Abstract
Background
Bipolar disorder (BD) is a highly heritable disease. While genome-wide association (GWA) studies have identified several genetic risk factors for BD, few of these studies have investigated the genetic etiology of specific disease subtypes. In particular, BD is positively associated with eating dysregulation traits such as binge eating behavior (BE), yet the genetic risk factors underlying BD with comorbid BE have not been investigated.
Methods
Utilizing data from the Genetic Association Information Network study of BD, which included 729,454 single nucleotide polymorphisms (SNPs) genotyped in 1001 European American bipolar cases and 1034 controls, we performed GWA analyses of bipolar subtypes defined by the presence or absence of BE history, and performed a case-only analysis comparing BD subjects with and without BE history. Association signals were refined using imputation, and network analysis was performed with Ingenuity Pathway Analysis software. Based on these results, candidate SNPs were selected for replication in an independent sample of 855 cases and 857 controls.
Results
Top ranking SNPs in the discovery set included rs6006893 in PRR5, rs17045162 in ANK2, rs13233490 near PER4, rs4665788 and rs10198175 downstream of APOB, rs2367911 in CACNA2D1, and rs7249968 near ZNF536. Rs10198175 in APOB also demonstrated evidence of association in the replication sample and a meta-analysis of the two samples.
Limitations
Without information of BE history in controls, it is not possible to determine whether the observed association with APOB reflects a risk factor for BE behavior in general or a risk factor for a subtype of BD with BE. Further longitudinal and functional studies are needed to determine the causal pathways underlying the observed associations.
Conclusions
This study identified new potential BD-susceptibility genes, highlighting the advantages of phenotypic sub-classification in genetic research and clinical practice.
Keywords: Bipolar disorder, Binge eating, Phenotypic subtypes, Network analysis, Genome-wide association study (GWAS), APOB
1. Introduction
Bipolar disorder (BD) is known to have a strong genetic component with estimated heritability between 60% and 85% (Lichtenstein et al., 2009; McGuffin et al., 2003). Multiple genome-wide association (GWA) studies of BD have been performed (Burton et al., 2007; Smith et al., 2009), identifying several BD susceptibility variants in genes such as ANK3, CACNA1C, and ODZ4 (Sklar et al., 2011; Ferreira et al., 2008). However, these genetic variants account for a small proportion of the heritability of BD, and the complex genetic etiology of BD remains largely unknown (Kendler, 2013). Detecting additional risk variants is hampered by low statistical power. While the primary strategy to improve power of genetic studies has relied on increasing the sample size, other avenues for improving statistical power (even with small sample sizes) need to be considered. Notably, incorporating important covariates or a more refined phenotype has the potential to substantially improve power by reducing phenotypic (and thus genetic) heterogeneity.
The BD phenotype is highly heterogeneous, with a number of important clinical comorbidities that constitute a wide range of disease subtypes. Because samples of BD cases are likely to be comprised of multiple subtypes controlled by different genetic mechanisms, the phenotypic heterogeneity of BD impedes the identification of genetic effects contributing to the disease (Alda et al., 2009; Alda, 2004). Definition of sub-phenotypes based on clinical factors known to be associated with BD may establish more refined subgroups of cases with distinct underlying genetic risk factors (Saunders et al., 2008). A similar strategy was successfully employed in a genetic study of BD that incorporated the effects of body mass index (Winham et al., 2013). However, with the exception of studies incorporating migraine and BD comorbidities defined by DSM diagnoses (Oedegaard et al., 2010; Kerner et al., 2011), few previous GWA studies have examined BD subtypes based on symptoms or diagnoses known to be associated with BD.
BD is associated with eating dysregulation phenotypes including binge eating behavior (BE) (McElroy et al., 2013; Wildes et al., 2007). Moreover, co-occurrence with BE is associated with greater bipolar illness burden (McElroy et al., 2013; Brietzke et al., 2011). However, despite the known associations between BD and BE, along with the heritability of BE, including broadly-defined BE (Hudson et al., 2006; Javaras et al., 2008; Klump et al., 2009; Thornton et al., 2011; Bulik et al., 1998; Sullivan et al., 1998), the genetic architectures of these traits have not been investigated simultaneously.
Given the relationship between BD and BE, pleiotropic effects and other commonalities in the genetic mechanisms underlying both diseases are plausible. In this study, we examined the genetic architecture of BD and BE in conjunction rather than isolation, enabling us to characterize an important subtype of BD and to advance our understanding of the genetic epidemiology of BD with comorbid BE. We utilized publically available data from a prior GWA study of BD conducted by the Genetic Association Information Network (GAIN) (Smith et al., 2009) to re-evaluate BD genetic associations with consideration of a subtype based on lifetime history of BE, and attempted replication of top findings in an independent cohort of BD cases and controls from Mayo Clinic. Although information on BE was collected in the GAIN study, this data was not incorporated in any prior GWA analyses. Our study utilizes this data to examine the genetic risk factors of BD with BE-related comorbidities, with validation in an independent sample.
2. Materials and methods
2.1. Study description
The data collected by the Bipolar Disorder Genome Study Consortium, part of the Genetic Association Information Network (GAIN), were accessed through dbGaP (Mailman et al., 2007). The data have been previously described, including descriptions of study subjects and genotyping and quality control procedures (Smith et al., 2009). After applying previously implemented quality control procedures (Smith et al., 2009), we performed GWA analyses of 729,454 single nucleotide polymorphism (SNP) markers using 1001 European American bipolar cases and 1034 mentally healthy European American controls, genotyped on the Affymetrix Genome-Wide Human SNP Array 6.0.
Bipolar subjects were enrolled at multiple institutions over a period of 18 years. Subjects recruited at different times received different psychiatric interviews based on the Diagnostic Interview for Genetic Studies 2, 3, or 4. All BD cases met the DSM IV criteria for bipolar I disorder. Control subjects completed a psychiatric questionnaire (separate from the Diagnostic Interview for Genetic Studies), and those meeting diagnostic criteria for depression or with a history of BD or psychosis were excluded. Controls were matched to cases for both gender and ethnicity (Smith et al., 2009).
All Diagnostic Interviews for Genetic Studies for BD subjects included the question “Has there ever been a time in your life when you went on food binges (i.e., rapid consumption of a large amount of food in a discrete period of time, usually less than two hours)?”, which was used to define history of BE (yes/no) in the cases (N=929). This is comparable to methods of assessing BE in twin studies. Binge eating information was not collected for control subjects.
