SUMMARY
The lateral line system of fishes and amphibians comprises two ancient sensory systems: mechanoreception and electroreception. Electroreception is found in all major vertebrate groups (i.e., jawless fishes, cartilaginous fishes and bony fishes); however, it was lost in several groups including anuran amphibians (frogs) and amniotes (reptiles, birds and mammals), as well as in the lineage leading to the neopterygian clade of bony fishes (bowfins, gars and teleosts). Electroreception is mediated by modified ‘hair cells’, which are collected in ampullary organs that flank lines of mechanosensory hair cell-containing neuromasts. In the axolotl (a urodele amphibian), grafting and ablation studies have shown a lateral line placode origin for both mechanosensory neuromasts and electrosensory ampullary organs (and the neurons that innervate them). However, little is known at the molecular level about the development of the amphibian lateral line system in general and electrosensory ampullary organs in particular. Previously, we identified Eya4 as a marker for lateral line (and otic) placodes, neuromasts and ampullary organs in a shark (a cartilaginous fish) and a paddlefish (a basal ray-finned fish). Here, we show that Eya4 is similarly expressed during otic and lateral line placode development in the axolotl (a representative of the lobe-finned fish clade). Furthermore, Eya4 expression is specifically restricted to hair cells in both neuromasts and ampullary organs, as identified by co-expression with the calcium-buffering protein Parvalbumin3. As well as identifying new molecular markers for amphibian mechanosensory and electrosensory hair cells, these data demonstrate that Eya4 is a conserved marker for lateral line placodes and their derivatives in all jawed vertebrates.
INTRODUCTION
The lateral line system of fishes and amphibians is comprised of mechanosensory neuromasts and electrosensory ampullary organs, which detect changes in local water flow and weak electric fields, respectively (Coombs and Montgomery 1999; Bullock et al. 2005). These sensory systems are important for behaviors such as prey/predator interactions, schooling, orientation, and obstacle avoidance. Neuromasts, which contain mechanosensory ‘hair cells’ (essentially identical to those found in the inner ear) and support cells, are arranged in lines along the head and trunk. Fields of electrosensory ampullary organs flank the neuromast lines. Ampullary organs, which also contain modified hair cells (electroreceptors) and support cells, are found recessed at the base of pores of varying length (depending on the species) filled with a jelly-like substance of low electrical resistance (Northcutt 1997; Jørgensen and Pickles 2002; Schlosser 2002b; Bodznick and Montgomery 2005; Jørgensen, 2005).
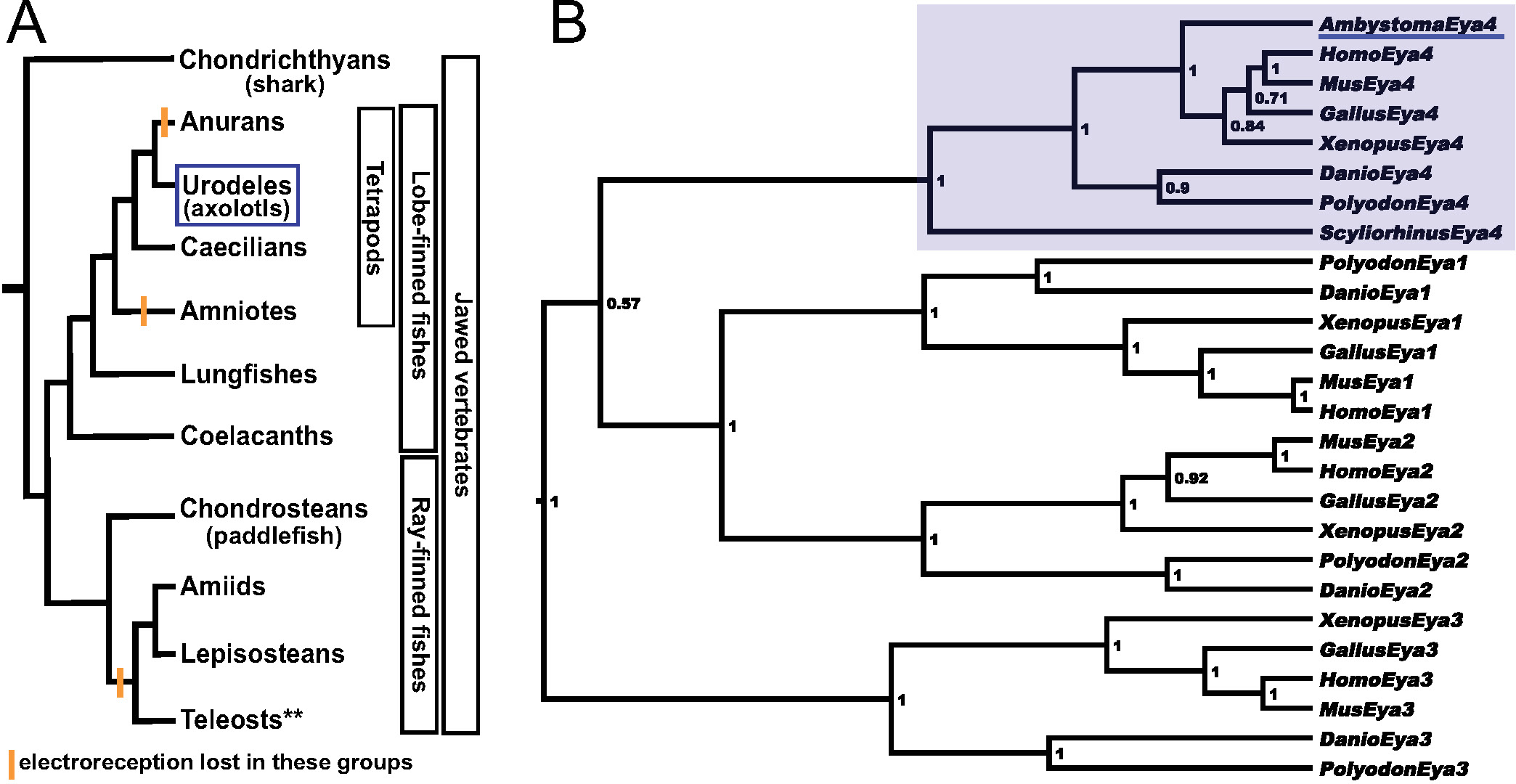
While the mechanosensory system is found in all fishes and larval and adult aquatic amphibians, the electrosensory division has been independently lost several times (Bullock et al. 1983; New 1997; Northcutt 1997; Schlosser 2002a) (Fig. 1A) suggesting that the development of these two sensory systems is, to some extent, independent. Electroreception is found in both the jawless fishes (lampreys, though not hagfishes) and the two groups that compose the jawed fishes, cartilaginous fishes (chondrichthyans) and bony fishes (osteichthyans). Bony fishes can be further subdivided into two groups: the lobe-finned fishes (sarcopterygians) and the ray-finned fishes (actinopterygians). Within the lobe-finned fish clade, electroreception was lost independently in the lineage leading to anuran frogs and the lineage leading to amniotes (presumably in the transition to land). Within the ray-finned fish clade, electroreception was lost in the lineage leading to the neopterygian clade, i.e., holostean fishes (bowfins and gars) (Grande 2010) and teleosts, although electroreception subsequently evolved independently at least twice in different teleost groups, e.g. catfishes (siluriforms), knifefishes (gymnotiforms) and elephantnose fishes (mormyriforms) (Bullock et al. 1983; New 1997; Northcutt 1997; Alves-Gomes 2001).
