Abstract
In 1990, the National Institutes of Health (NIH) gave some organisms special status as designated model organisms. This article documents publication trends for these NIH-designated model organisms over the past 40 years. We find that being designated a model organism by the NIH does not guarantee an increasing publication trend. An analysis of model and nonmodel organisms included in GENETICS since 1960 does reveal a sharp decline in the number of publications using nonmodel organisms yet no decline in the overall species diversity. We suggest that organisms with successful publication records tend to share critical characteristics, such as being well developed as standardized, experimental systems and being used by well-organized communities with good networks of exchange and methods of communication.
BY the end of the 20th century, “model organisms” were one of the centerpieces of biomedical research. Or so goes the usual narrative (Davis 2004). The historical origins of the term model organism are difficult to trace, but it is clear that the concept of a model organism took a firm hold in the 1960s and 1970s in part due to the rise of the techniques of molecular biology. The development of experimental systems around organisms, such as Drosophila, mice, and maize, has an even longer history in genetic research (Kohler 1994; Rader 2004), and it is defensible to claim that the concepts associated with model organisms long predate the actual use of the terminology. Contemporary model organisms tend to be species (or, more precisely, strains of these species) that are relatively simple and hence experimentally tractable. They were developed as resource materials in order to study particular biological phenomena exhaustively or in great detail, usually including genetic and developmental processes. Although in many cases the organism under study was of interest in its own right to those who did research with it, the underlying expectations were that discoveries made in these organisms would be useful or in some sense applicable for understanding other organisms, including humans, or even fundamental mechanisms shared by many or all living entities. (For an overview of this history and of the concept of a model organism, see Ankeny and Leonelli 2011, as well as earlier commentaries in Gest 1995; Ankeny 2001a.)
Some model organisms were selected because a certain subsystem or process was particularly accessible in the given species, such as the development of the nervous system in the nematode Caenorhabditis elegans (de Chadarevian 1998; Ankeny 2000, 2001b); others because techniques and information were already available from previous research work, for example, Drosophila melanogaster (Kohler 1994; Weber 2007), various mouse and rat strains (Rader 2004; Logan 2002, 2005; Leonelli et al. 2014); and still others such as zebrafish were chosen explicitly to be developed in detail although they had not previously been extensively studied (Grunwald and Eisen 2002). The criteria usually cited as justifying the use of some species as a model organism include a rapid life cycle that permits the growth of large populations in short periods of time and increases the likelihood of spontaneous genetic mutations, relatively simple reproductive cycles and genomes, and relatively small body sizes and physical robustness under laboratory conditions, such that large, standardized populations can be bred and maintained. A wide range of organisms were utilized to investigate fundamental biological processes during the 20th century, but it was not until the planning and implementation of the genome mapping and sequencing projects of the 1990s that a canonical set of model organisms was named by the National Institutes of Health (NIH)for biomedical research (NIH 1999).
The designation of a relatively limited set of organisms as model organisms by the NIH has proven controversial in some quarters. For instance, critics argue that many of these model organisms were chosen without attention to phylogenetic similarities and based on assumptions of conservation of various processes that were not warranted given the available evidence. They question the built-in assumptions of universality (or near universality), which they claim led to a lack of focus on variation and relatively little comparative work, as well as limitations on the study of development in it own right as well as in the context of evolution (e.g., Bolker 1995, 2012; Gest 1995; Jenner and Wills 2007; Gilbert 2009; Sedivy 2009; Sommer 2009; for a historical perspective, see Laubichler and Geison 2001). Further, the very processes that contribute to the standardization of model organisms within the laboratory, in fact, may render them insensitive to environmental variation and hence atypical in a problematic sense. Therefore, these skeptics censure research centered on model organisms for placing undue emphasis on molecular-level processes and paying inadequate attention to higher-level environmental and other epigenetic factors. In addition, disputes have occurred between the communities who work on particular model organisms as to what constitutes success or effectiveness in model organism research. Members of the wider biological community, particularly those who do not work on one of the NIH-designated model organisms, have complained about the “swamping out” of basic biological research due to the funneling of grant monies into research on these organisms (cf. Davies 2007, whose analysis shows no dominance of particular organisms within certain publications in developmental biology), while advocates of model organism approaches (e.g., Ledford 2010) note that continued funding is necessary to achieve the long-term goals inherent in this type of research.
