Abstract
Sex determination can be robustly genetic, strongly environmental, or genetic subject to environmental perturbation. The genetic basis of sex determination is unknown for zebrafish (Danio rerio), a model for development and human health. We used RAD-tag population genomics to identify sex-linked polymorphisms. After verifying this “RAD-sex” method on medaka (Oryzias latipes), we studied two domesticated zebrafish strains (AB and TU), two natural laboratory strains (WIK and EKW), and two recent isolates from nature (NA and CB). All four natural strains had a single sex-linked region at the right tip of chromosome 4, enabling sex genotyping by PCR. Genotypes for the single nucleotide polymorphism (SNP) with the strongest statistical association to sex suggested that wild zebrafish have WZ/ZZ sex chromosomes. In natural strains, “male genotypes” became males and some “female genotypes” also became males, suggesting that the environment or genetic background can cause female-to-male sex reversal. Surprisingly, TU and AB lacked detectable sex-linked loci. Phylogenomics rooted on D. nigrofasciatus verified that all strains are monophyletic. Because AB and TU branched as a monophyletic clade, we could not rule out shared loss of the wild sex locus in a common ancestor despite their independent domestication. Mitochondrial DNA sequences showed that investigated strains represent only one of the three identified zebrafish haplogroups. Results suggest that zebrafish in nature possess a WZ/ZZ sex-determination mechanism with a major determinant lying near the right telomere of chromosome 4 that was modified during domestication. Strains providing the zebrafish reference genome lack key components of the natural sex-determination system but may have evolved variant sex-determining mechanisms during two decades in laboratory culture.
Keywords: sex determination, RAD (restriction-site associated DNA)-sex, zebrafish, WZ, sex chromosome, genome-wide association study (GWAS)
…researchers using standard lines of zebrafish that have long been maintained in laboratories are often plagued by severe sex ratio distortions…. C. Lawrence, J. P. Ebersole, and R. V. Kesseli (2008)
CONSIDERING the fundamental importance of sex for species propagation, it is surprising that primary sex-determining mechanisms are not strongly conserved among animal taxa (Bull 1983; Charlesworth 1996; Ming et al. 2011; Bachtrog et al. 2014). Closely related species or even populations of the same species can have different sex-determining mechanisms (Takehana et al. 2007; Ross et al. 2009; Kobayashi et al. 2013; Heule et al. 2014; Larney et al. 2014). Zebrafish (Danio rerio) is a popular model for studies of vertebrate development, behavior, physiology, evolution, disease, and human health (Mills et al. 2007; Seth et al. 2013; Braasch et al. 2014; Ota and Kawahara 2014; Wilkinson et al. 2014), but researchers struggle with highly variable and distorted sex ratios, and investigations into the genetic nature of zebrafish sex determination are conflicting. To help understand these issues, we conducted a population genomic study of sex determination in multiple zebrafish strains.
Zebrafish exhibit a juvenile ovary phase in which the gonad contains meiotic oocytes in all individuals: in some juveniles, oocytes survive and the individual becomes a female, but in others, oocytes die from about 19 to 27 days postfertilization and the fish becomes a male (Takahashi 1977; Uchida et al. 2002; Wang et al. 2007; Rodriguez-Mari et al. 2010). In the absence of germ cells (Slanchev et al. 2005; Siegfried and Nusslein-Volhard 2008) and in mutants in which oocytes undergo apoptosis (Rodríguez-Marí et al. 2005, 2010; Rodríguez-Marí and Postlethwait 2011), gonads develop as testes and individuals become males. Remarkably, mutants that produce oocytes early and reproduce as females can transform into fertile males after oocyte depletion, showing that oocytes are necessary both for primary sex determination and for maintenance of female phenotype in adult zebrafish (Dranow et al. 2013).
As expected from the hypothesis that oocyte death is a major feature of zebrafish sex determination, harsh environmental conditions tend to shift sex ratios in favor of males; such factors include gamma rays, hypoxia, high density, high temperature, altered thermocycles, and poor nutrition (Walker-Durchanek 1980; Shang et al. 2006; Lawrence et al. 2008; Abozaid et al. 2011, 2012; Liew et al. 2012; Villamizar et al. 2012). Zebrafish does not, however, have a typical environmental sex-determination (ESD) mechanism like some sauropsids for which temperature is a cue (Charnier 1966; Lang and Andrews 1994; Merchant-Larios and Diaz-Hernandez 2013; Mork et al. 2014). It is more probable that zebrafish is like medaka (Oryzias latipes) in having a genetic sex-determination mechanism that is sensitive to environmental conditions (Hattori et al. 2007; Sato et al. 2005; Selim et al. 2009; Hayashi et al. 2010).
Zebrafish not only lacks classic ESD, but most stocks investigated do not have cytogenetically detectable chromosomal sex determination (Schreeb et al. 1993; Pijnacker and Ferwerda 1995; Daga et al. 1996; Gornung et al. 1997; Amores and Postlethwait 1999; Gornung et al. 2000; Sola and Gornung 2001; Traut and Winking 2001; Phillips et al. 2006). In contrast, the only investigation of zebrafish taken directly from nature in India concluded that zebrafish females are the heterogametic sex (Sharma et al. 1998). The disagreement in cytogenetic results suggests that different zebrafish strains may have different karyotypic bases for sex determination.
Not only is the chromosomal nature of zebrafish sex unresolved, but its genetic sex-determination (GSD) mechanism remains elusive. Repeated matings of zebrafish pairs from AB, TU (Tuebingen), and Toh strains produce consistent sex ratios, but different pairs can give quite different sex ratios (Liew et al. 2012). This result is expected if zebrafish has a genetic basis for sex determination with polygenic control that differs among strains (Liew et al. 2012). Furthermore, recent studies of sex genetics in different zebrafish strains identified sex-associated loci, confirming a genetic component to sex determination, but different studies identified different sex-associated loci (Orban et al. 2009; Siegfried 2010; Tong et al. 2010; Bradley et al. 2011; Anderson et al. 2012; Liew et al. 2012; Howe et al. 2013; Liew and Orban 2014). A cross between a female of the NA (Nadia) natural strain by a male of the AB laboratory strain identified a single sex-linked locus on zebrafish chromosome 4 (Chr4) while the reciprocal mating (female-AB-by-male-NA) showed sex-associated loci on both Chr4 and Chr3 (Anderson et al. 2012). In contrast, analysis of a female-AB-by-male-IN (India) cross identified sex-linked loci on Chr5 and Chr16 but none on Chr3 or Chr4 (Bradley et al. 2011). An F2 map constructed from a gynogenetic doubled-haploid TU female and a gynogenetic doubled-haploid AB male (i.e., both parents had only female-derived chromosomes) identified a sex-linked region on Chr16 that does not overlap with the one observed in the AB-by-IN mating (Bradley et al. 2011; Howe et al. 2013). Together, these results have been interpreted to support a polygenic sex-determination mechanism (Liew and Orban 2014), but they also support the hypothesis that different zebrafish strains utilize different genetic mechanisms to determine sex.
Mapping crosses like those cited above that mate two zebrafish strains that differ in their major genetic sex-determining mechanisms might give difficult to interpret or even spurious results due to epistatic interactions between loci. Furthermore, because environmental factors can influence zebrafish sex ratios, an individual’s phenotypic sex may not match its genotypic sex. Thus, a traditional F2 mapping cross may not identify a major sex-determining locus if one of the P0 or F1 individuals is by chance sex reversed. In addition, brother-by-sister matings to make F2 families and gynogenesis protocols lead to inbreeding, which results in strongly male-biased sex ratios (Brown et al. 2012a).
To help resolve the confusing state of zebrafish sex genetics, we conducted a genome-wide association study (GWAS) based on a population genomic analysis of RAD-tags (Baird et al. 2008) to identify SNPs found differentially in males or females (the “RAD-sex” method). This approach analyzes genotypes without regard to parentage and can identify loci of major effect (Atwell et al. 2010). To identify sex-specific SNPs in RAD-tags, we utilized Stacks, a program that infers genotypes from short-read sequences (Catchen et al. 2011, 2013). We investigated six zebrafish strains, including domesticated strains made lethal free for mutagenesis (AB and TU), natural strains cultured for a few years in the laboratory without deliberate genetic manipulations [EKW (EkkWill) and WIK (Wild India Kolkata)], and strains acquired directly from the wild in India [NA and CB (Cooch Behar)].
Analysis of >25,000 SNPs in each of these six strains identified alleles differentially associated with sex phenotype. In all four natural strains, results identified a single sex-associated locus at the end of the long (right) arm of Chr4 (Chr4R), a locus previously identified as sar4 (sex-associated region Chr4) (Anderson et al. 2012). Results showed that the distribution of sar4 alleles in populations and their inheritance patterns in crosses were consistent with natural zebrafish populations having a WZ/ZZ sex-determination system, as previously suggested (Tong et al. 2010), and are in accord with the reported WZ/ZZ karyotype inferred for a zebrafish population taken directly from the wild (Sharma et al. 1998). Surprisingly, our experiments failed to detect any sex-linked loci in either AB or TU, two strains domesticated for mutagenesis experiments and sequenced for the zebrafish reference genome. Domestication led either to the evolution of new methods of sex determination during recent decades of selection by zebrafish researchers or to the unveiling of preexisting minor genetic sex-determining mechanisms.
Materials and Methods
Fish strains
Zebrafish (D. rerio) were raised in the University of Oregon Zebrafish Research Facility under standard conditions (Westerfield 2007). Strains included the following:
AB (ZFIN ID: ZDB-GENO-960809-7), originating from a mating of strain A and strain B purchased at two different times from a pet shop in Albany, Oregon, in the late 1970s and screened for making large numbers of lethal-free embryos by in vitro fertilization and subsequently bottlenecked through 21 gynogenetic half-tetrad individuals produced by early pressure treatment to establish the current AB strain (Walker-Durchanek 1980; Streisinger et al. 1981; Chakrabarti et al. 1983; C. Walker, personal communication)
TU (Tuebingen, ZFIN ID: ZDB-GENO-990623-3), originating from a German pet store and selected to be lethal free ca. 1990 from multiple single pair crosses (Mullins et al. 1994)
NA (Nadia, ZFIN ID: ZDB-GENO-030115-2, Anderson et al. 2012), the eighth generation of animals taken from nature in Nadia, India, in 2000
WIK (Wild India Kolkata, ZFIN ID: ZDB-GENO-010531-2), which “derives from a wild catch in India” (Rauch et al. 1997), presumably Kolkata, about 140 km south of Nadia, originating from a single pair mating
EKW (EkkWill, ZFIN ID: ZDB-GENO-990520-2), zebrafish of unknown origin maintained for many years in large populations at EkkWill Waterlife Resources (Ruskin, FL), which has supplied fish to the pet store trade since 1962 (http://www.ekkwill.com/aboutekkwill.html) and obtained from M. Carvan (University of Wisconsin—Milwaukee) (Loucks and Carvan 2004)
CB (Cooch Behar), a new strain derived from fish collected in 2012 from Cooch Behar, India, ∼500 km north of Nadia, and purchased from Eugene Research Aquatics, LLC.