Fig. 1. Vertebrate phylogeny showing the distribution of electroreception and the molecular phylogenetic analysis of Eya4..
(A) Simplified phylogeny of the jawed vertebrates (gnathostomes). Electroreception is found in all major groups including both major clades of bony fishes, the lobe-finned fishes and the ray-finned fishes, but was lost in lineages indicated by orange bars. **Within teleosts, several groups independently evolved electroreception. (B) Phylogenetic tree showing the orthology of the Ambystoma mexicanum Eya4 cDNA fragment with other vertebrate Eya4 homologues (Eya4 clade is shaded in light blue). A. mexicanum Eya4 is underlined in blue. Numbers at nodes represent the Bayesian posterior probability of each clade. Species abbreviations are as follows: Ambystoma, Ambystoma mexicanum (axolotl); Danio, Danio rerio (zebrafish); Gallus, Gallus gallus (chicken); Homo, Homo sapiens (human); Mus, Mus musculus (mouse); Polyodon, Polyodon spathula (North American paddlefish); Scyliorhinus, Scyliorhinus canicula (lesser-spotted dogfish/catshark); Xenopus, Xenopus tropicalis (Western/tropical clawed frog).
Given the distribution of electroreception across jawed vertebrates (Fig. 1A), several groups stand out as being in phylogenetically important positions for understanding the development and evolution of electroreception: chondrichthyans (e.g. sharks, skates and rays), electroreceptive sarcopterygian derivatives such as the urodele amphibians (salamanders), and basal (non-teleost) electroreceptive actinopterygians such as the chondrostean fishes (paddlefishes and sturgeons). Indeed, studies within these groups have already provided a greater understanding of the developmental origin of ampullary organs. Fate-mapping studies in the axolotl (salamander) Ambystoma mexicanum (Northcutt et al. 1995) and the paddlefish Polyodon spathula (Modrell et al. 2011a) have conclusively demonstrated that both neuromasts and ampullary organs develop from lateral line placodes (i.e. thickened patches of cranial ectoderm) in these species, strongly supporting the hypothesis that these organs are lateral line placode-derived in all bony fishes. A placodal origin for chondrichthyan neuromasts has been suggested based on descriptive studies (Johnson 1917); however, a neural crest origin for both organ types has also been proposed based on gene expression data (Freitas et al. 2006). Thus, conclusive experimental data for the origin of chondrichthyan ampullary organs are still needed to confirm these hypotheses.
Relatively little is known about ampullary organ development at the molecular level. Recently, we found that Eya4 (a member of the Eyes absent/Eya family of transcription co-factors), whose expression was previously reported in neuromasts of the teleost zebrafish (Schönberger et al. 2005; Wang et al. 2008), is specifically expressed in lateral line (and otic) placodes, ampullary organs and neuromasts in both the shark Scyliorhinus canicula (O’Neill et al. 2007) and the paddlefish P. spathula (Modrell et al. 2011a). Here, we show that Eya4 is expressed in lateral line (and otic) placodes, ampullary organs and neuromasts in the axolotl, A. mexicanum, Furthermore, using a hair cell-specific antibody, we show that Eya4 expression is restricted to the mature sensory receptor cells in both ampullary organs and neuromasts. Taken together with the data from shark (a chondrichthyan) and paddlefish (a basal actinopterygian), our results from the axolotl (a sarcopterygian) demonstrate that Eya4 is a conserved marker for the developing lateral line system in all jawed vertebrates.
MATERIALS AND METHODS
Animal collection
Fixed Ambystoma mexicanum embryos were kindly provided by A. Johnson (University of Nottingham). Larval stages were purchased from the Ambystoma stock center (http://www.ambystoma.org/genetic-stock-center). Embryos were fixed in 4% paraformaldehyde overnight at 4°C, followed by dehydration into 100% methanol. Larvae were fixed in modified Carnoy’s fixative (6 volumes of 100% ethanol: 3 volumes of 37% formaldehyde: 1 volume of glacial acetic acid) overnight at 4°C (Etchevers et al. 2001). Samples were then washed with 75% ethanol in phosphate-buffered saline (PBS) followed by three washes in 100% ethanol, prior to storage at −20°C.
Cloning and phylogenetic analysis of Eya4
RNA from stage 45 embryos was extracted using Trizol reagent (Invitrogen). Single strand cDNA was generated using the Superscript III First Strand Synthesis kit (Invitrogen), as per the manufacturer’s instructions. Primers were designed based on sequence fragments obtained from the Ambystoma EST database (http://www.ambystoma.org/genome-resources). The following primers were used: Eya4F: ATGTTTCAGAGCCTCCAAAATTCC; Eya4R: TTTGCAAGTGCAGGGATAAGTTGCG. PCR fragments were cloned into the pDRIVE vector (Qiagen). Clones were isolated and sequenced by the Department of Biochemistry DNA Sequencing Facility, University of Cambridge. Sequences were analyzed using Sequencher (Gene Codes) and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) prior to phylogenetic analysis. The GenBank accession number for AmEya4 is JQ014149. Most amino acid sequences used in phylogenetic analyses were obtained from the National Center for Biotechnology Information (NCBI). Scyliorhinus canicula full-length Eya4 sequence was provided by Andrew Gillis (University of Cambridge). Danio rerio Eya4 sequence was obtained from the Zebrafish Vega database (http://vega.sanger.ac.uk/index.html). The following GenBank sequences were used in the analysis: Eya1: Danio rerio AAI54188; Gallus gallus XP_001231772; Homo sapiens NP_742056; Mus musculus AAH66860; Polyodon spathula AEQ28349, Xenopus tropicalis XP_002937115; Eya2: Danio rerio NP_001004558; Gallus gallus AAC98479; Homo sapiens CAA71310; Mus musculus AAB48018; Polyodon spathula AEQ28350; Xenopus tropicalis AAI35286; Eya3: Danio rerio AAI65887; Gallus gallus XP_417715; Homo sapiens CAA71311; Mus musculus AAB42068; Polyodon spathula AEQ28351; Xenopus tropicalis AAI61558; Eya4: Gallus gallus XP_419735; Homo sapiens CAA76636; Mus musculus CAA76637; Polyodon spathula AEQ28352; Xenopus tropicalis XP_002935856. Phylogenetic analyses were performed under a Bayesian-coalescence framework, implemented in BEAST v1.6.1 (Drummond and Rambaut 2007). The analysis was performed as previously described (Modrell et al. 2011b).
DASPEI staining
Live embryos were immersed in a solution of 1 mM DASPEI (2-(4-(dimethylamino)styryl)-N-ethylpyridinium iodide) (Invitrogen) in water for 10-20 minutes as previously described (Balak et al. 1990), then immediately rinsed in three changes of water. Embryos were anaesthetized with tricaine (ethyl m-aminobenzoate, MS-222, Sigma) prior to imaging.