Against the backdrop of this history, we set out to analyze publication trends in biological research using the organisms included on the NIH canonical list of model organisms as compared to other organisms. We focus on publications because these data are publicly accessible and fairly straightforward to analyze, once the onerous task of gathering the data is completed. Our goal was to elicit broad, general trends of publication, to map these trends onto more general historical trends in the lives of these model organisms. Such an investigation is useful for several reasons. First, it provides concrete data regarding the growth of model organism research over the past century. Second, it permits the identification of points in time when organisms become more or less successful, at least in terms of the quantity of literature published, arguably an important measure of success of a research community. Finally, it allows us to begin to assess the impacts of the NIH endorsement of particular organisms as model organisms.
Publication Trends for NIH-Designated Model Organisms
We initially extracted publication information from publicly available databases that have been developed within each organism-based community, as we thought it reasonable to assume that what was included in these databases reflected what the community considered to be “its” research. Within these databases, we excluded a range of publications that we concluded did not fit the conventional definition of research, including abstracts, dissertations, personal communication, supplemental material, letters, poems, book reviews, and sequence accessions. We are well aware that publishing norms may differ from community to community, and it was impossible to control for differential community norms in terms of the types of data published, the frequency with which investigators published, the quantity of data published (e.g., all articles regardless of length and content were counted as the same), data held back or only published electronically, and so on. However, it quickly became apparent that different databases used different procedures for collecting publication data and that a variety of types of publications were included. C. Robertson McClung published a publication chart that one of us (M.R.D.) prepared for Drosophila, Arabidopsis, maize, and rice using the research articles collected in databases for each organism: Drosophila, FlyBase, http://www.flybase.org; Arabidopsis, The Arabidopsis Information Resource, http://www.arabidopsis.org/index.jsp; maize, Maize Genetics and Genomics Database, http://www.maizegdp.org/; rice, Oryzabase, http://www.shigen.nig.ac.jp/rice/oryzabase/top/top.jsp (McClung 2008a,b). However, MaizeGDB and the Maize Genetics Executive Committee (2008) responded that the data on maize underestimated maize publications because they had stopped curating publications in 2003. The rice data were also reported to be an underestimate. McClung withdrew both maize and rice from the figure in the online ASPB News (MaizeGDB and the Maize Genetics Executive Committee 2008).
Moreover, curatorial standards for references in these databases are not standardized and so comparisons across these databases must be treated with caution. The Biocurator Discussion Wiki (2014) had been urging journal editors to ask for organism data to help standardize reporting of organism use. Thus, to compare trends across organisms using the same standards for reference curation, we searched titles in Web of Science for each organism using the expanded science database. This title search returned fewer publications per year than those reported in the organism databases. Expanding the search within Web of Science to search by the topic category produces many more results, but the data contain a substantial jump in publication numbers in 1992 when Web of Science began to include abstracts. Searching by title circumvents this artifact of the database. So, while title searches underreport the actual number of publications using model organisms, they are the only way of obtaining a consistent result from the Web of Science database.
Data for our analysis of publication trends was extracted from ISI Web of Knowledge: Science Citation Index Expanded (SCI-EXPANDED) 1900–present using a title search. All searches were conducted on August 15, 2010 and August 1, 2014. Search terms were as follows: rat (rat or Rattus or Rattus rattus), mouse (mouse or Mus musculus or M. musculus), Drosophila (Drosophila), Arabidopsis (Arabidopsis), chicken (chicken or Gallus or Gallus gallus or G. gallus), zebrafish (zebrafish or Danio rerio or D. rerio), Saccharomyces cerevisiae (Saccharomyces cerevisiae or S. cerevisiae), Xenopus (Xenopus), Schizosaccharomyces pombe (Schizosaccharomyces pombe or S. pombe), Neurospora (Neurospora or Neurospora crassa or N. crassa), Daphnia (Daphnia), C. elegans (Caenorhabditis elegans or C. elegans), and Dictyostelium (Dictyostelium discoideum or D. discoideum).