Cooch Behar individuals taken directly from the wild gave small clutches and showed greatly reduced sex dimorphism. Dissection of CB fish taken directly from nature revealed 28 females, 4 hermaphrodites that contained both ovary and testis tissue, 1 fish with translucent tissue at the location expected for gonads, and 8 males (20% males), suggesting disrupted sex development by stress, endocrine-disrupting substances during development, or differential survival during acquisition of these animals. Supporting Information, Figure S4 shows the location of origin of the wild fish stocks discussed here.
Group crosses of adults from each strain generated populations of fish from which we arbitrarily selected individuals for RAD-sex analysis. AB fish (UO stock no. S22191) came from stocks that the University of Oregon Zebrafish Research Facility maintains for shared use involving 10 crosses, each with 2 males and 2 females. Our TU population (S23232) came from in vitro fertilization of eggs from 4 females mixed with sperm pooled from 12 males. NA fish (S23847) derived from a natural cross of an unknown number males and females. WIK fish came from two different generations, one (S23069) a natural cross of 1 female by 2 males and the other (S24746) derived from multiple natural crosses. EKW fish were maintained in three tanks of 60 unsexed fish that were bred en masse by natural matings. CB individuals were from natural matings of the first generation offspring of 28 females and 8 males captured in India. The sex of each animal was determined by microscopic observation of dissected gonads. Individuals of undetermined sex were excluded from analysis.
A population of dwarf danio (D. nigrofasciatus), a close relative of zebrafish (Mayden et al. 2007; Tang et al. 2010), was raised in the University of Oregon fish facility under standard zebrafish culture conditions (Westerfield 2007). A population of medaka (O. latipes) from the Carbio strain (WLC#2674) was raised at the University of Würzburg according to standard laboratory practices (http://shigen.lab.nig.ac.jp/medaka/medakabook/index.php)(Kirchen and West 1976). Male and female medaka were selected at random from the standing aquarium population, which is maintained by natural matings of ∼50 males and 50 females each generation. The phenotypic sex of medaka was first determined from secondary sex characteristics (shape of dorsal and anal fins, spines on male anal fin rays) and confirmed by macroscopic inspection of dissected gonads. The genotypic sex of medaka was identified from the presence or absence of the dmrt1bY gene by PCR from fin clip DNA essentially as described (Nanda et al. 2003) using allele-specific primers: DMT1k (5′ CAA CTT TGT CCA AAC TCT GA 3′) and DMT1l (5′ AAC TAA TTC ATC CCC ATT CC 3′) at an annealing temperature of 56°.
RAD-tag genotyping
Genomic DNA was isolated from caudal fin clips and muscle with a Qiagen DNeasy blood and tissue kit. DNA was digested with high-fidelity SbfI restriction enzyme (New England Biolabs, no. R3642S). Barcode adapters five or six nucleotides long were ligated to each sample. Restriction-site associated DNA (RAD) libraries were prepared as described (Baird et al. 2008; Amores et al. 2011; Anderson et al. 2012) and were sequenced on an Illumina HiSequation 2000 or 2500 using 100-nucleotide single-end reads. We used Stacks software (http://creskolab.uoregon.edu/stacks/) to organize reads into loci and to identify polymorphisms (Catchen et al. 2011, 2013). We RAD-sequenced 20 males and 20 females from AB, 24 males and 24 females from TU, 25 males and 25 females from NA, 21 males and 37 females from EKW, 39 males and 28 females from WIK (of which 34 males and 27 females were retained for analysis), and 49 males and 28 females from CB. Illumina sequences were quality filtered with the process_radtags program of Stacks (Catchen et al. 2011) and then aligned to the zebrafish genome (v. Zv9)(Howe et al. 2013) using GSNAP (Wu and Nacu 2010), allowing nine mismatches. Medaka sequences were aligned to the O. latipes genome (v. MEDAKA1) (Kasahara et al. 2007). Sequences that aligned to multiple sites in reference genomes were discarded. Genotypes were called from aligned reads using the refmap.pl Stacks pipeline, requiring a minimum stack depth of 10 (−m 10). Reads that did not align to the genome were analyzed with the denovo_map.pl Stacks pipeline using the following parameters: a minimum stack depth of 10 (–m 10), up to three differences when merging stacks into loci (–M 3), and up to two differences between loci when building the catalog (–n 2).
Zebrafish linkage group numbers assigned based on genetic length (Postlethwait et al. 1994; Johnson et al. 1996) are given the name “chromosome” in the Zv9 version of the zebrafish reference genome; here we abbreviate “linkage group” as Chr according to zebrafish nomenclature conventions (https://wiki.zfin.org/display/general/zfin+zebrafish+nomenclature+guidelines). Cytogeneticists numbered zebrafish chromosomes based on physical length (Pijnacker and Ferwerda 1995; Daga et al. 1996; Gornung et al. 1997, 2000; Amores and Postlethwait 1999; Sola and Gornung 2001), so zebrafish genetic linkage group number and cytogenetic chromosome number are generally not the same. We assigned genetic linkage groups to physical chromosomes by fluorescent in situ hybridization (Phillips et al. 2006), which showed that, for example, cytogenetic chromosome 3 is linkage group 4, called chromosome 4 in the reference genome and Chr4 in this work.
Sequences for zebrafish, medaka, and dwarf danio RAD-tags were archived under accession no. SRP044635 and mitochondrial sequences under KM196113–KM196120.
Statistical analysis
Haplotypes were exported from the Stacks web interface requiring a minimum stack depth of three reads, and a blacklist of overmerged tags was generated with a custom python script (File S1). The Stacks ref_map pipeline identifies RAD-tags based on position in the genome. Overmerged tags arise when different regions of the genome have highly similar sequences, so that genomically separate RAD-tags align to the same location, resulting in a stack of RAD-tags with a biologically impossible number of alleles. We defined overmerged tags as those that had more than two alleles in more than one fish, and excluded these tags from further analysis.
Polymorphic RAD-tags were exported from Stacks in genomic format using Stacks’ populations program (Catchen et al. 2013). RAD-tags were required to be present in 75% of the individuals of each sex, and blacklisted tags were excluded. For each polymorphic SNP, the program SNPstats1 (http://webpages.uidaho.edu/hohenlohe/software.html) (Hohenlohe et al. 2010) calculated a G-test statistic comparing genotypes in males and females. False discovery rate (FDR) was calculated using the Qvalue R package (Storey and Tibshirani 2003) according to the method of Benjamini and Hochberg (1995). Markers that were significantly associated with sex after the first round of analysis were then inspected by hand. An arbitrarily selected subset of five individuals was evaluated in the Stacks web interface for all loci significantly associated with sex to ensure that Stacks had called genotypes correctly. If an error was identified in any of these five individuals, then all fish in the panel were checked at that locus and corrected by hand; for example, if three or more reads of an undersequenced allele were present in a stack, the genotype was corrected to a heterozygote in the genomic output file and statistics were recalculated.
Four WIK males were excluded from analysis because they had numerous genotypes throughout the genome that were not present in the vast majority of other fish from the population, suggesting migrants. One male and one female WIK were excluded because they had numerous alleles that were undersequenced and uncalled by Stacks, likely due to barcode contamination.
Mapping unassembled scaffolds
Despite the high quality of the zebrafish reference genome (Howe et al. 2013), many scaffolds remain unassembled, especially in areas rich in repeats, such as the heterochromatic and late-replicating right arm of zebrafish cytogenetic chromosome 3 (genetic linkage group 4, Ensembl Chr4) (Pijnacker and Ferwerda 1995; Daga et al. 1996; Amores and Postlethwait 1999; Traut and Winking 2001; Phillips et al. 2006; Anderson et al. 2012; Howe et al. 2013). Some RAD-tags that were strongly associated with sex reside on scaffolds that are either unassembled or are on Chr14 in Zv9. To test for misassembly, we used a previously published data set of SbfI-based RAD-tags mapped on the zebrafish HS (heat shock) meiotic recombination mapping panel (Kelly et al. 2000; Postlethwait et al. 2000; Woods et al. 2000, 2005; Catchen et al. 2011). Filtered raw reads from RAD-tagging the HS panel were aligned to the Zv9 assembly with GSNAP, allowing five mismatches. Because these gynogenetic fish were homozygous at all loci, we relaxed Stacks parameters and required a minimum stack depth of two reads (−m 2) to call genotypes. RAD-tags that aligned to Chr4 and Chr14 in the assembled genome and to all unassembled contigs and scaffolds were used to generate meiotic linkage maps for Chr4 and Chr14.
Markers were initially grouped in JoinMap 4.1 using the Independence LOD parameter under population grouping with a minimum LOD value of 8.0. Subsequent grouping was performed at a minimum LOD value of 6.0. Marker ordering was performed using the maximum-likelihood algorithm in JoinMap 4.1 with default parameters. The expected recombination count feature in JoinMap4.1 was used to identify individuals with a higher than expected number of recombination events and visual inspection of marker order was performed. When needed, marker order was optimized manually after visual inspection of the colorized graphical genotypes in JoinMap 4.1. If moving a marker or group of markers reduced the total number of recombination events, the marker was manually moved to the new position. This analysis resulted in the positioning of many unplaced contigs and scaffolds across the entire Zv9 reference genome.
Nadia sex-genotyping primers
A pair of primers (NA_sx.F_5-CCGGCCCTCAAGGACCGAAA-3 and NA_sx.R_5-GGTTGCTCAAGTGTTGGTGAGA-3) was designed within the sequence of a RAD-tag that aligned to Chr14:37,865,815–37,865,909 and included a sex-specific indel in the NA strain (see Figure S1). GoTaq Flexi DNA polymerase (Promega, M8298) was used to amplify the product with the following PCR protocol: denaturation at 94° for 6 min, 40 cycles of 94° denaturation, 55° annealing, and 72° extension followed by a final extension step of 72° for 10 min.
Phylogenetic reconstruction
To infer the history of zebrafish strains, we sampled RAD-tag sequences from 10 females and 10 males arbitrarily selected from each strain and identified 9,442 RAD-tag loci that were present in all 120 samples. To root the tree, we included orthologous RAD-tags from 1 male and 1 female D. nigrofasciatus for the 4,765 RAD-tags for which orthology could be inferred across species. RAD-tag loci were considered orthologous across strains and species if they mapped to the same unique location in the zebrafish reference genome Zv9. Tags with more than one best mapping location were excluded. We aligned sequences from each RAD-tag locus using Muscle (v. 3.8.31) with default parameters. Aligned loci were concatenated into a single 887.5-kb alignment. Using this alignment, we inferred phylogenetic trees and performed bootstrap replicates with RAxML (v. 7.2.8) using maximum parsimony and maximum likelihood under a GTR+I+Γ model with D. nigrofasciatus as the outgroup (Stamatakis 2014).
For zebrafish strains, we assembled sequences of the mitochondrial cytochrome-b gene from Illumina reads arising from small amounts of contaminating mitochondrial DNA. All reads used for RAD-tag analysis in Stacks have 6 bases left by the SbfI enzyme digestion (TGCAGG); reads without this sequence are from contaminating nuclear or mitochondrial genomic DNA. Reads with a correct barcode, but lacking a TGCAGG motif were identified with the process_radtags program from Stacks (Catchen et al. 2011) and aligned to the zebrafish genome using GSNAP (February 20, 2014, release) (Wu and Nacu 2010).