Immunohistochemistry
The polyclonal anti-Parvalbumin-3 antibody (a kind gift from A. J. Hudspeth, Rockefeller University) was used in whole-mount at 1:30,000, while the monoclonal anti-HuC/D (Invitrogen) was used in sections at 1:500 (overnight incubation at 4°C in block solution [10% sheep serum in PBS/0.1% Triton-X100]). Secondary antibodies were a horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson Labs.) used at 1:600 or an AlexaFluor594 goat anti-rabbit IgG or goat anti-mouse IgG (Invitrogen) used at 1:1000 (overnight incubation at 4°C in block solution). For the histochemical reaction, embryos were incubated in 600 μl of DAB + Ni solution (0.3 mg/ml 3,3′-diaminobenzidine [Sigma] in PBS/0.05% Tween-20 plus 5 μl of an 8% NiCl2 solution) for 5 minutes, before adding H2O2 to a final concentration of 0.03% to start the reaction. The reaction was allowed to proceed for up to 15 minutes and stopped using PBS/0.1% Tween-20 (PTw). Embryos were washed overnight in PTw, then cleared through a glycerol series into 70% glycerol/PBS.
In situ hybridization
Antisense RNA probes were synthesized using T7 or SP6 polymerases (Roche) and digoxigenin dUTPs (Roche). Samples (embryos and larvae) were first rehydrated through a stepwise series into PBS/0.1% Tween-20 (PTw). Pigmentation was removed by incubating the samples under strong white light in 0.5% saline-sodium citrate buffer (SSC: 3M NaCl, 0.3M sodium citrate, pH 5), 10% H2O2, 5% formamide for 20-40 minutes (depending on stage). Following bleaching, samples were rinsed several times in PTw before treating for 10-20 minutes (depending on stage) at room temperature with proteinase K (14-22μg/ml of Roche proteinase K). Samples were rinsed twice in PTw and then fixed again for 1 hour in 4% paraformaldehyde with 0.02% glutaraldehyde, followed by four rinses in PTw. Samples were graded into hybridization solution (1× salt solution [0.2 M NaCl, 10 mM Tris, pH 7.5, 5 mM NaH2PO4 .H2O, 5 mM Na2HPO4, 5 mM EDTA], 50% formamide, 10% dextran sulfate, 1 mg/ml yeast tRNA, 1× Denhardt’s solution). After pre-hybridizing for at least 3 hours (typically overnight) at 60°C, samples were incubated for at least 18 hours at 60°C with probe diluted in hybridization solution. Samples were then washed four times for 30-60 minutes each in wash solution (50% formamide, 1× SSC, 0.1% Tween-20) at 60°C, before being graded into MABT (1× Maleic acid buffer with 0.1% Tween-20) [10× MAB: 1M Maleic acid, 1.5M NaCl, pH 7.5]. After four 30-minute washes in MABT, samples were blocked in MABT with 20% filtered sheep serum and 2% Boehringer blocking reagent (Roche) for two to four hours at room temperature. Anti-DIG-AP antibody (Roche) at 1:2000 in block solution was applied and samples were incubated overnight at 4°C. On the following day, samples were given six 1-hour washes in MABT at room temperature, and then left overnight at 4°C in MABT. Samples were rinsed three times for 15 minutes each in NTMT buffer (0.1M NaCl, 0.1M Tris, pH9.5, 50mM MgCl2, 0.1% Tween-20) before incubation in 0.02% NBT/BCIP (nitro-blue tetrazolium/5-bromo-4-chloro-3′-indolyphosphate) stock solution (Roche) in NTMT until desired signal strength was attained. Following the coloration reaction, samples were rinsed in PBS and post-fixed with 4% paraformaldehyde. Sample were rinsed in PBS and then graded into 70% glycerol/PBS for whole-mount imaging.
Sectioning embryos after whole-mount in situ hybridization
Following whole-mount imaging, embryos were graded back into PBS from 70% glycerol/PBS through a glycerol series, incubated in 5% sucrose in PBS for several hours at room temperature, and then in 15% sucrose in PBS overnight at 4°C. Embryos were transferred into a prewarmed (37°C) 7.5% gelatin in 15% sucrose in PBS solution and incubated overnight at 37°C before embedding in plastic molds. Molds were then immersed in a dry ice/isopentane solution for one minute to freeze the gelatin. Embryos were cryosectioned at 20 μm intervals and allowed to dry for one hour to overnight at room temperature. The gelatin was removed by incubating the slides in prewarmed PBS (37°C) for 5 minutes before counterstaining with the nuclear marker DAPI (1 ng/ml) for 5 minutes and mounting in Fluoromount G (Southern Biotech.).
RESULTS
Cloning Eya4 from the axolotl, A. mexicanum
The eyes absent/Eya family of transcriptional co-activators (reviewed in Jemc and Rebay 2007), of which there are four members in vertebrates, act together with members of the sine oculis/Six and dachshund/Dac families of transcription factors and co-factors in multiple developmental contexts, including cranial placode formation (reviewed in Streit 2007; Schlosser 2010). Using the Ambystoma EST database, we generated gene-specific primers for axolotl Eya4. We made cDNA from stage 45 embryos and used the Eya4-specific primers to clone a 1630 bp cDNA fragment, which included the entire N-terminal transactivation domain and 190 amino acids of the C-terminal Eya domain.
We analyzed the orthology/paralogy of Ambystoma Eya4 under a Bayesian-coalescence framework, as implemented in the software package BEAST (Bayesian Evolutionary Analysis Sampling Trees) (Drummond and Rambaut 2007). For each parameter, this type of search results in a set of posterior probability distributions and credibility intervals, including the root and the nodes of each tree. An advantage of this method, based on the use of relaxed molecular clocks and the coalescent as a model to reconstruct the genealogical history of genes, is that outgroups are not necessary in order to root the phylogenetic tree: this is useful as it is not trivial to choose appropriate outgroups when determining gene (or gene family) orthology or paralogy. Phylogenetic analysis confirmed the orthology of Ambystoma Eya4 with other vertebrate Eya4 homologues (Fig. 1B).
Parvalbumin3 is a conserved marker for electrosensory and mechanosensory hair cells in bony fishes
The development of lateral line neuromasts and ampullary organs from lateral line placodes has been described in detail in axolotl using histology, scanning electron microscopy and fate-mapping techniques (Northcutt 1992; Northcutt et al. 1994; Northcutt and Brändle 1995; Northcutt et al. 1995). Using the vital fluorescent dye DASPEI, a general mitochondrial stain that eventually labels all cells but which selectively labels functional mechanosensory hair cells after a short incubation (Balak et al. 1990), we found that functional neuromasts were already present by stage 40 (Fig. 2A) and rapidly continued to form between stages 40 and 44 (Fig. 2B, C). Ampullary organs have formed by stage 44 (Northcutt et al. 1994) but are not labelled by DASPEI.
Fig. 2. Spatiotemporal development of sensory hair cells of axolotl lateral line organs.