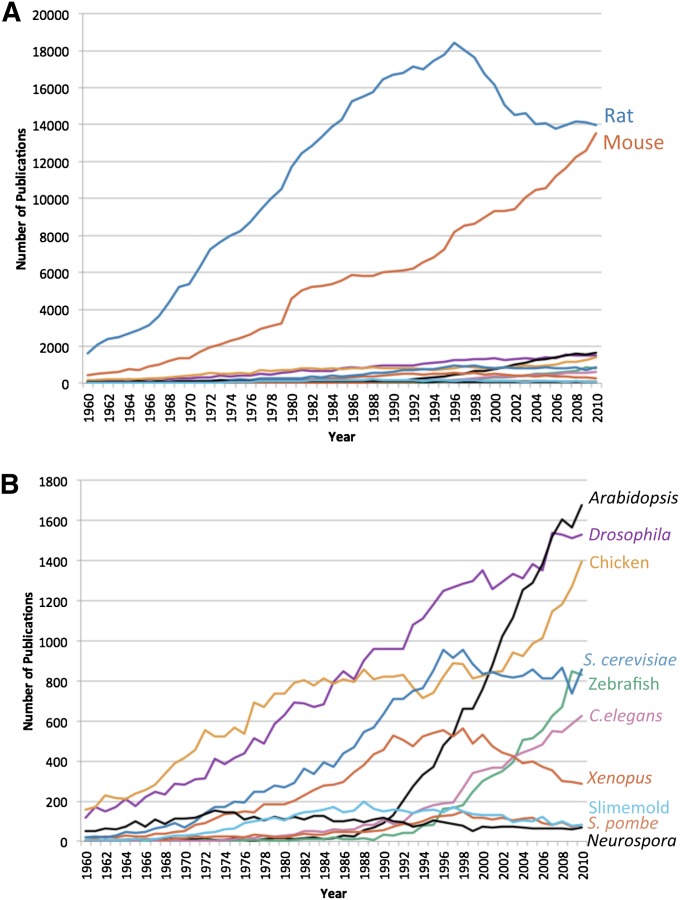
Comparison of the publication trends over the past 30 years reveals that the mammalian NIH-designated model organisms (rat and mouse) have dominated biological research publications (see Figure 1). They begin to diverge from the other organisms in the 1940s; rat in particular begins to grow very rapidly in the 1950s and 1960s. As other model organisms have begun to be used in the past 20 years, the number of rat publications has declined, although it still has the highest publication rate. The data more generally reveal that there are greater numbers of research publications on those higher-level organisms that prima facie are more analogous to human beings, which is not surprising against the backdrop of a contemporaneous push toward more translational research.
Figure 1.
General publication trends for NIH-designated model organisms, 1960–2010. (A) Numbers of publications per year by organism. (B) Numbers of publications per year without trends for rat and mouse.
Newer experimental organisms among those designated as NIH-model organism, such as Arabidopsis, zebrafish, and C. elegans, have shown dramatic increases in relatively short periods of time. The average rate of growth across all publications in the Science Citation Index Expanded database between 1997 and 2006 was 2.7% (Larsen and Von Ins 2010).1 During the same time period, organisms, such as Arabidopsis, zebrafish, and C. elegans, had publication growth rates of 160, 236, and 149%, respectively. Based on the timing of these increases in publication rates, it is clear that many of these have been more successful in terms of publications after being named as formal model organisms by NIH. In the case of Arabidopsis, this may be the result of an infusion of funding from the National Science Foundation (NSF), which has allowed the development of numerous resources essential to supporting model organism research, such as cyberinfrastructure and strain centers (see below for further discussion, and Leonelli 2007a,b; Leonelli and Ankeny 2011; Leonelli and Ankeny 2012; Leonelli and Ankeny 2013).
However, becoming a designated model organism does not guarantee a rise in publications. Xenopus has been in a declining trend since the late 1990s; its average growth rate between 1997 and 2006 is −29%. S. pombe, Neuropora, Dictyostelium, and even S. cerevisiae have been either relatively constant or declining in terms of rates of publication since being designated model organisms by the NIH in its 1999 Non-Mammalian Model Organisms conference and report (National Institutes of Health 1999). Merely designating something as a model organism is neither necessary nor sufficient for achieving high rates of publication, one of marks of success of such research.
Comparing Model and Nonmodel Organism Research
While designation by the NIH as a model organism does not consistently guarantee the same level of publication success, it may still provide a bump relative to organisms not designated as models by the NIH. To assess the relative standing of model and nonmodel organisms, we compared publication trends in Genetics. With limited space and high standards, publication in Genetics was necessarily competitive. To borrow terminology from ecology, did the designation of certain organisms as NIH model organisms lead to the competitive exclusion of research on other organisms (at least in the pages of Genetics)?
To contrast publication trends, we tabulated the organisms used in articles published in the journal Genetics every 5 years from 1960 to 2010. Every journal article in the sampled issue was read to determine which organisms had been used. Each organism was listed on our spreadsheet, and a tally of uses per year was tabulated. Organisms were sorted according to whether they appeared on the NIH-designated model organism list, which includes Drosophila, mice, rat, chicken, S. cerevisiae, S. pombe, C. elegans, Daphnia, Xenopus, zebrafish, Neurospora, and slimemolds (see Figure 2.).