2.2. Population stratification
The GAIN study of BD included data from both European American (N=2035) and African American (N=1015) subjects. To avoid potential population stratification, the current study used only data from the European American subjects. We also performed principle components (PC) analysis to correct for population stratification (Price et al., 2006). We evaluated the top 4 PCs to determine whether they were associated with potential phenotypes, including BD and the presence of BE history. The top 4 PCs were not associated with BD; however, the first PC was significantly associated with history of BE (p=0.03). Quantile–quantile (QQ) plots were closely examined in each analysis to monitor the degree of inflation, and the top association results were corrected for the first PC to evaluate the effect of population stratification.
2.3. Statistical analysis
A series of GWA analyses were conducted to characterize the genetic associations with BD and BE. Because questions regarding BE were absent from the control questionnaire, data for BE history were available only in case subjects, precluding a case-control analysis of gene–BE interaction to investigate the differential effect of SNPs on BD risk in the presence of BE history. Instead, we performed GWA analyses stratified by bipolar subtype: mentally healthy controls were separately compared to BD cases with the presence (N=206) or absence (N=723) of BE. Single SNP association tests were conducted using logistic regression for all 729,454 available SNP markers, assuming a log-additive genetic model. This stratified analysis allows the identification of genetic variants that may be associated with the subtype of BD in the absence of BE as well as the more severe subtype of BD with comorbid BE (BD+BE) (Brietzke et al., 2011).
We also performed a BD case-only GWA analysis, where we investigated SNP association with BE history only in subjects with BD. Under the assumption of independence between SNPs and BE, the case-only analysis is a powerful method for detecting gene-environment (i.e., SNP–BE) interaction effects (Kraft et al., 2007). Thus, the case-only analysis comparing BD subjects with BE history vs. BD subjects with no BE history may identify SNPs associated differentially with BD depending on the presence of BE. However, associations observed in this analysis can also reflect association of a SNP with BE (rather than SNP–BE interaction effects).
All analyses were performed in PLINK version 1.07 (Purcell et al., 2007). Reported p-values are not corrected for multiple testing; statistical significance was determined based on a Bonferroni-corrected significance threshold of p<6.85E–8.
2.4. Network analysis
After performing genome-wide analyses, we used Ingenuity Pathway Analysis (IPA) software to perform network analysis to facilitate interpretation of our results (www.ingenuity.com). IPA software was applied to the combined results of the analyses comparing BD cases with BE to controls and the BD case-only analysis, where SNP p-values within a gene were combined using the minimum p-value approach. SNPs were assigned to genes within 10 kb (or the nearest gene), and were pre-filtered to p≤0.05, resulting in ~41,000 top SNPs mapped to genes that were subjected to network and pathway analysis.
IPA used peer-reviewed, published scientific literature within the curated Ingenuity Knowledge Base to establish new networks of direct and indirect interactions between genes and molecules identified in our data based on a functional analysis algorithm. Furthermore, using a gene p-value threshold of p<0.001, IPA identified genes from our dataset that were associated with predefined processes such as biological functions, disease states, and canonical pathways of signaling/metabolic activation. Potential associations between our data and such predefined processes were evaluated using a 2 × 2 contingency right-tailed Fisher's Exact Test to compare the proportion of genes meeting the significance threshold (i.e., focus genes) in a particular process with the proportion of genes meeting the threshold among the remaining genes not associated with the process.
2.5. Genome-wide imputation
To determine whether stronger associations may exist with SNPs that were not genotyped, we imputed non-genotyped SNPs and repeated the analyses in the imputed data. Following phasing using SHAPEIT (Delaneau et al., 2013), imputation was performed on the phased data with Impute2.2.2 (Howie et al., 2012), using the 1000 Genomes Project Phase 1 Data (all populations) as the reference dataset. SNPs with poor imputation quality (dosage R2<0.3) or low minor allele frequency (MAF<0.01) were removed, resulting in 8,466,825 SNPs for analysis.
2.6. Replication sample
We used an independent sample from Mayo Clinic Biobanks to investigate possible replication of the top ranking SNPs. Cases consisted of 855 subjects from the Mayo Clinic Individualized Medicine Bipolar Biobank with a confirmed diagnosis of BD based on DSM-IV-TR criteria. The Mayo Clinic Bipolar Biobank is a collaborative effort across four sites (Mayo Clinic, Rochester, MN; Mayo Clinic Health System, Austin Medical Center, MN; Lindner Center of HOPE, Mason, OH; and University of Minnesota, Minneapolis, MN). BE was defined as having a lifetime history of binge eating disorder as determined by structured clinical interview (McElroy et al., 2013). Controls consisted of 857 mentally healthy subjects from the Mayo Clinic Community Biobank (Olson et al., 2013). Subjects who had a prior diagnosis of BD, schizophrenia, major depression, Down syndrome, autism, attention deficit hyperactivity disorder or other psychiatric condition, and those that reported having a first degree relative with BD, were excluded. Potential controls were also excluded if their answers to questions related to psychological well-being suggested the possibility of depression or mania. Eligible control subjects were matched to cases on age, sex, and race/ethnicity. All subjects were at least 18 years of age, and gave written informed consent. This genetic association study was approved by the Mayo Clinic Institutional Review Board.
Candidate genes and SNPs were selected for replication based on high rankings in the discovery sample combined with biological plausibility, and included SNPs in the following genes: PRR5, RNASE4, ANK2, APOB, PER4, CACNA2D1, and ZNF536. In addition to 9 candidate SNPs selected from the top ranking results, a set of tag SNPs was selected for each gene. Cases and controls were genotyped using the Illumina GoldenGate Platform. A CEPH trio was included on each plate and 20 study subjects were genotyped in duplicate, with genotype concordance rate >99.9%. Subjects that failed genotyping, had call rates <95%, or were not of European ancestry were excluded from analysis, resulting in 828 cases (including 70 with BE) and 832 controls that passed quality control. Five SNPs failed genotyping, and 2 SNPs that were out of Hardy–Weinberg Equilibrium (p<10−7) were removed from analysis; this resulted in 209 SNP in 7 genes after quality control (17 in PRR5, 14 in APOB, 63 in ANK2, 7 in the PER4 region, 73 in CACNA2D1, 8 in RNASE4, and 27 in ZNF536).
Analyses of the replication sample were performed as described above for the discovery sample. In particular, for each SNP, genetic associations with BE were examined within only the BD cases, and BD cases with BE history were compared to healthy controls. A fixed effects meta-analysis was conducted to combine results from the discovery and replication samples for each SNP. The meta-analyses were conducted using R statistical software (version 2.14.0) and the package ‘rmeta’ (http://cran.us.r-project.org/).