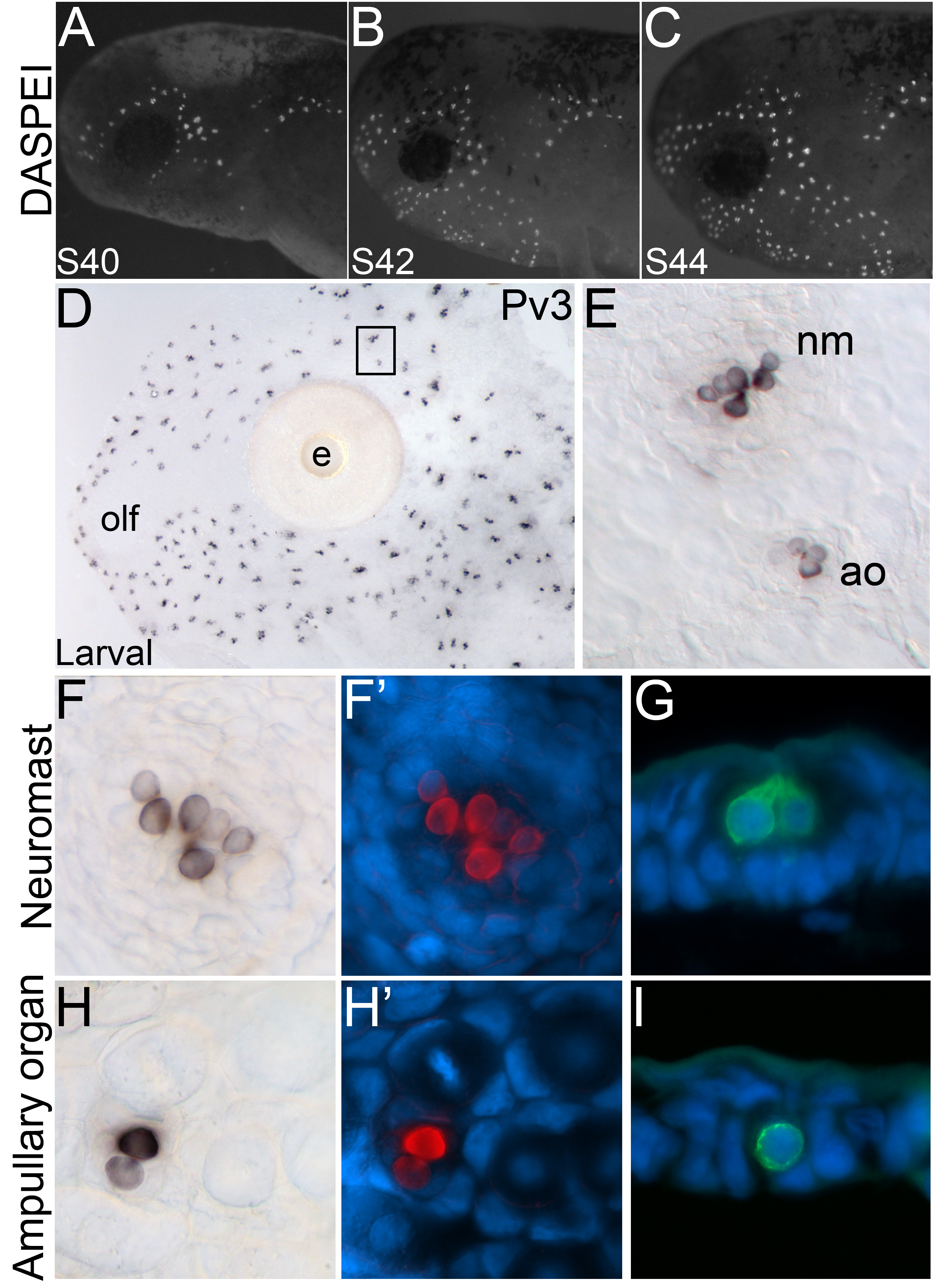
Anterior to the left; lateral views. (A-C) Fluorescent images superimposed on darkfield images of stage 40 through 44 embryos stained with the vital dye DASPEI (white), which selectively labels functional neuromasts after a short incubation. (A) Stage 40 embryo. Neuromasts derived from the anteriormost lateral line placodes are already present. (B) Stage 42 embryo. (C) Stage 44 embryo. (D-I) (D) Skinmount from a larva (1-2 cm in length) immunostained for Pv3, showing expression in mature lateral line organs. (E) Higher power view of the boxed area in A. Hair cells in both the mechanosensory neuromasts and electrosensory ampullary organs are labelled by Pv3. (F) Surface view of a single neuromast showing Pv3-positive hair cells (black) in a brightfield image and (F’) in a false color overlay (Pv3, red) with the nuclear marker DAPI (blue). In this view, approximately seven individual hair cells can be identified. (G) Transverse section through a neuromast showing Pv3-positive hair cells (green) and DAPI (blue). (H) Surface view of a single ampullary organ showing Pv3-positive hair cells (black) in a brightfield image and (H’) in a false color overlay (Pv3, red) with DAPI (blue). Ampullary organs contain fewer hair cells per organ than neuromasts: in this example, there are only two hair cells. (I) Transverse section through an ampullary organ showing a single Pv3-positive hair cell (green) with DAPI (blue). Abbreviations: ao, ampullary organ; e, eye; nm, neuromast; olf, olfactory organ.
Parvalbumin-3 (Pv3), a calcium-buffering protein, is expressed in mechanosensory hair cells in both the inner ear and lateral line neuromasts (Heller et al. 2002); we recently showed that Pv3 protein is also expressed in electrosensory hair cells in paddlefish ampullary organs (Modrell et al. 2011a). At larval stages in the axolotl, both neuromasts and ampullary organs are well developed and have erupted to the surface (Northcutt et al. 1994). Using whole-mount immunohistochemistry, we observed Pv3 expression in the sensory receptor cells of both neuromasts and ampullary organs (Fig. 2D), just as we observed in paddlefish (Modrell et al. 2011a). Closer examination of a skin-mount preparation revealed the difference in size and number of sensory receptor cells between the neuromasts and ampullary organs (Fig. 2E). Neuromasts are larger and contain more sensory receptor cells per organ (Fig. 2F-G) than do the ampullary organs (Fig. 2H-I), as observed in surface and transverse views, respectively. Taking these data together with our previous paddlefish data (Modrell et al. 2011a), we conclude that Pv3 is a conserved marker for electrosensory as well as mechanosensory hair cells in bony fishes.
Eya4 is expressed during lateral line placode development
We characterized the expression of Eya4 during lateral line development using a whole-mount in situ hybridization protocol that worked much better in our hands than other previously described protocols for axolotls. Our protocol uses a dextran sulfate-based hybridization buffer (see Methods) and a modified Carnoy’s fix (Etchevers et al. 2001) at larval stages instead of paraformaldehyde. (Paraformaldehyde works using this protocol for embryonic stages, but not for larval stages.) We found that embryos and larvae retain their original size and morphology better in this hybridization buffer, and that the signal to noise ratio was much stronger compared to the other protocols we tried.
During early stages of development, beginning around stage 28, Eya4 expression was only observed in the otic vesicle and weakly in the developing somites (Fig. 3A). By stage 33, expression of Eya4 was also observed in the developing lateral line placodes (Fig. 3B), but not in other cranial placodes. A transverse section through the anterodorsal lateral line placode revealed Eya4 in the thickened placodal epithelium (Fig. 3C). By stages 38-39, Eya4 was observed in the developing lines of neuromasts as well as in the lateral line ganglia (Fig. 3D). A section through the anterior-most lateral line ganglia, the anterodorsal and anteroventral lateral line ganglia, showed the restricted expression of Eya4 in only those ganglia (Fig. 3E). Immunostaining on the same sections with the pan-neuronal antibody HuC/D (which detects RNA-binding proteins expressed in all neurons; Wakamatsu and Weston, 1997) showed the close association of the anteroventral lateral line ganglion with the ganglion of the facial (VII) nerve, which did not express Eya4 (Fig. 3F). By stage 41, expression of Eya4 continued to expand as the lateral line organs further differentiated (Fig. 3G), including the neuromasts of the main trunk line, which were also Eya4-positive (Fig. 3H).