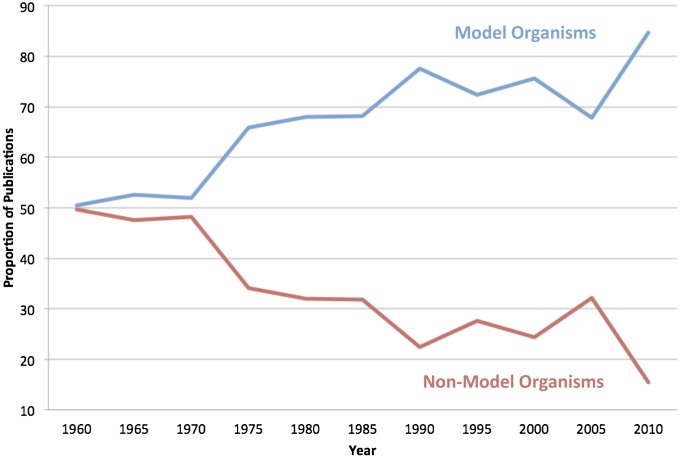
Figure 2.
Proportion of publications for NIH-designated model and other organisms in Genetics, 1960–2005.
Our analysis reveals that since around 1990 there has been a substantial and consistent difference in the proportion of articles using NIH-model and nonmodel organisms. Approximately 75% of the articles used NIH-designated model organisms, while ∼25% used other organisms. Looking back, however, it is clear that this was not always so. Throughout the 1960s and into the 1970s, the proportion of publications with those later designated by NIH as model organisms and those that were not was almost 50:50. This approximate balance begins to shift dramatically by 1975, when there are 38 more publications using what became NIH-designated model organisms and a decline in publications with other species of organisms. The increase in publications focused on NIH-designated model organisms occurred mainly as a result of a large increase in publications using yeast. The consistent divergence in the proportion of publications certainly speaks to the relative success of what became the NIH-designated model organisms, but the steady presence of other species of organisms does not support claims about the competitive exclusion of non-NIH-designated model organisms from publications in Genetics.
While articles on non-NIH-designated model organisms continued to grace the pages of Genetics in the period analyzed here, we wished to investigate whether their scarcity corresponded with a reduced representation of biological diversity in the journal. To quantify the diversity of species of organisms used in articles published in Genetics between 1960 and 2005, we modified measures of species richness and diversity utilized in ecology. Where ecologists calculate species richness and diversity in a geographic area, we calculated organism richness and diversity present in a year of publications in Genetics. For present purposes, species richness (R) is the total number of different organisms used in the articles in Genetics that year. While richness captures the change in number of different species used, diversity captures the relative abundance of each species. We calculated species diversity using Simpson’s diversity index,
where N is the total number of organisms, and ni is the number of individual organisms within each type (Simpson 1949). The value of Ds ranges from 0 to 1. Higher values of Ds reflect higher diversity. This index can be interpreted as the probability that two organisms will be selected from different species when drawn randomly from publications in a given year of Genetics. (It is critical to note that “diversity” in this sense does not incorporate any measure of representativeness in biological terms, and hence this measure does not provide data relevant to the key criticisms of those who question the relevance of model organisms for work in evo-devo, which were discussed previously. This measure of diversity is relative only to the species in the sample from Genetics, which is clearly far from representative of the actual biodiversity present in the world.)