Results
Verifying the RAD-sex method: Medaka
To verify that a RAD-tag-based population genomics approach would identify a major sex-determination locus if one exists, we first tested a species in which the sex chromosome and major sex-determination locus has already been identified. Although Japanese medaka has a strong XY sex-determination system based on the sex-determining gene dmrt1bY located on chromosome Ola1 (O. latipes chromosome 1) (Matsuda et al. 2002; Nanda et al. 2002; Kondo et al. 2006, 2009), environmental temperature can override the system and cause genotypic females to develop as phenotypic males (Sato et al. 2005; Hattori et al. 2007; Selim et al. 2009; Hayashi et al. 2010). We analyzed 21,909 RAD-tags (Table 1) from 30 female and 31 male medaka and conducted a G-test for genotypes that are significantly associated with male or female sex at 36,115 SNPs. Although the medaka Y chromosome is fully assembled, including dmrt1bY in the middle of Ola1 (NCBI accession nos. AP006150–AP006153 (Kondo et al. 2006)), the male-specific region is not assembled well in the reference genome sequence (http://www.ensembl.org/Oryzias_latipes/Info/Index) and dmrt1bY itself is on the unassembled scaffold1535, which contains no SbfI site and hence no RAD-tags, thus excluding dmrt1bY from our analyses. Nevertheless, results revealed SNPs between 14.3 and 32.5 Mb on Ola1 that were strongly associated with sex (Figure 1A; see Table S1). Only six sex-linked RAD-tags failed to align to the reference genome, none of which were as strongly associated with sex as the best hits within the genome (Table S2). In the medaka reference genome, a region of 18.2 Mb around the position of the sex-determining locus dmrt1bY contained polymorphisms that are highly associated with sex, and 3.5 Mb of this region showed 100% correlation with the genotypic sex that had been determined previously by PCR, consistent with recombination suppression over the region showing strongly sex-specific RAD-tags. PCR genotyping and RAD-sex analysis both agreed that 2 of 31 medaka individuals with a male phenotype had a female genotype, showing that despite occasional sex reversal, the RAD-sex method is robust enough to identify sex-linked markers. Sex-linked SNPs were also identified in single RAD-tags on Ola3, Ola13, and Ola17. These SNPs are in linkage disequilibrium with the sex-linked SNPs on Ola1 (Figure 1, B and C), a result expected if these parts of Ola3, Ola13, and Ola17 were misassembled in the medaka reference genome. These experiments show that a RAD-tag-based GWAS study is an effective method for identifying a major sex-determining region despite occasional sex reversal.
Table 1. Number of RAD-tags analyzed for each population tested.
| Total no. RAD-tags in population | Tags present in at least 75% of Individuals | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total no. RAD-tags | Polymorphic RAD-tags | SNPs analyzed | ||||||||||
| Population | Denovo | Refmap | Total | Denovo | Refmap | Total | Denovo | Refmap | Total | Denovo | Refmap | Total |
| Medaka | ||||||||||||
| Carbio | 8054 | 65849 | 73903 | 2802 | 51309 | 54111 | 1034 | 20875 | 21909 | 1588 | 34527 | 36115 |
| Zebrafish | ||||||||||||
| AB | 6455 | 54389 | 60844 | 1789 | 33542 | 35331 | 440 | 15099 | 15539 | 709 | 30293 | 31002 |
| Tuebingen | 6408 | 80284 | 86692 | 1983 | 46844 | 48827 | 469 | 19535 | 20004 | 770 | 39621 | 40391 |
| Nadia | 12020 | 44314 | 56334 | 1430 | 20705 | 22135 | 536 | 11828 | 12364 | 977 | 25314 | 26291 |
| WIK | 15792 | 138963 | 154755 | 3223 | 44481 | 47704 | 751 | 19143 | 19894 | 1202 | 38054 | 39256 |
| EkkWill | 14440 | 119701 | 134141 | 2303 | 40218 | 42521 | 772 | 22396 | 23168 | 1328 | 47854 | 49182 |
| Cooch Behar | 21712 | 75353 | 97065 | 1335 | 33149 | 34484 | 1161 | 28494 | 29655 | 3158 | 91339 | 94497 |
Figure 1.
RAD-sex for the medaka (O. latipes) assembled genome sequence. (A) The -log10P of a G-test of genotypes associated with male or female phenotype plotted against position in the 24 medaka linkage groups, with odd-numbered linkage groups having a white background and even-numbered linkage groups having a gray background. The solid horizontal line represents a q-value of 0.01, and the dashed line represents a q-value of 0.05. The analysis identified a broad peak of sex-associated SNPs on chromosome 1, (Ola1) the medaka sex chromosome. Isolated single SNPs on chromosomes 3, 13, and 17 were also highly linked to sex. (B) Medaka females were homozygous at SNPs strongly linked to sex, as expected from an XX karyotype, on Ola1 (only two of which are shown), and Ola3, Ola13, and Ola17. (C) Most medaka males were heterozygous at SNPs strongly linked to sex, as expected from an XY sex-determination system. Phenotypic males M1 and M4 had a female genotype. SNPs on Ola3, Ola13, and Ola17 were in linkage disequilibrium with sex-linked SNPs on Ola1, a result that would occur if these regions are on Ola1 in the fish genome but have been misassembled in the medaka reference genome sequence.
Identifying a major zebrafish sex-determination locus
To identify loci linked to sex phenotype in zebrafish, we initially analyzed SbfI-associated RAD-tags (Table 1) from the widely used AB and TU strains. We tested 20 males and 20 females of the AB strain and 24 males and 24 females of the TU strain and compared results to 25 males and 25 females from NA, a natural strain from India. We analyzed 31,002 SNPs in AB and 40,391 SNPs in TU but found no markers that were significantly linked to sex (Figure 2, A and B). In contrast, analysis of 26,291 SNPs in NA fish revealed several sharp peaks of association with phenotypic sex. Sex-linked SNPs in NA were located at the right end of Chr4 between position 61.1 and 62.1 Mb, in the middle of Chr14 between 37.87 and 37.90 Mb, and on the “Not Assembled” contigs NA482 and NA683 (Figure 2C and Table S3).
Figure 2.
RAD-sex results for zebrafish strains show plots of the −log10P of a G-test of genotypes associated with male or female phenotype in zebrafish plotted against the 25 linkage groups of the assembled zebrafish genome Zv9, with odd-numbered linkage groups having a white background and even-numbered linkage groups having a gray background. Solid lines represent a q-value of 0.01, and dashed lines represent a q-value of 0.05. No SNPs were significantly associated with sex in (A) AB or (B) Tuebingen strains. In contrast SNPs significantly associated with sex were identified on Chr4 and Chr14 in (C) Nadia and (D) WIK, while only loci on Chr4 were associated with sex in (E) EkkWill and (F) Cooch Behar. In addition to the assembled genome, sex-associated SNPs appeared on unassembled contigs NA482 and NA683 for WIK and NA, NA851 for WIK and EKW, scaffold3519 for WIK, EKW, and CB, and scaffold3545 for WIK.
Puzzled by the differences between AB and TU fish vs. NA fish, we analyzed two additional commonly used wild-type strains: WIK and EKW. We analyzed 39,256 polymorphic sites in 34 male and 27 female WIK fish and identified sex-associated SNPs located between 61.4 and 62.1 Mb at the end of Chr4, between 37.87 and 37.90 Mb on Chr14, and on the unassembled contigs NA482 and NA683 as found for the NA strain; in addition, sex-associated SNPs for WIK appeared on contig NA851, scaffold3519, and scaffold3545 (Figure 2D and Table S3). Analysis of 49,182 SNPs in 21 male and 37 female EKW fish again identified sex-associated SNPs on Chr4 between 61.1 and 61.2 Mb, and the unassembled fragments NA851 and scaffold3519, both of which were also identified in the WIK RAD-sex results (Figure 2E and Table S3). No locus on Chr14 was associated with sex in EKW.
To verify the discrepancy between domesticated and natural stocks, we obtained zebrafish directly from a natural population in Cooch Behar in India. We bred wild-caught individuals in the lab to establish the CB strain and used some of the offspring of wild-caught individuals for RAD-sex analysis. We analyzed 94,497 polymorphic sites in 49 males and 28 females (this strain showed much higher levels of heterozygosity than any of the other stocks examined; Figure S2) and identified sex-linked SNPs on Chr4:60.6 Mb and on scaffold3519, which was also found to harbor sex-linked SNPs in WIK (Figure 2F and Table S3).
DNA for the zebrafish reference genome sequence initially came from several thousand TU embryos (Howe et al. 2013), some of which would have become males and others females, despite our finding that TU had no loci strongly linked to sex phenotype. If the reference genome lacked one of the sex chromosomes, then many sex-linked RAD-tags from wild stocks would fail to align to it. Analysis, however, showed that only one sex-linked RAD-tag in NA, one in EKW, and four in WIK failed to align to the zebrafish reference genome (Table S4). SNPs in all nonaligning sex-linked RAD-tags were less strongly associated with sex than those that actually aligned to assembled or unassembled portions of the Zv9 reference genome. This result suggests that the assembly does not lack substantial amounts of any sex chromosome or that the major sex determinant is in a genomic region or unassembled contig with no SbfI sites, as is true for medaka.
Sex-linked unassembled scaffolds map to the sex-associated region at the right tip of Chr4R
While all four natural populations we studied had a strong sex-associated region near the right telomere of Chr4, several strains showed sex-associated loci at other locations, including a small portion of Chr14 and a number of unassembled contigs and scaffolds (Table S5). Suspecting that the Chr14 locus and unassembled contigs and scaffolds belong on Chr4R, we utilized a previously published data set of RAD-tags generated from the zebrafish HS meiotic mapping panel (Kelly et al. 2000; Postlethwait et al. 2000; Woods et al. 2000; Woods et al. 2005; Catchen et al. 2011) to identify the position of these loci on a genetic map and hence to correct possible misassembly of these regions in Zv9. Results showed that nonassembled contigs NA683 and NA851 and scaffold3519, which are linked to sex in multiple natural populations (Table S5), all localized to the end of Chr4 near other sex-linked tags in the meiotic mapping panel (Figure 3). Sex-associated fragments NA482 and scaffold3545 lacked any RAD-tags that were polymorphic in the meiotic mapping panel and so could not be mapped. Two sex-associated RAD-tags that had aligned to Chr14 at positions Chr14:37,844,151 and Chr14:37,879,718 also mapped to Chr4R near other sex-linked RAD-tag polymorphisms. The finding that all mappable sex-linked RAD-tags in four natural strains (NA, WIK, EKW, and CB) occupy a single 1.5-Mb region at the right tip of Chr4 supports the conclusion that Chr4 represents a sex chromosome in natural populations of zebrafish. The failure to detect any sex-linked loci in AB and TU in these analyses suggests that a wild sex determinant was lost or greatly modified in the domestication of zebrafish for laboratory work and that other mechanisms have since taken the place of the natural wild genetic sex-determination system.