3. Results
3.1. Discovery sample
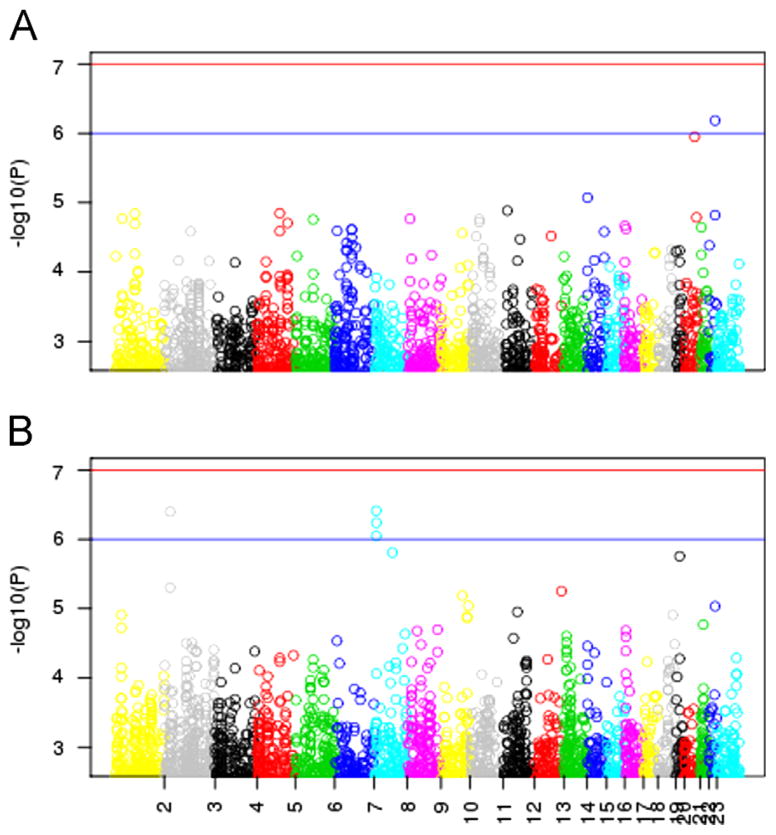
Of the BD cases, 206 subjects (22.2%) reported a history of BE, whereas 723 (77.8%) had no BE history. None of the SNP associations were significant at a genome-wide significance level after correction for multiple testing. The top ranking SNP in the case-only GWA analysis of BE history was rs6006893 in an intron of the PRR5 gene (Fig. 1A and Table 1, p=6.5E–7). Rs17045162 in an intron of ANK2 was also ranked highly in the case-only analysis of BE (p=1.4E–5).
Fig 1.
Manhattan plots for GWA analyses. For each SNP, −log10(p-value) (y-axis) is plotted against chromosomal position (x-axis) for (A) the BD case-only analysis comparing cases with and without a history of BE and (B) the analysis comparing the subtype of BD cases with BE to healthy controls.
Table 1.
Top 10 SNP results for analysis of BE history in BD cases only.
| SNP | CHR | Nearest gene | Minor allele | MAF BD+BE | MAF BD−BE | OR | p-Value |
|---|---|---|---|---|---|---|---|
| rs6006893 | 22 | PRR5 | T | 0.175 | 0.089 | 2.25 | 6.53E–07 |
| rs13037732 | 20 | LOC79015 | T | 0.135 | 0.060 | 2.52 | 1.12E–06 |
| rs5019558 | 14 | RNASE4 | G | 0.392 | 0.489 | 0.59 | 8.46E–06 |
| rs1003077 | 11 | COPB1 | T | 0.213 | 0.126 | 1.88 | 1.30E–05 |
| rs17045162 | 4 | ANK2 | G | 0.083 | 0.032 | 2.83 | 1.42E–05 |
| rs17414905 | 1 | SLC44A3 | T | 0.142 | 0.243 | 0.51 | 1.44E–05 |
| rs8135269 | 22 | PRR5-ARHGAP8 | T | 0.174 | 0.100 | 1.96 | 1.51E–05 |
| rs6068048 | 20 | ZFP64 | A | 0.187 | 0.104 | 1.95 | 1.63E–05 |
| rs7545742 | 1 | C1orf94 | C | 0.157 | 0.083 | 2.11 | 1.69E–05 |
| rs2189887 | 8 | MCPH1 | A | 0.191 | 0.112 | 1.96 | 1.71E–05 |
Abbreviations: SNP, single nucleotide polymorphism; CHR, chromosome location; MAF, minor allele frequency; OR, odds ratio.
In the stratified analysis, when comparing BD cases with BE to controls (Fig. 1B and Table 2), the strongest signal was rs13233490–603 kb upstream of the pseudogene PER4 (p=3.9E–7). SNP rs17158578, which is 578 kb upstream of PER4 and in high linkage disequilibrium with rs13233490, was also highly ranked (p=8.9E–7). SNPs rs4665788 and rs10198175, both downstream of APOB, showed a trend of association with the BD+BE subtype (p=4.0E–7, and p=5.0E–6, respectively). SNP rs2367911 in an intron in CACNA2D1 also demonstrated some evidence of association with BE+BD as compared to controls (p=1.6E–6). Also, rs7249968 in an intergenic region near ZNF536 may be associated with an increased odds of BE+BD (p=1.8E–6).
Table 2.
Top 10 SNP results for analysis of BD stratified by BE history.
| SNP | CHR | Nearest gene | Minor allele | MAF BD+BE | MAF BD−BE | MAF controls | BD+BE vs. controls N=206 vs. N=1034 |
BD−BE vs. controls N=723 vs. N=1034 |
||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | p-Value | OR | p-Value | |||||||
| rs13233490 | 7p21.3 | PER4 | G | 0.042 | 0.018 | 0.007 | 6.37 | 3.87E–07 | 2.61 | 0.0045 |
| rs4665788 | 2p24-p23 | APOB | T | 0.377 | 0.278 | 0.248 | 1.79 | 4.01E–07 | 1.16 | 0.0542 |
| rs10224816 | 7p21.3 | NDUFA4 | G | 0.174 | 0.103 | 0.088 | 2.16 | 5.78E–07 | 1.18 | 0.1465 |
| rs17158578 | 7p21.3 | PER4 | G | 0.046 | 0.022 | 0.009 | 5.13 | 8.95E–07 | 2.35 | 0.0041 |
| rs2367911 | 7p21-q22 | CACNA2D1 | A | 0.071 | 0.039 | 0.024 | 3.29 | 1.55E–06 | 1.69 | 0.0096 |
| rs7249968 | 19q12 | ZNF536 | G | 0.310 | 0.223 | 0.200 | 1.79 | 1.76E–06 | 1.15 | 0.1109 |
| rs10198175 | 2p24-p23 | APOB | T | 0.163 | 0.097 | 0.086 | 2.09 | 5.00E–06 | 1.15 | 0.2441 |
| rs17448220 | 12q24.31 | LOC400084 | C | 0.270 | 0.197 | 0.172 | 1.81 | 5.61E–06 | 1.19 | 0.0578 |
| rs10491807 | 9q32 | TAL2 | T | 0.159 | 0.087 | 0.086 | 2.07 | 6.51E–06 | 1.01 | 0.9070 |
| rs10988393 | 9q34.11 | C9orf50 | A | 0.323 | 0.234 | 0.223 | 1.75 | 9.14E–06 | 1.06 | 0.4825 |
Results for the stratified analysis comparing BD subjects without history of BE to controls were similar to the unadjusted results for European Americans reported by Smith et al. (2009) (Supplementary Table S3).