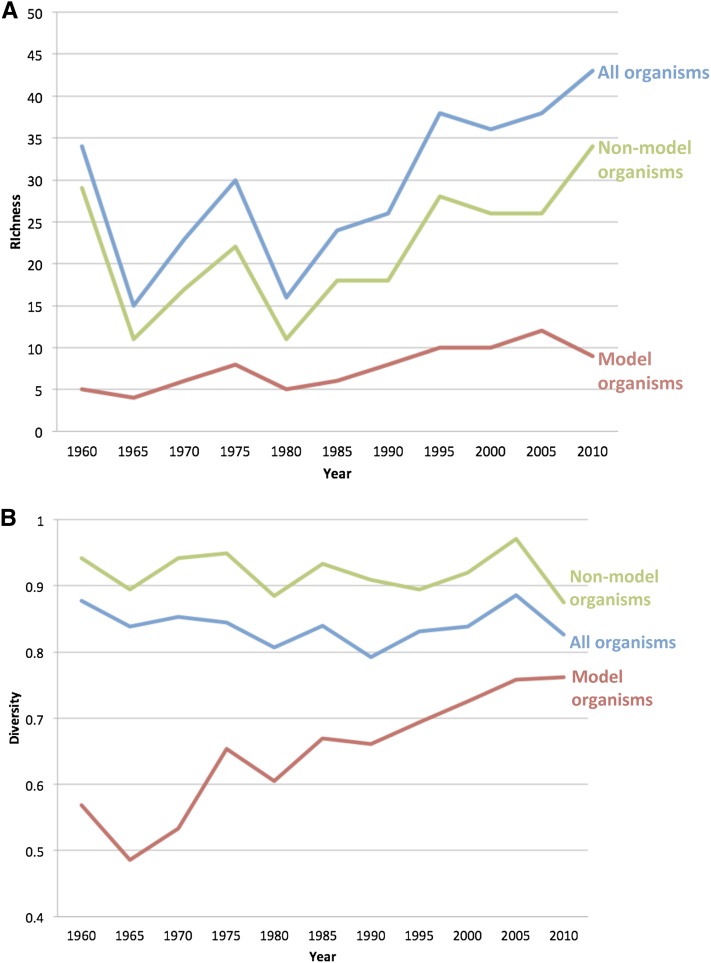
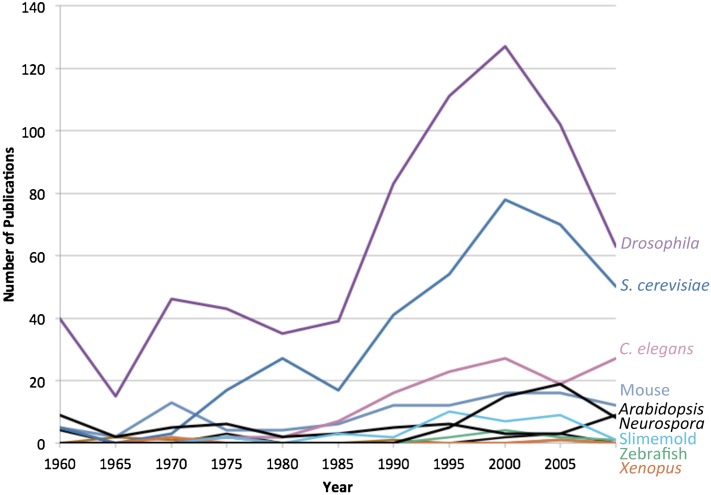
Even when non-NIH designated model organisms represented ∼25% of the publications in Genetics, they still had a higher species richness than model organism species (see Figure 3). There were more non-NIH designated model species included in the journal than there were NIH-designated model species. The rise of model species richness reflects the inclusion and rise of newly developed model organisms, such as Arabidopsis. Similarly, the diversity of non-NIH designated model organisms is consistently high. While some nonmodel species, such as maize and Escherichea coli, had relatively high number of publications per year in Genetics, most nonmodel organisms were represented in fewer than two or three publications per year. The resulting evenness of the distribution of organisms across species produced a high diversity index. The consistently lower diversity index for NIH-designated model organisms reflects the dominance of publications featuring Drosophila and S. cerevisiae (see Figure 4). The recent rise in the number of publications for C. elegans, zebrafish, and Arabidopsis resulted in the rise in the diversity index for model organisms since 1990. Overall, the number of species represented in the pages of Genetics has increased since 1990, after a period of decline during the 1970s and 1980s. The number of NIH-designated model species has generally increased since 1990, but most of the increase in species richness is the result of the inclusion of publications using non-NIH-designated model species. So, even as the proportion of articles utilizing organisms not designated as models by the NIH hovers ∼25%, the number of these species represented in those pages has increased. Part of the rise in species richness is the result of more articles being published per year in Genetics. When species richness is represented proportionally, non-NIH-designated model organisms represent 85% of the total species richness in 1960, but that number drops to 75% by 1965, moving between a low of 70% in 1980 and high of 80% in 2010. Overall, then, the 25% of the publications that include nonmodel organisms account for ∼75% of the species richness in Genetics. Nonmodel organisms have not been excluded, although their proportion has declined, and the number of species represented in its pages in 2010 exceeded that of 1960. Yet, the fact remains that NIH-designated model organisms occupy the vast majority of articles in Genetics.
Figure 3.
Species richness and diversity in Genetics, 1960–2010. (A) Species richness measured every five years from 1960 to 2010 for NIH-designated model and nonmodel organisms. (B) Species diversity measured every 5 years from 1960 to 2010 for NIH-designated model and nonmodel organisms.
Figure 4.
Number of publications for NIH-designated model organisms in Genetics, 1960–2010.