Figure 3.
All sex-associated RAD-tags map to Chr4. Sex-associated SNPs were mapped on the HS meiotic mapping cross panel (Kelly et al. 2000; Postlethwait et al. 2000; Woods et al. 2000; Catchen et al. 2013). The sar4 region is indicated by shading. RAD-tags that aligned to Chr14 at 37.8 Mb and to the unassembled scaffolds NA683, NA851, scaffold3519, scaffold3462, and scaffold3536 (indicated in boldface italic type) all mapped to the distal tip of Chr4R on the HS panel.
Wild zebrafish populations have a ZW/ZZ sex chromosome system
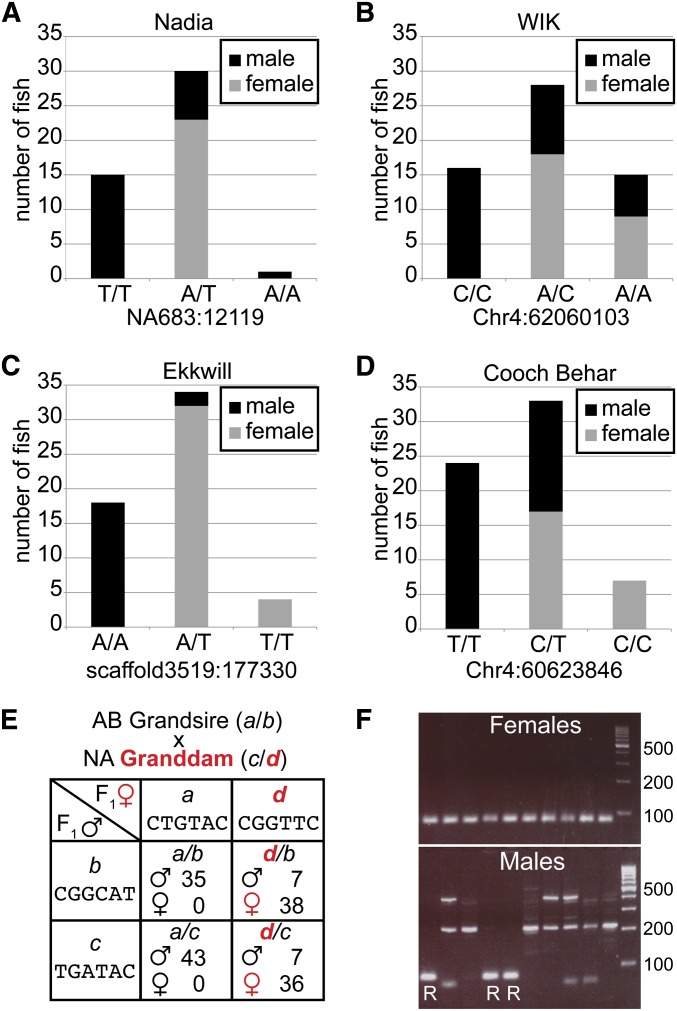
To determine whether natural zebrafish utilize an XY or ZW sex-determination system, we scrutinized the SNP with the strongest statistical support for linkage to sex phenotype in each of the four natural populations. In NA, the SNP most strongly associated with sex was an A > T (nucleotide A in the reference genome vs. T in NA) polymorphism on the unassembled contig NA683 at nucleotide position 12,119 with support of −log10P = 6.8. All 15 fish with the homozygous T/T genotype were males, while 77% (23/30) of A/T heterozygotes developed as females. No individual developed as a female that did not have at least one A allele at this SNP. Only one individual was homozygous A/A, and it, rather surprisingly, developed as a male (Figure 4A). An A/A male could result from sex reversal or from recombination events that separated the sex-linked RAD-tag locus from the causative sex-determination locus. The rather small size of the zebrafish sex-associated region, about 1.5 Mb (60.6–62.1 Mb when considering all four natural strains and including all SNPs with a q-value <0.01) compared to the large region we detected in medaka (14.3–32.5 Mb, or 18.2 Mb using the same q-value) suggests that recombination suppression is stronger around the medaka sex locus than around the zebrafish sex locus.
Figure 4.
Genotypes for SNPs linked to sex phenotype with the highest statistical significance for four natural populations of zebrafish. (A) Nadia SNP at NA683:12,119 (nonassembled contig:nucleotide position), A > T, −log10P = 6.8. All T/T fish were male; 77% of A/T fish were female; the only A/A fish was male. (B) WIK SNP Chr4:62,060,103, A > C, −log10P = 5.4. All C/C fish were male; 64% of A/C and 60% of A/A fish were females. (C) EkkWill SNP at scaffold3519:177,330, A > T, −log10P = 12.5. All A/A fish were male; 94% of A/T and all T/T fish were females. (D) Cooch Behar SNP Chr4:60,623,846, C > T, −log10P = 8.5. All T/T fish were male; 52% of T/C and all seven C/C fish became female. In each of the four populations, all individuals homozygous for the “male allele” (T in NA, C in WIK, A in EKW, and T in CB) developed as males, heterozygotes (A/T in NA, C/A in WIK, T/A in EKW, and C/T in CB) developed mostly as females, and homozygotes for the “female” allele (A in NA, A in WIK, T in EKW, and C in CB) were rare and usually female. In each strain, fish with the homozygous “male allele” all became males and no individuals without the “non-male allele” (the “female allele”) developed into a female. Individuals homozygous for the “female allele” were obtained much less frequently than expected from random mating, which would be expected either if there were few matings between a male and a female both of which had at least one “female” allele or if homozygotes for the “female allele” were less likely to survive. These patterns would be expected of a WZ female/ZZ male sex-determination system with some female genotypes sex reversing to become males. (E) Analysis of the AB × NA F2 sex mapping cross (Anderson et al. 2012). RAD tag 32204, for example, which aligns to Chr4:61,934,186–61,934,280, has allele d that is present in the granddam, the F1 female, and all F2 females as expected if it resides on the W of a WZ/ZZ sex-determination system. (F) Sex-genotyping primers for NA. In the NA strain, the “female allele” in RAD-tag 4086 (which aligned to Chr14:37,865,815–37,865,909 in Zv9 but mapped to the right tip of Chr4 on the HS meiotic mapping panel, Figure 3) has a 12-nt deletion relative to the reference sequence. Primers with the sequence of the female allele in this RAD-tag gave an 85-bp fragment in females but produced an alternative amplification pattern in males. Sex-reversed individuals (as determined by this and other sex-linked RAD-tags) are indicated with an R.
These data for NA show that: (1) the A allele of this SNP in NA is linked to a dominant factor that is necessary but not sufficient for development of a female phenotype; (2) the NA strain has a heterozygous female/homozygous male (e.g., ZW female/ZZ male) sex-determination system; (3) some A/T and A/A “genetic females” are sex reversed to a male phenotype due to the effects of the environment or to segregating minor genetic modifiers that our protocol could not detect; and (4) the small number of individuals with an A/A genotype could be due to the infrequent mating of a normal heterozygous A/T genotypic female to a sex-reversed heterozygous genotypic A/T male, that the homozygous A genotype is semilethal, or that these genotypes come from a recombination event between the RAD-tag and the functional sex locus.
This pattern was repeated in the other natural populations. In WIK, an A > C polymorphism at Chr4:62,060,103 showed the strongest statistical support for sex linkage (−log10P = 5.4). All 16 homozygous C/C WIK fish were male, but 64% (18/28) of A/C heterozygotes and 60% (9/15) of A/A homozygotes were females (Figure 4B). In EKW, an A > T polymorphism at scaffold3519:177,330 was statistically most strongly linked to sex (−log10P = 12.5). All 18 A/A homozygotes developed as males, while 94% (32/34) of A/T heterozygotes and all four T/T homozygotes became females (Figure 4C). In CB, a C > T polymorphism at Chr4:60,623,846 was statistically most strongly linked to sex (−log10P = 8.5). All 24 homozygous T/T fish were male, while 52% (17/33) of heterozygous T/C individuals and all seven homozygous C/C fish were female (Figure 4D). While the CB result could be interpreted as supporting a balancing sex-determination system, with T/T fish developing as males, C/C fish developing as females, and heterozygotes developing as either sex, a more parsimonious explanation that mirrors other native strains is that the C allele is linked to a locus that is necessary but not sufficient for female sex development and that homozygous C/C fish come from the mating of a normal T/C female to a sex-reversed phenotypic T/C male. Taken together, data from all four wild stocks provide strong support for a female-heterogametic sex-determination system.
Segregation in a mapping cross supports a ZW/ZZ sex chromosome system in wild zebrafish
Reanalysis of a female-NA-by-male-AB F2 mapping panel (Anderson et al. 2012) supports female heterogamety. For RAD-tag 32204 (Anderson et al. 2012), all 78 F2 individuals that inherited the grandsire’s a allele became males, but a is not a male-specifying allele because it was inherited through the F1 female. The grandsire’s b allele and the granddam’s c allele were about equally likely to be found in males and females in the F2 (52 and 58% males, respectively), so they are unrelated to sex differentiation. In contrast, 84% (74/88) of F2 fish with the granddam’s d allele, inherited through the F1 female, themselves became females and no fish without the d allele became a female (Figure 4E). These results show that a sequence linked to the d allele is necessary but not sufficient for female development in this female-NA-by-male-AB cross, independent of the other allele, consistent with a dominant, environmentally sensitive, female-determining locus at sar4 and female heterogamety.
Genotyping sex in NA
One of the RAD-tags (4086) that was tightly linked to sex in NA (−log10P = 6.0) contained a 12-bp indel with the insertion present in the female-linked allele. Although this RAD-tag aligned uniquely to Chr14:37,865,815–37,865,909 in Zv9, it mapped to the right tip of Chr4 on the HS meiotic mapping panel (Figure 3). Within the RAD-tag, we designed a forward primer containing four female-specific SNPs and a reverse primer with one female-specific SNP that amplify a single 85-bp band in NA females and a 73-bp band or a nonspecific banding pattern in NA males (Figure 4F). We tested all 50 individuals from our NA population genomic analyses, of which Stacks had genotyped 24 phenotypic females and 22 phenotypic males at this locus. PCR verified the genetic sex determined by RAD-tag analysis for all individuals, including those experiencing female-to-male sex reversal. These primers also accurately identified the sex of individuals that Stacks did not genotype at this locus (due to insufficient read depth) but were genotyped for sex at nearby SNPs.
After verifying primers on the RAD-sex population, we tested other NA fish. Of eight phenotypic females tested, all had the female genotype and of eight phenotypic males tested, seven had the male genotype; the other phenotypic male had a female genotype, consistent with occasional female-to-male sex reversal observed in other natural stocks. These results show that this primer pair can identify NA fish that will definitely become males or that have a high probability of becoming females. These sex-genotyping primers will be useful for identifying genetic sex long before phenotypic sex becomes evident, which will facilitate the study of developmental mechanisms.