The genome-wide analyses did not exhibit effects of population stratification, even though the first PC was significantly associated with history of BE. The QQ plots (Supplementary Figs. S1–S3) demonstrated little inflation of the association test statistics (λ =1.02, 1.03, and 1.02 for the tests evaluating BE history in BD cases only, BD+BE compared to controls, and BD cases without BE compared to controls, respectively), and for all three genome-wide analyses, the five top ranking SNPs for each unadjusted analysis remained in the top 10 PC-adjusted rankings. Although the rankings were preserved, some SNPs demonstrated slightly attenuated effect sizes after PC-adjustment, particularly for the analysis comparing BD cases with BE to controls (Supplementary Tables S1–S3).
3.2. Network analysis
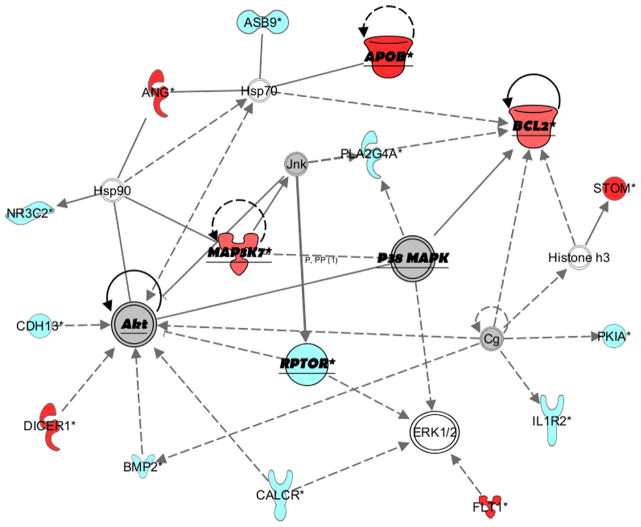
Two top networks/interactomes were estimated with IPA software based on our data: one involving APOB and the other CACNA2D1 (Fig. 2 and Supplementary Fig. S4). The APOB network also linked with AKT, MAPK, DICER1, ERK, BCL2, and STOM (highly ranked in our analysis). The CACNA2D1 network involved other voltage-dependent calcium channel genes such as CACNA1C and CACNB2, as well as metabolic and inflammatory genes. The pre-defined canonical pathways of calcium signaling and glucocorticoid receptor signaling were highly ranked, with 12 out of 189 (6.3%) and 16 out of 277 (5.8%) pathway molecules identified (p<0.001) in our analyses, respectively.
Fig 2.
Gene network involving APOB generated with IPA. The color of genes/molecules indicates the degree of association in our analyses (by p-value): red=high rank, light blue=moderate rank, gray=low rank, white=did not pass filter of p<0.001. Shapes indicate the gene/molecule type (cytokine, transporter, kinase, etc.). Solid lines indicate direct relationships and dashed lines/arrows indicate indirect relationships. Genes/molecules emphasized in underlined/bold text are discussed in the manuscript. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.3. Imputation
Results that include imputed SNPs are similar to the results for the observed genotypes, with denser peaks in the top-ranking regions (Supplementary Figs. S5 and S6). In the BE case-only analysis of imputed SNPs, a clear peak is evident in the PRR5 region, and a new high ranking variant in an intergenic region on chromosome 1 is now apparent (Supplementary Table S4). The BD+BE vs. controls analysis of imputed data resulted in dense peaks near the observed top-ranking SNPs in APOB, PER4, and ZNF536, and additional high-ranking imputed variants on chromosomes 4 and 10 (Supplementary Table S5). However, these new peaks represent single SNPs with low imputation quality scores, so should be interpreted with caution.
3.4. Replication sample
In the analysis comparing BD cases with and without BE, the top ranking SNP from the discovery set, rs6006893 in PRR5, showed a consistent effect direction, although the effect size was attenuated and not statistically significant in the replication analysis (OR=1.41, p=0.21). Most interestingly, the effect estimates for rs10198175 in APOB were nearly identical between the discovery and replication sets (OR=1.84 vs. 1.80), with nominally significant evidence of replication (p=0.017). Little evidence of replication was obtained for the remaining candidate SNPs, with many of the effect size estimates being in opposite directions in the discovery and replication samples (Table 3). Similarly, in the analysis comparing the subtype of BD cases with BE to healthy controls, little evidence of replication was observed besides rs10198175 in APOB (Table 4), which again displayed a similar effect size between the discovery and replication sets (OR=2.09 vs. 1.57) with marginally significant evidence of replication (p=0.06).
Table 3.
Results for associations with BE within BD cases for the 9 candidate SNPs identified from the discovery set; results are presented for the discovery set, replication set, and the meta-analysis.
| SNP | CHR | Nearest gene | Discovery set
|
Replication set
|
Meta-analysis
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | P | MAF BD+BE | MAF BD−BE | OR | p | OR | p | |||
| rs10198175 | 2 | APOB | 1.84 | 2.98E–04 | 0.171 | 0.103 | 1.80 | 0.017 | 1.83 | 1.48E–05 |
| rs13233490 | 7 | PER4 | 2.55 | 2.85E–03 | 0.007 | 0.019 | 0.48 | 0.463 | 2.20 | 8.53E–03 |
| rs17045162 | 4 | ANK2 | 2.83 | 1.42E–05 | 0.029 | 0.043 | 0.93 | 0.885 | 2.34 | 9.87E–05 |
| rs17158578 | 7 | PER4 | 2.27 | 6.07E–03 | 0.007 | 0.019 | 0.48 | 0.463 | 2.00 | 1.54E–02 |
| rs2367911 | 7 | CACNA2D1 | 1.95 | 6.12E–03 | 0.043 | 0.036 | 0.86 | 0.803 | 1.75 | 1.40E–02 |
| rs4665788 | 2 | APOB | 1.54 | 2.38E–04 | 0.321 | 0.253 | 1.36 | 0.118 | 1.49 | 7.70E–05 |
| rs5019558 | 14 | RNASE4 | 0.59 | 8.46E–06 | 0.493 | 0.479 | 1.13 | 0.513 | 0.71 | 6.31E–04 |
| rs6006893 | 22 | PRR5 | 2.25 | 6.53E–07 | 0.132 | 0.108 | 1.41 | 0.213 | 1.99 | 9.09E–07 |
| rs7249968 | 19 | ZNF536 | 1.60 | 2.61E–04 | 0.207 | 0.231 | 0.82 | 0.379 | 1.37 | 5.64E–03 |
Table 4.