Behind the Trends
No simple set of causes is available to explain the complex factors and circumstances underlying trends noted during 50 years of research in biology, or even within that biological research that happened to be published in Genetics. We have shown that NIH designation, for instance, does not seem to have a consistent effect on publication rates, contrary to many claims by critics. This does not mean that it has had no effect, but simply that it did not have the same impact on every designated organism. In addition to NIH designation, a number of other factors may have informed the publication trends for different model organisms and certain non-NIH-designated model organisms, particularly those that have been more successful. These explanatory factors include funding levels within the United States and globally, from public agencies, private foundations, and industry; the relative standardization of the organism and other experimental factors; the degree of community organization including organism-specific societies, conferences, and training courses, the creation of stock centers, organism databases, and other networks of exchange and communication, in concert with the size of the community and its relative maturity; and having an available genome sequence or sequences. Our analysis has necessarily been limited, as we have focused on publication as a proxy for success. Tracing a detailed history of each of these factors is a task for future research, but we offer some brief examples to illustrate how these factors may have operated.
Gone are the days when Thomas Hunt Morgan could start a Drosophila lab with old milk bottles and a bunch of bananas (Kohler 1994). Today funding is essential for biological research, and funding levels can have a considerable impact on research outputs for any given organism. The impact of funding on model organisms is most obvious in the case of Arabidopsis. Using the NSF’s reported data, we compared funding levels per organism at 5-year intervals. In 1995, Arabidopsis research received $10.3 million in funding. By 2000, NSF funding for Arabidopsis rose to $67 million and by 2005, $87.1 million. Publications using Arabidopsis rose dramatically from 814 in 2000 to 3481 in 2010. No other organism showed such a sharp rise in publication numbers. Using funding data from the NIH Reporter database, we found a similar pattern for correlated funding and publication increases in zebrafish and C. elegans. NIH funding for zebrafish rose from $57.2 million in 2005 to $353.6 million in 2010, while publications in the same interval increased from 214 to 834. In C. elegans, NIH funding climbed from $93.7 million in 2005 to $264 million in 2010 with publications increased from 392 to 803 in the same period. Other funders also likely contributed, including non-U.S. sources, although our analysis documents that the NIH dominates funding of organismal research in the period examined. While it should not be surprising that funding is correlated with published results, funding levels are not the only factor influencing publication rates.
Model organism research also has depended critically on building infrastructure around each model organism, including both stock/strain centers and cyberinfrastructure, such as community databases for communication of results within and across communities of researchers. Many of the NIH-designated model organisms had received funding to support “community resources” such as stock centers (e.g., C. elegans and Arabidopsis) and community databases such as FlyBase, The Arabidopsis Information Resource (TAIR), and WormBase (Leonelli and Ankeny 2012). Organismal standardization was a vital aspect of these community-building resources by allowing materials and techniques to travel with some security. Standardization of organisms is a multifaceted process that creates and documents stable features of different strains (see Ankeny 2000). For organisms, such as Drosophila, which arose as an experimental system for genetic research, standardization included the documentation of mutants, special experimental lines, and techniques for everything from building growth chambers to mixing fly food (Kohler 1994). For organisms used in developmental work, such as the chicken, documenting the normal stages of development was essential for clear communication among researchers (Hamburger and Hamilton 1951). Without cyberinfrastructure and communal access to standardized specimens, the exchange of information about model organisms and their use for comparative purposes would be impossible to realize on the appropriate scale, given the large-scale integrative goals of many contemporary biological research programs (Leonelli and Ankeny 2012). Due to their capacities for bringing results, people, and specimens together, community databases and stock centers have come to play crucial roles in defining what counts as knowledge of organisms in the postgenomic era (Rosenthal and Ashburner 2002). Those who participate actively in model organism communities are expected to contribute material to the stock centers, support the community databases by providing data and other information, and perhaps even assist in the curation of such databases, in exchange for being critically dependent on the specialized information and resources available through the database and the strain center (on these processes in Arabidopsis, see Leonelli 2007a,b).
Training biologists to use and contribute to community resources is also a central feature of successful organismal communities. In the case of research on yeast (S. cerevisiae), the Cold Spring Harbor (CHS) courses played a pivotal role in popularizing the routine usage of yeast. The first yeast course began in 1970 and built off the tradition of phage genetics at Cold Spring Harbor (Hall 1993). The summer yeast courses led to the Molecular Biology of Yeast Meetings, the first with 166 members in 1976. Attendance at the meetings was, from the start, well represented by major molecular biology labs and alumni of the phage and yeast courses, with an impressive doubling of attendance every year. The meetings adopted an egalitarian approach, replicating the model used in phage conferences in which abstract submission was open and all talks were limited to an allotted time (Hall 1993). Both the course and the conference helped create a sense of community that helped establish yeast as a model organism in the 1990s.
