides DN. Observation of accelerating airy beams. Phys Rev Lett. 2007;99:213901. doi: 10.1103/PhysRevLett.99.213901. [DOI] [PubMed] [Google Scholar]
- 3.Siviloglou GA, Christodoulides DN. Accelerating finite energy Airy beams. Opt Lett. 2007;32:979–981. doi: 10.1364/ol.32.000979. [DOI] [PubMed] [Google Scholar]
- 4.Zhang P, et al. Nonparaxial Mathieu and Weber Accelerating Beams. Phys Rev Lett. 2012;109:193901. doi: 10.1103/PhysRevLett.109.193901. [DOI] [PubMed] [Google Scholar]
- 5.Kaminer I, Bekenstein R, Nemirovsky J, Segev M. Nondiffracting accelerating wave packets of Maxwell's equations. Phys Rev Lett. 2012;108:163901. doi: 10.1103/PhysRevLett.108.163901. [DOI] [PubMed] [Google Scholar]
- 6.Baumgartl J, Mazilu M, Dholakia K. Optically mediated particle clearing using Airy wavepackets. Nature Photon. 2008;2:675–678. [Google Scholar]
- 7.Polynkin P, Kolesik M, Moloney JV, Siviloglou GA, Christodoulides DN. Curved Plasma Channel Generation Using Ultraintense Airy Beams. Science. 2009;324:229–232. doi: 10.1126/science.1169544. [DOI] [PubMed] [Google Scholar]
- 8.Zhang P, et al. Plasmonic Airy beams with dynamically controlled trajectories. Opt Lett. 2011;36:3191–3193. doi: 10.1364/OL.36.003191. [DOI] [PubMed] [Google Scholar]
- 9.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 10.Huang B, Babcock H, Zhuang X. Breaking the diffraction barrier: super-resolution imaging of cells. Cell. 2010;143:1047–1058. doi: 10.1016/j.cell.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rust MJ, Bates M, Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat Methods. 2006;3:793–796. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 13.Hess ST, Girirajan TPK, Mason MD. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juette MF, et al. Three-dimensional sub-100 nm resolution fluorescence microscopy of thick samples. Nat Methods. 2008;5:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 16.Pavani SR, et al. Three-dimensional, single-molecule fluorescence imaging beyond the diffraction limit by using a double-helix point spread function. Proc Natl Acad Sci USA. 2009;106:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu K, Babcock HP, Zhuang X. Dual-objective STORM reveals three-dimensional filament organization in the actin cytoskeleton. Nat Methods. 2012;9:185–188. doi: 10.1038/nmeth.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aquino D, et al. Two-color nanoscopy of three-dimensional volumes by 4Pi detection of stochastically switched fluorophores. Nat Methods. 2011;8:353–359. doi: 10.1038/nmeth.1583. [DOI] [PubMed] [Google Scholar]
- 19.Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci USA. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang J, Akerboom J, Vaziri A, Looger LL, Shank CV. Near-isotropic 3D optical nanoscopy with photon-limited chromophores. Proc Natl Acad Sci USA. 2010;107:10068–10073. doi: 10.1073/pnas.1004899107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Broky J, Siviloglou GA, Dogariu A, Christodoulides DN. Self-healing properties of optical Airy beams. Optics express. 2008;16:12880–12891. doi: 10.1364/oe.16.012880. [DOI] [PubMed] [Google Scholar]
- 22.Huang B, Jones SA, Brandenburg B, Zhuang X. Whole-cell 3D STORM reveals interactions between cellular structures with nanometer-scale resolution. Nat Methods. 2008;5:1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bates M, Blosser TR, Zhuang X. Short-range spectroscopic ruler based on a single-molecule optical switch. Phys Rev Lett. 2005;94:108101. doi: 10.1103/PhysRevLett.94.108101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heilemann M, Margeat E, Kasper R, Sauer M, Tinnefeld P. Carbocyanine dyes as efficient reversible single-molecule optical switch. J Am Chem Soc. 2005;127:3801–3806. doi: 10.1021/ja044686x. [DOI] [PubMed] [Google Scholar]
- 25.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:193901. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HL, Sahl SJ, Lew MD, Moerner WE. The double-helix microscope super-resolves extended biological structures by localizing single blinking molecules in three dimensions with nanoscale precision. Applied physics letters. 2012;100:153701–1537013. doi: 10.1063/1.3700446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quirin S, Pavani SR, Piestun R. Optimal 3D single-molecule localization for superresolution microscopy with aberrations and engineered point spread functions. Proc Natl Acad Sci USA. 2012;109:675–679. doi: 10.1073/pnas.1109011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gebhardt JC, et al. Single-molecule imaging of transcription factor binding to DNA in live mammalian cells. Nat Methods. 2013;10:421–426. doi: 10.1038/nmeth.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grover G, Quirin S, Fiedler C, Piestun R. Photon efficient double-helix PSF microscopy with application to 3D photo-activation localization imaging. Biomed Opt Express. 2011;2:3010–3020. doi: 10.1364/BOE.2.003010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vaughan JC, Jia S, Zhuang X. Ultrabright photoactivatable fluorophores created by reductive caging. Nat Methods. 2012;9:1181–1184. doi: 10.1038/nmeth.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egner A, Hell SW. In: Handbook of Biological Confocal Microscopy. Pawley J, editor. Springer; New York: 2006. p. 404. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.