Results for comparison between BD cases with BE to healthy controls for the 9 candidate SNPs identified from the discovery set; results are presented for the discovery set, replication set, and the meta-analysis.
| SNP | CHR | Nearest gene | Discovery set
|
Replication set
|
Meta-analysis
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR | p | MAF BD+BE | MAF controls | OR | p | OR | p | |||
| rs10198175 | 2 | APOB | 2.09 | 5.00E–06 | 0.171 | 0.118 | 1.57 | 0.061 | 1.92 | 1.34E–06 |
| rs13233490 | 7 | PER4 | 6.37 | 3.87E–07 | 0.007 | 0.012 | 0.59 | 0.607 | 4.89 | 3.95E–06 |
| rs17045162 | 4 | ANK2 | 2.38 | 5.85E–05 | 0.029 | 0.043 | 0.66 | 0.422 | 1.98 | 6.46E–04 |
| rs17158578 | 7 | PER4 | 5.13 | 8.95E–07 | 0.007 | 0.011 | 0.62 | 0.644 | 4.21 | 5.76E–06 |
| rs2367911 | 7 | CACNA2D1 | 3.29 | 1.55E–06 | 0.043 | 0.036 | 1.19 | 0.687 | 2.56 | 1.27E–05 |
| rs4665788 | 2 | APOB | 1.79 | 4.01E–07 | 0.321 | 0.274 | 1.25 | 0.235 | 1.62 | 7.54E–07 |
| rs5019558 | 14 | RNASE4 | 0.63 | 3.48E–05 | 0.493 | 0.490 | 1.01 | 0.943 | 0.72 | 4.80E–04 |
| rs6006893 | 22 | PRR5 | 1.95 | 9.28E–06 | 0.132 | 0.113 | 1.20 | 0.491 | 1.73 | 2.86E–05 |
| rs7249968 | 19 | ZNF536 | 1.79 | 1.76E–06 | 0.207 | 0.206 | 1.01 | 0.978 | 1.56 | 3.02E–05 |
Most SNP associations for top-ranking SNPs in the discovery sample were not strengthened when the replication sample was combined with the discovery sample via meta-analysis, with the exception of rs10198175 in APOB, for which the BD+BE subtype association was strengthened over that observed in the discovery sample alone (p=2.98E–4 vs. p=1.48E–5, Table 3; p=5.00E–6 vs. p=1.92E–6, Table 4).
The tag SNPs for the seven candidate genes genotyped in the replication sample also did not yield significant findings after correction for multiple testing. The SNPs with smallest p-values included rs10418356 in ZNF536 (p=0.0029 in the analysis comparing BD cases with and without BE and p=0.012 in the analysis comparing BD+BE cases with controls) and rs6533680 in ANK2 (p=0.0055 in the analysis comparing BD cases with and without BE and p=0.019 in the analysis comparing BD+BE cases with controls). These results are not statistically significant after correction for the number of tag SNPs analyzed in the replication study.
4. Discussion
In the current study, we examined genome-wide associations with BE as a comorbidity of BD, in order to uncover novel susceptibility variants of BD with this eating dysregulation sub-phenotype. Our top-ranking SNPs include previously unidentified variants in genes with functions that are biologically relevant to BD and BE. In particular, in the stratified analysis comparing the subtype of BD cases with BE to healthy controls, we identified biologically plausible genetic associations with SNPs in APOB. In the discovery set, we identified two SNPs within the APOB gene associated with the subtype of BD with BE. In our replication set, the minor allele of rs10198175 displayed trends towards increased risk of BE among BD cases only and increased risk of the BD subtype with BE compared to controls. Similarly, in the meta-analysis of the discovery and replication sets, evidence of the involvement of rs10198175 in risk for BD and BE was strengthened in both the case-only and subtype analyses. APOB is a protein-coding gene for apolipoprotein B, the main component of chylomicrons and low density lipoproteins (LDL), implicated in the pathogenesis of atherosclerosis (Park et al., 2011), cardiovascular disease (Willer et al., 2008), cerebral β-amyloidosis, cognitive decline (Ramirez et al., 2011) and blood–brain barrier disruption (Pallebage-Gamarallage et al., 2012). Individual reports have implicated APOB mutations with suicide and violent behavior (Ramirez et al., 2011); on the other hand, mixed evidence has emerged, regarding a possible role in the interaction between depressive symptoms and atherogenic metabolic profiles (Hummel et al., 2011); yet its brain functional role is not completely understood (Elliott et al., 2010).
The BD case-only analysis of BE suggested a possible association between a variant in PRR5 and BE history in subjects with BD; although this variant was not statistically significant in our replication sample, the effect size estimates in the replication were consistent with those seen in the discovery sample. The PRR5 gene encodes a subunit of the mammalian target of rapamycin (mTOR) complex 2 (mTORC2). mTOR is a serine/threonine kinase expressed in many tissues, including the brain, where it participates in food intake regulation, acting as an energy sensor in the hypothalamus (Cota et al., 2006), neuronal development and synaptic plasticity (Weber and Gutmann, 2012). Deficits in mTOR expression appear to be related to depression pathophysiology (Jernigan et al., 2011) and mTOR inhibition has an anorexic and pro-depressant effect (Russo et al., 2012; Zhou et al., 2010). Consistently, its activation is one of the mechanisms of ketamine antidepressant action (Li et al., 2010). Thus the potential role of PRR5 in risk of BD with comorbid BE deserves further investigation.