Conclusion
A select group of model organisms were designated by the NIH as part of the push toward the Human Genome Project. Each of the initial NIH-designated model organisms was slated to have its genome sequenced. Each of the organisms selected in both phases of the NIH process had a history of research success and a developed community with shared resources that supported research. An available genome sequence was understood as a valued resource that made these organisms amenable to analysis with a growing body of tools and techniques. Indeed, within the Xenopus community, their 2009 white paper acknowledged the “substantial and continuing” investment by the NIH (“427 grants for a total cost of $127,583,776 for FY08 and FY09”), but noted that “Despite this investment in individuals’ research, the Xenopus community lacks many resources that are considered entirely essential for other model systems, including a complete genome sequence, stock and training centers, and a comprehensive model organism database” (Xenopus Community White Paper 2009, their emphasis). The Xenopus community now enjoys these resources with its research and training center at the Marine Biological Laboratory at Woods Hole. However, for our purposes, what is significant about their 2009 statement is its identification of a combination of factors essential for success as a model organism system. According to them, publication success depends on funding, combined with informatics resources (genomic and otherwise), stock centers for the distribution of material, and training centers. To this list of requirements for success, we would add standardization of the organism and associated techniques, as well as community-based mechanisms that promote communication and create a sense of an organism-centered scientific community. In the end, publication success for any organism, model or otherwise, depends critically on the social and scientific organization of research as much as designation by the NIH as a canonical model organism, the biology of the particular organism, or even levels of funding.
Acknowledgments
We are grateful for the constructive commentary provided by C. Robertson McClung, James Collins, and participants in the Model Organism Workshop hosted by Jason Robert, as well as for advice on trends associated with Arabidopsis and general feedback from Sabina Leonelli.
Footnotes
Other databases had higher growth rates for the same period (up to 13.5%). See Larsen and von Ins (2010) for a detailed discussion.
Communicating editor: A. S. Wilkins
Literature Cited
- Ankeny R. A., 2000. Fashioning descriptive models in biology: of worms and wiring diagrams. Philos. Sci. 67: S260–S272. [Google Scholar]
- Ankeny R. A., 2001a Model organisms as models: understanding the ‘lingua franca’ of the Human Genome Project. Philos. Sci. 68: S251–S261. [Google Scholar]
- Ankeny R. A., 2001b The natural history of Caenorhabditis elegans research. Nat. Rev. Genet. 2: 474–479. [DOI] [PubMed] [Google Scholar]
- Ankeny R. A., Leonelli S., 2011. What’s so special about model organisms? Stud. Hist. Philos. Sci. 42: 313–323. [Google Scholar]
- Biocurator Discussion Wiki, 2014 Available at: http://wiki.geneontology.org/index.php/BioCurator_Discussion_Topics. Accessed: June 18, 2014.
- Bolker J., 1995. Model systems in developmental biology. BioEssays 17: 451––455.. [DOI] [PubMed] [Google Scholar]
- Bolker J., 2012. Model organisms: there’s more to life than rats and flies. Nature 491: 31–33. [DOI] [PubMed] [Google Scholar]
- Davies J. A., 2007. Developmental biologists’ choice of subjects approximates to a power law, with no evidence for the existence of a special group of “model organisms.” BMC Dev. Biol. 7: 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H., 2004. The age of model organisms. Nat. Rev. Genet. 5: 69–76. [DOI] [PubMed] [Google Scholar]
- de Chadarevian S., 1998. Of worms and programmes: Caenorhabiditis elegans and the study of development. Stud Hist Phil Biol Biomed Sci 29: 81–105. [Google Scholar]
- Gest H., 1995. Arabidopsis to zebrafish: a commentary on “Rosetta stone” model systems in the biological sciences. Perspect. Biol. Med. 39: 77–85. [Google Scholar]