Fig. 3. Eya4 expression in lateral line placodes and ganglia.
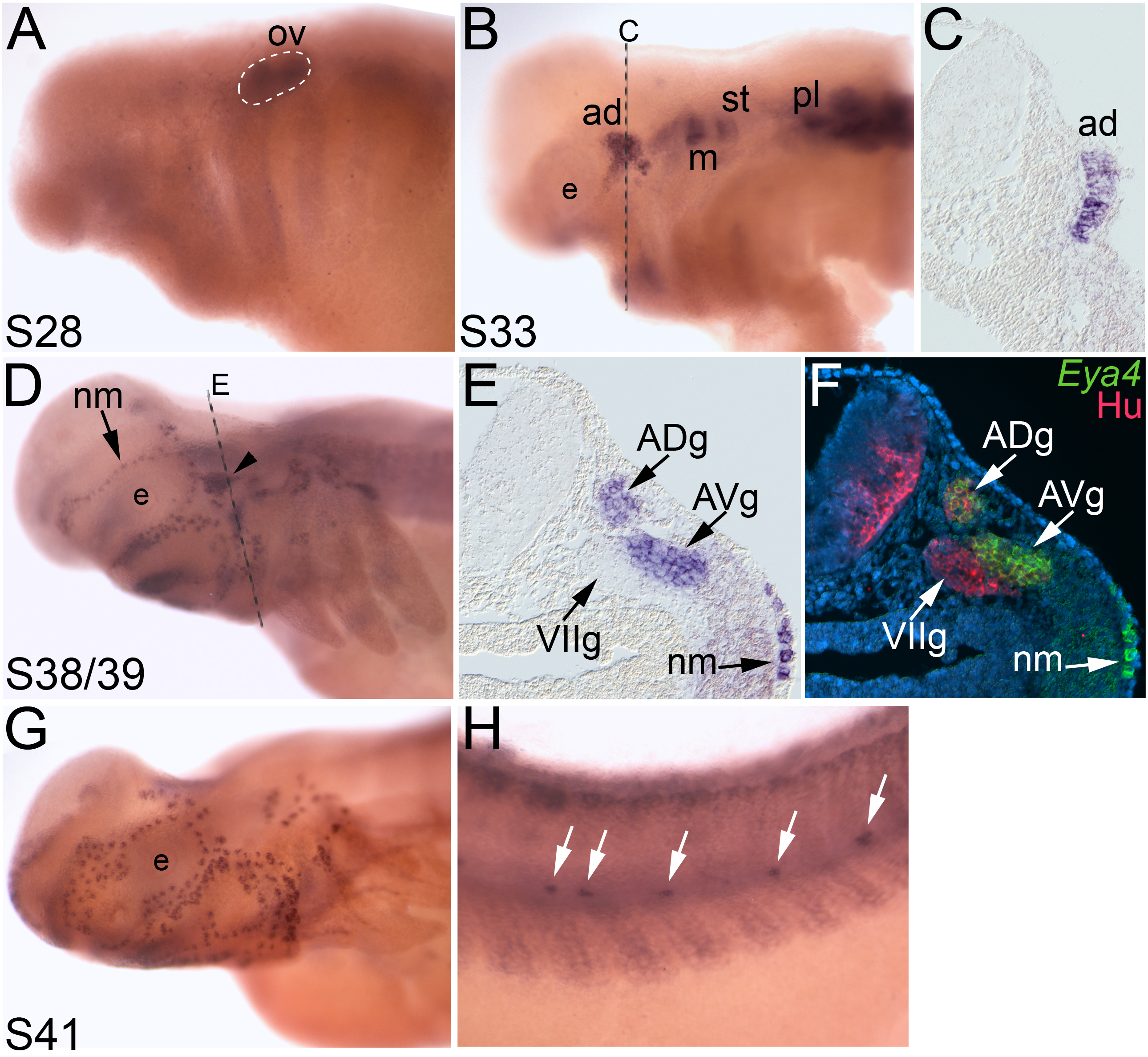
In situ hybridizations of whole-mount axolotl embryos showing the localization of Eya4. Anterior to the left; lateral views unless otherwise noted. (A) Stage 28 embryo. Eya4 expression is restricted to the otic vesicle (outlined) and weakly in the developing somites. (B) By stage 33, several lateral line placodes have begun to express Eya4. (C) Transverse section through the anterodorsal lateral line placode (ad) indicated in B by the dashed line. Eya4 expression is restricted to the thickened epithelium (i.e. the placode). (D) At stages 38-39, Eya4 expression is observed in neuromasts (arrow) and lateral line ganglia (arrowhead). Dashed line indicates plane of section shown in E. (E) Transverse section through the region indicated in D by the dashed line. Eya4 is expressed in the anterodorsal and anteroventral lateral line ganglia, and in developing neuromasts beneath the surface ectoderm. (F) The same transverse section shown in E, immunostained for the panneuronal marker HuC/D (pink), with a false color overlay of the brightfield image in E, showing Eya4 (green) and the nuclear marker DAPI (blue). Eya4 is restricted to the lateral line ganglia and is not observed in other cranial ganglia, such as the ganglion of the facial (VII) nerve. (G) Stage 41 embryo. Neuromast expression of Eya4 continues as more organs differentiate. (H). Trunk of the stage 41 embryo shown in G. The posterior trunk neuromasts also express Eya4 (arrows). Abbreviations: ad, anterodorsal lateral line placode; ADg, ganglion of the anterodorsal lateral line nerve; AVg, ganglion of the anteroventral lateral line nerve; e, eye; m, middle lateral line placode; nm, neuromast; ov, otic vesicle; pl, posterior lateral line placode; st, supratemporal lateral line placode; VIIg, ganglion of the facial (VII) nerve.
Eya4 expression is restricted to sensory receptor cells in mature ampullary organs and neuromasts
Using in situ hybridization on larval stage samples (~ 1 to 2 cm total length), when more ampullary organs have erupted, we found that Eya4 was also expressed in mature neuromasts and ampullary organs (Fig. 4A). The in situ hybridization pattern in whole-mount specimens suggested that Eya4 transcripts might be restricted to a subset of cells within the organs (Fig. 4B, C). To confirm whether these Eya4-positive cells were sensory receptor cells or support cells (or a subset of both), we immunostained the embryos for the calcium-buffering protein Pv3 (Fig. 2). In skinmounts, we observed the localization of Pv3 in the nuclei and Eya4 in the cytoplasm specifically within the sensory receptor cells in both neuromasts (Fig. 4B’, B”) and ampullary organs (Fig. 4C’, C”). The support cells and surrounding cells lacked Eya4 expression.
Fig. 4. Eya4 is restricted to the hair cells in mature neuromasts and ampullary organs.