Phylogenomics of sex determinant loss
To determine whether our four natural strains represent separate accessions from the wild and to understand their historical relationships to the two strains that lack the Chr4R sex determinant, we conducted a phylogenetic analysis using RAD-sex sequence data. We collected RAD-tag genomic data from dwarf danio (D. nigrofasciatus), one of the closest extant relatives of zebrafish, for use as outgroup (Mayden et al. 2007; Tang et al. 2010). Because D. nigrofasciatus is native to the Sittang basin in Myanmar, which is far outside the range of D. rerio (Figure S4), the two species are not sympatric (Engeszer et al. 2007; Whiteley et al. 2011). Analysis utilized 888 kb of sequence from 9442 RAD-tags present in all six D. rerio strains, including 58,282 variable positions of which 51,444 SNPs were parsimony informative.
Analysis of the maximum-likelihood tree (Figure 5) and the maximum-parsimony tree (see Figure S3) provided several conclusions:
Figure 5.
Maximum-likelihood phylogeny of six D. rerio strains. The phenotypic sex of each individual is abbreviated as F (female) or M (male). Branch labels reflect bootstrap support. Each zebrafish strain represents a distinct clade.
Maximum-likelihood and maximum-parsimony both gave strong support for the same phylogenetic relationships among strains. In contrast, relationships between individuals within a strain varied between the two approaches and across maximum-likelihood bootstrap replicates.
Analyses consistently recovered the same relationships among the three terminal taxa [EKW (AB,TU)] across bootstrap replicates under a GTR+I+Γ model. Because AB and TU occupied a monophyletic clade that was sister to EKW, phylogenomics does not resolve the question of whether the lack of the Chr4R sex-determination locus in AB and TU is due to two independent events during their separate and independent routes to domestication in Oregon and Germany or to a single event that occurred either in nature or in the pet trade before the divergence of these two populations.
Recent extractions from East India (NA, WIK, CB) were genetically distinct from the [EKW (AB,TU))]clade, although relationships among NA, WIK, and CB varied across bootstrap replicates, as evidenced by poor support of the respective internal nodes (41 and 43%, Figure 5). The inability to resolve relationships among these strains even with a large amount of available sequence data are due to variation in the placement of the root of the tree. The length of the branch between zebrafish and dwarf danio is orders of magnitude longer than the length of the internal nodes separating strains because dwarf danio lacks zebrafish-specific RAD-tags. Unfortunately, outgroups closer to zebrafish are as yet unknown.
CB appears to have two subclades, suggesting population substructure within that isolate. In addition, CB harbors more genetic variation than the other strains as evidenced by the depth of the root of that strain and analysis of heterozygosity (Figure 5 and Figure S2). This result is expected for the offspring of individuals taken directly from nature.
Males and females do not group separately within strains, showing that standing genetic variation in the bulk of the genome masks any sex-specific differences in the phylogenetic analyses. In sum, phylogenomics showed that each of the six strains used is a distinct population, that the most recently accessed natural strains lie basal in the tree, and that the domesticated strains AB and TU are rather closely related among the strains we tested.
To determine whether our strains cover the broad diversity of zebrafish natural variation, we investigated cytb, encoded by the mitochondrial genome. Zebrafish populations collected widely across India fall into three major clades: haplogroup 1 in Northern India and Eastern and Western Nepal, haplogroup 2 in Bangladesh and Southern India, and haplogroup 3 in Central Nepal (Whiteley et al. 2011). Infrequent mitochondrial genome contamination in RAD-tag libraries allowed us to reconstruct the cytb locus and assign populations to mitochondrial clades (accession nos. KM196113–KM196120). Results (see Figure S5) identified 14 SNPs that were shared across all our strains and all fish of haplogroup 1 but were absent from all haplogroups 2 and 3, showing that NA, WIK, EKW, and CB all derived from haplogroup 1 (Whiteley et al. 2011). Among the six strains we used and the 16 populations in the Whiteley et al. (2011) study, only AB appeared in both studies, and in both cases was in haplogroup 1. We conclude that our samples represent a single branch of the broad diversity of mitochondrial lineages across the zebrafish species.
Discussion
A major genetic sex determinant is linked to the distal tip of Chr4R in natural zebrafish populations
RAD-sex analysis identified a 1.5-Mb interval containing a major genetic sex determinant near the telomere of zebrafish Chr4R in two natural laboratory strains (WIK and EKW) and in two recent accessions from the wild (NA and CB). These four natural strains have not been extensively manipulated for domestication and were derived from natural populations in India (NA, WIK, and CB) or are of unknown, but likely unmanipulated, origin (EKW). Results also detected sex-linked loci within a small portion of Chr14 and on several unassembled contigs and scaffolds, as might be expected if zebrafish sex phenotype is polygenic (Tong et al. 2010; Bradley et al. 2011; Anderson et al. 2012; Liew et al. 2012; Howe et al. 2013; Liew and Orban 2014). To determine whether these several sex-linked sequences represented a polygenic sex-determination system or whether they were due to errors in the genome assembly, we mapped the unassembled and Chr14 sex-linked loci on a meiotic mapping panel. Because results showed that all sex-linked sequences map to the right tip of Chr4 in Zv9, we conclude that natural wild zebrafish have a single major sex-determining region within a short segment of Chr4R, sar4.
In natural zebrafish populations, Chr4 is likely a sex chromosome
The discovery of a strongly sex-linked locus only in Chr4R in natural zebrafish raises the question of whether Chr4 is a sex chromosome. Genetically defined Chr4 (Postlethwait et al. 1994; Johnson et al. 1996) corresponds to cytogenetic chromosome 3 (Phillips et al. 2006), the right arm of which is the only arm that is late replicating and heterochromatic (Pijnacker and Ferwerda 1995; Gornung et al. 1997; Amores and Postlethwait 1999; Gornung et al. 2000; Sola and Gornung 2001; Traut and Winking 2001; Phillips et al. 2006). Chr4R is impoverished in protein-coding genes, contains most of the genome’s 5S–RNA genes, is enriched in satellite repeats, and has high GC content (Anderson et al. 2012; Howe et al. 2013); these properties are shared with sex chromosomes in other species (Peichel et al. 2004; Charlesworth et al. 2005). Finding sex-linked markers only on the sole chromosome arm with cytogenetic properties of sex chromosomes is consistent with the hypothesis that Chr4 is a sex chromosome in zebrafish.
Females are the heterogametic sex in natural zebrafish populations
The conclusion that Chr4 is a sex chromosome raises the question of whether zebrafish has a female-XX/male-XY, a female-WZ/male-ZZ, or other type of chromosomal sex mechanism. Our population genomics and meiotic mapping both support the conclusion from testosterone-treated AB × WIK individuals (Tong et al. 2010) that females are the heterogametic sex (WZ) in natural populations of zebrafish. For the SNP that is statistically associated most strongly to sex phenotype in each natural population: (1) all individuals that were homozygous for one allele (e.g., m/m), which could be on the Z chromosome, developed as males; (2) most individuals that were heterozygous (e.g., m/f), which could represent a WZ karyotype, became females; and (3) fish that were homozygous for the allele that is not homozygous in most males (e.g., f/f fish, presumably WW) usually developed as females and were fewer than expected by random mating and equal viability. These fish would occur from the mating of a genetic (WZ) female with a, sex-reversed (WZ) phenotypic male. And (4) although some fish with a female genotype (f/m) developed as males, no individual with a male genotype (m/m) became a female. This result suggests that (5), an allele (f) is necessary but not sufficient to make a female phenotype, or the Z version of the native Chr4 has a dosage-sensitive locus for which two doses guarantees male development and one dose favors, but does not assure, female development.
The dominant-female-allele hypothesis and the two-dose-male–one-dose-female hypothesis have both been described in vertebrates. A W-linked, dominantly acting truncated copy of DMRT1 triggers ovary development in the frog Xenopus laevis, likely acting as a dominant-negative inhibitor of the normal DMRT1 gene (Yoshimoto et al. 2008; Yoshimoto et al. 2010; Yoshimoto and Ito 2011). DMRT1 is also involved in sex determination in birds, but the agent is a Z-linked copy that affects sex development by a dosage-sensitive mechanism (Smith et al. 2009). The Z, but not the W, in half-smooth tongue sole has a functional copy of dmrt1, consistent with the dosage-sensitive hypothesis (Chen et al. 2014). A variant of dmrt1 is also the major Y-linked sex determinant of Japanese medaka O. latipes (Matsuda et al. 2002; Nanda et al. 2002; Kondo et al. 2006, 2009). In some species of Oryzias, variants of different genes in the sex-determination pathway, including gsdf and sox3,are at the top of the sex-determination hierarchy (Takehana et al. 2007, 2014; Kondo et al. 2009; Myosho et al. 2012; Kikuchi and Hamaguchi 2013), while in other species of fish, other genes are at the top of the hierarchy, including a variant of irf9 in trout and amhr in fugu (Kamiya et al. 2012; Yano et al. 2012). The molecular genetic nature of the zebrafish sex determinant is as yet unknown. About 80% of Chr4R genes have no apparent human ortholog and are highly duplicated, with, for example, 109 genes encoding NOD-like receptors and zinc finger proteins (Howe et al. 2013), and as discussed below, the strains from which the Zv9 zebrafish reference genome was derived lack or have a greatly modified Chr4-linked sex-determination system, making it likely that the reference genome does not contain the normal wild sex determinant.
The only karyotypes known to involve zebrafish collected directly from nature (Mansar Lake, Jammu) tentatively support the WW/WZ chromosomal sex system (Sharma et al. 1998). The relevance of the Mansar Lake fish to our populations, however, is unknown. Mansar Lake is 2000 km west of Nadia, Kolkata, and Cooch Behar and the phylogenetic relationship of its zebrafish to our populations is unknown. Furthermore, the Z chromosome in Mansar Lake fish was cytogenetically much larger than the W chromosome (Sharma et al. 1998). Assuming that SbfI sites are distributed with approximately the same density across Z and W chromosomes, a large proportion of sex-linked RAD-tags should be homozygous in ZZ males and heterozygous in WZ females. In our experiments, however, only a small fraction of Chr4-linked RAD-tags fit these criteria, suggesting that our Indian populations, while maintaining a WZ/ZZ system, might have a different sex chromosome karyotype than the one suggested by published analyses of the Mansar Lake population.
Domesticated zebrafish strains lack a single strong sex-linked locus
In contrast to natural strains, RAD-sex failed to detect any sex-linked loci in AB and TU, which is surprising given that published studies involving these strains identified sex-biasing loci. For the SATmap study, we made fully homozygous (doubled haploid) gynogenetic AB and TU fish, some of which became males and others females despite all fish having only female-derived chromosomes (Howe et al. 2013). Crossing a fully homozygous female TU fish to a fully homozygous male AB fish produced a clutch of genetically identical F1 heterozygotes, some of which became males and others females (Howe et al. 2013), showing that genetic differences are not essential for zebrafish sex determination. Apparently, in the absence of genetic differences, other forces influence zebrafish sex determination; for example, stochastic differences might cause some eggs to have less yolk than others or environmental differences might arise if late hatching larvae have less access to food; both situations would lead to poorer nutrition, slower growth, and a greater likelihood of developing as a male (Lawrence et al. 2008).