In subjects with BD, we observed an association between a variant near ANK2 and history of BE. ANK2 codes for ankyrin 2, a gene related to known BD susceptibility gene ANK3. Although a plausible gene for susceptibility to BD, we did not observe evidence for ANK2 in our replication sample or meta-analysis. A variant within CACNA2D1 (in the same family of voltage-dependent calcium channels as CACNA1C) was also a top ranking SNP in our stratified analysis of BD cases with BE compared to controls. Notably, the same CACNA2D1 variant was previously reported in a meta-analysis including the GAIN data (Smith et al., 2011), but was not reported in the original analysis. In fact the association with CACNA2D1 was not identified until the sample size was increased to 2191 cases and 1434 controls; in the present study, we elicited the same variant with a reduced sample size based on a refined subtype, emphasizing the potential increase in power to detect relevant genetic association as a result of phenotypic refinement and reduced trait variation.
IPA analysis revealed two main networks showing high gene/ molecule interaction. These networks involved two of our top ranked genes: APOB and CACNA2D1. Interestingly, in the same network as APOB, growth factor downstream signaling cascade molecules Akt, ERK, MAPK and Bcl-2 were linked; these molecules have been related to BD pathophysiology and are involved in mood stabilization mechanisms (Soeiro-de-Souza et al., 2012). Lithium acts by disinhibiting Akt (Beaulieu et al., 2008), while ketamine activates Akt and ERK (Li et al., 2010). Lithium and valproate increase levels of ERK in the hippocampus, and its inhibition is correlated with manic-like effects in rodents (Einat et al., 2003). Lower Bcl-2 expression with subsequent alterations of calcium homeostasis (Schloesser et al., 2012) and glutamate dysregulation (Soeiro-de-Souza et al., 2013), have been identified as part of BD pathophysiology (Chong et al., 2012); its neuroprotective role therefore has been suggested as a potential target for future mood pharmacotherapy (Li et al., 2012).
The top ranking SNPs based on our analyses differ from those previously reported for European Americans in the GAIN data (Smith et al., 2009) based on analyses that did not incorporate BE history. However, rs7690204 in GYPA and rs10193871 in NCKAP5 (a.k.aNAP5) reported by Smith et al. were also highly ranked in our analyses, with more extreme odds ratio estimates in our analyses (Supplementary Table S6). Furthermore, our analyses integrating additional information into the investigation of genome-wide associations with BD (through restriction to a BE subtype) generally pointed to variants with greater biological relevance, as well as results of greater significance (including lower p-values and higher odds ratios) than those reported in the original analyses. This is striking, considering that our analysis utilized a smaller sample size due to stratification and the restriction to those with available BE data. This highlights the potential advantage in increased power to detect genetic effects through the reduction of heterogeneity. By considering phenotypic subtypes, potentially important genetic factors became more readily apparent.
5. Limitations
While power may have improved by reducing heterogeneity through the use of more refined phenotypic subtypes, such refinement resulted in a reduction in sample size to only N=206 BD cases with BE in the discovery sample – a distinct limitation of this study. To overcome such a small sample size, we performed replication analysis for candidate SNPs within an independent sample, and increased sample size via meta-analysis with the data from GAIN, providing stronger evidence for the involvement of the APOB gene in the risk of BD with BE.
Because rs10198175 in APOB demonstrated evidence of a relationship with BD and BE risk in both analyses, this SNP may be associated with BE behavior in general, a subtype of BD with BE, or may reflect a SNP–BE interaction effect on BD susceptibility; however these potential effects cannot be untangled without relevant information on history of BE in control subjects. Further genetic association studies in well-characterized human samples, as well as functional and longitudinal studies, may help to determine the causal pathways underlying the observed associations with this APOB sequence variant.
Additional study limitations should be noted. Because subjects were recruited over a long time period, subsets of BD cases were given different diagnostic interviews, which were also different from those given to controls. This restricted our analyses to subsets of the full sample, precluding a thorough assessment of the relationship between BD, BE, and SNP effects and limiting the interpretation of results. For example, the BD case-only analysis of BE behavior can be used to investigate potential SNP–BE interactions, under the assumption that BE is independent of a given genetic effect; however, for some SNPs in the genome, this assumption is likely to be false, since BE is thought to be under genetic influence and may in fact share genetic factors with BD (Hudson et al., 2008). The lack of data on BE for mentally healthy control subjects without BD impedes this investigation, because we cannot determine whether associated SNPs reflect an interaction effect on BD or whether the SNP is associated with BE independent of BD. Similarly, for variants associated with the BD+BE subtype, we are unable to determine whether the association is driven by BD, BE, or both. However, our analyses allow us to study an underlying construct that may be responsible for both mood disorders and eating behaviors; the details of the mechanism of action of identified risk factors on these phenotypes needs to be examined in subsequent studies. A final limitation is that the definition of BE used in the discovery sample was broader than that in the replication sample.
6. Conclusion
In this study, we were able to identify a number of variants with biologically plausible roles in BD and BE, and through an independent replication sample and meta-analysis, provided additional evidence for variants within the APOB gene. However, it remains to be determined whether the variants identified in these analyses have functional relationships to BD, BE behavior, or both; further investigation is necessary to determine the specific role of APOB in BD susceptibility. Nevertheless, this study demonstrates the importance of phenotypic sub-classification in genetic studies of complex psychiatric traits. It lays the groundwork for further genome-wide investigation of eating dysregulation and obesity-related measures in bipolar patients, and more broadly, further examination of potential genetic effects underlying other phenotypic subtypes.
Supplementary Material
Acknowledgments
Role of funding source
This study was supported by funding from the Marriott Family Foundation, Mayo Clinic's Center for Individualized Medicine, and NIH K12 HD65987 (SJW). Funding support for the Whole Genome Association Study of Bipolar Disorder was provided by the National Institute of Mental Health (NIMH) and the genotyping of samples was provided through the Genetic Association Information Network (GAIN).