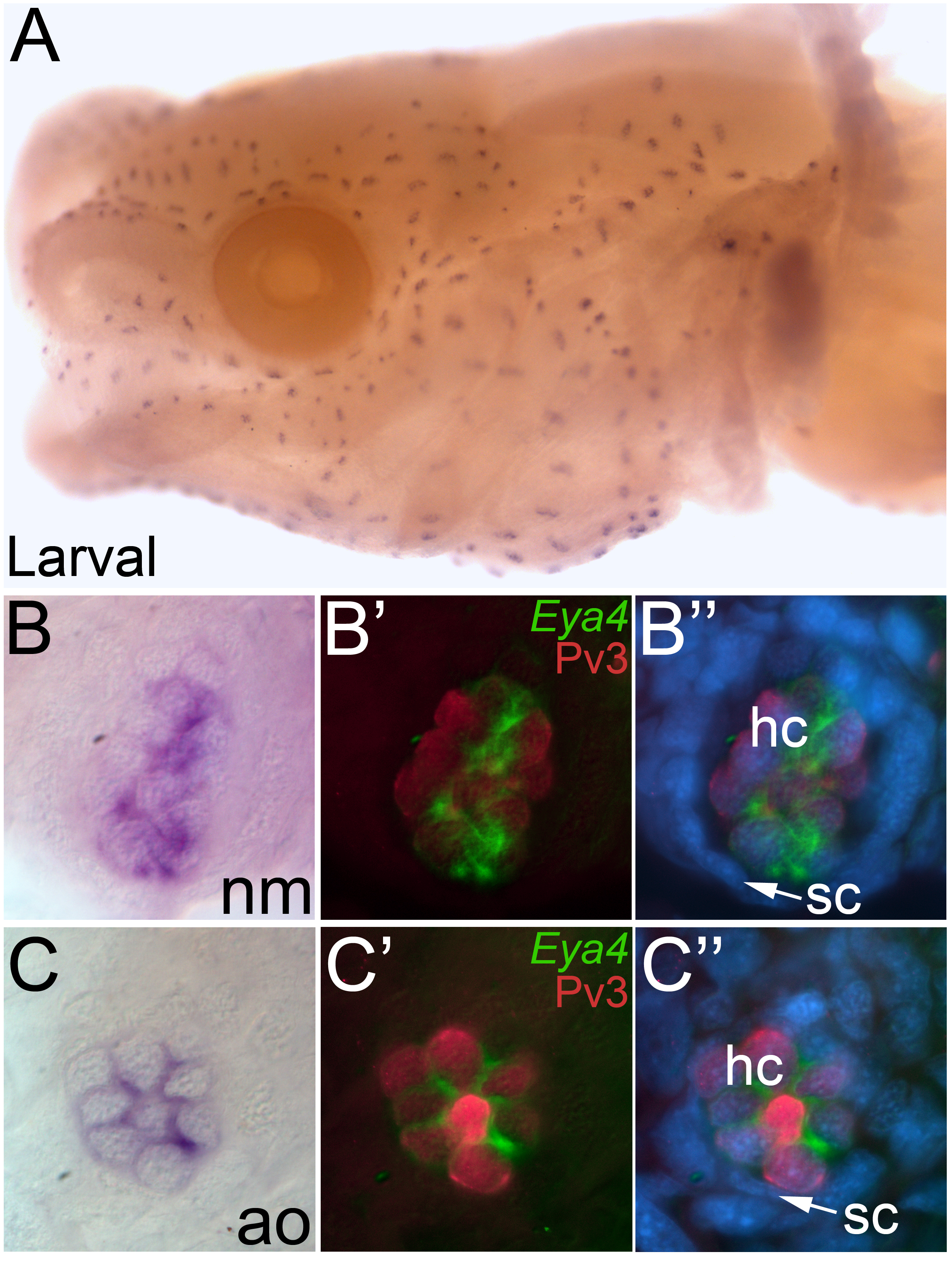
Whole-mount in situ hybridization for Eya4 in larval axolotls. (A) Head view of a larval axolotl. Anterior to the left, lateral view. Eya4 expression is observed in both the neuromasts and ampullary organs. (B) Higher power view of a single neuromast, showing the restricted localization of Eya4. (B’, B”) Fluorescent image of the neuromast in B, immunostained for the hair cell marker Pv3 (red) with Eya4 as a false colored overlay (green) plus (in B”) the nuclear marker DAPI (blue). (C) Higher power view of a single ampullary organ showing the restricted localization of Eya4. (C’, C”) Fluorescent image of the ampullary organ in C, immunostained for the hair cell marker Pv3 (red) with Eya4 as a false colored overlay (green), plus (in C”) the nuclear marker DAPI (blue). In both neuromasts and ampullary organs, Eya4 transcripts are localized within the cytoplasm of the Pv3-positive hair cells. Abbreviations: ao, ampullary organ; hc, hair cell; nm, neuromast; sc, support cell.
Overall, taken together with our previous data from a chondrichthyan (shark, O’Neill et al. 2007) and a basal actinopterygian (paddlefish, Modrell et al. 2011a), these results confirm that Eya4 is a conserved marker for the development of lateral line placodes, neuromasts and ampullary organs in jawed vertebrates.
DISCUSSION
Eya4 is one of the four vertebrate homologues of the eyes absent/Eya gene family, which are involved in a diverse array of developmental processes including mechanosensory hair cell development (reviewed in Rebay et al. 2005; Jemc and Rebay 2007). Eya4 function has been mostly examined in humans and mice, in which mutations in Eya4 result in a variety of inner ear and/or hearing defects (Wayne et al. 2001; Pfister et al. 2002; Schönberger et al. 2005). In zebrafish, Eya4 is expressed in the otic vesicle as well as in lateral line neuromasts (Schönberger et al. 2005; Wang et al. 2008). Morpholino (MO)-mediated knockdown of Eya4 in zebrafish results in malformed otic vesicles and fewer neuromasts, as well as fewer hair cells per neuromast (Wang et al. 2008). Eya4 MO knockdown has also been reported to lead to apoptosis in the anterior lateral line ganglion and neuromasts (Formella 2008) suggesting that Eya4 is required for proper patterning of lateral line placode-derived neurons and neuromasts, as well as the otic vesicle. However, zebrafish lack the electrosensory division of the lateral line system, thus cannot shed light on the potential role of Eya4 in ampullary organs.
Previously, we identified Eya4 as a marker for lateral line (and otic) placodes, neuromasts and ampullary organs in a shark (chondrichthyan) and a paddlefish (basal actinopterygian) (O’Neill et al. 2007; Modrell et al. 2011a). Here, we have shown that Eya4 is similarly expressed during otic and lateral line placode development in the axolotl (a sarcopterygian), confirming that Eya4 is a conserved marker for lateral line placodes and their derivatives in all jawed vertebrates. Our data also show that the calcium-buffering protein Pv3 (Heller et al. 2002) is a conserved marker for electrosensory as well as mechanosensory hair cells in bony fishes (axolotl, this paper; paddlefish, Modrell et al. 2011a). Finally, since Eya4 is specifically co-expressed with Pv3 in mature neuromasts and ampullary organs, we have identified Eya4 as a new molecular marker for amphibian mechanosensory and electrosensory hair cells.