- Gilbert, S., 2009 The adequacy of model systems for evo-devo: modeling the formation of organisms/modeling the formation of society, pp. 57–68, 155–169 in Mapping the future of biology, edited by A. Barberousse, M. Morange, and T. Pradeu. Springer, Dordrecht. [Google Scholar]
- Grunwald D. J., Eisen J. S., 2002. Headwaters of the zebrafish: emergence of a new model vertebrate. Nat. Rev. Genet. 3: 717–723. [DOI] [PubMed] [Google Scholar]
- Hall M. N. (Editor), 1993. The Early Days of Yeast Genetics. Cold Spring Harbor Press, Cold Spring Harbor, NY. [Google Scholar]
- Hamburger, V., and Hamilton, H. L. 1951 A series of normal stages in the development of the chick embryo J. Morphol. 88(1): 49–92. [PubMed]
- Jenner R. A., Wills M. A., 2007. The choice of model organisms in evo-devo. Nat. Rev. Genet. 8: 311–319. [DOI] [PubMed] [Google Scholar]
- Kohler R., 1994. Lords of the Fly: Drosophila Genetics and the Experimental Life. Univ. of Chicago Press, Chicago. [Google Scholar]
- Larsen P. O., von Ins M., 2010. The rate of growth in scientific publications and the decline in coverage provided by Science Citation Index. Scientometrics 84: 575–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laubichler M. D., Geison G., 2001. The varied lives of organisms: variation in the historiography of the biological sciences. Stud. Hist. Philos. Biol. Biomed. Sci. 32: 1–29. [Google Scholar]
- Ledford H., 2010. Plant biologists fear for cress project. Nature 464: 154. [DOI] [PubMed] [Google Scholar]
- Leonelli S., 2007a What is in a model?, pp. 15–36 in Modeling Biology: Structures, Behaviours, Evolution, edited by Laubichler M., Muller G. B. MIT Press, Cambridge, MA. [Google Scholar]
- Leonelli S., 2007b Growing weed, producing knowledge: an epistemic history of Arabidopsis thaliana. Hist. Philos. Life Sci. 29: 55–87. [PubMed] [Google Scholar]
- Leonelli S., Ankeny R. A., 2012 Re-thinking organisms: the impact of databases on model organism biology. Studies in History and Philosophy of Biological and Biomedical Sciences 43: 29–36. [DOI] [PubMed] [Google Scholar]
- Leonelli S., Ankeny R. A., 2013 What makes a model organism? Endeavour 37: 209–12. [DOI] [PubMed] [Google Scholar]
- Leonelli S., Ankeny R. A., Nelson N. C., Ramsden E., 2014 Making organisms model human behavior: situated models in North American alcohol research, 1950–onwards. Sci. Context 27: 485–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C., 2002. Before there were standards: the role of test animals in the production of scientific generality in physiology. J. Hist. Biol. 35: 329–363. [DOI] [PubMed] [Google Scholar]
- Logan C., 2005. The legacy of Adolf Meyer’s comparative approach: Worcester rats and the strange birth of the animal model. Integr. Physiol. Behav. Sci. 40: 169–181. [DOI] [PubMed] [Google Scholar]
- MaizeGDB and the Maize Genetics Executive Committee, 2008 Letter to Rob McClung ASPB News 35. http://newsletter.aspb.org/2008/septoct08.pdf.
- McClung, R., 2008a A model citizen ASPB News 35. http://newsletter.aspb.org/2008/julaug08.pdf. Last accessed 2014.
- McClung, R., 2008b Turn! Turn! Turn! ASPB News 35. http://newsletter.aspb.org/2008/septoct08.pdf. Last accessed 2014.
- National Institutes of Health, 1999 Non-mammalian models workshop. http://www.nih.gov/science/models/nmm/. Last accessed June 18, 2014.
- Rader, K., 2004 Making Mice: Standardizing Animals for American Biomedical Research, 1900–1955. Princeton Univ. Press, Princeton.
- Rosenthal N., Ashburner M., 2002. Taking stock of our models: the function and future of stock centres. Nat. Rev. Genet. 3: 711–717. [DOI] [PubMed] [Google Scholar]
- Sedivy J. M., 2009. How to learn new and interesting things from model systems based on ‘exotic’ biological species. Proc. Natl. Acad. Sci. USA 106: 19207–19208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson E. H., 1949. Measurement of diversity. Nature 163: 688. [Google Scholar]
- Sommer R. J., 2009. The future of evo-devo: model systems and evolutionary theory. Nature 10: 416–422. [DOI] [PubMed] [Google Scholar]
- Weber M., 2007. Redesigning the fruit fly: the molecularization of Drosophila, pp. 23–45 in Science without Laws: Model Systems, Cases, Exemplary Narratives, edited by Creager A. N. H., Lunbeck E., Wise M. N. Duke University Press, Durham, NC. [Google Scholar]
- Xenopus Community White Paper, 2009 http://www.xenbase.org/community/static/xenopuswhitepaper/2009/XWP_final.jsp. Last accessed June 18, 2014.