The datasets used for the analyses described in this manuscript were obtained from the database of Genotypes and Phenotypes (dbGaP) found at 〈http://www.ncbi.nlm.nih.gov/gap〉 through dbGaP accession number phs000017.v3.p1. Samples and associated phenotype data for the Collaborative Genomic Study of Bipolar Disorder were provided by the NIMH Genetics Initiative for Bipolar Disorder. Data and biomaterials were collected in four projects that participated in NIMH Bipolar Disorder Genetics Initiative. From 1991–1998, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, U01 MH46282, John Nurnberger, M.D., Ph.D., Marvin Miller, M.D., and Elizabeth Bowman, M.D.; Washington University, St. Louis, MO, U01 MH46280, Theodore Reich, M.D., Allison Goate, Ph.D., and John Rice, Ph.D.; Johns Hopkins University, Baltimore, MD U01 MH46274, J. Raymond DePaulo, Jr., M.D., Sylvia Simpson, M.D., MPH, and Colin Stine, Ph.D.; NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, MD, Elliot Gershon, M.D., Diane Kazuba, B.A., and Elizabeth Maxwell, M.S. W. Data and biomaterials were collected as part of ten projects that participated in the NIMH Bipolar Disorder Genetics Initiative. From 1999–2003, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, M.D., Ph.D., Marvin J. Miller, M.D., Elizabeth S. Bowman, M.D., N. Leela Rau, M.D., P. Ryan Moe, M.D., Nalini Samavedy, M.D., Rif El-Mallakh, M.D. (at University of Louisville), Husseini Manji, M.D. (at Wayne State University), Debra A. Glitz, M.D. (at Wayne State University), Eric T. Meyer, M.S., Carrie Smiley, R.N., Tatiana Foroud, Ph.D., Leah Flury, M.S., Danielle M. Dick, Ph.D., Howard Edenberg, Ph.D.; Washington University, St. Louis, MO, R01 MH059534, John Rice, Ph.D, Theodore Reich, M.D., Allison Goate, Ph.D., Laura Bierut, M.D.; Johns Hopkins University, Baltimore, MD, R01 MH59533, Melvin McInnis M.D., J. Raymond DePaulo, Jr., M.D., Dean F. MacKinnon, M.D., Francis M. Mondimore, M. D., James B. Potash, M.D., Peter P. Zandi, Ph.D., Dimitrios Avramopoulos, and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini M. D., Ph.D.; University of California at Irvine, CA, R01 MH60068, William Byerley M.D., and Mark Vawter M.D.; University of Iowa, IA, R01 MH059548, William Coryell M. D., and Raymond Crowe M.D.; University of Chicago, IL, R01 MH59535, Elliot Gershon, M.D., Judith Badner Ph.D., Francis McMahon M.D., Chunyu Liu Ph.D., Alan Sanders M.D., Maria Caserta, Steven Dinwiddie M.D., Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, John Kelsoe, M.D., Rebecca McKinney, B.A.; Rush University, IL, R01 MH059556, William Scheftner M.D., Howard M. Kravitz, D.O., M.P.H., Diana Marta, B.S., Annette Vaughn-Brown, MSN, RN, and Laurie Bederow, MA; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, Francis J. McMahon, M.D., Layla Kassem, PsyD, Sevilla Detera-Wadleigh, Ph.D, Lisa Austin, Ph.D, Dennis L. Murphy, M.D.
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.jad.2014.04.026.
Footnotes
Conflict of interest
Dr. McElroy is a consultant to or member of the scientific advisory boards of Alkermes, Bracket, Corcept, MedAvante, Shire, and Teva. She is a principal or co-investigator on studies sponsored by the Agency for Healthcare Research & Quality (AHRQ), Alkermes, AstraZeneca, Cephalon, Eli Lilly and Company, Forest, Marriott Foundation, National Institute of Mental Health, Orexigen Therapeutics, Inc., Pfizer, Shire, Takeda Pharmaceutical Company Ltd., and Transcept Pharmaceutical, Inc. Dr. McElroy has consulted for: F. Hoffman LaRoche and Naurex. She is also an inventor on United States Patent No. 6,323,236 B2, Use of Sulfamate Derivatives for Treating Impulse Control Disorders, and along with the patient's assignee, University of Cincinnati, Cincinnati, Ohio, has received payments from Johnson & Johnson, which has exclusive rights under the patent.
Dr. Crow has received research grants from Shire, Alkermes, and National Institute of Diabetes and Digestive and Kidney Diseases, United States (P30DK50456).
Dr Frye has received grant support from Assurex Health, Myriad, Pfizer, National Institute of Mental Health (RO1 MH079261), National Institute of Alcohol Abuse and Alcoholism (P20AA017830), Mayo Foundation; has been a consultant to Janssen Global Services, LLC, Mitsubishi Tanabe Pharma Corporation, Myriad, Sunovion, and Teva Pharmaceuticals; has received CME/Travel Support/presentation from CME Outfitters Inc. and Sunovian.
All other authors have no conflicts of interest to declare.
References
- Alda M. The phenotypic spectra of bipolar disorder. Eur Neuropsychopharmacol. 2004;14:S94–S99. doi: 10.1016/j.euroneuro.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Alda M, et al. Treatment of bipolar disorder: new perspectives. Ann Med. 2009;41 (3):186–196. doi: 10.1080/07853890802409489. [DOI] [PubMed] [Google Scholar]
- Beaulieu JM, et al. A beta-arrestin 2 signaling complex mediates lithium action on behavior. Cell. 2008;132 (1):125–136. doi: 10.1016/j.cell.2007.11.041. [DOI] [PubMed] [Google Scholar]
- Brietzke E, et al. Clinical correlates of eating disorder comorbidity in women with bipolar disorder type I. J Affect Disord. 2011;130 (1–2):162–165. doi: 10.1016/j.jad.2010.10.020. [DOI] [PubMed] [Google Scholar]
- Bulik CM, Sullivan PF, Kendler KS. Heritability of binge-eating and broadly defined bulimia nervosa. Biol Psychiatry. 1998;44 (12):1210–1218. doi: 10.1016/s0006-3223(98)00280-7. [DOI] [PubMed] [Google Scholar]
- Burton PR, et al. Genome-wide association study of 14,000 cases of seven common diseases and 3000 shared controls. Nature. 2007;447 (7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong ZZ, et al. A critical kinase cascade in neurological disorders: PI 3-K, Akt, and mTOR. Future neurol. 2012;7 (6):733–748. doi: 10.2217/fnl.12.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cota D, et al. Hypothalamic mTOR signaling regulates food intake. Science. 2006;312 (5775):927–930. doi: 10.1126/science.1124147. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10 (1):5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- Einat H, et al. The role of the extracellular signal-regulated kinase signaling pathway in mood modulation. J Neurosci. 2003;23 (19):7311–7316. doi: 10.1523/JNEUROSCI.23-19-07311.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott DA, Weickert CS, Garner B. Apolipoproteins in the brain: implications for neurological and psychiatric disorders. Clin Lipidol. 2010;51 (4):555–573. doi: 10.2217/CLP.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40 (9):1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, et al. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44 (8):955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JI, et al. Binge-eating disorder as a distinct familial phenotype in obese individuals. Arch Gen Psychiatry. 2006;63 (3):313–319. doi: 10.1001/archpsyc.63.3.313. [DOI] [PubMed] [Google Scholar]
- Hudson JI, et al. A structural approach to the familial coaggregation of disorders. Epidemiology. 2008;19 (3):431–439. doi: 10.1097/EDE.0b013e31816a9de7. [DOI] [PubMed] [Google Scholar]