Several other members of the Eya family of transcription co-factors, as well as of the Six1/2 and Six4/5 families, are known to be involved in lateral line placode and neuromast development (reviewed in Streit 2007; Schlosser 2010). In particular, Eya1, Six1 and Six4 are all expressed during lateral line placode formation and/or in developing neuromasts in Xenopus and zebrafish (Sahly et al. 1999; Kobayashi et al. 2000; Pandur and Moody 2000; David et al. 2001; Bessarab et al. 2004; Schlosser and Ahrens 2004). Furthermore, we recently showed that all four Eya genes, as well as Six1, Six2 and Six4, are expressed in lateral line placodes, neuromasts and ampullary organs in the paddlefish (Modrell et al. 2011a). Hence, Eya and Six family members are likely to constitute a highly conserved gene network during the development of lateral line placodes and the sensory organs derived from them.
Although much is known about the molecular cues involved in cranial placode development in the frog Xenopus laevis (Schlosser and Northcutt 2000; Schlosser and Ahrens 2004; Schlosser 2010; Pieper et al. 2011), little is known at the molecular level about amphibian lateral line organ development after placode formation, in particular the ampullary organs (which are lacking in Xenopus). To our knowledge, the homeobox genes Msx2 and Dlx3 are the only previously described transcription factors expressed during neuromast and ampullary organ development in the axolotl: in the mature sense organs, both genes were reported to be expressed in the sensory receptor cells and support cells, though expression data were only shown for neuromasts (Metscher et al. 1997). However, neither of these genes is expressed during neuromast or ampullary organ development in the paddlefish (M.S.M., unpublished data), suggesting they may not be conserved markers for lateral line sense organs in bony fishes.
Until now, the only reported electrosensory hair cell-specific marker in amphibians was miR-183 (Pierce et al. 2008), a member of the miR-183 family (miR-96, miR-182 and miR-183) of microRNAs (highly conserved, small non-coding RNAs that can post-transcriptionally regulate gene expression). miR-183 family members are expressed in ciliated neurosensory cell types in both invertebrates and vertebrates (Weston et al. 2006; Pierce et al. 2008; Li et al. 2010; Weston et al. 2011), including in vertebrate mechanosensory hair cells (of both the inner ear and neuromasts) (Wienholds et al. 2005; Pierce et al. 2008; Li et al. 2010). Recently, the SoxB1-group family member Sox2 was validated as a target for the miR-183 family during mouse mechanosensory hair cell development (Weston et al. 2011), consistent with a role for miR-183 family members in repressing Sox2 specifically in mechanosensory hair cells (Soukup 2009). The expression of Sox2 in axolotls is currently unknown; however, in paddlefish, Sox2 is expressed in both neuromasts and ampullary organs (M.S.M., unpublished data). Recently, we also identified the related SoxB1-group family member Sox3 as a marker for both neuromasts and ampullary organs in the paddlefish (Modrell et al. 2011b).
In conclusion, we have identified Eya4 as a conserved marker for lateral line (and otic) placodes, ampullary organs and neuromasts in all jawed vertebrates, Pv3 as a conserved marker for electrosensory (as well as mechanosensory) hair cells in bony fishes, and both Eya4 and Pv3 as novel markers for hair cells (both electrosensory and mechanosensory) in mature amphibian lateral line organs.
Acknowledgments
We thank David Buckley for assistance with the phylogenetic analysis, Andrew Gillis for the full-length S. canicula Eya4 sequence, Andrew Johnson for fixed axolotl embryos and Jim Hudspeth for the anti-Parvalbumin3 antibody.
REFERENCES
- Alves-Gomes JA. The evolution of electroreception and bioelectrogenesis in teleost fish: a phylogenetic perspective. J. Fish Biol. 2001;58:1489–1511. [Google Scholar]
- Balak KJ, Corwin JT, Jones JE. Regenerated hair cells can originate from supporting cell progeny: evidence from phototoxicity and laser ablation experiments in the lateral line system. J. Neurosci. 1990;10:2502–2512. doi: 10.1523/JNEUROSCI.10-08-02502.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessarab DA, Chong SW, Korzh V. Expression of zebrafish six1 during sensory organ development and myogenesis. Dev. Dyn. 2004;230:781–786. doi: 10.1002/dvdy.20093. [DOI] [PubMed] [Google Scholar]
- Bodznick D, Montgomery JC. The physiology of low-frequency electrosensory systems. In: Bullock TH, Hopkins CD, Popper AN, Fay RR, editors. Electroreception. Springer; New York: 2005. pp. 132–153. [Google Scholar]
- Bullock TH, Hopkins CD, Popper AN, Fay RR, editors. Electroreception. Springer; New York: 2005. [Google Scholar]
- Bullock TH, Bodznick DA, Northcutt RG. The phylogenetic distribution of electroreception: evidence for convergent evolution of a primitive vertebrate sense modality. Brain Res. 1983;287:25–46. doi: 10.1016/0165-0173(83)90003-6. [DOI] [PubMed] [Google Scholar]
- Coombs S, Montgomery JC. The enigmatic lateral line system. In: Popper AN, Fay RR, editors. Comparative Hearing: Fishes and Amphibians. Springer-Verlag; New York: 1999. pp. 319–362. [Google Scholar]
- David R, Ahrens K, Wedlich D, Schlosser G. Xenopus Eya1 demarcates all neurogenic placodes as well as migrating hypaxial muscle precursors. Mech. Dev. 2001;103:189–192. doi: 10.1016/s0925-4773(01)00355-0. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchevers HC, Vincent C, Le Douarin NM, Couly GF. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128:1059–1068. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- Formella I. Identification and characterisation of three novel eyes absent genes in the cranial sensory placodes, the developing inner ear and lateral line in the zebrafish Danio rerio. PhD dissertation. Technische Universität Darmstadt; Darmstadt, Germany: 2008. Retrieved from http://tuprints.ulb.tu-darmstadt.de/id/eprint/986. [Google Scholar]
- Freitas R, Zhang G, Albert JS, Evans DH, Cohn MJ. Developmental origin of shark electrosensory organs. Evol. Dev. 2006;8:74–80. doi: 10.1111/j.1525-142X.2006.05076.x. [DOI] [PubMed] [Google Scholar]
- Grande L. An Empirical Synthetic Pattern Study of Gars (Lepisosteiformes) and Closely Related Species, Based Mostly on Skeletal Anatomy: The Resurrection of Holostei. Vol. 6. American Society of Ichthyologists and Herpetologists Special Publication; 2010. [Google Scholar]
- Heller S, Bell AM, Denis CS, Choe Y, Hudspeth AJ. Parvalbumin 3 is an abundant Ca2+ buffer in hair cells. J. Assoc. Res. Otolaryngol. 2002;3:488–498. doi: 10.1007/s10162-002-2050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemc J, Rebay I. The eyes absent family of phosphotyrosine phosphatases: properties and roles in developmental regulation of transcription. Annu. Rev. Biochem. 2007;76:513–538. doi: 10.1146/annurev.biochem.76.052705.164916. [DOI] [PubMed] [Google Scholar]