To produce the F2 SATmap population, a heterozygous AB/TU F1 male was crossed to his genetically identical sister (Howe et al. 2013). Analysis identified a single significant peak linked to sex at Chr16:19 Mb–23 Mb. Homozygotes for the granddam (TU) allele were 70% likely to become female but homozygotes for the grandsire (AB) allele were only 26% likely to become female, while heterozygotes were about equally likely to be male or female (Howe et al. 2013). This result shows that a locus on Chr16 has female-favoring allele(s) in TU and/or male-favoring alleles in AB. This result does not address the question of whether sex-determination alleles are polymorphic within each strain, just that alleles differ between strains. For example, AB fish might have partially deleterious alleles at this locus so that AB homozygotes have fewer primordial germ cells, are slower developing, or are less successful at obtaining nutrition and thus more likely to become males relative to individuals homozygous for TU alleles (Lawrence et al. 2008; Siegfried and Nusslein-Volhard 2008; Rodriguez-Mari et al. 2010; Rodríguez-Marí and Postlethwait 2011; Dranow et al. 2013). In contrast, RAD-sex detects sex-linked polymorphisms segregating within a strain, not between strains. If the Chr16 sex-biasing factor that varies between AB and TU is not polymorphic within AB or within TU, our protocol would not find it. Furthermore, the failure to find sar4 in the SATmap experiments supports our finding that AB and TU lack sar4.
Modification of the Chr4R sex determinant in domesticated strains
AB and TU both experienced selection to remove preexisting mutations for mutagenesis experiments (Walker-Durchanek 1980; Streisinger et al. 1981; Mullins et al. 1994). C. Walker produced AB from 21 females derived by half-tetrad gynogenesis (Streisinger et al. 1981), which produces offspring that are homozygous except for regions distal to a recombination event; thus, half-tetrad gynogenesis might or might not eliminate heterozygosity at the distally located sar4 locus. Because homozygosity can result in male bias due to loss of fitness (Brown et al. 2012a,b), some half-tetrad animals might sex reverse to male development despite heterozygosity at sar4. The mating of a WW (or ZZ) gynogenetic female (or male) to a sex-reversed WW (or ZZ) gynogenetic male (or female) might result in the loss of the Z (or W) chromosome making a homozygous WW (or ZZ) strain. Sex-biasing alleles or environmental factors could cause some WW (or ZZ) individuals to become males (or females), allowing the strain to propagate. Researchers might select for any preexisting male- or female-biasing alleles as they set up mating pairs; eventually a new genetic sex-determining mechanism might evolve in the domesticated strain, perhaps similar to the rapid evolution of new sex chromosomes in cichlids (Roberts et al. 2009; Ser et al. 2010; Parnell and Streelman 2013).
TU originated from a pet store in Germany about 1990. Mullins et al. (1994) used multiple single-pair matings to make a lethal-free strain without gynogenesis and with a conscious effort to maintain genetic variation. A hypothesis is that the major wild male or female sex determinant was linked to lethal or deleterious alleles, leading to the loss of the sex determinant along with lethal allele loss. In the absence of male or female alleles of the major sex determinant, occasional sex reversal would allow for stock maintenance, and researchers would strongly select for alleles favoring the development of both sexes. If this hypothesis were true, then the evolution of new sex determinants in TU provides a unique opportunity to study the evolution of new sex-determining mechanisms.
Despite the absence of a strong sex-linked locus on Chr4, AB and TU may have retained some components of the wild sex-determination mechanism:
A female-AB-by-male-IN cross (MGH cross) identified sex-linked loci at Chr5:44.5–46.6 Mb and Chr16:13–17 Mb that together account for just 16% of the variance with regard to sex (Knapik et al. 1996, 1998; Bradley et al. 2011). If the AB parental female lacked a strong female sex determinant as in our AB fish, then the MGH cross might miss sar4 even if IN females, which were not involved in the cross, do possess the natural wild sex determinant.
Our female-AB-by-male-NA cross identified sex-linked loci in addition to sar4, specifically, one on Chr3, although the reciprocal cross identified only sar4 (Anderson et al. 2012).
As discussed above, the female-TU-by-male-AB SATmap cross identified sex-linked loci on Chr16, but in a different location than in the MGH cross (Howe et al. 2013).
The identification of autosomal sex-associated loci (Chr3, Chr5, Chr16) could represent either the unmasking of weak sex determinants given the loss of the sar4, or the rapid evolution of new sex-determination systems after the divergence of EKW, which has the Chr4R sex-determination system, from the common ancestor of AB and TU.
Female-to-male sex reversal
What causes male genotypes to develop only as males but female genotypes to sometimes develop as sex-reversed phenotypes? Answers likely lie in environmental factors and background genetic features that affect the strength of a meiotic oocyte-derived pro-female signal that inhibits oocyte apoptosis, probably by maintaining aromatase production (Slanchev et al. 2005; Houwing et al. 2007; Wang et al. 2007; Siegfried and Nusslein-Volhard 2008; Rodriguez-Mari et al. 2010, 2011; Rodríguez-Marí and Postlethwait 2011; Pradhan et al. 2012; Dranow et al. 2013). In general, harsh conditions, including high density and poor nutrition, tend to promote male development (Walker-Durchanek 1980; Pelegri and Schulte-Merker 1999; Shang et al. 2006; Lawrence et al. 2008; Abozaid et al. 2011, 2012; Liew et al. 2012; Villamizar et al. 2012). These harsh factors may act to decrease the pro-female signal by depressing the pool of meiotic oocytes, either by inhibiting primary germ-cell proliferation or entry into meiosis or by promoting oocyte apoptosis. Sex reversals can happen even in species with a strong genetic sex determinant. In medaka, high temperature and hypoxia can cause female-to-male sex reversal, accompanied by increased cortisol and depressed aromatase (Sato et al. 2005; Hattori et al. 2007; Selim et al. 2009; Kitano et al. 2012; Cheung et al. 2014). It remains to be tested whether stressful conditions cause sex reversal in zebrafish by a similar mechanism.
In addition to environmental factors, background genetic factors might decrease the strength of sar4 activity in domesticated stocks. Although our experiments showed that WIK has a strong sex determinant, inbreeding, which results in homozygosis of partially deleterious alleles, biases WIK fish toward male development (Brown et al. 2012a). Especially likely would be an interaction of background genotype with environmental factors that could override the influence of sar4 on phenotypic sex.
Population diversity and the modification of sex-determining mechanisms in domesticated strains
Did AB and TU lose sar4 independently or was this feature present in the last common ancestor of the two strains? Phylogenomic analyses showed that AB and TU occupy a monophyletic sister clade to EKW, suggesting that AB and TU may have been derived from EKW-related fish—AB from a pet store in Oregon and TU from a pet store in Germany. By parsimony, these results do not rule out the possibility that the lack of sex-linked loci in Chr4R was a shared trait derived from the last common ancestor of AB and TU. Nevertheless, due to their independent domestication in Eugene and Tuebingen, and the presence of sar4 in EKW, we suspect that the loss of a strong sex determinant occurred independently during the separate domestication of the two lineages, a conclusion that is not incompatible with the phylogenomic data.
Analysis of the mitochondrial genome-encoded cytb gene showed that our strains all derived from mitochondrial haplogroup 1, suggesting that a broader understanding of the genetics of sex determination across the full diversity of zebrafish will require investigations of haplogroups 2 and 3 (populations in Nepal and Southern India, respectively), which diverged from haplogroup 1 about 3 million years ago (Whiteley et al. 2011). Investigations of Danio species closely related to zebrafish will also add to our understanding, although the sar4 sex-determination locus may not be widely conserved with other danios. The closest lineage to zebrafish (Cypriniformes; Cyprinidae; Danio) that has a genome-wide analysis of conserved syntenies is the gudgeon (Cypriniformes; Cyprinidae; Gnathopogon) (Kakioka et al. 2013), whose lineage separated from the zebrafish lineage about 117 million years ago (Saitoh et al. 2011). Analyses showed strong conservation of syntenies among all zebrafish chromosomes except Chr4R (Kakioka et al. 2013).
Conclusions
Results presented here show that zebrafish in nature has a strong sex determinant linked to the right tip of Chr4, the only chromosome arm with cytogenetic features frequently found in sex chromosomes; that the determinant is necessary but not sufficient for female development; and that in natural populations, females are WZ and males are ZZ. In contrast, domesticated strains cleaned of background mutations for mutagenesis experiments lack or have greatly weakened versions of the Chr4 sex-determination system.
These conclusions have several important implications:
Because domesticated strains make males and females (often with widely fluctuating sex ratios) even without the full complement of natural genetic sex determinants, these strains apparently have a functional sex-determining mechanism, perhaps due to polygenic sex determination unmasked by the evolutionary strengthening of weak sex-ratio modifiers under the heavy hand of “unnatural selection” wielded by zebrafish researchers, or the unmasking of latent but preexisting environmental sex-determination mechanisms.
Some studies investigating the biology of zebrafish sex determination using TU and AB fish should be revisited using sex-genotyped animals from a strain that possesses the natural genetic sex determinant.
Although mechanisms downstream of sar4 are likely to be the same in all zebrafish stocks, future work that includes studies of natural strains containing the wild sex determinants would provide richer understanding.
The zebrafish genome sequence, which was derived mainly from TU with input from AB and substantial correction using the SATmap, is unlikely to contain strong alleles of both the male and the female alternatives of the major natural sex-determining gene.
We need to concentrate efforts to identify the molecular genetic basis of wild sex in zebrafish.
Supplementary Material
Acknowledgments
We thank Bill Trevarrow (Eugene Research Aquatics, LLC) for obtaining Nadia and Cooch Behar fish and Yann Guiguen and William Cresko for helpful discussions. Research was supported by National Institutes of Health grants R01GM085318 and R01OD011116 to J.H.P. and a Major Research Instrumentation grant from the National Science Foundation, Office of Cyber Infrastructure “MRI-R2: Acquisition of an Applied Computational Instrument for Scientific Synthesis (ACISS) ,” Grant no. OCI-0960354.
Footnotes
Supporting information is available online at www.genetics.org/content/early/2014/09/18/genetics.114.169284/suppl/DC1.
Sequences for zebrafish, medaka, and dwarf danio RAD-tag reads from this study have been archived at NCBI under accession no. SRP044635. Mitochondrial sequence accession numbers: KM196113–KM196120.
Communicating editor: D. Parichy
IRB number: IACUC Protocol no. 14-08, University of Oregon.