- Hummel J, et al. Serum lipoproteins improve after successful pharmacologic antidepressant treatment: a randomized open-label prospective trial. J Clin Psychiatry. 2011;72 (7):885–891. doi: 10.4088/JCP.09m05853blu. [DOI] [PubMed] [Google Scholar]
- Javaras KN, et al. Familiality and heritability of binge eating disorder: results of a case-control family study and a twin study. Int J Eat Disord. 2008;41 (2):174–179. doi: 10.1002/eat.20484. [DOI] [PubMed] [Google Scholar]
- Jernigan CS, et al. The mTOR signaling pathway in the prefrontal cortex is compromised in major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35 (7):1774–1779. doi: 10.1016/j.pnpbp.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS. What psychiatric genetics has taught us about the nature of psychiatric illness and what is left to learn. Mol Psychiatry. 2013;18 (10):1058–1066. doi: 10.1038/mp.2013.50. [DOI] [PubMed] [Google Scholar]
- Kerner B, Lambert CG, Muthen BO. Genome-wide association study in bipolar patients stratified by co-morbidity. PLoS One. 2011;6 (12):e28477. doi: 10.1371/journal.pone.0028477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klump KL, et al. Genetic and environmental influences on disordered eating: an adoption study. J Abnorm Psychol. 2009;118 (4):797–805. doi: 10.1037/a0017204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft P, et al. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63 (2):111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- Li N, et al. mTOR-dependent synapse formation underlies the rapid antidepressant effects of NMDA antagonists. Science. 2010;329 (5994):959–964. doi: 10.1126/science.1190287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Frye MA, Shelton RC. Review of pharmacological treatment in mood disorders and future directions for drug development. Neuropsychopharmacol: Off Publ Am Coll Neuropsychopharmacol. 2012;37 (1):77–101. doi: 10.1038/npp.2011.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373 (9659):234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39 (10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy SL, et al. Clinical phenotype of bipolar disorder with comorbid binge eating disorder. J Affect Disord. 2013;150 (3):981–986. doi: 10.1016/j.jad.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, et al. The heritability of bipolar affective disorder and the genetic relationship to unipolar depression. Arch Gen Psychiatry. 2003;60 (5):497–502. doi: 10.1001/archpsyc.60.5.497. [DOI] [PubMed] [Google Scholar]
- Oedegaard KJ, et al. A genome-wide association study of bipolar disorder and comorbid migraine. Genes Brain Behav. 2010;9 (7):673–680. doi: 10.1111/j.1601-183X.2010.00601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JE, et al. The Mayo Clinic Biobank: a building block for individualized medicine. Mayo Clin Proc. 2013;88 (9):952–962. doi: 10.1016/j.mayocp.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallebage-Gamarallage M, et al. Restoration of dietary-fat induced blood–brain barrier dysfunction by anti-inflammatory lipid-modulating agents. Lipids Health Dis. 2012;11:117. doi: 10.1186/1476-511X-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, et al. High levels of apolipoprotein B/AI ratio are associated with intracranial atherosclerotic stenosis. Stroke: J Cereb Circ. 2011;42 (11):3040–3046. doi: 10.1161/STROKEAHA.111.620104. [DOI] [PubMed] [Google Scholar]
- Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38 (8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81 (3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez C, et al. ApoB100/LDLR−/− hypercholesterolaemic mice as a model for mild cognitive impairment and neuronal damage. PLoS One. 2011;6 (7):e22712. doi: 10.1371/journal.pone.0022712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo E, et al. mTOR inhibition modulates epileptogenesis, seizures and depressive behavior in a genetic rat model of absence epilepsy. Neuropharmacology. 2012;69:25–36. doi: 10.1016/j.neuropharm.2012.09.019. [DOI] [PubMed] [Google Scholar]
- Saunders EH, et al. Familiality and diagnostic patterns of subphenotypes in the National Institutes of Mental Health bipolar sample. Am J Med Genet B: Neuropsychiatr Genet. 2008;147B (1):18–26. doi: 10.1002/ajmg.b.30558. [DOI] [PubMed] [Google Scholar]
- Schloesser RJ, Martinowich K, Manji HK. Mood-stabilizing drugs: mechanisms of action. Trends Neurosci. 2012;35 (1):36–46. doi: 10.1016/j.tins.2011.11.009. [DOI] [PubMed] [Google Scholar]
- Sklar P, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43 (10):977–983. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, et al. Genome-wide association study of bipolar disorder in European American and African American individuals. Mol Psychiatry. 2009;14 (8):755–763. doi: 10.1038/mp.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, et al. Genome-wide association of bipolar disorder suggests an enrichment of replicable associations in regions near genes. PLoS Genet. 2011;7 (6):e1002134. doi: 10.1371/journal.pgen.1002134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeiro-de-Souza MG, et al. Translating neurotrophic and cellular plasticity: from pathophysiology to improved therapeutics for bipolar disorder. Acta Psychiatr Scand. 2012;126 (5):332–341. doi: 10.1111/j.1600-0447.2012.01889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soeiro-de-Souza MG, et al. Bcl-2 rs956572 polymorphism is associated with increased anterior cingulate cortical glutamate in euthymic bipolar I disorder. Neuropsychopharmacol. 2013;38 (3):468–475. doi: 10.1038/npp.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Bulik CM, Kendler KS. Genetic epidemiology of binging and vomiting. Br J Psychiatry: J Ment Sci. 1998;75–79173:75–79. doi: 10.1192/bjp.173.1.75. [DOI] [PubMed] [Google Scholar]
- Thornton LM, Mazzeo SE, Bulik CM. The heritability of eating disorders: methods and current findings. Curr Top Behav Neurosci. 2011;6:141–156. doi: 10.1007/7854_2010_91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Gutmann DH. Deconvoluting mTOR biology. Cell Cycle. 2012;11 (2):236–248. doi: 10.4161/cc.11.2.19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildes JE, Marcus MD, Fagiolini A. Eating disorders and illness burden in patients with bipolar spectrum disorders. Compr Psychiatry. 2007;48 (6):516–521. doi: 10.1016/j.comppsych.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40 (2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winham SJ, et al. Genome-wide association study of bipolar disorder accounting for effect of body mass index identifies a new risk allele in TCF7L2. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.159. (online publication, 10 December 2013; http://dx.doi.org/10.1038/mp.2013.159) [DOI] [PubMed]
- Zhou H, Luo Y, Huang S. Updates of mTOR inhibitors. Anticancer Agents Med Chem. 2010;10 (7):571–581. doi: 10.2174/187152010793498663. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.