- Johnson SE. Structure and development of the sense organs of the lateral canal system of Selachians (Mustelus canis and Squalus acanthias) J. Comp. Neurol. 1917;28:1–74. [Google Scholar]
- Jørgensen JM. Morphology of electroreceptive sensory organs. In: Bullock TH, Hopkins CD, Popper AN, Fay RR, editors. Electroreception. Springer; New York: 2005. pp. 47–67. [Google Scholar]
- Jørgensen JM, Pickles JO. The lateral line canal sensory organs of the epaulette shark (Hemiscyllium ocellatum) Acta Zool. (Stockh.) 2002;83:337–343. [Google Scholar]
- Kobayashi M, Osanai H, Kawakami K, Yamamoto M. Expression of three zebrafish Six4 genes in the cranial sensory placodes and the developing somites. Mech. Dev. 2000;98:151–155. doi: 10.1016/s0925-4773(00)00451-2. [DOI] [PubMed] [Google Scholar]
- Li H, Kloosterman W, Fekete DM. MicroRNA-183 family members regulate sensorineural fates in the inner ear. J. Neurosci. 2010;30:3254–3263. doi: 10.1523/JNEUROSCI.4948-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metscher BD, Northcutt RG, Gardiner DM, Bryant SV. Homeobox genes in axolotl lateral line placodes and neuromasts. Dev. Genes Evol. 1997;207:287–295. doi: 10.1007/s004270050116. [DOI] [PubMed] [Google Scholar]
- Modrell MS, Bemis WE, Northcutt RG, Davis MC, Baker CVH. Electrosensory ampullary organs are derived from lateral line placodes in bony fishes. Nat. Commun. 2011a;2:496. doi: 10.1038/ncomms1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrell MS, Buckley D, Baker CVH. Molecular analysis of neurogenic placode development in a basal ray-finned fish. Genesis. 2011b;4:278–294. doi: 10.1002/dvg.20707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- New JG. The evolution of vertebrate electrosensory systems. Brain Behav. Evol. 1997;50:244–252. doi: 10.1159/000113338. [DOI] [PubMed] [Google Scholar]
- Northcutt RG. Distribution and innervation of lateral line organs in the axolotl. J. Comp. Neurol. 1992;325:95–123. doi: 10.1002/cne.903250109. [DOI] [PubMed] [Google Scholar]
- Northcutt RG. Evolution of gnathostome lateral line ontogenies. Brain Behav. Evol. 1997;50:25–37. doi: 10.1159/000113319. [DOI] [PubMed] [Google Scholar]
- Northcutt RG, Brändle K. Development of branchiomeric and lateral line nerves in the axolotl. J. Comp. Neurol. 1995;355:427–454. doi: 10.1002/cne.903550309. [DOI] [PubMed] [Google Scholar]
- Northcutt RG, Brändle K, Fritzsch B. Electroreceptors and mechanosensory lateral line organs arise from single placodes in axolotls. Dev. Biol. 1995;168:358–373. doi: 10.1006/dbio.1995.1086. [DOI] [PubMed] [Google Scholar]
- Northcutt RG, Catania KC, Criley BB. Development of lateral line organs in the axolotl. J. Comp. Neurol. 1994;340:480–514. doi: 10.1002/cne.903400404. [DOI] [PubMed] [Google Scholar]
- O’Neill P, McCole RB, Baker CVH. A molecular analysis of neurogenic placode and cranial sensory ganglion development in the shark. Scyliorhinus canicula. Dev. Biol. 2007;304:156–181. doi: 10.1016/j.ydbio.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur PD, Moody SA. Xenopus Six1 gene is expressed in neurogenic cranial placodes and maintained in the differentiating lateral lines. Mech. Dev. 2000;96:253–257. doi: 10.1016/s0925-4773(00)00396-8. [DOI] [PubMed] [Google Scholar]
- Pfister M, Toth T, Thiele H, Haack B, Blin N, Zenner HP, Sziklai I, Nurnberg P, Kupka S. A 4-bp insertion in the eya-homologous region (eyaHR) of EYA4 causes hearing impairment in a Hungarian family linked to DFNA10. Mol. Med. 2002;8:607–611. [PMC free article] [PubMed] [Google Scholar]
- Pieper M, Eagleson GW, Wosniok W, Schlosser G. Origin and segregation of cranial placodes in Xenopus laevis. Dev. Biol. 2011;360:257–275. doi: 10.1016/j.ydbio.2011.09.024. DOI: 10.1016/j.ydbio.2011.09.024. [DOI] [PubMed] [Google Scholar]
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol. Dev. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebay I, Silver SJ, Tootle TL. New vision from Eyes absent: transcription factors as enzymes. Trends Genet. 2005;21:163–171. doi: 10.1016/j.tig.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Sahly I, Andermann P, Petit C. The zebrafish eya1 gene and its expression pattern during embryogenesis. Dev. Genes Evol. 1999;209:399–410. doi: 10.1007/s004270050270. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Making senses: development of vertebrate cranial placodes. Int. Rev. Cell Mol. Biol. 2010;283:129–234. doi: 10.1016/S1937-6448(10)83004-7. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Development and evolution of lateral line placodes in amphibians I. Development. Zoology (Jena) 2002a;105:119–146. doi: 10.1078/0944-2006-00058. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Development and evolution of lateral line placodes in amphibians. II. Evolutionary diversification. Zoology (Jena) 2002b;105:177–193. doi: 10.1078/0944-2006-00062. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Ahrens K. Molecular anatomy of placode development in Xenopus laevis. Dev. Biol. 2004;271:439–466. doi: 10.1016/j.ydbio.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Northcutt RG. Development of neurogenic placodes in Xenopus laevis. J. Comp. Neurol. 2000;418:121–146. [PubMed] [Google Scholar]
- Schönberger J, Wang L, Shin JT, Kim SD, Depreux FF, Zhu H, Zon L, Pizard A, Kim JB, Macrae CA, Mungall AJ, Seidman JG, Seidman CE. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat. Genet. 2005;37:418–422. doi: 10.1038/ng1527. [DOI] [PubMed] [Google Scholar]
- Soukup GA. Little but loud: small RNAs have a resounding affect on ear development. Brain Res. 2009;1277:104–114. doi: 10.1016/j.brainres.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streit A. The preplacodal region: an ectodermal domain with multipotential progenitors that contribute to sense organs and cranial sensory ganglia. Int. J. Dev. Biol. 2007;51:447–461. doi: 10.1387/ijdb.072327as. [DOI] [PubMed] [Google Scholar]
- Wakamatsu Y, Weston JA. Sequential expression and role of Hu RNA-binding proteins during neurogenesis. Development. 1997;124:3449–3460. doi: 10.1242/dev.124.17.3449. [DOI] [PubMed] [Google Scholar]
- Wang L, Sewell WF, Kim SD, Shin JT, Macrae CA, Zon LI, Seidman JG, Seidman CE. Eya4 regulation of Na+/K+-ATPase is required for sensory system development in zebrafish. Development. 2008;135:3425–3434. doi: 10.1242/dev.012237. [DOI] [PubMed] [Google Scholar]
- Wayne S, Robertson NG, DeClau F, Chen N, Verhoeven K, Prasad S, Tranebjarg L, Morton CC, Ryan AF, Van Camp G, Smith RJ. Mutations in the transcriptional activator EYA4 cause late-onset deafness at the DFNA10 locus. Hum. Mol. Genet. 2001;10:195–200. doi: 10.1093/hmg/10.3.195. [DOI] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev. Dyn. 2011;240:808–819. doi: 10.1002/dvdy.22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA gene expression in the mouse inner ear. Brain Res. 2006;1111:95–104. doi: 10.1016/j.brainres.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]