Literature Cited
- Abozaid H., Wessels S., Horstgen-Schwark G., 2011. Effect of rearing temperatures during embryonic development on the phenotypic sex in zebrafish (Danio rerio). Sex Dev. 5: 259–265. [DOI] [PubMed] [Google Scholar]
- Abozaid H., Wessels S., Horstgen-Schwark G., 2012. Elevated temperature applied during gonadal transformation leads to male bias in zebrafish (Danio rerio). Sex Dev. 6: 201–209. [DOI] [PubMed] [Google Scholar]
- Amores A., Postlethwait J. H., 1999. Banded chromosomes and the zebrafish karyotype. Methods Cell Biol. 60: 323–338. [DOI] [PubMed] [Google Scholar]
- Amores A., Catchen J., Ferrara A., Fontenot Q., Postlethwait J. H., 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188: 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. L., Rodriguez-Mari A., Braasch I., Amores A., Hohenlohe P., et al. , 2012. Multiple sex-associated regions and a putative sex chromosome in zebrafish revealed by RAD mapping and population genomics. PLoS ONE 7: e40701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S., Huang Y. S., Vilhjalmsson B. J., Willems G., Horton M., et al. , 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D., Mank J. E., Peichel C. L., Kirkpatrick M., Otto S. P., et al. , 2014. Sex determination: Why so many ways of doing it? PLoS Biol. 12: e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird N. A., Etter P. D., Atwood T. S., Currey M. C., Shiver A. L., et al. , 2008. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 3: e3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc., B 57: 289–300. [Google Scholar]
- Braasch I., Guiguen Y., Loker R., Letaw J. H., Ferrara A., et al. , 2014. Connectivity of vertebrate genomes: paired-related homeobox (Prrx) genes in spotted gar, basal teleosts, and tetrapods. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 163: 24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, K. M., J. P. Breyer, D. B. Melville, K. W. Broman, E. W. Knapik et al., 2011 An SNP-based linkage map for zebrafish reveals sex determination loci. G3 (Bethesda) 1: 3–9. [DOI] [PMC free article] [PubMed]
- Brown A. R., Bickley L. K., Ryan T. A., Paull G. C., Hamilton P. B., et al. , 2012a Differences in sexual development in inbred and outbred zebrafish (Danio rerio) and implications for chemical testing. Aquat. Toxicol. 112–113: 27–38. [DOI] [PubMed] [Google Scholar]
- Brown K. H., Dobrinski K. P., Lee A. S., Gokcumen O., Mills R. E., et al. , 2012b Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc. Natl. Acad. Sci. USA 109: 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull J. J., 1983. Evolution of Sex Determining Mechanisms. Benjamin/Cummings, Menlo Park, CA. [Google Scholar]
- Catchen, J. M., A. Amores, P. Hohenlohe, W. Cresko, and J. H. Postlethwait, 2011 Stacks: building and genotyping Loci de novo from short-read sequences. G3 (Bethesda) 1: 171–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J., Hohenlohe P. A., Bassham S., Amores A., Cresko W. A., 2013. Stacks: an analysis tool set for population genomics. Mol. Ecol. 22: 3124–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti S., Streisinger G., Singer F., Walker C., 1983. Frequency of gamma-ray induced specific locus and recessive lethal mutations in mature germ cells of the zebrafish, Brachydanio rerio. Genetics 103: 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B., 1996. The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 6: 149–162. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G., 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95: 118–128. [DOI] [PubMed] [Google Scholar]
- Charnier M., 1966. Action of temperature on sex-ratio in agama agamas embryo (Agamidae lacertilien). C. R. Seances Soc. Biol. Fil. 160: 620–622. [PubMed] [Google Scholar]
- Chen S., Zhang G., Shao C., Huang Q., Liu G., et al. , 2014. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nat. Genet. 46: 253–260. [DOI] [PubMed] [Google Scholar]
- Cheung C. H., Chiu J. M., Wu R. S., 2014. Hypoxia turns genotypic female medaka fish into phenotypic males. Ecotoxicology 23: 1260–1269. [DOI] [PubMed] [Google Scholar]
- Daga R. R., Thode G., Amores A., 1996. Chromosome complement, C-banding, Ag-NOR and replication banding in the zebrafish Danio rerio. Chromosome Res. 4: 29–32. [DOI] [PubMed] [Google Scholar]
- Dranow D. B., Tucker R. P., Draper B. W., 2013. Germ cells are required to maintain a stable sexual phenotype in adult zebrafish. Dev. Biol. 376: 43–50. [DOI] [PubMed] [Google Scholar]
- Engeszer R. E., Patterson L. B., Rao A. A., Parichy D. M., 2007. Zebrafish in the wild: a review of natural history and new notes from the field. Zebrafish 4: 21–40. [DOI] [PubMed] [Google Scholar]
- Gornung E., Gabrielli I., Cataudella S., Sola L., 1997. CMA3-banding pattern and fluorescence in situ hybridization with 18S rRNA genes in zebrafish chromosomes. Chromosome Res. 5: 40–46. [DOI] [PubMed] [Google Scholar]
- Gornung E., De Innocentiis S., Annesi F., Sola L., 2000. Zebrafish 5S rRNA genes map to the long arms of chromosome 3. Chromosome Res. 8: 362. [DOI] [PubMed] [Google Scholar]
- Hattori R. S., Gould R. J., Fujioka T., Saito T., Kurita J., et al. , 2007. Temperature-dependent sex determination in Hd-rR medaka Oryzias latipes: gender sensitivity, thermal threshold, critical period, and DMRT1 expression profile. Sex Dev. 1: 138–146. [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Kobira H., Yamaguchi T., Shiraishi E., Yazawa T., et al. , 2010. High temperature causes masculinization of genetically female medaka by elevation of cortisol. Mol. Reprod. Dev. 77: 679–686. [DOI] [PubMed] [Google Scholar]
- Heule C., Salzburger W., Bohne A., 2014. Genetics of sexual development: an evolutionary playground for fish. Genetics 196: 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohenlohe P. A., Bassham S., Etter P. D., Stiffler N., Johnson E. A., et al. , 2010. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 6: e1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houwing S., Kamminga L. M., Berezikov E., Cronembold D., Girard A., et al. , 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129: 69–82. [DOI] [PubMed] [Google Scholar]
- Howe K., Clark M. D., Torroja C. F., Torrance J., Berthelot C., et al. , 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. L., Gates M. A., Johnson M., Talbot W. S., Horne S., et al. , 1996. Centromere-linkage analysis and consolidation of the zebrafish genetic map. Genetics 142: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakioka R., Kokita T., Kumada H., Watanabe K., Okuda N., 2013. A RAD-based linkage map and comparative genomics in the gudgeons (genus Gnathopogon, Cyprinidae). BMC Genomics 14: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiya T., Kai W., Tasumi S., Oka A., Matsunaga T., et al. , 2012. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8: e1002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M., Naruse K., Sasaki S., Nakatani Y., Qu W., et al. , 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447: 714–719. [DOI] [PubMed] [Google Scholar]
- Kelly P. D., Chu F., Woods I. G., Ngo-Hazelett P., Cardozo T., et al. , 2000. Genetic linkage mapping of zebrafish genes and ESTs. Genome Res. 10: 558–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi K., Hamaguchi S., 2013. Novel sex-determining genes in fish and sex chromosome evolution. Dev. Dyn. 242: 339–353. [DOI] [PubMed] [Google Scholar]
- Kirchen R. V., West W. R., 1976. The Japanese Medaka: Its Care and Development. Carolina Biological Supply Co., Burlington, NC. [Google Scholar]
- Kitano T., Hayashi Y., Shiraishi E., Kamei Y., 2012. Estrogen rescues masculinization of genetically female medaka by exposure to cortisol or high temperature. Mol. Reprod. Dev. 79: 719–726. [DOI] [PubMed] [Google Scholar]
- Knapik E. W., Goodman A., Atkinson O. S., Roberts C. T., Shiozawa M., et al. , 1996. A reference cross DNA panel for zebrafish (Danio rerio) anchored with simple sequence length polymorphisms. Development 123: 451–460. [DOI] [PubMed] [Google Scholar]
- Knapik E. W., Goodman A., Ekker M., Chevrette M., Delgado J., et al. , 1998. A microsatellite genetic linkage map for zebrafish (Danio rerio). Nat. Genet. 18: 338–343. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Nagahama Y., Nakamura M., 2013. Diversity and plasticity of sex determination and differentiation in fishes. Sex Dev. 7: 115–125. [DOI] [PubMed] [Google Scholar]
- Kondo M., Hornung U., Nanda I., Imai S., Sasaki T., et al. , 2006. Genomic organization of the sex-determining and adjacent regions of the sex chromosomes of medaka. Genome Res. 16: 815–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Nanda I., Schmid M., Schartl M., 2009. Sex determination and sex chromosome evolution: insights from medaka. Sex Dev. 3: 88–98. [DOI] [PubMed] [Google Scholar]
- Lang J. W., Andrews H. V., 1994. Temperature-dependent sex determination in crocodilians. J. Exp. Zool. 270: 28–44. [Google Scholar]
- Larney C., Bailey T. L., Koopman P., 2014. Switching on sex: transcriptional regulation of the testis-determining gene Sry. Development 141: 2195–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C., Ebersole J. P., Kesseli R. V., 2008. Rapid growth and out-crossing promote female development in zebrafish (Danio rerio). Environ. Biol. Fishes 81: 239–246. [Google Scholar]
- Liew W. C., Orban L., 2014. Zebrafish sex: a complicated affair. Brief Funct Genomics 13: 172–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew W. C., Bartfai R., Lim Z., Sreenivasan R., Siegfried K. R., et al. , 2012. Polygenic sex determination system in zebrafish. PLoS ONE 7: e34397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loucks E., Carvan M. J., 2004. Strain-dependent effects of developmental ethanol exposure in zebrafish. Neurotoxicol. Teratol. 26: 745–755. [DOI] [PubMed] [Google Scholar]
- Matsuda M., Nagahama Y., Shinomiya A., Sato T., Matsuda C., et al. , 2002. DMY is a Y-specific DM-domain gene required for male development in the medaka fish. Nature 417: 559–563. [DOI] [PubMed] [Google Scholar]
- Mayden R. L., Tang K. L., Conway K. W., Freyhof J., Chamberlain S., et al. , 2007. Phylogenetic relationships of Danio within the order cypriniformes: a framework for comparative and evolutionary studies of a model species. J. Exp. Zoolog. B Mol. Dev. Evol. 308B: 642–654. [DOI] [PubMed] [Google Scholar]
- Merchant-Larios H., Diaz-Hernandez V., 2013. Environmental sex determination mechanisms in reptiles. Sex Dev. 7: 95–103. [DOI] [PubMed] [Google Scholar]
- Mills M. G., Nuckels R. J., Parichy D. M., 2007. Deconstructing evolution of adult phenotypes: genetic analyses of kit reveal homology and evolutionary novelty during adult pigment pattern development of Danio fishes. Development 134: 1081–1090. [DOI] [PubMed] [Google Scholar]
- Ming R., Bendahmane A., Renner S. S., 2011. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 62: 485–514. [DOI] [PubMed] [Google Scholar]
- Mork L., Czerwinski M., Capel B., 2014. Predetermination of sexual fate in a turtle with temperature-dependent sex determination. Dev. Biol. 386: 264–271. [DOI] [PubMed] [Google Scholar]
- Mullins M. C., Hammerschmidt M., Haffter P., Nusslein-Volhard C., 1994. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Curr. Biol. 4: 189–202. [DOI] [PubMed] [Google Scholar]
- Myosho T., Otake H., Masuyama H., Matsuda M., Kuroki Y., et al. , 2012. Tracing the emergence of a novel sex-determining gene in medaka, Oryzias luzonensis. Genetics 191: 163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I., Kondo M., Hornung U., Asakawa S., Winkler C., et al. , 2002. A duplicated copy of DMRT1 in the sex-determining region of the Y chromosome of the medaka, Oryzias latipes. Proc. Natl. Acad. Sci. USA 99: 11778–11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda I., Hornung U., Kondo M., Schmid M., Schartl M., 2003. Common spontaneous sex-reversed XX males of the medaka Oryzias latipes. Genetics 163: 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban L., Sreenivasan R., Olsson P. E., 2009. Long and winding roads: testis differentiation in zebrafish. Mol. Cell. Endocrinol. 312: 35–41. [DOI] [PubMed] [Google Scholar]
- Ota S., Kawahara A., 2014. Zebrafish: a model vertebrate suitable for the analysis of human genetic disorders. Congenit. Anom. 54: 8–11. [DOI] [PubMed] [Google Scholar]
- Parnell N. F., Streelman J. T., 2013. Genetic interactions controlling sex and color establish the potential for sexual conflict in Lake Malawi cichlid fishes. Heredity 110: 239–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peichel C. L., Ross J. A., Matson C. K., Dickson M., Grimwood J., et al. , 2004. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr. Biol. 14: 1416–1424. [DOI] [PubMed] [Google Scholar]
- Pelegri F., Schulte-Merker S., 1999. A gynogenesis-based screen for maternal-effect genes in the zebrafish, Danio rerio. Methods Cell Biol. 60: 1–20. [DOI] [PubMed] [Google Scholar]
- Phillips R. B., Amores A., Morasch M. R., Wilson C., Postlethwait J. H., 2006. Assignment of zebrafish genetic linkage groups to chromosomes. Cytogenet. Genome Res. 114: 155–162. [DOI] [PubMed] [Google Scholar]
- Pijnacker L. P., Ferwerda M. A., 1995. Zebrafish chromosome banding. Genome 38: 1052–1055. [DOI] [PubMed] [Google Scholar]
- Postlethwait J. H., Johnson S., Midson C. N., Talbot W. S., Gates M., et al. , 1994. A genetic linkage map for the zebrafish. Science 264: 699–703. [DOI] [PubMed] [Google Scholar]
- Postlethwait J. H., Woods I. G., Ngo-Hazelett P., Yan Y. L., Kelly P. D., et al. , 2000. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10: 1890–1902. [DOI] [PubMed] [Google Scholar]
- Pradhan A., Khalaf H., Ochsner S. A., Sreenivasan R., Koskinen J., et al. , 2012. Activation of NF-kappaB protein prevents the transition from juvenile ovary to testis and promotes ovarian development in zebrafish. J. Biol. Chem. 287: 37926–37938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch G. J., Granato M., Haffter P., 1997. A polymorphic zebrafish line for genetic mapping using SSLPs on high-percentage agarose gels. Tech. Tips Online 2: 148–150. [Google Scholar]
- Roberts R. B., Ser J. R., Kocher T. D., 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 326: 998–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Marí A., Postlethwait J. H., 2011. The role of Fanconi anemia/BRCA genes in zebrafish sex determination. Methods Cell Biol. 105: 461–490. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Marí A., Yan Y. L., Bremiller R. A., Wilson C., Cañestro C., et al. , 2005. Characterization and expression pattern of zebrafish anti-Müllerian hormone (amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr. Patterns 5: 655–667. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Mari A., Canestro C., R. A. BreMiller, A. Nguyen-Johnson, K. Asakawa et al, 2010. Sex reversal in zebrafish fancl mutants is caused by Tp53-mediated germ cell apoptosis. PLoS Genet. 6: e1001034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Mari A., Wilson C., Titus T. A., Canestro C., R. A. BreMiller et al, 2011. Roles of brca2 (fancd1) in oocyte nuclear architecture, gametogenesis, gonad tumors, and genome stability in zebrafish. PLoS Genet. 7: e1001357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. A., Urton J. R., Boland J., Shapiro M. D., Peichel C. L., 2009. Turnover of sex chromosomes in the stickleback fishes (gasterosteidae). PLoS Genet. 5: e1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh K., Sado T., Doosey M. H., Bart H. L., Inoue J. G., et al. , 2011. Evidence from mitochondrial genomics supports the lower Mesozoic of South Asia as the time and place of basal divergence of cypriniform fishes (Actinopterygii: Ostariophysi). Zool. J. Linn. Soc. 161: 633–662. [Google Scholar]
- Sato T., Endo T., Yamahira K., Hamaguchi S., Sakaizumi M., 2005b Induction of female-to-male sex reversal by high temperature treatment in Medaka, Oryzias latipes. Zoolog. Sci. 22: 985–988. [DOI] [PubMed] [Google Scholar]
- Schreeb K. H., Groth G., Sachsse W., Freundt K. J., 1993. The karyotype of the zebrafish (Brachydanio rerio). J. Exp. Anim. Sci. 36: 27–31. [PubMed] [Google Scholar]
- Selim K. M., Shinomiya A., Otake H., Hamaguchi S., Sakaizumi M., 2009. Effects of high temperature on sex differentiation and germ cell population in medaka, Oryzias latipes. Aquaculture 289: 340–349. [Google Scholar]
- Ser J. R., Roberts R. B., Kocher T. D., 2010. Multiple interacting loci control sex determination in lake Malawi cichlid fish. Evolution 64: 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth A., Stemple D. L., Barroso I., 2013. The emerging use of zebrafish to model metabolic disease. Dis. Model. Mech. 6: 1080–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang E. H., Yu R. M., Wu R. S., 2006. Hypoxia affects sex differentiation and development, leading to a male-dominated population in zebrafish (Danio rerio). Environ. Sci. Technol. 40: 3118–3122. [DOI] [PubMed] [Google Scholar]
- Sharma K. K., Sharma O. P., Tripathi N. K., 1998. Female heterogamety in Danio rerio (Cypriniformes: Cyprinidae). Proc. Nat. Acad. Sci. India 68B: 123–126. [Google Scholar]
- Siegfried K. R., 2010. In search of determinants: gene expression during gonadal sex differentiation. J. Fish Biol. 76: 1879–1902. [DOI] [PubMed] [Google Scholar]
- Siegfried K. R., Nusslein-Volhard C., 2008. Germ line control of female sex determination in zebrafish. Dev. Biol. 324: 277–287. [DOI] [PubMed] [Google Scholar]
- Slanchev K., Stebler J., de la Cueva-Mendez G., Raz E., 2005. Development without germ cells: the role of the germ line in zebrafish sex differentiation. Proc. Natl. Acad. Sci. USA 102: 4074–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A., Roeszler K. N., Ohnesorg T., Cummins D. M., Farlie P. G., et al. , 2009. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature 461: 267–271. [DOI] [PubMed] [Google Scholar]
- Sola L., Gornung E., 2001. Classical and molecular cytogenetics of the zebrafish, Danio rerio (Cyprinidae, Cypriniformes): an overview. Genetica 111: 397–412. [DOI] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J., Tibshirani R., 2003. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Walker C., Dower N., Knauber D., Singer F., 1981. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 291: 293–296. [DOI] [PubMed] [Google Scholar]
- Takahashi H., 1977. Juvenile hermaphroditism in the zebrafish, Brachydanio rerio. Bull. Fac. Fish. Hokkaido Univ. 28: 57–65. [Google Scholar]
- Takehana Y., Matsuda M., Myosho T., Suster M. L., Kawakami K., et al. , 2014. Co-option of Sox3 as the male-determining factor on the Y chromosome in the fish Oryzias dancena. Nat. Commun. 5: 4157. [DOI] [PubMed] [Google Scholar]
- Takehana Y., Naruse K., Hamaguchi S., Sakaizumi M., 2007. Evolution of ZZ/ZW and XX/XY sex-determination systems in the closely related medaka species, Oryzias hubbsi and O. dancena. Chromosoma 116: 463–470. [DOI] [PubMed] [Google Scholar]
- Tang K. L., Agnew M. K., Hirt M. V., Sado T., Schneider L. M., et al. , 2010. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol. Phylogenet. Evol. 57: 189–214. [DOI] [PubMed] [Google Scholar]
- Tong S. K., Hsu H. J., Chung B. C., 2010. Zebrafish monosex population reveals female dominance in sex determination and earliest events of gonad differentiation. Dev. Biol. 344: 849–856. [DOI] [PubMed] [Google Scholar]
- Traut W., Winking H., 2001. Meiotic chromosomes and stages of sex chromosome evolution in fish: zebrafish, platyfish and guppy. Chromosome Res. 9: 659–672. [DOI] [PubMed] [Google Scholar]
- Uchida D., Yamashita M., Kitano T., Iguchi T., 2002. Oocyte apoptosis during the transition from ovary-like tissue to testes during sex differentiation of juvenile zebrafish. J. Exp. Biol. 205: 711–718. [DOI] [PubMed] [Google Scholar]
- Villamizar N., Ribas L., Piferrer F., Vera L. M., Sanchez-Vazquez F. J., 2012. Impact of daily thermocycles on hatching rhythms, larval performance and sex differentiation of zebrafish. PLoS ONE 7: e52153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker-Durchanek, R. C., 1980 Induction of germ line mutations by gamma-irradiation of zebrafish embryos, p. 96. Ph.D. thesis, Department of Biology, University of Oregon, Eugene, OR. [Google Scholar]
- Wang X., Bartfai R., Sleptsova-Freidrich I., Orban L., 2007. The timing and extent of ‘juvenile ovary’ phase are highly variable during zebrafish testis differentiation. J. Fish Biol. 70: 33–44. [Google Scholar]
- Westerfield M., 2007. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish Danio (Brachydanio) rerio, Ed. 5 M. Westerfield, Eugene, OR. [Google Scholar]
- Whiteley A. R., Bhat A., Martins E. P., Mayden R. L., Arunachalam M., et al. , 2011. Population genomics of wild and laboratory zebrafish (Danio rerio). Mol. Ecol. 20: 4259–4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson R. N., Jopling C., van Eeden F. J., 2014. Zebrafish as a model of cardiac disease. Prog. Mol. Biol. Transl. Sci. 124: 65–91. [DOI] [PubMed] [Google Scholar]
- Woods I. G., Kelly P. D., Chu F., Ngo-Hazelett P., Yan Y. L., et al. , 2000. A comparative map of the zebrafish genome. Genome Res. 10: 1903–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods I. G., Wilson C., Friedlander B., Chang P., Reyes D. K., et al. , 2005. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 15: 1307–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. D., Nacu S., 2010. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 26: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano A., Guyomard R., Nicol B., Jouanno E., Quillet E., et al. , 2012. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22: 1423–1428. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S., Ito M., 2011. A ZZ/ZW-type sex determination in Xenopus laevis. FEBS J. 278: 1020–1026. [DOI] [PubMed] [Google Scholar]
- Yoshimoto S., Okada E., Umemoto H., Tamura K., Uno Y., et al. , 2008. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl. Acad. Sci. USA 105: 2469–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto S., Ikeda N., Izutsu Y., Shiba T., Takamatsu N., et al. , 2010. Opposite roles of DMRT1 and its W-linked paralogue, DM-W, in sexual dimorphism of Xenopus laevis: implications of a ZZ/ZW-type sex-determining system. Development 137: 2519–2526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.